Abstract
Of the three critical enhancer elements that mediate β-cell-specific and glucose-responsive expression of the insulin gene, only the identity of the transcription factor binding to the RIPE3b element (RIPE3b1) has remained elusive. Using a biochemical purification approach, we have identified the RIPE3b1 factor as a mammalian homologue of avian MafA/L-Maf (mMafA). The avian MafA is a cell-type determination factor that expressed ectopically can trigger lens differentiation program, but no mammalian homologue of avian MafA has previously been identified. Here, we report cloning of the human mafA (hMafA) and demonstrate that it can specifically bind the insulin enhancer element RIPE3b and activate insulin-gene expression. In addition, mMafA has a very restrictive cellular distribution and is selectively expressed in pancreatic β but not in α cells. We suggest that mMafA has an essential role in the function and differentiation of β-cells and thus may be associated with the pathophysiological origins of diabetes.
Pancreatic β-cells synthesize and secrete insulin, which is essential for the maintenance of normal metabolism. Thus, a reduction in the functional mass of pancreatic β-cells results in diabetes. β-cell-specific expression of the insulin gene is regulated by transcription factors binding to three conserved insulin enhancer elements [E1 (−100 to −91 bp), A3 (−201 to −196 bp) and RIPE3b (−126 to −101 bp)] (1–5). Two of the three factors binding to these elements, PDX-1 and BETA2, have been identified and found to have profound roles in regulating pancreatic development and the differentiation of β-cells (6–13). Thus, transcription factors regulating insulin gene expression are key mediators of development, differentiation, and function of β-cells. Hence, identification and characterization of insulin gene transcription factors is critical for understanding the pathophysiology of diabetes.
Pancreatic β-cell-specific insulin gene expression results from the expression of a unique combination of PDX-1, BETA2, and RIPE3b1 factors in this cell type. The transcription factor PDX-1 is expressed in pancreatic β-cells and has a heterogeneous expression pattern in other pancreatic cell types and in the duodenum (6–10, 14). BETA2 is expressed in all pancreatic endocrine cell types, some intestinal endocrine cells, and the brain (11–13). The cellular distribution of RIPE3b-binding activity has been characterized by electrophoretic mobility-shift assay (EMSA) with nuclear extracts from both insulin-producing and non-insulin-producing cell lines (2, 3, 15, 16). Two specific RIPE3b-binding complexes have been identified:(i) RIPE3b1 detected only in pancreatic β-cell lines, and (ii) the RIPE3b2-binding complex detected in all cell lines examined. The binding of these transcription factors to the A3, E1, and RIPE3b elements also regulates glucose-mediated alterations in insulin gene expression (3, 17–19). Our earlier studies demonstrated that the binding activity of the RIPE3b1 activator was induced in response to acute changes in glucose concentration (3), while under glucotoxic conditions, the RIPE3b1 and PDX-1-binding activities were inhibited (20, 21). These observations suggest that the RIPE3b1 transcription factor is critical for maintaining normal β-cell functions in response to alterations in glucose levels.
Here we report the identification and cloning of the transcription factor RIPE3b1, which binds to the third and remaining critical insulin enhancer element, as the mammalian homologue of an important avian regulator of cellular differentiation, MafA/L-Maf (22, 23). Expression of avian MafA is highly restricted to lens cells, and it regulates expression of multiple lens-specific genes (22, 23). We demonstrate that the mammalian MafA (mMafA) also has a very restrictive cellular distribution and, furthermore, is capable of binding the RIPE3b element and activating insulin gene expression. We expect that mMafA will have an equally important role in regulating development, differentiation, and function of pancreatic β-cells based on such roles for other β-cell-specific transcription factors and the role of avian MafA in lens differentiation.
Materials and Methods
EMSAs.
Wild-type and the mutant rat insulin II oligonucleotides used in the study have been described before (24). The RIPE3b1 and RIPE3b2 factors can bind rat insulin II oligonucleotides RIPE3b, −139 to −101 bp, −114.113m, −125.124m, but not the oligonucleotides −139 to −101 bp, −110.109m, and −125.124m (24). Other oligonucleotides used in the EMSA are wild type (5′-ACGTAGCATTCCAGCTGCTGACGGTGCAGCCTCTCCCCCGAG-3′), or mutant αA-crystallin (5′-ACGTAGCATTCCAGCTGCTGCCGGTGCAGCCTCTCCCCCGAG-3′) (25). Competition experiments were performed by simultaneous addition of radiolabeled probe and 50-fold excess unlabeled competitors to the binding reaction. For antibody supershift experiments, antibodies were preincubated with the nuclear extract for 20 min at room temperature, followed by another 20-min incubation in the presence of the radiolabeled probe and analysis by EMSA. Joel Habener (Massachusetts General Hospital, Boston) generously provided the anti-IDX-1 (PDX-1) antibody, while anti-cMaf and anti-nucleolin antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA).
Purification of the RIPE3b1.
For large-scale purification, 20 ml of nuclear extract (26) (about 90 mg of total protein containing at least 100 pmol of the RIPE3b1 factor) from hamster insulinoma cell line HIT T-15 was loaded onto HiTrap heparin column (Amersham Pharmacia-Pharmacia Biotech), and fractions were eluted with binding buffer containing increasing amounts of salt. Fractions containing the RIPE3b1 binding activity were pooled into one heparin-pooled (HP) fraction. For affinity purification, annealed 5′-biotinylated (top strand) RIPE3b (−126 to −101 bp) dimer oligonucleotide (5′-TGA CTG GAA ACT GCA GCT TCA GCC CCT CTG GAA ACT GCA GCT TCA GCC CCA C-3′) was incubated for 1 h with the HP fraction followed by addition of 3 ml of streptavidin-agarose (Pierce). The mixture was packed into a column and washed with RIPE3b1-buffer containing increasing concentration of salt. Proteins present in the affinity-purified fractions were concentrated by using the Orgosol protein concentration kit (GenoTech, St. Louis). The concentrated fraction was run on an SDS/PAGE 10% gel, and protein bands were detected using a silver stain kit (Bio-Rad).
To identify that the protein band contained the RIPE3b1 factor, SDS/PAGE fractionation was performed (27). An unheated purified fraction was run in duplicate on an SDS/PAGE 10% gel. The gel was allowed to renature for 30 min and was then transferred onto a poly(vinylidene difluoride) (PVDF) membrane. The membrane was cut into two, and one half was stained with colloidal gold stain (Bio-Rad); the protein bands were cut from the other half and proteins were eluted in a buffer containing Triton X-100.
Concentrated purified protein fractions were run on an SDS/PAGE gel, and the 47-kDa protein band (approximately 45 pmol) was cut out. In-gel reduction, alkylation, and trypsin digestion were performed. Approximately 10% of the digested sample was used to determine the amino acid sequence of the tryptic peptides by microcapillary reverse-phase HPLC nano-electrospray tandem mass spectrometry (μLC/MS/MS) with a Finnigan LCQ quadrupole ion trap mass spectrometer at Harvard Microchemistry Facility.
Cloning of the Human MafA.
Degenerate oligonucleotide primers corresponding to peptide sequences SDDQLV (WSIGAYGAYCARYTIGTIWS) and LYKEKY (TCRTAYTTYTCYTTRTAIAR) of the 47-kDa protein were used in an reverse-transcription PCR with RNAs from insulin-producing (HIT T-15 and MIN-6) and glucagon-producing (αTC 1.6) cell lines. PCR product from HIT T-15 RNA was isolated, cloned, sequenced, and used to design specific internal oligonucleotide primers (5′ primer GATGTCGGTGCGGGAGCTGAACC and 3′ primer CCCACCTCCAGCTTCAGCTGCTC); cyclophilin primers have been described (28). To clone the complete coding region of human MafA (hMafA), genomic DNA (CLONTECH) was used as a template with the 5′ primer (ATGGCCGCGGAGCTGGCGATG) and 3′ primer (CAGGAAGAAGTCGGCCGTGCCCT) in a PCR.
Northern Blot Analysis.
Approximately 10 μg of total RNA from mouse islets, insulin-producing (MIN6 and HIT-T15) and non-insulin-producing αTC1.6 cell lines were used for Northern blot analysis. N-terminal fragment (base pairs 1–473) of human MafA was labeled using Strip-EZ kit (Ambion, Austin, TX) for Northern blot analysis.
Determination of Insulin Gene Expression.
HeLa cells were transfected with 2 μg of luciferase reporter plasmid (−238 WT LUC or −122.121m LUC) (24), 0.5 μg of pcDNA3.1 vector or full-length hMafA or an N-terminal deletion derivative of hMafA lacking the first 138 aa (ΔN-hMafA) cloned into pcDNA3.1, and 0.5 μg of pSV-β-galactosidase plasmid (Promega). Luciferase and β-galactosidase activities were determined 48 h after transfection. For immunostaining, HeLa cells were transfected with 1 μg of rat insulin I:GFP reporter (−410 rINSI:GFP; GFP indicates green fluorescent protein) and 1.5 μg of hMafA, ΔN-hMafA or pcDNA3.1 expression plasmids. Forty-eight hours after transfection, HeLa cells were fixed with 10% buffered formalin, and fusion proteins were detected by using αHSV.tag monoclonal antibody and a secondary Texas red-conjugated anti-mouse antibody.
Results and Discussion
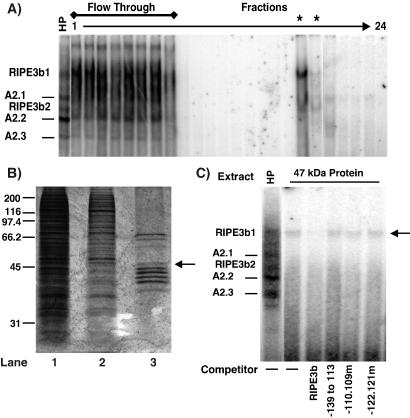
Attempts to clone the RIPE3b1 factor by genetic approaches have been unsuccessful but resulted in the isolation of Rip-1 (15). The Rip-1 factor is a likely constituent of the ubiquitous RIPE3b2 complex, with a negligible effect on insulin gene expression (15). Thus, a two-step purification approach was established to clone the β-cell-specific RIPE3b1 activator from hamster insulinoma cell line HIT T-15. As a first step, HIT T-15 nuclear extract was partially purified by using a HiTrap heparin column (Amersham Pharmacia-Pharmacia Biotech). Purified fractions containing RIPE3b1-binding activity were pooled into one HP fraction. This HP fraction contained about 85% of the recovered RIPE3b1 factor with a 2.5-fold purification (data not shown). The RIPE3b-binding factor was then affinity purified from the HP fraction by using a 5′-biotinylated RIPE3b oligonucleotide and streptavidin-agarose. The purified fractions were analyzed for binding to a rat insulin II −139 to −101 bp probe by EMSA [Fig. 1A (24)]. The −139 to −101 bp probe contains the RIPE3b and overlapping A2 elements. Five distinct factors (RIPE3b1, RIPE3b2, and three A2-specific complexes) can bind the −139 to −101 bp probe. Thus the −139 to −101 bp probe serves as a good indicator of the purification process. The RIPE3b1-binding activity was eluted in two fractions and was the predominant DNA-binding activity in these fractions. We were able to accomplish about 95-fold purification of RIPE3b1-binding activity as compared with the HP fraction and over 200-fold overall. The purified fraction was next analyzed on an SDS/PAGE 10% gel along with the starting HIT T-15 nuclear extract and HP fraction. Results showed a significant purification after the affinity column, although the purified fraction still contained at least 10 bands as detected by silver staining (Fig. 1B). The regions corresponding to the individual protein bands were cut, the proteins were then eluted and analyzed for the RIPE3b1-binding activity. One of the eluted proteins (≈47-kDa) formed a RIPE3b-binding complex with identical electrophoretic mobility and competition profile as the RIPE3b1 complex [Fig. 1C (24)], demonstrating that the 47-kDa protein is highly enriched in the RIPE3b1 factor.
Figure 1.
A 47-kDa protein constitutes the RIPE3b1 factor. (A) Small aliquots from affinity purification fractions were incubated with 32P-radiolabeled rat insulin II −139 to −101 bp probe and binding activity was analyzed by EMSA. Asterisks denote fractions containing the RIPE3b1 factor and HP denotes partially purified heparin-pooled fraction used as the starting material for the affinity purification. Positions of RIPE3b- and A2-binding complexes are shown. (B) To determine the degree of purification, the concentrated and pooled purified fraction (0.4 μg, lane 3) was run on an SDS/PAGE 10% gel along with the starting HIT T-15 nuclear extract (4.0 μg, lane1) and HP fraction (0.8 μg, lane 2) and Silver stained. The positions and molecular masses (×103 kDa) of protein standards are indicated; the arrow marks the position of the 47-kDa band. (C) The 47-kDa protein band was eluted by using SDS/PAGE fractionation; the eluted protein and HP fraction were analyzed for binding to the −139 to −101 bp probe. Binding reactions were performed in the absence (−) or presence of 50-fold excess of the indicated competitors. Positions of the various binding complexes are shown.
Amino acid sequences of the 47-kDa protein were determined by using HPLC tandem mass spectrometry at Harvard Microchemistry Facility. Results from this analysis identified the chicken transcription factor L-Maf (GenBank accession no. AF034570) as sharing identity with five tryptic peptides of the 47-kDa protein. L-Maf was originally cloned from a chicken embryonic lens cDNA library (22) but has also been cloned from quail and termed MafA (23). The transcription factor MafA/L-Maf regulates lens cell-specific expression of crystallin genes and triggers the lens differentiation program. Thus far, no mammalian homologue of MafA/L-Maf has been identified. Maf proteins are subdivided into two classes, “large” (236–370 aa; c-Maf, MafB, NRL, and MafA/L-Maf), and “small” (149–162 aa; MafF, MafG, and MafK) (22, 25, 29, 30). All large Maf proteins have an N-terminal serine/proline/threonine-rich acidic activation domain that is absent in the “small-Maf” proteins (29, 31). However, both large and small Maf proteins have a high degree of similarity in the C-terminal basic and leucine zipper domains. Although all five tryptic peptides are from the C-terminal domain of MafA, several of the amino acids in these peptides discriminate this factor from the other Maf factors, suggesting that the RIPE3b1 factor is a previously unrecognized mammalian homologue of MafA (mMafA).
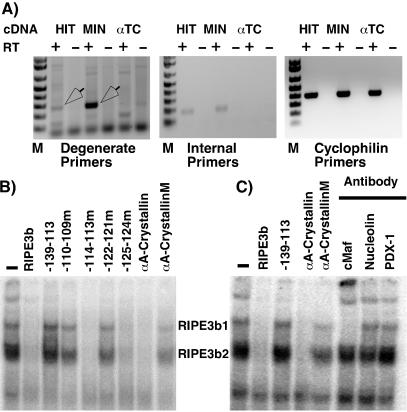
To confirm that RIPE3b1 factor is mMafA, we determined the expression of mMafA in insulin-producing cells and demonstrated that the RIPE3b1 factor can specifically bind a Maf-binding site. Furthermore we showed that the endogenous RIPE3b1 complex is recognized by a large Maf-specific antibody (Fig. 2). Degenerate oligonucleotides were designed from the amino acid sequences of two tryptic peptides, such that the PCR product would contain the sequence corresponding to the remaining three peptides. Since RIPE3b1-binding activity is detected only in pancreatic β cells and not α cells, cDNAs from the insulinoma cell lines HIT T-15 and MIN-6, as well as from the glucagon-producing cell line αTC1.6, were used with degenerate oligonucleotide primers in a PCR. An approximately 300-bp-long PCR product was amplified in this reaction from insulin-producing cell lines but not from αTC1.6 cells (Fig. 2A); this PCR product was cloned and sequenced. Then specific internal primers were designed and used in similar PCRs, again demonstrating expression of MafA only in insulin-producing cells. The deduced amino acid sequence obtained from the PCR product demonstrated the presence of the expected internal peptides, establishing that the avian MafA homologue is selectively expressed in insulin-producing cells.
Figure 2.
The RIPE3b1 factor is mammalian MafA. (A) Ethidium-bromide-agarose-gel-electrophoretic analysis of PCR products amplified by using cDNAs from insulin-producing (HIT T-15 and MIN-6) and glucagon-producing (αTC 1.6) cell lines. Degenerate oligonucleotide primers corresponding to peptide sequences of 47-kDa protein selectively amplify a PCR product (arrows) from cDNAs from insulin-producing cells. Gel-electrophoretic analysis of PCR products amplified by using internal primers and cyclophilin primers demonstrates that mMafA is expressed only in insulin-producing cells. HIT T-15 nuclear extract was incubated with radiolabeled RIPE3b probe (B) in the absence or presence of 50-fold excess of indicated unlabeled αA-crystallin wild type or mutant αA-crystallin competitors, or rat insulin II oligonucleotides; and (C) in the absence or presence of αc-Maf, αnucleolin, or αPDX-1 antibodies. Binding reactions were analyzed by EMSA, and the positions of the RIPE3b1 and RIPE3b2 complexes are indicated. Wild-type αA-crystallin competitor successfully competed for both the RIPE3b1 and RIPE3b2-binding activities. However, αc-Maf antibody that detects only large Maf family members recognized only the RIPE3b1 complex.
MafA can bind chicken αA-crystallin Maf element (which is nearly identical to mouse αA-crystallin element), and mutations in this element inhibit DNA binding of MafA and transcriptional activation of the αA-crystallin gene (22, 25). To demonstrate that the RIPE3b1 factor can specifically bind the Maf-binding site (Fig. 2B), nuclear extract from HIT T-15 cells was incubated with the RIPE3b probe in the absence or presence of wild type or mutant RIPE3b oligonucleotides (24), with wild type or mutant Maf-binding oligonucleotides from mouse αA-crystallin gene as unlabeled competitors (22, 25). The RIPE3b1 and RIPE3b2 factors can bind rat insulin II oligonucleotides RIPE3b, −139 to −101 bp, −114.113m, −125.124m, but not the oligonucleotides −139 to −101 bp, −110.109m, and −125.124m (24), and thus showed the expected competition profile (Fig. 2). Importantly, formation of both the RIPE3b1 and RIPE3b2 complexes was successfully blocked by competition by the wild type but not by an oligonucleotide containing a single-base substitution mutation in the Maf-binding site (Fig. 2B). Similar results were obtained in a converse experiment using the Maf-binding site from the αA-crystallin gene as a probe (data not shown). The Maf family of factors recognize the extended DNA element TGC(N)6–7GCA but show a significant variation in DNA-binding specificity for individual family members (29, 30, 32). The RIPE3b element shows modest homology with the MAF-binding site of the mouse αA-crystallin gene (CTGN6CAGCC) and the MAF consensus sequence (TGCN7GC). Conserved GC nucleotides between Maf and RIPE3b element (−118, −117, and −109, −108 bp in rat insulin II gene) are important for the binding of RIPE3b1 and RIPE3b2 factors (24). Interestingly, nucleotides upstream of the conserved region (−122 and −121 bp) are also critical for binding of these factors (24). These results suggest that the RIPE3b element shares a reasonable homology with the consensus MAF-binding element and that the RIPE3b1 and RIPE3b2 factors belong to the Maf family of transcription factors.
To confirm that the RIPE3b-binding factors belong to the Maf family of transcription factors, DNA-binding reactions were preincubated in the presence of anti-c-Maf (which recognizes only the large Maf family members), anti-nucleolin, and anti-PDX-1 antibodies (Fig. 2C). Although, as shown in Fig. 2B, both RIPE3b1 and RIPE3b2 complexes specifically competed with wild-type but not mutant αA-crystallin oligonucleotides, anti-c-Maf antibody specifically recognized only the RIPE3b1 complex and not the RIPE3b2 complex. This result demonstrates that the RIPE3b1 factor is a member of large Maf family of transcription factor and that the RIPE3b2 factor is most likely a small Maf factor. Of the four large-Maf family members, MafB and c-Maf have broad cellular distributions, while NRL and MafA have more restrictive expression (22, 25, 29, 31). By reverse transcription PCR, we could detect expression of MafB in pancreatic α and β cells and that of c-Maf in α cells. However, expression of NRL could not be detected in either of the pancreatic cell types (data not shown). mMafA is the only large-Maf family member that is selectively expressed in pancreatic β cells, and our amino acid sequencing identifies it as the 47-kDa RIPE3b-binding factor; therefore, we suggest that RIPE3b1 factor is mMafA.
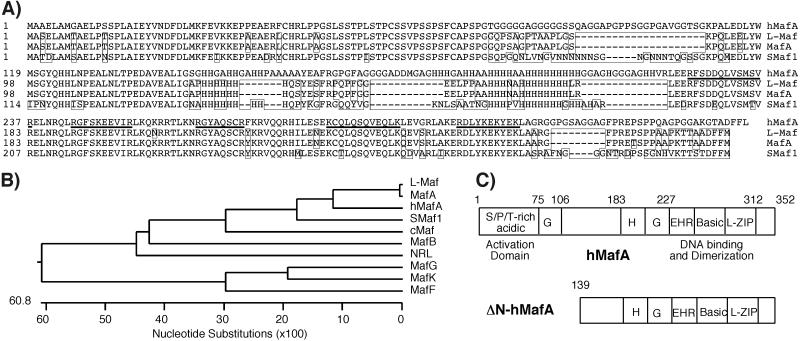
To clone the full-length gene, we compared the nucleotide sequence of the PCR fragment amplified by using degenerate oligonucleotide primers and the public human genome database. Results from blast analysis showed significant homology to all known Maf genes but the highest homology with a “non-Maf” region of the human genome on chromosome 8q24. Although there was significant sequence homology between human genomic contig and our fragment, annotation of the human genome (build 24) did not identify the human mafA (hmafA) gene in this region. Additionally, the publicly available Celera genomic assembly (GA_X54KRCEUARB) contained a gap in the N-terminal region of the hmafA gene. Oligonucleotide primers based on the human genome sequence were designed to amplify the sequence around the region of homology, resulting in identification of an intronless ORF (352 aa) corresponding to hmafA (Fig. 3). The coding region corresponding to hMafA was PCR-amplified by using human genomic DNA as a template. The predicted molecular mass of hMafA is approximately 36,850 Da, which is less than the purified protein band (47,000 Da). We suggest that the apparent difference is due to posttranslational modification of the hMafA. Quail MafA, a 286-aa protein with approximate molecular mass of 32,450 Da, is highly phosphorylated and has multiple isoforms ranging in size from 35 to 43 kDa (31). Hence, it is quite likely that hMafA (352 aa), which is larger than quail MafA, can have an isoform with a molecular mass of 47 kDa. Amino acid sequence alignment of hMafA with other human large and small-Maf family members, chicken L-Maf, quail MafA, and zebrafish Smaf1, clearly shows that RIPE3b1 factor is more closely related to the chicken and quail protein than to other human Maf factors (Fig. 3 A and B). hMafA is a member of the large-Maf family of transcription factors containing an N-terminal activation domain and C-terminal bZIP domain (Fig. 3C). These observations demonstrate that a mammalian Maf factor, mMafA, is selectively expressed in insulin-producing cells.
Figure 3.
Cloning of a novel mammalian MafA factor. (A) Multiple amino acid sequence alignments of the human MafA with quail MafA, chicken L-Maf, zebrafish Smaf1, and known human large and small Mafs were performed with sequence alignment program MEGALIGN using CLUSTAL W method with BLOSUM matrix (DNASTAR, Madison, WI). Sequence alignments of hMafA (GenBank accession no. AY083269) with quail MafA, chicken L-Maf, and zebrafish-Smaf1 are shown, and boxed amino acids depict sequences different from hMafA. Amino acid sequences of five tryptic peptides of the 47-kDa protein are underlined. (B) Phylogenetic tree derived from this alignment demonstrates that the hmafA is a human/mammalian homologue of avian MafA/L-Maf factor. (C) Schematic of deduced hMafA amino acid sequence shows various structural [serine/proline/threonine (S/P/T)-rich acidic domain, glycine (G)- and histidine (H)-rich regions, extended homology region (EHR); basic and leucine zipper (L-ZIP)] and functional (transcriptional activation, DNA-binding, and protein dimerization) domains. Schematic of ΔN-hMafA shows lack of the putative transcriptional activation domain.
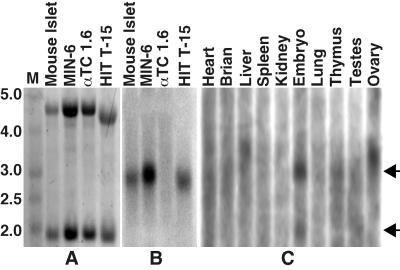
Sequence analysis of the human genomic contig NT_023684 using programs FGENESH 1.0 and FGENES-M 1.5 (http://genomic.sanger.ac.uk) indicated the presence of possible transcription start and stop sites, predicting a full-length hMafA mRNA to be ≈2.7 kb in size. To demonstrate that the hMafA homologue is expressed in other mammalian systems, approximately 10 μg of total RNA from insulin-producing and non-insulin-producing cells was used in Northern analysis with the N-terminal fragment of the hMafA as a probe. An ≈2.8-kb message from insulin-producing cells hybridized with the hMafA probe (Fig. 4B), but no such signal was detected in the lane from glucagon-producing αTC1.6 cells, even though ethidium bromide staining showed equal amounts of RNA (Fig. 4A). To further characterize the expression profile of mMafA, mouse [2 μg of poly(A) RNA] multiple tissue Northern blot (Ambion) was hybridized with the same N-terminal hMafA probe (Fig. 4C). RNA from E14 mouse embryo showed two faint bands corresponding to 1.9- and 2.8-kb transcripts. The presence of 1.9-kb transcript in E14 embryo may result from an alternative transcription start site suggesting an interesting regulation of this gene during embryonic development. In addition to expression in E14 embryo, a faint 2.8 Kb band was also seen in RNA from thymus, but no signal was detected from other tissues (heart, brain, liver, spleen, kidney, lung, testes, and ovary). Results from this analysis demonstrate that, like its avian counterpart, mMafA has a very restrictive cellular expression profile.
Figure 4.
mMafA mRNA is expressed in insulin-producing cells. (A) Total RNA from the indicated samples was run on an agarose/formaldehyde gel and stained with ethidium bromide to show 18S and 28S rRNA bands. (B) Northern blot of the gel shown in A, hybridized with a hMafA probe (15-h exposure). (C) A mouse multiple tissue Northern blot (Ambion) containing 2 μg of poly(A) RNA from 10 tissues hybridized to the same probe (72 h exposure). Arrows denote the positions of 1.9- and 2.8-kb transcripts.
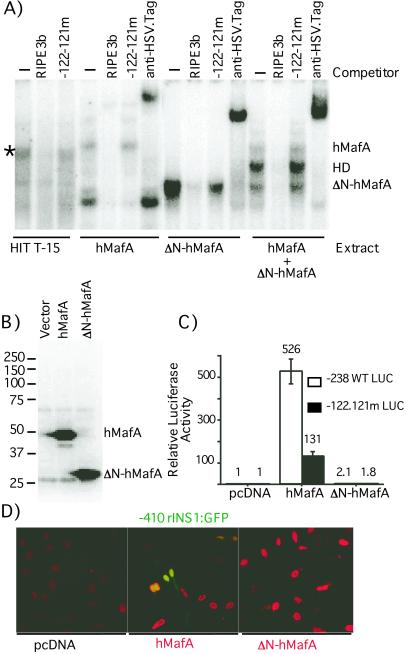
To confirm that RIPE3b1 factor is mMafA, we determined the ability of hMafA to bind the RIPE3b element and activate insulin gene expression (Fig. 5). Both the full-length factor and an N-terminal deletion derivative (ΔN) that lacks the first 138 aa and the activation domain [based on comparison with other large Maf family members (29)] were cloned in-frame with HSV.tag in expression vector pETBlue-2. The presence of the tag sequence increased the molecular size of full-length and ΔN constructs (by ≈3.5 kDa) to 40.4 and 26.8 kDa, respectively. Interestingly, in vitro transcribed and translated full-length hMafA, using a TnT Kit (Promega), has an approximate molecular size of 50 kDa, while the ΔN-hMafA protein runs at 28 kDa (Fig. 5B). These observations demonstrate that the expressed mMafA protein is significantly larger than that predicted from the primary amino acid sequence. Furthermore, most of the posttranslational modification that alters the molecular size of the protein appears to affect the N-terminal 138 aa. These results support our observation that 36.8-kDa mMafA is present as a 47-kDa protein in HIT T-15 nuclear extract. To determine whether the hMafA can bind the RIPE3b element, rabbit reticulocyte lysates from in vitro translated full-length and ΔN-hMafA constructs were used in EMSA (Fig. 5A). Both full-length and ΔN-hMafA bound the RIPE3b probe with DNA-binding specificity identical to that of the RIPE3b1 factor [Fig. 5A, data not shown (24)], and specific DNA-binding complexes were recognized by αHSV.tag antibody. By running the EMSA gel for an extended length of time, we could demonstrate a slight difference in the migration of the full-length DNA-binding complex and that of the endogenous RIPE3b1 factor present in HIT T-15 nuclear extract. We suggest that the difference in the migration of these complexes is possibly due to the presence of the sequence tag. Interestingly, the DNA-binding reaction with cotranslated hMafA and ΔN-hMafA resulted in formation of three DNA-binding complexes: two corresponding to the full-length and ΔN-hMafA, and one with an intermediate migration. Similarly, a combination of translated ΔN-hMafA and HIT T-15 nuclear extract formed RIPE3b-binding complexes that migrated between the endogenous RIPE3b1 complex and the faster-migrating ΔN-hMafA complex (data not shown). These results clearly demonstrate that hMafA most likely binds the RIPE3b element as a homodimer.
Figure 5.
hMafA can bind the RIPE3b element and activate insulin gene expression. (A) HIT T-15 nuclear extract and lysates from indicated in vitro transcription-translation reactions were incubated with RIPE3b probe in the absence (−) or presence of indicated unlabeled wild-type RIPE3b oligonucleotide or one with mutations at positions −122 and −121 as competitor. In addition, binding reactions were also incubated with an αHSV.tag antibody. EMSA was performed as described before with minor modifications; the gels were run for an additional 30 min to resolve binding complexes. Position of the RIPE3b1 complex is denoted by an asterisk (*), and the positions of hMafA, ΔN-hMafA, and heterodimer (HD) complexes are indicated. (B) Equal volume of in vitro transcribed and translated lysates from hMafA, ΔN-hMafA, or pETBlue-2 vector were run on an SDS/PAGE 10% gel, expression of specific proteins was determined by immunoblotting with αHSV.tag monoclonal antibody (Novagen) and bands were detected by ECL kit (Amersham Pharmacia-Pharmacia Biotech). Positions and molecular masses (×103 kDa) of protein standards are indicated. (C) Non-insulin-producing HeLa cells were transfected with luciferase reporter plasmid (-238 WT LUC or -122.121m LUC) (24) and indicated expression plasmid (hMafA, ΔN-hMafA, or pcDNA3.1). Luciferase activity was determined 48 h after transfection. Results are presented relative to -238 WT luciferase activity ± SE (n = 4), after normalization for internal control. (D) HeLa cells were transfected with rat insulin I:GFP reporter (-410 rINSI:GFP) and indicated expression plasmids. Forty-eight hours after transfection, HeLa cells were fixed with buffered 10% formalin, and fusion proteins were detected by using αHSV.tag monoclonal antibody and a secondary Texas red-conjugated anti-mouse antibody. Expression of both hMafA and ΔN-hMafA was localized to the nuclei (red) of the transfected cells. However, only hMafA was able to activate GFP expression from the insulin reporter construct (green), as seen in some hMafA-transfected cells with both GFP and Texas red fluorescence.
To demonstrate that mMafA can activate insulin gene expression, full-length and ΔN-hMafA constructs were cotransfected with the wild-type (−238 WT LUC) and mutant (−122.121m LUC) insulin promoter:luciferase reporter constructs (24) in non-insulin-producing HeLa cells (Fig. 5C). Alone, insulin reporters had very low-level expression in HeLa cells, and the mutation in the insulin enhancer at base pairs −122 and 121, which prevents binding of the RIPE3b1 factor, had no effect on the basal luciferase expression. Cotransfection of the full-length hMafA induced luciferase gene expression by several hundredfold over the −238 WT LUC construct alone. However, coexpression of hMafA with the mutant-reporter construct activated reporter-gene expression by only 15–20% of that of the WT construct. Interestingly, coexpression of ΔN-hMafA did not activate either the WT or mutant constructs. These observations were visually confirmed by using an insulin:GFP reporter construct (−410 rINS I:GFP) and immunostaining with αHSV.tag antibody to detect expression of the full-length and ΔN-hMafA. Fig. 5D shows that both full-length and ΔN-hMafA proteins are expressed in HeLa cells and have nuclear localization but that only the full-length hMafA construct activated insulin gene expression. These results demonstrate that mMafA can specifically bind the insulin enhancer element RIPE3b and activate expression of insulin gene, thus establishing mMafA as the insulin gene transcription factor RIPE3b1.
Maf factors play important roles in cell determination and control of cellular differentiation. The mafB gene is important for the segmentation of the hindbrain (33) and contributes to monoblast and macrophage differentiation (34). The transcription factor c-Maf has been implicated in lymphopoiesis and lens development (35, 36). Expression of NRL (mouse) and MafA/L-Maf (quail/chicken) is crucial for lens development (22, 25, 31). Our demonstration of the insulin gene transcription factor RIPE3b1 as the mMafA reveals a role for the Maf family of transcription factors in regulating pancreatic β cell function. Based on the important developmental role of the avian MafA factor, we suggest that mMafA (RIPE3b1) regulates the development and differentiation of pancreatic β cells. In addition, earlier results showed that the RIPE3b1 factor regulates β cell-specific and glucose-responsive expression of insulin; thus cloning of the RIPE3b1/mMafA should lead to better understanding of β cell function during the development of diabetes.
Acknowledgments
We thank Drs. Susan Bonner-Weir and Gordon Weir for constructive criticism of the manuscript, Dr. Bonner-Weir for her help with immunostaining, Robert Harrington and Alan Guilbault for technical assistance, Jack O'Neal and Jennifer Lock for islets, and Dr. Joel Habener for IDX-1 antibody, Dr. Ed Leiter (The Jackson Laboratory) for αTC1.6 cell line, and Dr. J. I. Miyazaki (Osaka University, Suito, Japan) for the MIN6 cell line. Funding for this study was provided by a Juvenile Diabetes Research Foundation Research Grant, and by a Career Development Award from American Diabetes Association (to A.S.). We appreciate the services provided by the DNA and Tissue Culture cores of Joslin Diabetes Center (supported by the Diabetes Endocrinology Research Center Grant NIH DK-36836), and Harvard Microchemistry Facility for amino acid sequencing.
Abbreviations
- EMSA
electophoretic mobility shift assay
- HP
heparin-pooled
- GFP
green fluorescent protein
- WT
wild type
Note Added in Proof.
While this manuscript was under review, a recent Human Genome build no. 28 indicated that the hMafA is located on chromosome 8q24 as well as on chromosome 11p15.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY083269).
References
- 1.Boam D S W, Docherty K. Biochem J. 1989;264:233–239. doi: 10.1042/bj2640233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shieh S-Y, Tsai M-J. J Biol Chem. 1991;266:16708–16714. [PubMed] [Google Scholar]
- 3.Sharma A, Stein R. Mol Cell Biol. 1994;14:871–879. doi: 10.1128/mcb.14.2.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karlsson O, Edlund T, Moss J B, Rutter W J, Walker M D. Proc Natl Acad Sci USA. 1987;84:8819–8823. doi: 10.1073/pnas.84.24.8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crowe D T, Tsai M-J. Mol Cell Biol. 1989;9:1784–1789. doi: 10.1128/mcb.9.4.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohlsson H, Karlsson K, Edlund T. EMBO J. 1993;12:4251–4259. doi: 10.1002/j.1460-2075.1993.tb06109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leonard J, Peers B, Johnson T, Ferreri K, Lee S, Montminy M R. Mol Endocrinol. 1993;10:1275–1283. doi: 10.1210/mend.7.10.7505393. [DOI] [PubMed] [Google Scholar]
- 8.Miller C P, McGehee R E, Habener J F. EMBO J. 1994;13:1145–1156. doi: 10.1002/j.1460-2075.1994.tb06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jonsson J, Carlsson L, Edlund T, Edlund H. Nature (London) 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 10.Offield M F, Jetton T L, Labosky P, Ray M, Stein R, Magnuson M, Hogan B L M, Wright C V E. Development (Cambridge, UK) 1996;122:983–985. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 11.Lee J E, Hollenberg S M, Snider L, Turner D L, Lipnick N, Weintraub H. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 12.Naya F J, Stellrecht C M M, Tsai M-J. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 13.Naya F J, Huang H P, Qiu Y, Mutoh H, DeMayo F J, Leiter A B, Tsai M J. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahlgren U, Jonsson J, Edlund H. Development (Cambridge, UK) 1996;122:1409–1416. doi: 10.1242/dev.122.5.1409. [DOI] [PubMed] [Google Scholar]
- 15.Shieh S Y, Stellrecht C M M, Tsai M J. J Biol Chem. 1995;270:21503–21508. doi: 10.1074/jbc.270.37.21503. [DOI] [PubMed] [Google Scholar]
- 16.Zhao L, Cissell M A, Henderson E, Colbran R, Stein R. J Biol Chem. 2000;275:10532–10537. doi: 10.1074/jbc.275.14.10532. [DOI] [PubMed] [Google Scholar]
- 17.MacFarlane W M, Read M L, Gilligan M, Bujalska I, Docherty K. Biochem J. 1994;303:625–631. doi: 10.1042/bj3030625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melloul D, Ben-Neriah Y, Cerasi E. Proc Natl Acad Sci USA. 1993;90:3865–3869. doi: 10.1073/pnas.90.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.German M S, Wang J. Mol Cell Biol. 1994;14:4067–4075. doi: 10.1128/mcb.14.6.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma A, Olson L K, Robertson R P, Stein R. Mol Endocrinol. 1995;9:1127–1134. doi: 10.1210/mend.9.9.7491105. [DOI] [PubMed] [Google Scholar]
- 21.Olson L K, Sharma A, Peshavaria M, Wright C V E, Towle H C, Robertson R P, Stein R. Proc Natl Acad Sci USA. 1995;92:9127–9131. doi: 10.1073/pnas.92.20.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogino H, Yasuda K. Science. 1998;280:115–118. doi: 10.1126/science.280.5360.115. [DOI] [PubMed] [Google Scholar]
- 23.Benkhelifa S, Provot S, Lecoq O, Pouponnot C, Calothy G, Felder-Schmittbuhl M P. Oncogene. 1998;17:247–254. doi: 10.1038/sj.onc.1201898. [DOI] [PubMed] [Google Scholar]
- 24.Harrington R H, Sharma A. J Biol Chem. 2001;276:104–113. doi: 10.1074/jbc.M008415200. [DOI] [PubMed] [Google Scholar]
- 25.Ring B Z, Cordes S P, Overbeek P A, Barsh G S. Development (Cambridge, UK) 2000;127:307–317. doi: 10.1242/dev.127.2.307. [DOI] [PubMed] [Google Scholar]
- 26.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silva C M, Tully D B, Petch L A, Jewell C M. Proc Natl Acad Sci USA. 1987;84:1744–1748. doi: 10.1073/pnas.84.7.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jonas J-C, Sharma A, Hasenkamp W, Ilkova H, Patane G, Laybutt R, Bonner-Weir S, Weir G C. J Biol Chem. 1999;274:14112–14121. doi: 10.1074/jbc.274.20.14112. [DOI] [PubMed] [Google Scholar]
- 29.Blank V, Andrews N C. Trends Biochem Sci. 1997;22:437–441. doi: 10.1016/s0968-0004(97)01105-5. [DOI] [PubMed] [Google Scholar]
- 30.Matsushima-Hibiya Y, Nishi S, Sakai M. Biochem Biophys Res Commun. 1998;245:412–418. doi: 10.1006/bbrc.1998.8447. [DOI] [PubMed] [Google Scholar]
- 31.Benkhelifa S, Provot S, Nabais E, Eychene A, Calothy G, Felder-Schmittbuhl M-P. Mol Cell Biol. 2001;21:4441–4452. doi: 10.1128/MCB.21.14.4441-4452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dlakic M, Grinberg A V, Leonard D A, Kerppola T K. EMBO J. 2001;20:828–840. doi: 10.1093/emboj/20.4.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manzanares M, Cordes S, Kwan C-T, Sham M H, Barsh G S, Krumlauf R. Nature (London) 1997;387:191–195. doi: 10.1038/387191a0. [DOI] [PubMed] [Google Scholar]
- 34.Sieweke M H, Tekotte H, Frampton J, Graf T. Cell. 1996;85:49–60. doi: 10.1016/s0092-8674(00)81081-8. [DOI] [PubMed] [Google Scholar]
- 35.Kim J I, Li T, Ho I-C, Grusby M J, Glimcher L H. Proc Natl Acad Sci USA. 1999;96:3781–3785. doi: 10.1073/pnas.96.7.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho I-C, Hodge M R, Rooney J W, Glimcher L H. Cell. 1996;85:973–983. doi: 10.1016/s0092-8674(00)81299-4. [DOI] [PubMed] [Google Scholar]