Abstract
The best studied nuclear export processes are mediated by classical leucine-rich nuclear export signals that specify recognition by the CRM1 export receptor. However, details concerning alternative nuclear export signals and pathways are beginning to emerge. Within the family of Herpesviridae, a set of homologous regulatory proteins that are exemplified by the ICP27 of herpes simplex virus were described recently as nucleocytoplasmic shuttling proteins. Here we report that pUL69 of the β-herpesvirus human cytomegalovirus is a nuclear protein that is able to shuttle between the nucleus and the cytoplasm independently of virus-encoded cofactors. In contrast to proteins containing a leucine-rich export signal, the shuttling activity of pUL69 was not affected by leptomycin B, indicating that pUL69 trafficking is not mediated by the export receptor CRM1. Importantly, we identified and characterized a novel type of transferable, leptomycin B-insensitive export signal that is distinct from other export signals described previously and is required for pUL69-mediated activation of gene expression. These data suggest that pUL69 is exported via a novel nuclear export pathway, based on a so far unique nuclear export signal of 28 amino acids.
Keywords: human cytomegalovirus/leptomycin B/nuclear export signal/nucleocytoplasmic shuttling
Introduction
A defining feature of eukaryotic cells is their division into nucleoplasm and cytoplasm. This segregation requires specific mechanisms for the continuous transport of large numbers of macromolecules between both compartments. Nucleocytoplasmic transport occurs through the nuclear pore complex (NPC) and is mediated by specific transport receptors. These receptors shuttle between the nucleus and the cytoplasm and are able to recognize and bind cargo molecules. According to the direction in which they carry a cargo, the receptors can be classified as importins or exportins (reviewed in Gorlich and Kutay, 1999; Nakielny and Dreyfuss, 1999).
Protein trafficking into and out of the nucleus occurs through direct interaction between a transport signal of the cargo protein and the corresponding interaction domain of the transport receptor. There are several types of nuclear localization signals (NLSs), which mediate nuclear import by direct binding to one or more importin receptors of the α or β type (reviewed in Nakielny and Dreyfuss, 1999). In contrast, the pathways and details of signal-mediated nuclear export are less defined. The best characterized nuclear export signal (NES) is the small, hydrophobic, leucine-rich NES, which was identified initially in the human immunodeficiency virus type 1 (HIV-1) Rev protein (Fischer et al., 1995) and the cellular kinase PKI (Wen et al., 1995). Functionally related export sequences resembling a leucine-rich NES have been detected since then in many cellular and viral proteins of diverse functions (Fukuda et al., 1996; Dobbelstein et al., 1997; Sandri-Goldin, 1998). The direct interaction with the importin-β-related export factor CRM1 (exportin1) is essential for the export of proteins containing a leucine-rich NES (Fornerod et al., 1997; Fukuda et al., 1997; Stade et al., 1997). This interaction can be inhibited specifically by the antibiotic leptomycin B (LMB), resulting in a block of the nuclear export of proteins with a leucine-rich NES (Wolff et al., 1997; Kudo et al., 1999).
Proteins containing both nuclear import and export signals have the capacity to shuttle between the nucleus and cytoplasm. However, nucleocytoplasmic shuttling can also be controlled by signals combining both functions. These sequences were termed bidirectional or nucleocytoplasmic shuttling (NS) signals (reviewed in Michael, 2000). The pathways mediating the nuclear export of proteins containing bidirectional signals are not sensitive to LMB inhibition and are therefore thought to differ from the classical protein export. Specific export receptors have not been identified yet (Nakielny and Dreyfuss, 1999; Michael, 2000).
Recently, within the family of Herpesviridae, a set of homologous polypeptides implicated in RNA processing and transport were described as NS proteins (Sandri-Goldin, 1998; Goodwin et al., 1999; Farjot et al., 2000). The most intensively studied member of this group is the post-transcriptional regulator ICP27 of the α-herpesvirus herpes simplex virus type 1 (HSV-1) (Sandri-Goldin and Mendoza, 1992). The ICP27 homolog of the β-herpesvirus human cytomegalovirus (HCMV) is the multifunctional nuclear phosphoprotein pUL69 (Winkler et al., 1994; Winkler and Stamminger, 1996). Both pUL69 and ICP27 were described as promiscuous transactivators of viral and cellular gene expression (Sandri-Goldin and Mendoza, 1992; Winkler et al., 1994). While it has been reported that ICP27 is exported from the nucleus via CRM1-dependent and -independent pathways, thereby promoting the nuclear export of viral RNAs (Soliman et al., 1997; Sandri-Goldin, 1998; Soliman and Silverstein, 2000), the mechanism of pUL69 transactivation is not entirely understood. We have shown recently that pUL69 is able to interact with the cellular factor hSPT6, which has been implicated in the regulation of chromatin structure and transcriptional elongation (Winkler et al., 2000). Furthermore, it was shown that pUL69 is involved in cell cycle control, since it induces infected or transfected cells to accumulate in the G1 phase of the cell cycle (Lu and Shenk, 1999; Hayashi et al., 2000). Despite the sequence homology between pUL69 and ICP27, there is evidence for major functional differences between the two polypeptides. For instance, while it has been reported that ICP27 impairs RNA splicing (Sandri-Goldin and Mendoza, 1992), pUL69 does not exert any measurable negative regulation that is dependent on the presence of introns (Winkler et al., 1994).
Based on recent results that defined ICP27 of HSV-1 as a nucleocytoplasmic shuttling protein, we speculated whether pUL69 of HCMV might share this function with ICP27 and initiated our studies on pUL69 nuclear trafficking. Here, we report that pUL69 of HCMV is a nuclear protein that is able to shuttle between the nucleus and the cytoplasm. Importantly, the shuttling activity of pUL69 was not affected by LMB, a finding which indicated that the trafficking of pUL69 does not involve binding to CRM1. We identified a novel type of short, transferable NES, comprising 28 amino acids, which is distinct from that of HIV-1 Rev and other shuttling proteins. These data suggest that the activity of pUL69 is linked to a novel, so far uncharacterized nuclear export pathway.
Results
The UL69 but not the IE1 protein shuttles between the nucleus and the cytoplasm in HCMV-infected human fibroblasts
The UL69 gene product of HCMV belongs to a family of regulatory proteins that are conserved among all herpesviruses, some of which share the capability to shuttle between the nucleus and the cytoplasm. Therefore, we addressed the question of whether pUL69 also has nucleocytoplasmic shuttling activity and performed an interspecies heterokaryon analysis as originally described by Pinol-Roma and Dreyfuss (1992). This assay has been used successfully to identify a series of cellular and viral shuttling proteins (Meyer and Malim, 1994; Michael et al., 1997; Caceres et al., 1998; Mears and Rice, 1998). HCMV-infected human fibroblasts were fused to non-permissive murine NIH 3T3 cells in order to produce heterokaryons. Two hours after fusion, the cells were fixed and the localization of viral nucleoproteins was assessed by indirect immunofluorescence (Figure 1). To differentiate human from murine nuclei, cells were counterstained with Hoechst 33258 dye; murine nuclei display a characteristic punctate pattern, whereas human nuclei are stained diffusely with this reagent (Figure 1A). In a quantitative microscopic examination of heterokaryons, pUL69 could be found in the vast majority (>95%) of those murine nuclei that were part of heterokaryons (Figure 1B). In contrast, IE1, which is an important regulatory protein of HCMV, was detected in murine nuclei only in a minority (<5%) of interspecies heterokaryons (Figure 1C). Neither pUL69 nor IE1 were detected in unfused murine nuclei. Taken together, this indicates that pUL69 migrated from infected human foreskin fibroblast (HFF) nuclei to the cytoplasm and subsequently entered the murine nuclei. Thus, in infected cells, pUL69 is able to shuttle across the nuclear membrane in both directions.

Fig. 1. Nucleocytoplasmic shuttling of pUL69 in HCMV-infected primary human fibroblasts. Heterokaryons were generated by fusion of HCMV-infected human fibroblasts and NIH 3T3 mouse cells. Protein synthesis was blocked with cycloheximide (50 µg/ml) 30 min prior to cell fusion and throughout the experiment. Two hours after fusion, cells were fixed and a double immunofluorescence analysis was performed with a polyclonal antiserum directed against pUL69 (B) and a monoclonal antibody against the IE1 protein (C). Staining with Hoechst 33258 (A) was used to differentiate between human and murine nuclei within the heterokaryon. Murine nuclei display a characteristic punctate pattern, whereas human nuclei are stained diffusely with this reagent; murine nuclei are indicated by arrows. (D) The phase contrast image of the heterokaryons.
Nucleocytoplasmic shuttling of pUL69 occurs in the absence of viral infection
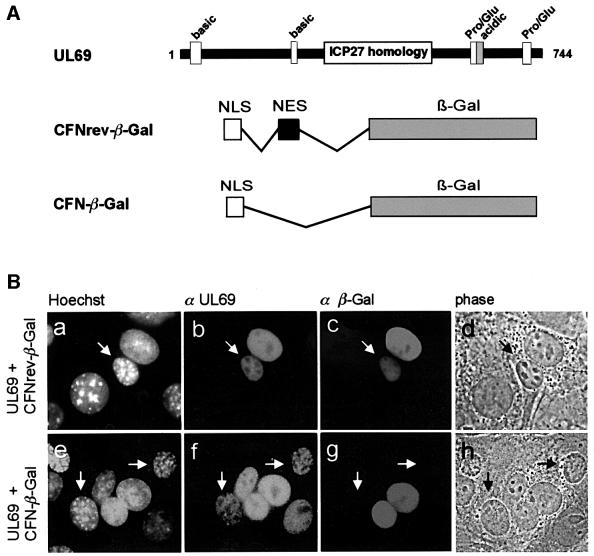
It has been reported that the shuttling activity of some viral proteins depends on virus-encoded cofactors, i.e. proteins or RNA (Dobbelstein et al., 1997; Soliman et al., 1997). Therefore, we examined whether pUL69 is able to shuttle in uninfected cells in the absence of additional viral factors. For this, HeLa cells were co-transfected with a pUL69 expression plasmid and either the internal control plasmid CFN-βGal or CFNrev-βGal (Figure 2A). CFNrev-βGal encodes β-galactosidase (β-Gal) fused to the NLS of the SV40 T-antigen and the 10 amino acid NES of the HIV-1 Rev protein. Thus, this fusion protein is able to shuttle rapidly between the nucleus and the cytoplasm in transfected cells. CFN-βGal expresses only the SV40 T-antigen NLS fused to β-Gal and is therefore restricted to nuclei (Roth and Dobbelstein, 1997). Transfected cells were allowed to synthesize pUL69 together with one of the control proteins and subsequently were subjected to heterokaryon formation. After fixation, cells were co-stained for pUL69 and β-Gal. In interspecies heterokaryons that co-expressed pUL69 and NLS-NES-βGal (as encoded by CFNrev-βGal), both proteins were observed in murine and human nuclei (Figure 2B, a–d). In contrast, when the localization of pUL69 and NLS-βGal (as encoded by CFN-βGal) was assessed, pUL69 alone was found to be present in the murine nuclei, while NLS-βGal was detected exclusively in the human nuclei (Figure 2B, e–h). In conclusion, the ability of pUL69 to shuttle between nucleus and cytoplasm does not depend on viral infection or other HCMV-encoded cofactors.
Fig. 2. Nucleocytoplasmic shuttling of pUL69 in transfected HeLa cells. (A) Schematic representation of the wild-type (wt) UL69 protein and β-Gal fusion proteins that were used as internal controls in this heterokaryon analysis. The region of highest homology to the HSV-1 ICP27 and the location of basic, acidic and proline/glutamine-rich amino acid clusters are indicated for pUL69. Plasmid CFNrev-βgal (expressing β-Gal fused to the NLS from SV40 T-Ag and the NES from HIV 1-Rev) served as a positive control for nucleocytoplasmic shuttling in heterokaryon analysis. Plasmid CFN-βGal, which lacks the Rev NES, served as a negative control (Roth and Dobbelstein, 1997). (B) HeLa cells were co-transfected with the UL69 expression plasmid pHM160 and one of the internal control plasmids as indicated (a–d, UL69 + CFNrev-βGal; e–f, UL69 + CFN-βGal). The transfected cells were subjected to the interspecies heterokaryon assay as described in Materials and methods. Double immunofluorescence analysis with polyclonal anti-pUL69 serum and a monoclonal antibody against β-Gal was performed in order to detect the expressed proteins. Hoechst, staining with Hoechst 33258 (a and e); α UL69, staining with anti-pUL69 antiserum (b and f); α β-Gal, staining with anti-β-galactosidase antibody; phase, phase contrast image of the heterokaryon; murine nuclei are indicated by arrows.
Inhibition of the CRM1-mediated nuclear export pathway does not affect pUL69 shuttling activity
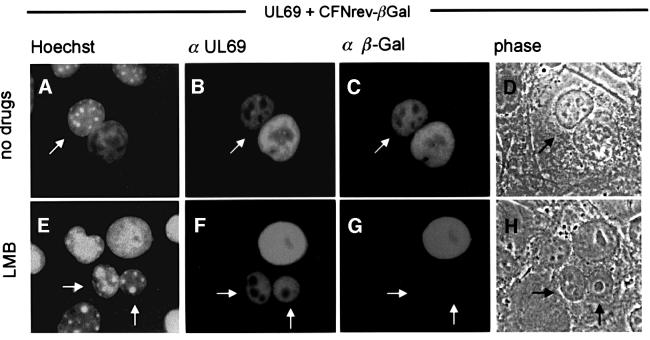
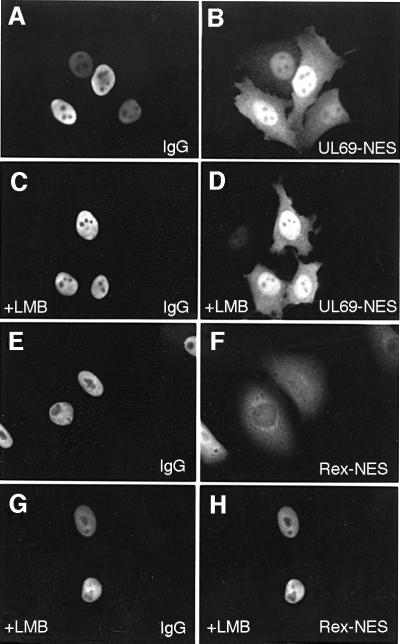
The vast majority of shuttling proteins described so far possess an NES similar to the leucine-rich NES of the HIV-1 Rev protein. Furthermore, this signal sequence has been reported to be sufficient to mediate the active nuclear export of fused proteins (Elfgang et al., 1999). Referring to this, an export receptor for the leucine-rich NES of the HIV-1 Rev type, termed CRM1, was identified (Fornerod et al., 1997; Fukuda et al., 1997; Stade et al., 1997). Previous studies demonstrated that LMB inhibits export of leucine-rich NES-containing proteins by a covalent modification of CRM1, thereby preventing its association with the NES substrate without directly affecting the other known nuclear transport pathways (Fornerod et al., 1997; Wolff et al., 1997; Kudo et al., 1998, 1999). Interestingly, the predicted amino acid sequence of pUL69 does not contain a leucine-rich, classical NES, suggesting that the pUL69 export is mediated either by a non-conventional but CRM1-dependent NES, or by a novel type of CRM1-independent NES. To address the question of whether nucleocytoplasmic shuttling of pUL69 is dependent on CRM1, transfected HeLa cells expressing pUL69 together with the NLS-NES-βGal control protein were pre- incubated with LMB at 2.5 ng/ml in culture medium for 3 h and throughout post-fusion incubation. The transfected cells were used to perform heterokaryon assays as described above. The nucleocytoplasmic shuttling of the NLS-NES-βGal fusion protein (CFNrev-βGal) containing the HIV-1 Rev NES was inhibited (Figure 3G), while, in contrast, the nuclear export of pUL69 was completely unaffected by LMB (Figure 3F). To rule out the possibility that a higher concentration of LMB might be required to inhibit the putative NES within pUL69, two additional experiments were performed. As a result of these, pUL69 shuttling could still be observed at 5 and 10 ng/ml of LMB (data not shown). These findings indicate that the pUL69 shuttling activity is not affected by LMB, and thus suggest that the pUL69 protein is not transported via the classical CRM1-dependent pathway.
Fig. 3. Effect of LMB on the shuttling of pUL69 in interspecies heterokaryon assays. HeLa cells were co-transfected with expression constructs for wild-type pUL69 together with plasmid CFNrev-βGal. Three hours prior to fusion and throughout the experiment, cells were incubated in the absence (A–D) or presence of 2.5 ng/ml LMB (E–H). Two hours after fusion, the proteins were detected by double-label immunofluorescence analysis as described in the legend of Figure 2.
A C-terminal region of pUL69 is required for nuclear export
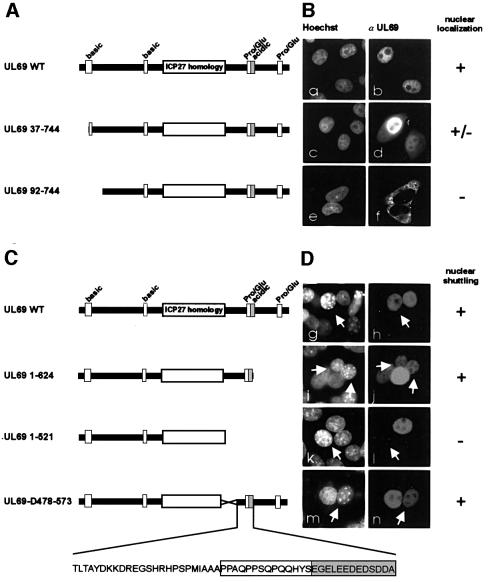
The identification of a nucleocytoplasmic shuttling protein exported via a CRM1-independent pathway raised the question of what was the corresponding export signal. In order to delineate the domain required for nuclear export of pUL69, we first constructed two N-terminal deletion mutants termed UL69 37–744 and UL69 92–744 (Figure 4A). In contrast to the wild-type pUL69, which showed a strictly nuclear localization, mutant UL69 92–744 could only be detected within the cytoplasm of transfected cells (Figure 4B, a, b, e and f). Sequence inspection of the deleted N-terminal domain revealed a motif with homology to a classical bipartite NLS (amino acids 21–45). Consequently, mutant UL69 37–744 disrupting this sequence showed an intermediate nuclear and cytoplasmic staining (Figure 4A, and B, c and d). Due to these results, which suggested the presence of a functional NLS at the N-terminus of pUL69, we constructed two additional C-terminal deletion mutants termed UL69 1–624 and UL69 1–521 (Figure 4C). Since these mutants were strictly nuclear after transfection (data not shown), they could be analyzed for their ability to enter murine nuclei in heterokaryon assays. As shown in Figure 4, the deletion mutant lacking 223 C-terminal amino acids (UL69 1–521) lost its export activity (Figure 4D, k and l), while the mutant lacking 120 C-terminal residues (UL69 1–624) was still able to shuttle (Figure 4D, i and j). Due to these results, we then tested the internal deletion mutant UL69-D478–573 for nucleocytoplasmic shuttling activity (Figure 4C). Since the protein encoded by UL69-D478–573 retained its shuttling activity, we mapped the location of the NES to the C-terminal region of pUL69 between amino acid residues 574 and 624 (Figure 4C, and D, m and n).
Fig. 4. Delineation of a putative NES within the C-terminus of pUL69 by deletion mutagenesis. (A) Schematic diagram illustrating N-terminal deletion mutants of pUL69. The N-terminal cluster of basic amino acids shows homology to a bipartite NLS. (B) HeLa cells transfected with plasmids expressing N-terminal pUL69 deletion mutants were analyzed for the subcellular localization of the indicated mutants via indirect immunofluorescence analysis. (C) Schematic diagram illustrating C-terminal or internal deletion mutants of pUL69; in the expanded section at the bottom of (C), the putative NES (amino acids 574–624) of pUL69 is shown. (D) HeLa cells transfected with plasmids expressing the pUL69 mutants as shown in (C) were subjected to heterokaryon analyses. Hoechst, DNA staining of transfected cells (B) or heterokaryons (D) with Hoechst 33258 (a, c, e, g, i, k and m); α UL69, transfected cells (B) or heterokaryons (D) stained for pUL69 are shown in b, d, f, h, j, l and n.
The UL69 protein contains a novel, short NES which can mediate nuclear export of a carrier protein in domain swap experiments
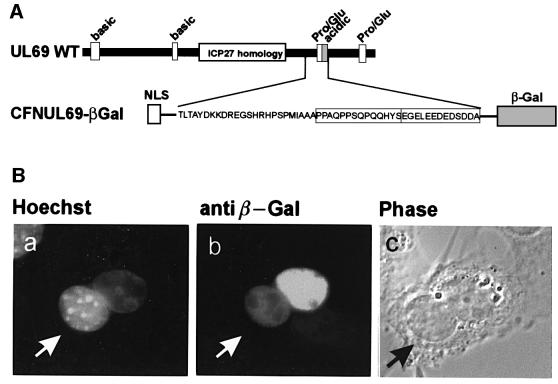
In general, the stable binding of export substrates to their cognate export receptors can be mediated by either large parts of the protein or short continuous (canonical) NES. Based on our finding that the stretch of amino acids 574–624 of the C-terminal part of pUL69 might be essential for its shuttling activity, this raised the possibility that this domain might contain a canonical NES. Therefore, we investigated whether the putative export signal of pUL69 is not only necessary, but also sufficient to allow nuclear export of a protein. This was investigated by domain swap experiments. For this purpose, the Rev NES within the artificial shuttling protein expressed by plasmid CFNrev-βGal was replaced by the pUL69 sequence containing the putative NES (Figure 5A). As shown in Figure 5B, the resulting β-Gal fusion protein could be detected in murine nuclei after fusion with transfected HeLa cells in heterokaryon analyses. This result indicates that the pUL69 signal sequence is transferable and can function as an autonomous NES.
Fig. 5. Nuclear export of a heterologous protein by the putative pUL69 NES. (A) Schematic representation of the wild-type UL69 coding region showing the putative NES which was inserted into plasmid CFNrev-βGal, thus replacing the NES of HIV-1 Rev. The resulting expression vector encoding β-Gal in fusion with the NLS of SV40 T-Ag and the putative NES of pUL69 was termed CFNUL69-βGal. (B) Heterokaryon experiments as described in the legend of Figure 2 were performed to visualize the nuclear export activity of the expressed β-Gal fusion protein.
Microinjection experiments confirm the autonomous NES activity and distinguish the pUL69 NES from a bidirectional signal
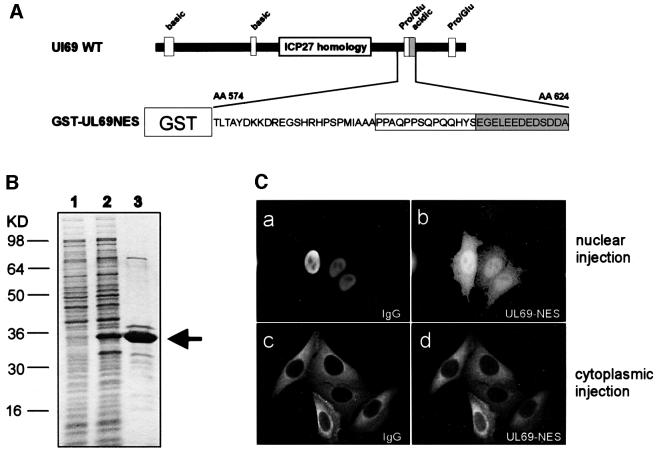
In order to confirm the results of domain swap experiments, we decided to perform microinjections as an alternative experimental approach. For this, we fused the amino acids 574–624 of pUL69 to the C-terminus of GST, which served as a carrier protein (Figure 6A). The fusion protein was purified from Escherichia coli (Figure 6B) and microinjected into the nuclei of HeLa cells along with rabbit IgG as a marker for the injection site (Figure 6C). One hour after injection, the cells were fixed and double immunostained for GST and the co-injected IgG control, respectively. As shown in Figure 6C (a and b), the injected rabbit IgG was detected exclusively within the nucleus, whereas the GST–UL69NES fusion protein had undergone significant relocalization from the nuclear injection site to the cytoplasm. In addition, we also injected unfused GST as an export-deficient control and observed that the native GST protein remained nuclear (data not shown). As outlined in the Introduction, some shuttling cargos have been shown to contain a bidirectional signal that confers both import and export. Therefore, we injected the GST– UL69NES fusion protein into the cytoplasm of HeLa cells in order to identify an intrinsic nuclear import activity (Figure 6C, c and d). No significant nuclear import of the fusion protein could be detected at 1 h after injection. Thus, pUL69 harbors an autonomous NES which does not confer bidirectional activity.
Fig. 6. Confirmation of the autonomous function of the pUL69 nuclear export sequence by microinjection experiments. (A) Schematic representation of the UL69 coding region showing the putative NES fused to the C-terminus of GST to produce GST–UL69NES. (B) Prokaryotic expression and purification of GST–UL69NES. Shown is a Coomassie Blue-stained gel: lane 1, extracts from E.coli cells grown without isopropyl-β-d-thiogalactopyranoside (IPTG); lane 2, extracts of E.coli cells grown in the presence of IPTG; lane 3, purified GST fusion protein. (C) HeLa cells were co-microinjected into the nucleus (a and b) or the cytoplasm (c and d) with GST–UL69NES and rabbit IgG (IgG, injection control). Cells were incubated for 1 h, fixed and analyzed for GST- and rabbit IgG-specific localization by double-label immunofluorescence microscopy. The injected GST fusion protein was visualized by using a GST-specific mouse monoclonal antibody followed by a Cy3-conjugated goat anti-mouse antibody (b and d). The co-microinjected rabbit IgG was detected with a Cy2-conjugated goat anti-rabbit polyclonal antibody (a and c).
The pUL69 NES mediates CRM1-independent nuclear export
In a second series of microinjection experiments, we investigated whether the nuclear export of the GST–UL69NES is, in agreement with results obtained with the full-length protein (shown in Figure 3), not affected by the CRM1 inhibitor LMB. Therefore, we pre-treated HeLa cell cultures with LMB (2.5 ng/ml) 2 h before microinjection of the indicated proteins (Figure 7). A GST fusion protein containing the classical leucine-rich NES of the human T-cell leukemia virus type 1 (HTLV-1) Rex transactivator served as control export cargo in these experiments (Figure 7E and F), since nuclear export of this cargo has been described previously to be LMB sensitive (Elfgang et al., 1999). Figure 7 summarizes the data, showing that LMB completely abolished the CRM1-dependent nuclear export of GST–Rex (Figure 7G and H). In contrast, nuclear export mediated by the pUL69 NES fused to GST was not affected by LMB (Figure 7C and D). However, there were differences in the kinetics of this export: the NES from HTLV-1 Rex could mediate complete export in <1 h, whereas the fusion construct containing the pUL69 NES was only exported to a limited degree of ∼50% in the given time (Figure 7B and F).
Fig. 7. Effect of LMB on nuclear export of GST–UL69NES in microinjection experiments. HeLa cells were co-microinjected into the nucleus with the indicated GST fusion protein and rabbit IgG and analyzed as described in the legend of Figure 6. Cell cultures in (C), (D), (G) and (H) were supplemented 2 h before microinjection and throughout the experiment with LMB at a concentration of 2.5 ng/ml. IgG, rabbit IgG, used as a control for nuclear injection; pUL69 NES, GST fusion protein with the putative NES of pUL69; Rex NES, GST fusion protein with the NES of Rex.
Fine mapping and competition experiments characterize the pUL69 NES as a unique export signal
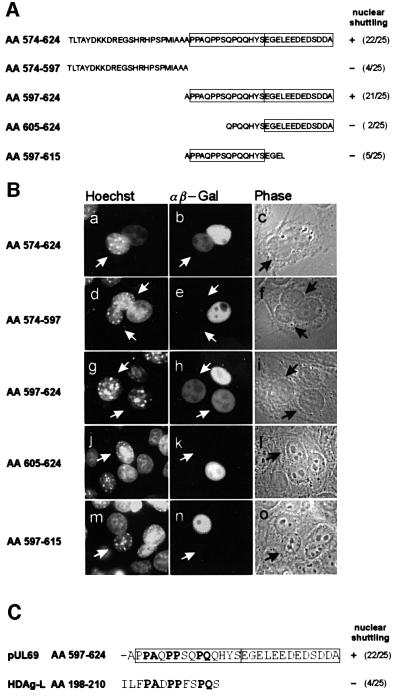
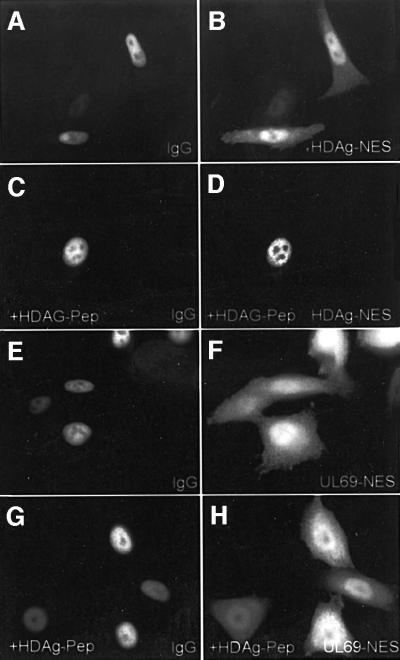
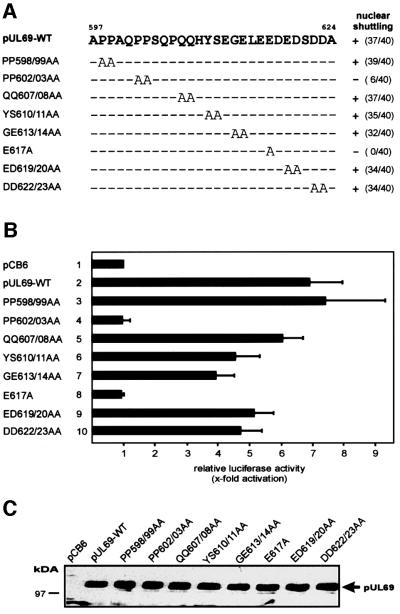
As described in Figure 4, we have localized the pUL69 NES by deletion mutagenesis of the wild-type protein to between amino acids 574 and 624. In order to narrow down the minimal sequence within this domain determining NES activity, we performed further deletion mutagenesis in the context of plasmid CFN-βGal (Figure 8A). The shuttling ability of the resulting fusion proteins was analyzed in heterokaryon assays. The results are summarized in Figure 8B, showing that amino acids 597–624 represent the smallest fragment which is sufficient to maintain NES activity. Removal of eight amino acids from the N-terminus of this fragment as well as removal of nine residues from the C-terminus inactivated the NES (Figure 8B, j–l and m–o). The identified motif corresponds to a sequence which we notified previously when inspecting the C-terminal part of pUL69, as a proline/glutamine cluster followed by an acidic domain. Interestingly, a proline-rich sequence within the large form of the hepatitis delta antigen (HDAg-L) recently has been reported to act as an LMB-insensitive NES (Lee et al., 2001). Since this sequence showed some similarity to the proline-rich part of the pUL69 NES, we compared the activity of both sequences in the context of plasmid CFN-βGal by heterokaryon experiments (Figure 8C). Surprisingly, when quantifying the number of positive heterokaryons which were analyzed routinely 2 h after cell fusion, only the pUL69 NES revealed significant nucleocytoplasmic shuttling, suggesting a lower export activity of the HDAg-L signal compared with the pUL69 NES (Figure 8C). Consistent with this, when GST fusion proteins containing either the HDAg-L NES or the pUL69 NES were microinjected into HeLa cell nuclei, we observed a weaker nuclear export mediated by the HDAg-L NES (see Figure 9A, B, E and F). Signal-mediated nuclear export previously has been shown to be saturable as well as susceptible to inhibition by competition (Meyer et al., 1996; Watanabe et al., 1999). In order to examine whether both signals access identical export receptors, we used a HDAg-L NES peptide conjugated to bovine serum albumin (BSA) as a competitor for the nuclear export of GST–UL69NES or GST–HDAg-L NES. The use of a BSA conjugate was considered advantageous since each carries ∼10 cross-linked peptides, thus allowing the use of a high molar excess in co-microinjection experiments (Meyer et al., 1996). Furthermore, it has been shown previously that a BSA–HDAg-L NES itself exits the nucleus (Lee et al., 2001). As shown in Figure 9C and D, co-micro injection of the peptide competitor together with the GST–HDAg-L NES resulted in a complete block of nuclear export in the majority of co-injected cells. Thus, the HDAg-L NES-mediated transport was clearly saturable. Importantly, however, nuclear export of GST– UL69NES was not affected by co-microinjection of the BSA conjugate (Figure 9G and H), indicating that the HDAg-L NES cannot compete for pUL69 NES-mediated nuclear export.
Fig. 8. Delineation of a minimal pUL69 export sequence. (A) Schematic representation of N- and C-terminal deletions of the pUL69 NES generated to identify a minimal domain that is able to mediate nuclear export. The shuttling activity of each pUL69 NES mutant is indicated by + or –. Numbers in parentheses indicate the number of heterokaryons showing positive staining/the number of heterokaryons examined. (B) pUL69 NES deletion mutants shown in (A) were inserted in plasmid CFNrev-βGal between the SV40 T-Ag NLS and β-Gal, thus replacing the HIV-1 Rev NES. HeLa cells were transfected and analyzed by interspecies heterokaryon analysis as described in the legend of Figure 2. (C) Comparison of nuclear shuttling mediated by the pUL69 NES and the HDAg-L NES by heterokaryon analysis. The HDAg-L NES was inserted in plasmid CFNrev-βGal and analyzed as described in (A) for the pUL69 NES.
Fig. 9. The HDAg-L NES is not able to compete for nuclear export mediated by the pUL69 NES. HeLa cells were co-microinjected into the nucleus with the respective GST fusion proteins, rabbit IgG and either with or without an excess amount of the HDAg-L NES peptide cross-linked to BSA (as indicated in the figure). Cells were incubated at 37°C for 2 h and analyzed by indirect immunofluorescence. The injected GST fusion proteins were visualized by using a GST-specific mouse monoclonal antibody followed by a Cy3-conjugated goat anti-mouse antibody (B, D, F and H). The co-microinjected rabbit IgG was detected with a Cy2-conjugated goat anti-rabbit polyclonal antibody (A, C, E and G). UL69-NES, GST fusion protein with the putative NES of pUL69; HDAg-NES, GST fusion protein with the NES of HDAg-L; HDAg-Pep, HDAg-L NES peptides covalently coupled to BSA.
Mutational analysis of the pUL69 NES reveals a correlation between nucleocytoplasmic shuttling and transactivation
To identify further critical amino acids within the newly defined pUL69 NES, we performed alanine replacement mutagenesis in the context of the wild-type UL69 protein. Nucleocytoplasmic shuttling of the resulting mutants was compared by heterokaryon analysis and quantification of positive heterokaryons. As summarized in Figure 10A, mutants PP602/03AA and E617A lost their shuttling activity. Since it has been shown for ICP27 of HSV-1 that nuclear export is required for its stimulatory effects on gene expression, we tested whether the UL69 mutants were still able to transactivate in co-transfection experiments using a pUL69-responsive luciferase reporter plasmid (Winkler et al., 2000). This revealed a perfect correlation between nucleocytoplasmic shuttling and pUL69-mediated transactivation since mutants PP602/03AA and E617A were no longer able to stimulate gene expression from a luciferase reporter plasmid under control of the IE1/2 enhancer-promoter of HCMV (Figure 10B). This effect was not due to instability of the respective mutant polypeptides since western blotting revealed an equal expression of all tested proteins (Figure 9C). Taken together, these results demonstrate that the nuclear export of pUL69 is mediated via a novel type of canonical NES and indicate that nucleocytoplasmic shuttling is essential for pUL69-mediated activation of gene expression.
Fig. 10. Identification of amino acids important for pUL69 export activity and transactivation. (A) A series of single or double alanine replacement mutations were generated in the context of wild-type pUL69 to identify the amino acids that are critical for pUL69 nuclear export activity. The resulting mutants were then tested in heterokaryon analyses for nuclear shuttling. The shuttling activity of each pUL69 NES mutant is indicated by + or –. Numbers in parentheses indicate the number of heterokaryons showing positive staining/the number of heterokaryons examined. (B) Luciferase analysis after co-transfection of a pUL69-responsive luciferase reporter construct carrying the IE1/2 enhancer-promoter together with expression plasmids for the pUL69 alanine replacement mutants. Lane 1, co-transfection was performed with the empty expression vector pCB6; lane 2, co-transfection was performed with an expression vector for wild-type pUL69; lanes 3–10, co-transfections were performed with expression vectors representing the mutants indicated in (A). Fold activation was calculated relative to the basal activity of the reporter construct after co-transfection with the empty vector pCB6. Each experiment was performed in triplicate and was repeated at least three times. (C) Western blot analysis of U373MG cell extracts after transfection of the pUL69 mutants indicated in (A) using the pUL69-specific monoclonal antibody 69-66.
Discussion
UL69 of HCMV encodes a nucleocytoplasmic shuttling protein
Recently, it has become apparent that some homologous proteins of pUL69 within the subfamilies of α- and γ-herpesviruses, including HSV-1 ICP27, Epstein–Barr virus (EBV) EB2, human herpesvirus type 8 (HHV-8) ORF57 and herpesvirus saimiri (HVS) ORF57, continuously shuttle between the nucleus and the cytoplasm (Soliman et al., 1997; Mears and Rice, 1998; Semmes et al., 1998; Bello et al., 1999; Goodwin et al., 1999). Although the physiological significance of nuclear shuttling is not exactly understood in all cases, most of these proteins have been reported to be implicated in RNA processing and transport. Thus, the HSV-1 shuttling protein ICP27 functions as a post-transcriptional transactivator which activates nuclear export of intronless RNA (Sandri-Goldin, 1998) and down-regulates expression of intron-containing genes (Hardy and Sandri-Goldin, 1994). Nuclear export of ICP27 has been reported to be mediated by a leucine-rich NES at the N-terminus of the protein (Sandri-Goldin, 1998), which could be blocked by the CRM1 inhibitor LMB (Soliman and Silverstein, 2000). As demonstrated in the present study, the HCMV homolog pUL69 differs from ICP27 in this respect, since LMB could not inhibit its nuclear export. Furthermore, pUL69 did not accumulate in the cytoplasm of cells in the presence of actinomycin D (data not shown). Treatment of cells with actinomycin D has been used as an experimental approach to demonstrate nucleocytoplasmic shuttling of several proteins including HSV-1 ICP27 (Phelan and Clements, 1997). However, cytoplasmic accumulation in the presence of actinomycin D is not a general feature of nucleocytoplasmic shuttling proteins, since a series of other polypeptides with nuclear export activity (e.g. ORF57 of HHV-8) show a similar lack of sensitivity (Bello et al., 1999). In these cases, the nuclear export of the respective proteins could be identified unequivocally by the use of heterokaryon analyses. The results of our interspecies heterokaryon experiments demonstrate that pUL69 can traffic efficiently from HCMV-infected human to murine nuclei. Furthermore, pUL69 shuttling activity was detectable upon transient expression in uninfected cells, indicating that its shuttling does not require viral infection or any other viral components. So far, it is unresolved whether pUL69 nucleocytoplasmic shuttling plays a role in transport of viral or cellular RNAs similar to that which has been described for ICP27. However, since we could demonstrate a correlation between pUL69-mediated stimulation of gene expression and nuclear export, the functional importance of pUL69 shuttling is strongly suggested.
Nuclear export of pUL69 is not mediated via the CRM1 pathway
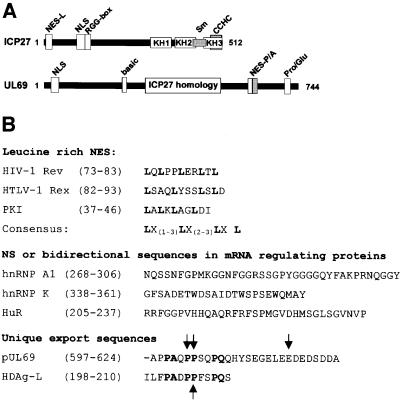
A nuclear shuttling protein requires an export machinery in order to be transported. Proteins of the importin-β superfamily have been described to function as transport receptors mediating many, although not all, transport pathways between nucleus and cytoplasm. Five family members from higher eukaryotes, namely CRM1 (Stade et al., 1997), CAS, exportin t, exportin 4 and importin 13, are known to function in export (Fornerod et al., 1997; Kutay et al., 1997, 1998; Lipowsky et al., 2000). While four of these receptors seem to be very specific for unique export substrates (e.g. CAS mediates the export of importin-α, exportin t that of tRNA, exportin 4 that of eIF5a, and importin 13 that of eIF1A), CRM1 accounts for export of the vast majority of published shuttling proteins containing a leucine-rich NES. LMB has been described specifically to abrogate the export of proteins containing a leucine-rich NES by inactivating the corresponding export receptor CRM1 through covalent binding to a cysteine residue in the central domain of the polypeptide (Wolff et al., 1997; Kudo et al., 1998, 1999). Interestingly, our characterization of the pUL69 export pathway by heterokaryon assays demonstrated that the nuclear export was not sensitive to LMB, while the Rev NES-dependent nuclear export was completely inhibited. Furthermore, we could show that the isolated pUL69-derived NES can direct LMB-insensitive nuclear export of a carrier protein. Recently, it has been proposed that CRM1 may be able to recognize alternative export signals via separate protein domains (Paraskeva et al., 1999; Kunzler et al., 2000). However, since it was shown that LMB modification of CRM1 induces a dramatic alteration of the three-dimensional protein structure (Kudo et al., 1999), we favor a mechanism whereby the pUL69 NES can circumvent the CRM1-dependent pathway by directing nuclear export of pUL69 to a so far uncharacterized pathway. It cannot be excluded entirely that the nuclear export of pUL69 is mediated via one of the alternative export receptors of the importin-β family described above. Alternatively, pUL69 nuclear export might possibly be linked to the pathway of cellular mRNA transport. This idea is supported by three observations: first, a group of mRNA-interacting proteins which are likely to regulate mRNA export such as hnRNP A1 (Nakielny and Dreyfuss, 1999), hnRNP K (Henderson and Eleftheriou, 2000) or TAP (Kang and Cullen, 1999) are also LMB-insensitive shuttling proteins. Secondly, while this paper was in revision, Koffa and colleagues reported that ICP27 of HSV-1 can also be exported in a CRM-1-independent manner utilizing the cellular factors REF and TAP/NXF1 that recently have been identified as mediators of cellular mRNA export (Stutz et al., 2000; Zhou et al., 2000; Koffa et al., 2001; Rodrigues et al., 2001). Thirdly, the pUL69 homolog of the γ-herpesvirus EBV, termed EB2, has been described previously to shuttle between the nucleus and the cytoplasm in an LMB-insensitive manner, thereby contributing to the cytoplasmic accumulation of intron-containing mRNA (Farjot et al., 2000). The NES mediating the CRM-1-independent export of ICP27 and EB2 have not yet been identified. We mapped the pUL69 NES to a unique C-terminal amino acid sequence which is characteristic for the β-herpesviral members of the ICP27 protein family since this domain is not conserved within the respective proteins encoded by α- or γ-herpesviruses (Figure 11A) (Winkler et al., 2000). Whether similar sequences within EB2 and ICP27 mediate nucleocytoplasmic shuttling requires further investigation.
Fig. 11. (A) Schematic representation of ICP27 and pUL69 showing the various protein domains as described by Bryant et al. (2001), Winkler et al. (2000) and this study. NES-L, leucine-rich nuclear export signal; NES-P/A, proline-rich/acidic nuclear export signal; NLS, nuclear localization signal; RGG box, arginine-rich RNA-binding region; KH1-3, hnRNP K homology domains; Sm, Sm protein homology domain; CCHC, cysteine–histidine zinc finger-like domain; basic, cluster of basic amino acids; Pro/Glu, cluster of prolines and glutamines; ICP27 homology, domain of pUL69 with high homology to ICP27. (B) Sequence comparison of various types of NES. Amino acid sequences of NES of the classical leucine-rich type are compared with bidirectional shuttling signals and unique NES. The NES of HIV-1 Rev (Fischer et al., 1995), HTLV-1 Rex (Palmeri and Malim, 1996), protein kinase inhibitor (PKI) (Wen et al., 1995) and a derived consensus NES (Henderson and Eleftheriou, 2000) are listed for representation of leucine-rich NES; hnRNP A1 (Michael et al., 1995), hnRNP K (Michael et al., 1997) and HuR (Fan and Steitz, 1998) represent proteins containing bidirectional signals; the hepatitis D antigen HDAg-L NES (Lee et al., 2001) recently has been described as a unique export sequence. Identical residues in the unique export sequences identified in HDAg-L and pUL69 are shown in bold. The residues that are essential for nuclear export in HDAg-L and pUL69 are indicated by arrows. Numbers refer to the positions of the amino acid sequences within each protein.
The identified NES of pUL69 is distinct from known types of export signals
Since we identified a novel, LMB-insensitive shuttling protein, it was important to address the composition of the corresponding export signal. In general, export cargos either bind their cognate exportins via a short transferable NES or large parts of the cargo molecules are required for stable binding, e.g. importin-α (Kutay et al., 1997) or snurportin 1 (Paraskeva et al., 1999). In our study, deletion analysis and gene fusion experiments demonstrated that 28 amino acids within the C-terminus of pUL69 are sufficient to direct LMB-insensitive nuclear export of reporter proteins. The observed slower export kinetics of a GST–UL69NES protein in comparison with a similar construct driven by a leucine-rich NES might be a consequence of a different nuclear export pathway used by the pUL69 NES; alternatively, this might be explained by a lack of flanking regulatory sequences in the fusion construct containing the minimal NES as described for MAPKAP kinase 2 (Engel et al., 1998). Another possibility for the weaker export observed after microinjection of a GST–UL69NES protein could be a bidirectional transport activity of the identified sequence, since the intracellular localization of a shuttling protein is determined in most cases by the rate of nuclear import and export. This idea is in line with the finding that the LMB-insensitive M9 or KNS shuttling signals of hnRNP A1 and hnRNP K (Michael et al., 1995, 1997) are clearly less active in mediating nuclear export than NES of the HIV-1 Rev type (Henderson and Eleftheriou, 2000). However, we could not detect a significant nuclear uptake of a GST–UL69NES fusion protein injected into the cytoplasm of HeLa cells. Yet, we cannot entirely exclude that the identified signal is a bidirectional signal with enhanced export activity, since the visualization of intrinsic import activity was complicated by the fact that nuclear export could not be inhibited by LMB.
The pUL69 NES represents an NES consisting of an N-terminal cluster of proline and glutamine residues and a C-terminal domain containing mainly acidic amino acids, thereby representing a novel type of short, transferable NES. Known types of NES are represented by (i) the leucine-rich NES which was identified in a series of proteins of diverse function; (ii) the divergent bidirectional signals in the mRNA regulating proteins hnRNP A1, Nup153 (M9 signal), hnRNP K (KNS signal) and HuR (HNS signal); and (iii) a recently described unique NES detected in the large form of HDAg-L (Figure 11B, and references in the figure legend). The pUL69 NES differs functionally from the leucine-rich NES in that it is not transported via CRM1, and from the bidirectional signals in that it is not coupled with NLS activities. Interestingly, the N-terminal part of the pUL69 export sequence shows some similarities to the proline-rich NES of HDAg-L. Alanine-scanning mutagenesis of this signal demonstrated that Pro205 is essential for nuclear export of HDAg-L, and this proline residue is conserved at a comparable position of the pUL69 NES and was also essential for NES function (Figure 11B). However, the HDAg-L NES lacks an acidic domain that was shown to be necessary for efficient nuclear export mediated by the pUL69 NES (Figure 8A and B). An essential function of the acidic domain is also supported by our observation that mutation of Glu617 inactivates nucleocytoplasmic shuttling of wild-type pUL69 (Figure 10A). When we compared nuclear export mediated by the pUL69 NES and the recently described HDAg-L NES, no significant activity was detected for the HDAg-L NES when heterokaryons were examined 2 h after fusion (Figure 8C). However, analysis of heterokaryons at 5 h after fusion as described by Lee et al. (2001) revealed NES activity of this signal (data not shown) and, compared with the pUL69 NES, a weaker export activity of the HDAg-L NES could be confirmed in microinjection experiments (Figure 9A and B). Furthermore, we could show in co-microinjection experiments using a large excess of the HDAg-L NES as a competitor that pUL69 NES-mediated nuclear export was not inhibited whereas HDAg-L NES-mediated transport was completely abolished. This suggests that distinct export receptors are used by both signals, thus supporting our assumption that the pUL69 NES is unique.
In summary, we have identified a novel type of NES mediating efficient CRM1-independent nuclear export within the pUL69 protein of HCMV. The identification of proteins that interact with this signal may help to characterize a novel nuclear export pathway.
Materials and methods
Oligonucleotides
Oligonucleotides were obtained from Eurogentec (Seraing, Belgium) and ARK (Darmstadt, Germany). The following oligonucleotides (5′ to 3′ sequences) were used for cloning and deletion mutagenesis: 5UL69Bam1720 (ATCGGGATCCACGGCACGCTGACAGCCTACGATAAAAAG), 3UL69Eco1869 (GATCGAATTCCCAGCGTCGTCACTGTCTTCGTCCTC), UL69-5′ (TATTAGGATCCCCATGGAGCTGCACTCAC), 69D2 (GGGAATTCTAGATATCGAAGGGACAGTACGCTAT), 5UL69XbaL1720 (AGCTTCTAGACCCGGGCCCGGGACGCTGACAGCCTACGATAAAAAG), 3UL69 AspL1872 (AGCTGGTACCCCCCGGGCCCGGGAGCGTCGTCACTGTCTTCGTCCTC), 5UL69XbaL1788 (AGCTTCTAGACCCGGGCCCGGGGCTCCTCCCGCCCAGCCACCGTCG), 3UL69AspL1790 (AGCTGGTACCCCCCGGGCCCGGGAGCGGCGGCGATCATTGGAGAGGG), 3UL69AspL1851 (AGCTGGTACCCCCCGGGCCCGGGAGCCTCTTCCAGCTCGCCCTCAGA), 5UL69XbaL1813 (AGCTTCTAGACCCGGGCCCGGGCAACCACAGCAGCACTACTCTGAG), HDAgNES-C (GATCCACGGCATACTCTTCCCAGCCGACCCTCCCTTCTCTCCCCAGAGTGGG), HDAgNES-nC (AATTCCCACTCTGGGGAGAGAAGGGAGGGTCGGCTGGGAAGAGTATGCCGTG), HDAgCFN-C (CTAGACCCGGGCCCGGGATACTCTTCCCAGCCGACCCTCCCTTCTCTCCCCAGAGTCCCGGGCCCGGGCG), HDAgCFN-nC (GTACCGCCCGGGCCCGGGACTCTGGGGAGAGAAGGGAGGGTCGGCTGGGAAGAGTATCCCGGGCCCGGGT).
Plasmid construction and in vitro mutagenesis
The eukaryotic expression plasmid pHM160 for wild-type pUL69, plasmid UL69-D478–573 (alternatively termed pHM937), expressing an internal UL69 deletion mutant, and the luciferase reporter construct pHM284, expressing luciferase under the control of the IE1/2 enhancer-promoter, were described previously (Winkler et al., 1994, 2000; Gebert et al., 1997). Plasmids pCFN-βGal and pCFNrev-βGal used as controls in interspecies heterokaryon assays were obtained from M.Dobbelstein (Roth and Dobbelstein, 1997). The N-terminal deletion mutants UL69 37–744 and UL69 92–744 were constructed by cloning the BamHI–HindIII fragments of pHM506 and pHM507 (Winkler et al., 2000) into the BglII–HindIII-digested vector pCB6 (Brewer, 1994). To construct the C-terminal deletion mutant UL69 1–521 (pHM475), the sequence of UL69 encoding amino acids 1–521 was amplified by PCR using primers UL69–5′ and 69D2, and pHM160 as a template. The amplified fragment was cleaved with MluI–EcoRI and inserted into the MluI–EcoRI-digested vector pHM160. UL69 1–624 (pHM1139) was generated by cloning of the BamHI–NheI fragment of pHM300 containing the UL69 reading frame (Winkler et al., 2000) into vector pcDNA3 (Invitrogen Corp., San Diego, CA). The prokaryotic vector GST–UL69NES (pHM1441) expressing amino acids 574–624 of pUL69 fused to GST was constructed by PCR amplification using oligonucleotides 5UL69Bam1720 and 3UL69Eco1869, followed by cleaveage with BamHI–EcoRI and ligation with the pGEX-3X vector (Pharmacia Biotech, Freiburg, Germany). Fusion mutants combining fragments of the SV40 T-Ag-NLS, pUL69 NES and βGal for the heterokaryon analysis were also constructed by PCR using the oligonucleotides 5UL69 XbaL1720/3UL69AspL1872 (CFNUL69-βGal), 5UL69XbaL1720/3UL69AspL1790 (AA574–597), 5UL69XbaL1788/ 3UL69AspL1872 (AA597–624), 5UL69XbaL1813/3UL69AspL1872 (AA605–624) and 5UL69XbaL1788/3UL69AspL1851 (AA597–615), respectively. The primers contained XbaI–Asp718 restriction sites followed by short spacer sequences coding for Gly/Pro/Gly/Pro. The amplified fragments were cleaved with XbaI–Asp718 followed by in-frame insertion into the NLS-βGal-expressing vector pCFNrev-βGal, thereby replacing the Rev NES (the resulting plasmids were termed pHM1473 to pHM1475 and pHM1542, pHM1543, respectively). The prokaryotic vector GST– HDAg-L NES (pHM1697) was constructed by inserting complementary oligonucleotides coding for amino acids 198–210 of the HDV HDAg-L into the pGEX-3X vector. The fusion mutant combining the SV40 T-Ag NLS, HDAg-L NES and βGal for heterokaryon analyses was also constructed by cloning complementary oligonucleotides coding for the HDAg-L NES into pCFNrev-βGal, thereby replacing the Rev NES (pHM1698). Site-directed mutagenesis within pHM160 was performed using a QuickChange site-directed mutagenesis kit as instructed by the manufacturer (Stratagene, Heidelberg, Germany) using complementary oligonucleotide primers that introduced the alanine replacement mutations indicated in Figure 9. The resulting plasmids were termed PP598/99AA (pHM1688), PP602/03AA (pHM1567), QQ607/08AA (pHM1689), YS610/11AA (pHM1690), GE613/14AA (pHM1655), E617A (pHM1577), ED619/20AA (pHM1610) and DD622/23AA (pHM1651). The DNA sequence of each plasmid construct was confirmed by automated sequence analysis (ABI, Weiterstadt, Germany).
Infection and transfection of cells, luciferase reporter assays and immunoblotting
Primary HFFs, HeLa, NIH 3T3 (murine fibroblast) and U373MG cells were cultured as described previously (Winkler et al., 1994). HFFs were infected with HCMV (strain AD169) at a m.o.i. of 1–2 p.f.u. per cell for 70 h. HeLa cells were transfected via the calcium phosphate co-precipitation procedure using N,N-bis(2-hydroxyethyl)-2-aminoethansulfonic acid as described earlier (Winkler et al., 1994). Heterokaryon assays were performed 36 h post-transfection. DEAE–dextran transfection of U373MG cells and luciferase reporter assays were carried out exactly as described previously (Winkler et al., 2000). Luciferase activity was determined using a luminometer (Berthold, Freiburg, Germany); each transfection was performed in triplicate and was repeated at least three times. For western blot analysis, transfected U373MG cells were lysed in SDS–Laemmli buffer and boiled at 94°C for 10 min. Samples were electrophoresed by SDS–PAGE on 15% polyacrylamide gels, and the proteins were transferred onto nitrocellulose membranes (Schleicher and Schuell, Dassel, Germany). Western blotting and chemiluminescence detection were performed according to the manufacturer’s protocol (ECL Western detection Kit; Amersham Pharmacia Europe, Freiburg, Germany). The monoclonal antibody 69-66 (Winkler et al., 2000) was used for detection of pUL69 wild-type and mutants. The anti-mouse horseradish peroxidase-conjugated secondary antibody was obtained from Dianova (Hamburg, Germany)
Heterokaryon assays and immunofluorescence analysis
Nucleocytoplasmic shuttling was detected by using a heterokaryon assay, as initially described by Pinol-Roma and Dreyfuss (1992). Infected HFFs or transfected HeLa cells (600 000) were seeded on glass coverslips in 6-well dishes together with an equal number of NIH 3T3 cells. Co-cultures of infected HFFs and NIH 3T3 cells were allowed to grow for 3 h at 37°C; co-cultures of transfected HeLa and NIH 3T3 cells were incubated overnight. Protein synthesis was blocked with 50 µg/ml cycloheximide 30 min prior to the fusion and throughout the experiment. If inhibition of CRM1-dependent protein export was required, cells were treated additionally with 2.5 ng/ml LMB (kindly provided by Dr Minoru Yoshida, Tokyo, Japan) 3 h before cell fusion. Subsequently, cells were washed in phosphate-buffered saline (PBS), and heterokaryons were formed by incubating the coverslips for 2 min in 50% polyethylene glycol 8000 in Dulbecco’s modified Eagle’s medium (without fetal calf serum). Following cell fusion, coverslips were washed extensively in PBS and returned to fresh medium containing 50 µg/ml cycloheximide and 2.5 ng/ml LMB when needed. After exactly 2 h, cells were fixed with 4% paraformaldehyde and indirect immunofluorescence analysis was performed according to a standard protocol as described previously (Hofmann et al., 2000). After the secondary antibody incubation, 5 µg/ml of Hoechst 33258 (Sigma, Taufkirchen, Germany) were added for 5 min and cells were mounted using Vectashield mounting medium (Vector Laboratories, Burlingame, CA). The polyclonal antiserum against pUL69 and the monoclonal antibody against the IE1 protein were described previously (Andreoni et al., 1989; Winkler et al., 1994). The monoclonal anti-β-galactosidase antibody, anti-mouse and anti-rabbit fluorescein isothiocyanate (FITC)- or tetramethyl rhodamine isothiocyanate (TRITC)-conjugated secondary antibodies were obtained from Roche (Mannheim, Germany) and Dianova (Hamburg, Germany), respectively. Images were analyzed using a Zeiss Axioplan-2 microscope and recorded with a cooled Spot color digital camera (Diagnostic Instruments, Burroughs, MI). The Meta-Imaging series and Adobe Photoshop package (Universal Imaging Corp., Brandywine, PA; Adobe Systems Inc.) were used for processing. At least 25 heterokaryons were analyzed for each experiment, and nuclear shuttling was only scored positive when a minimum of 80% of the heterokaryons showed a positive staining of the investigated protein.
Purification of GST–NES fusion proteins and peptides
The purification of a GST–Rex fusion protein was described in detail elsewhere (Elfgang et al., 1999). The putative NES of pUL69 and the HDAg-L NES were expressed in fusion with GST using E.coli strain BL21. The fusion proteins were purified from crude lysates by affinity chromatography with glutathione–Sepharose 4B according to the specifications of the manufacturer (Pharmacia Biotech, Germany). Eluted proteins were analyzed by SDS–PAGE and Coomassie Blue staining. Fractions were pooled, concentrated by ultrafiltration with a Nanosep filter (Pall-Filtron, Northborough, MA) and stored at –70°C. Peptides representing the HDAg-L NES (CILFPADPPFSPQS) covalently coupled to BSA at a ratio of 1:10 were synthesized by Eurogentec (Seraing, Belgium) and used for competition in microinjection experiments (HDAg-Pep).
Microinjection
HeLa cells were co-microinjected into the nucleus or the cytoplasm with either GST–UL69NES (1.5 mg/ml), GST–Rex (1.0 mg/ml) or GST–HDAg-L NES (1.5 mg/ml) in combination with rabbit IgG (1.0 mg/ml) (injection control) by using a CompiC INJECT computer-assisted injection system (Cellbiology Trading, Hamburg, Germany). For competition experiments, HDAg-L NES–BSA conjugates (25 mg/ml) were mixed with the respective GST fusion proteins and used for co-microinjection as indicated. Cells were fixed at given times post-injection with paraformaldehyde, and the injected proteins were visualized by indirect immunofluorescence analysis. The primary mouse anti-GST monoclonal antibody (Serotec, Oxford, UK) was used at a 1:50 dilution. The secondary antibodies, Cy2-conjugated goat anti-rabbit IgG and Cy3-conjugated goat anti-mouse IgG, were used at a 1:200 dilution and were obtained from Dianova (Hamburg, Germany).
Acknowledgments
Acknowledgements
We thank Dr Minoru Yoshida (Tokyo, Japan) for his generous gift of LMB and Dr M.Dobbelstein for providing plasmids pCFN-βGal and pCFNrev-βGal. We are grateful to Dr Manfred Marschall for participation in the generation of the manuscript. This work was supported by the DFG (SFB473) and the IZKF Erlangen.
References
- Andreoni M., Faircloth,M., Vugler,L. and Britt,W.J. (1989) A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. J. Virol. Methods, 23, 157–167. [DOI] [PubMed] [Google Scholar]
- Bello L.J., Davison,A.J., Glenn,M.A., Whitehouse,A., Rethmeier,N., Schulz,T.F. and Barklie,C.J. (1999) The human herpesvirus-8 ORF 57 gene and its properties. J. Gen. Virol., 80, 3207–3215. [DOI] [PubMed] [Google Scholar]
- Brewer C.B. (1994) Cytomegalovirus plasmid vectors for permanent lines of polarized epithelial cells. Methods Cell Biol., 43, 233–245. [DOI] [PubMed] [Google Scholar]
- Bryant H.E., Wadd,S.E., Lamond,A.I., Silverstein,S.J. and Clements,J.B. (2001) Herpes simplex virus IE63 (ICP27) protein interacts with spliceosome-associated protein 145 and inhibits splicing prior to the first catalytic step. J. Virol., 75, 4376–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres J.F., Screaton,G.R. and Krainer,A.R. (1998) A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev., 12, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelstein M., Roth,J., Kimberly,W.T., Levine,A.J. and Shenk,T. (1997) Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J., 16, 4276–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfgang C., Rosorius,O., Hofer,L., Jaksche,H., Hauber,J. and Bevec,D. (1999) Evidence for specific nucleocytoplasmic transport pathways used by leucine-rich nuclear export signals. Proc. Natl Acad. Sci. USA, 96, 6229–6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel K., Kotlyarov,A. and Gaestel,M. (1998) Leptomycin B-sensitive nuclear export of MAPKAP kinase 2 is regulated by phosphorylation. EMBO J., 17, 3363–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X.C. and Steitz,J.A. (1998) HNS, a nuclear–cytoplasmic shuttling sequence in HuR. Proc. Natl Acad. Sci. USA, 95, 15293–15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farjot G., Buisson,M., Duc,D.M., Gazzolo,L., Sergeant,A. and Mikaelian,I. (2000) Epstein–Barr virus EB2 protein exports unspliced RNA via a crm-1-independent pathway. J. Virol., 74, 6068–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U., Huber,J., Boelens,W.C., Mattaj,I.W. and Luhrmann,R. (1995) The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell, 82, 475–483. [DOI] [PubMed] [Google Scholar]
- Fornerod M., Ohno,M., Yoshida,M. and Mattaj,I.W. (1997) CRM1 is an export receptor for leucine-rich nuclear export signals. Cell, 90, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Fukuda M., Gotoh,I., Gotoh,Y. and Nishida,E. (1996) Cytoplasmic localization of mitogen-activated protein kinase kinase directed by its NH2-terminal, leucine-rich short amino acid sequence, which acts as a nuclear export signal. J. Biol. Chem., 271, 20024–20028. [DOI] [PubMed] [Google Scholar]
- Fukuda M., Asano,S., Nakamura,T., Adachi,M., Yoshida,M., Yanagida,M. and Nishida,E. (1997) CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature, 390, 308–311. [DOI] [PubMed] [Google Scholar]
- Gebert S., Schmolke,S., Sorg,G., Floss,S., Plachter,B. and Stamminger,T. (1997) The UL84 protein of human cytomegalovirus acts as a transdominant inhibitor of immediate-early-mediated transactivation that is able to prevent viral replication. J. Virol., 71, 7048–7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin D.J., Hall,K.T., Stevenson,A.J., Markham,A.F. and Whitehouse,A. (1999) The open reading frame 57 gene product of herpesvirus saimiri shuttles between the nucleus and cytoplasm and is involved in viral RNA nuclear export. J. Virol., 73, 10519–10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D. and Kutay,U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell. Dev. Biol., 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Hardy W.R. and Sandri-Goldin,R.M. (1994) Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J. Virol., 68, 7790–7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M.L., Blankenship,C. and Shenk,T. (2000) Human cytomegalovirus UL69 protein is required for efficient accumulation of infected cells in the G1 phase of the cell cycle. Proc. Natl Acad. Sci. USA, 97, 2692–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson B.R. and Eleftheriou,A. (2000) A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp. Cell Res., 256, 213–224. [DOI] [PubMed] [Google Scholar]
- Hofmann H., Floss,S. and Stamminger,T. (2000) Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J. Virol., 74, 2510–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y. and Cullen,B.R. (1999) The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev., 13, 1126–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koffa M.D., Clements,J.B., Izaurralde,E., Wadd,S., Wilson,S.A., Mattaj,I.W. and Kuersten,S. (2001) Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J., 20, 5769–5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N., Wolff,B., Sekimoto,T., Schreiner,E.P., Yoneda,Y., Yanagida,M., Horinouchi,S. and Yoshida,M. (1998) Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res., 242, 540–547. [DOI] [PubMed] [Google Scholar]
- Kudo N., Matsumori,N., Taoka,H., Fujiwara,D., Schreiner,E.P., Wolff,B., Yoshida,M. and Horinouchi,S. (1999) Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl Acad. Sci. USA, 96, 9112–9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunzler M., Gerstberger,T., Stutz,F., Bischoff,F.R. and Hurt,E. (2000) Yeast Ran-binding protein 1 (Yrb1) shuttles between the nucleus and cytoplasm and is exported from the nucleus via a CRM1 (XPO1)-dependent pathway. Mol. Cell. Biol., 20, 4295–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U., Bischoff,F.R., Kostka,S., Kraft,R. and Gorlich,D. (1997) Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell, 90, 1061–1071. [DOI] [PubMed] [Google Scholar]
- Kutay U., Lipowsky,G., Izaurralde,E., Bischoff,F.R., Schwarzmaier,P., Hartmann,E. and Gorlich,D. (1998) Identification of a tRNA-specific nuclear export receptor. Mol. Cell, 1, 359–369. [DOI] [PubMed] [Google Scholar]
- Lee C.H., Chang,S.C., Wu,C.H. and Chang,M.F. (2001) A novel chromosome region maintenance 1-independent nuclear export signal of the large form of hepatitis delta antigen that is required for the viral assembly. J. Biol. Chem., 276, 8142–8148. [DOI] [PubMed] [Google Scholar]
- Lipowsky G., Bischoff,F.R., Schwarzmaier,P., Kraft,R., Kostka,S., Hartmann,E., Kutay,U. and Gorlich,D. (2000) Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J., 19, 4362–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M. and Shenk,T. (1999) Human cytomegalovirus UL69 protein induces cells to accumulate in G1 phase of the cell cycle. J. Virol., 73, 676–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears W.E. and Rice,S.A. (1998) The herpes simplex virus immediate-early protein ICP27 shuttles between nucleus and cytoplasm. Virology, 242, 128–137. [DOI] [PubMed] [Google Scholar]
- Meyer B.E. and Malim,M.H. (1994) The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev., 8, 1538–1547. [DOI] [PubMed] [Google Scholar]
- Meyer B.E., Meinkoth,J.L. and Malim,M.H. (1996) Nuclear transport of human immunodeficiency virus type 1, visna virus, and equine infectious anemia virus Rev proteins: identification of a family of transferable nuclear export signals. J. Virol., 70, 2350–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael W.M. (2000) Nucleocytoplasmic shuttling signals: two for the price of one. Trends Cell Biol., 10, 46–50. [DOI] [PubMed] [Google Scholar]
- Michael W.M., Choi,M. and Dreyfuss,G. (1995) A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell, 83, 415–422. [DOI] [PubMed] [Google Scholar]
- Michael W.M., Eder,P.S. and Dreyfuss,G. (1997) The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J., 16, 3587–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S. and Dreyfuss,G. (1999) Transport of proteins and RNAs in and out of the nucleus. Cell, 99, 677–690. [DOI] [PubMed] [Google Scholar]
- Palmeri D. and Malim,M.H. (1996) The human T-cell leukemia virus type 1 posttranscriptional trans-activator Rex contains a nuclear export signal. J. Virol., 70, 6442–6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskeva E., Izaurralde,E., Bischoff,F.R., Huber,J., Kutay,U., Hartmann,E., Luhrmann,R. and Gorlich,D. (1999) CRM1-mediated recycling of snurportin 1 to the cytoplasm. J. Cell Biol., 145, 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan A. and Clements,J.B. (1997) Herpes simplex virus type 1 immediate early protein IE63 shuttles between nuclear compartments and the cytoplasm. J. Gen. Virol., 78, 3327–3331. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S. and Dreyfuss,G. (1992) Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature, 355, 730–732. [DOI] [PubMed] [Google Scholar]
- Rodrigues J.P., Rode,M., Gatfield,D., Blencowe,B., Carmo-Fonseca,M. and Izaurralde,E. (2001) REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl Acad. Sci. USA, 98, 1030–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J. and Dobbelstein,M. (1997) Export of hepatitis B virus RNA on a Rev-like pathway: inhibition by the regenerating liver inhibitory factor IκBα. J. Virol., 71, 8933–8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri-Goldin R.M. (1998) ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev., 12, 868–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri-Goldin R.M. and Mendoza,G.E. (1992) A herpesvirus regulatory protein appears to act post-transcriptionally by affecting mRNA processing. Genes Dev., 6, 848–863. [DOI] [PubMed] [Google Scholar]
- Semmes O.J., Chen,L., Sarisky,R.T., Gao,Z., Zhong,L. and Hayward,S.D. (1998) Mta has properties of an RNA export protein and increases cytoplasmic accumulation of Epstein–Barr virus replication gene mRNA. J. Virol., 72, 9526–9534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman T.M. and Silverstein,S.J. (2000) Herpesvirus mRNAs are sorted for export via Crm1-dependent and -independent pathways. J. Virol., 74, 2814–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman T.M., Sandri-Goldin,R.M. and Silverstein,S.J. (1997) Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J. Virol., 71, 9188–9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K., Ford,C.S., Guthrie,C. and Weis,K. (1997) Exportin 1 (Crm1p) is an essential nuclear export factor. Cell, 90, 1041–1050. [DOI] [PubMed] [Google Scholar]
- Stutz F., Bachi,A., Doerks,T., Braun,I.C., Seraphin,B., Wilm,M., Bork,P. and Izaurralde,E. (2000) REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA, 6, 638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Fukuda,M., Yoshida,M., Yanagida,M. and Nishida,E. (1999) Involvement of CRM1, a nuclear export receptor, in mRNA export in mammalian cells and fission yeast. Genes Cells, 4, 291–297. [DOI] [PubMed] [Google Scholar]
- Wen W., Meinkoth,J.L., Tsien,R.Y. and Taylor,S.S. (1995) Identification of a signal for rapid export of proteins from the nucleus. Cell, 82, 463–473. [DOI] [PubMed] [Google Scholar]
- Winkler M. and Stamminger,T. (1996) A specific subform of the human cytomegalovirus transactivator protein pUL69 is contained within the tegument of virus particles. J. Virol., 70, 8984–8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler M., Rice,S.A. and Stamminger,T. (1994) UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J. Virol., 68, 3943–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler M., aus Dem,S.T. and Stamminger,T. (2000) Functional interaction between pleiotropic transactivator pUL69 of human cytomegalovirus and the human homolog of yeast chromatin regulatory protein SPT6. J. Virol., 74, 8053–8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff B., Sanglier,J.J. and Wang,Y. (1997) Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol., 4, 139–147. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Luo,M.J., Straesser,K., Katahira,J., Hurt,E. and Reed,R. (2000) The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature, 407, 401–405. [DOI] [PubMed] [Google Scholar]