Abstract
The pathogenesis of tuberculosis involves multiple phases and is believed to involve both a carefully deployed series of adaptive bacterial virulence factors and inappropriate host immune responses that lead to tissue damage. A defined Mycobacterium tuberculosis mutant strain lacking the sigH-encoded transcription factor showed a distinctive infection phenotype. In resistant C57BL/6 mice, the mutant achieved high bacterial counts in lung and spleen that persisted in tissues in a pattern identical to those of wild-type bacteria. Despite a high bacterial burden, the mutant produced a blunted, delayed pulmonary inflammatory response, and recruited fewer CD4+ and CD8+ T cells to the lung in the early stages of infection. In susceptible C3H mice, the mutant again showed diminished immunopathology and was nonlethal at over 170 days after intravenous infection, in contrast to isogenic wild-type bacilli, which killed with a median time to death of 52 days. Complete genomic microarray analysis revealed that M. tuberculosis sigH may mediate the transcription of at least 31 genes directly and that it modulates the expression of about 150 others; the SigH regulon governs thioredoxin recycling and may be involved in the maintenance of intrabacterial reducing capacity. These data show that the M. tuberculosis sigH gene is dispensable for bacterial growth and survival within the host, but is required for the production of immunopathology and lethality. This phenotype demonstrates that beyond an ability to grow and persist within the host, M. tuberculosis has distinct virulence mechanisms that elicit deleterious host responses and progressive pulmonary disease.
Tuberculosis is one of the leading infectious causes of death and claims ≈2 million lives annually (1). There is controversy over whether the disease is primarily a dysfunctional immunologic reaction to a persistent microbe or whether the bacteria themselves produce tissue damage; there is evidence that both host and bacterial factors play key roles in disease severity. In susceptible mouse strains, such as C3H, the pathogen elicits a dysregulated, necrotizing host immune response leading to tissue destruction and further bacterial replication. In resistant mice such as C57BL/6, Mycobacterium tuberculosis survives in high numbers for many months and is contained in organized granulomatous lesions in the lung without progressive lung damage. Thus whereas mycobacteria survive in both genetic backgrounds, disease progression is delayed in resistant animals (2–4).
On the bacterial side, M. tuberculosis virulence has been associated with its initial survival within macrophages and resistance to reactive oxygen and nitrogen intermediates (ROIs and RNIs) (5–8). Tubercle bacilli demonstrate inducible responses to oxidative stresses, and several M. tuberculosis genes, including katG (catalase peroxidase), ahpC (alkylhydroperoxide reductase), and sodA and sodC (superoxide dismutases) have been implicated in protection from the macrophage oxidative burst (9–11). Another potential oxidative defense system possessed by M. tuberculosis is a thioredoxin system composed of thioredoxin (TrxA) and thioredoxin reductase (TrxB) which requires NADH as a cofactor. The reduced form of thioredoxin is a general protein disulfide reductant, which can reactivate proteins that have been oxidized by H2O2 (12, 13). The ability of the thioredoxin system to reduce reactive oxygen species and thereby protect the cell against oxidative stress has been established in several systems (14, 15). Other genes, not associated with resistance to oxidants, have also been implicated as bacterial virulence factors and reveal reduced bacterial counts in different stages of disease (16–18). Although reported in genetically undefined strain variants, attenuated M. tuberculosis mutants that survive with normal colony-forming unit (cfu) counts in animal tissues have not been extensively characterized (19).
Studies using in vitro stresses such as reactive oxygen intermediates (20) and studies using macrophage infection models have demonstrated that several M. tuberculosis σ factors are strongly induced by conditions associated with infection (21). M. tuberculosis SigH belongs to the extracytoplasmic function (ECF) family of alternative σ70 RNA polymerase subunits (22). Expression of Mycobacterium smegmatis sigH is induced by both heat shock and redox stress in vitro (23), and M. tuberculosis sigH expression has been shown to be up-regulated at several time points during macrophage infection (21). ECF σ factors are believed to control the expression of gene products associated with the cell envelope and/or secreted functions of bacteria. Hence, such σ factors of M. tuberculosis might be expected to play a role in surface antigen expression and the secretion of immunomodulatory molecules by the pathogen.
Materials and Methods
Mutant Construction.
The targeted disruption strategy was based on the vector pCK0686, which contains a hygromycin-resistance cassette flanked by unique multiple cloning sites (MCS). A left flank PCR product containing 1,722 bp proximal to the sigH gene and a right flank PCR product containing 2,059 bp distal to the sigH gene were cloned into the MCSs. The sigH specific flanks were nonoverlapping so that a central region of the sigH gene was deleted from the targeting vector, leaving 82 bp of the 5′ end and 113 bp of the 3′ end of the sigH gene in place. The resulting plasmid pCK0708 was then used to transform M. tuberculosis CDC1551 in a two-step selection process using Southern blotting to confirm allelic exchange. A complemented strain of the sigH mutant (pCK0758) was generated by cloning a 2,862-bp M. tuberculosis CDC1551 genomic fragment encompassing the coding as well as regulatory region of the sigH gene plus the complete coding sequences of three distal genes (MT3319, MT3318, and MT3317) into an L5 phage-based integration-proficient vector for mycobacteria, pMH94 (24), and then transforming the M. tuberculosis ΔsigH mutant with this vector.
Mouse Virulence Assays.
For mouse organ cfu assays, C57BL/6 mice were inoculated by the aerosol route. The liquid inoculum placed in the nebulizer was determined empirically for each bacterial strain to result in the implantation of 100 bacteria per lung at day 1 of infection. Groups of six mice were killed at day 1 and weeks 4, 8, 12, and 16. Lung and spleen tissues were homogenized in their entirely in PBS/0.05% Tween 80, and colonies were enumerated on 7H10 plates grown at 37°C for 3–4 weeks. Intermediate lobes of the right lung of infected mice were saved for histologic analysis (25). For time-to-death assays C3H:HeJ and CB17-severe combined immunodeficient (CB17-SCID) mice were infected intravenously with 106 and 104 bacteria, respectively, and C57BL/6J mice were infected by the aerosol route as described above.
Fluorescence-Activated Cell Sorting (FACS) Analysis and Intracellular Cytokine (ICC) Staining.
Single-cell suspensions of lung and spleen cells were prepared, washed, and plated as described (26–28) and were stimulated with anti-CD3 (0.1 μg/ml) and anti-CD28 (1 μg/ml) antibodies (Ab) in the presence of 3 μM monensin for 5 h. Cells were washed and stained with anti-CD4 (0.2 μg of anti-CD4-CyChrome Ab per 106 cells) and anti-CD8 (0.2 μg of anti-CD8-FITC Ab per 106 cells), washed, and fixed. For ICC, cells were permeabilized with saponin and stained for interferon γ (IFN-γ) (0.4 μg of anti-IFN-γ-PE Ab per 106 cells) or tumor necrosis factor α (TNF-α) (0.4 μg of anti-TNF-α-PE Ab per 106 cells) (PharMingen, San Diego) and analyzed by FACS (Becton Dickinson) as described (26–28). Cells were gated on the lymphocyte population based on cell size. Isotype controls for each Ab were used and uninfected control mice were tested in each experiment.
Microarrays.
Gene-specific PCR primers were designed to amplify internal fragments from a total of 4,016 ORFs from the annotated sequences of M. tuberculosis CDC1551 and H37Rv (29). Individual purified PCR products were spotted in duplicate on high contact angle slides (Corning, Corning, NY) by using a 96-well format IAS arrayer (Intelligent Automation Systems, Cambridge, MA). Bacterial RNA prepared by the Trizol method was reverse transcribed and labeled with Cy3 or Cy5 probes (Amersham Pharmacia) by using the aminoallyl labeling method. The slides were scanned with an Axon Genepix scanner, and the TIGR (The Institute for Genomic Research) Spotfinder and Array Viewer software systems were used to define and quantify spot intensities. For each bacterial growth point, two independent RNA preparations from wild type and mutant were prepared, and for each RNA sample pair reverse labeling was performed (four hybridizations for each growth point). As each amplicon was spotted in duplicate, eight relative hybridization values were available for each gene for each growth point.
Results
Construction of the sigH Mutant and Complemented Strain.
To study the role of M. tuberculosis SigH in the pathogenesis of tuberculosis, we used allelic exchange to delete and replace the sigH gene (MT3320, Rv3223c) (29) with a hygromycin-resistance gene cassette (ΔsigH mutant) in M. tuberculosis strain CDC1551 (wild type) (30). We also complemented the sigH mutation by cloning a complete copy of the sigH gene and its promoter into the single-copy integrating plasmid pMH94. The ΔsigH mutant, the complemented strain, and the wild-type strain showed identical growth rates in Middlebrook 7H9 medium (Difco) in vitro.
Proliferation and Survival in Mouse Tissues.
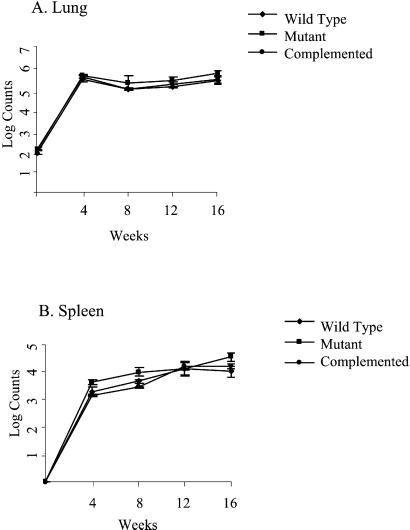
We studied the ability of these three M. tuberculosis strains to survive in the tissues of resistant C57BL/6 mice by cfu analysis after aerosol exposure. Resistant C57BL/6 mice were selected for this analysis because they restrict the tissue proliferation of virulent M. tuberculosis and hence would be expected to amplify survival defects in attenuated strains. After aerosol exposure using inocula that implanted 100 bacilli in the lungs (as assessed by cfu counts the day after infection), mice were monitored for 16 weeks. As may be seen in Fig. 1 A and B, the pattern of mouse organ burden of M. tuberculosis seen with infection by the wild-type, ΔsigH mutant, and complemented strains did not differ. All three strains proliferated in lungs and spleen for 4 weeks, and then persisted in both tissues between 4 and 16 weeks with approximately 106 organisms in the lungs and 104 bacilli in the spleen.
Figure 1.
Lung (A) and spleen (B) cfu counts in C57BL/6 mice infected with wild-type M. tuberculosis (♦), the ΔsigH mutant (■), and the complemented strain (●) by the aerosol route. Groups of six mice infected were evaluated at each time point.
Histopathologic Analysis.
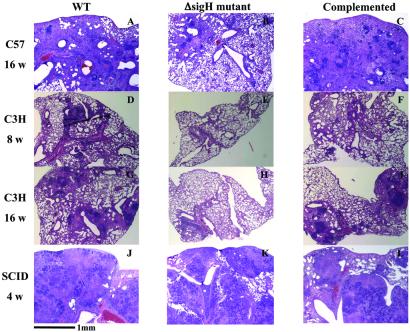
Despite the apparent full virulence of the ΔsigH mutant by organ cfu analysis, histopathologic assessments of the organs from C57BL/6 mice demonstrated an attenuated phenotype in mutant-infected lungs. As may be seen in Fig. 2 A–C, whereas the wild-type and complemented-mutant strains produced progressive granulomatous inflammation and loss of functional alveoli attributable to enlarging and coalescing lesions, the mutant infection showed smaller, less abundant granulomas, and these lesions progressed less rapidly. After 16 weeks, mouse lungs infected by the ΔsigH mutant revealed minimal interstitial mononuclear cell infiltrates and peribronchial inflammation without large inflammatory wedges or granulomatous lesions (Fig. 2B) that were clearly observed in the corresponding wild-type and complemented strain infections (Fig. 2 A and C). A similar decrease in intensity of granulomatous inflammation and a delay in its evolution was observed in the lungs of C3H mice infected with the ΔsigH mutant for 8 or 16 weeks (Fig. 2 E and H) in comparison with infections by the wild-type and complemented strains (Fig. 2 D and F, and G and I). In contrast, the lungs of SCID mice infected with the three different M. tuberculosis strains did not show histopathologic differences at 4 weeks (Fig. 2 J–L), indicating that intact cellular immunity is required for the differential effect. Interestingly, analysis of spleens of all three strains of mice (C57BL/6, C3H, and SCID) indicated a similar extent of granuloma formation and tissue damage with mutant and wild-type bacterial strains (data not shown), suggesting that the disease delay phenotype in the ΔsigH mutant may be lung specific.
Figure 2.
The ΔsigH mutant (B, E, H, and K) produces less tissue pathology in lungs of infected C57BL/6 and C3H mice compared with wild-type (A, D, G, and J) and complemented strains (C, F, I, and L). C57BL/6 mice infected by the aerosol route (as in Fig. 1) were evaluated at 16 weeks (A–C). C3H mice infected intravenously with 106 bacilli were analyzed at 8 weeks (D–F) and 16 weeks (G–I). In contrast, SCID mice infected intravenously with 104 bacilli and analyzed at 4 weeks (J–L) showed no significant difference in the degree of tuberculous pneumonia. (×40; hematoxylin and eosin staining.)
Cellular Immune Responses in Mouse Infections.
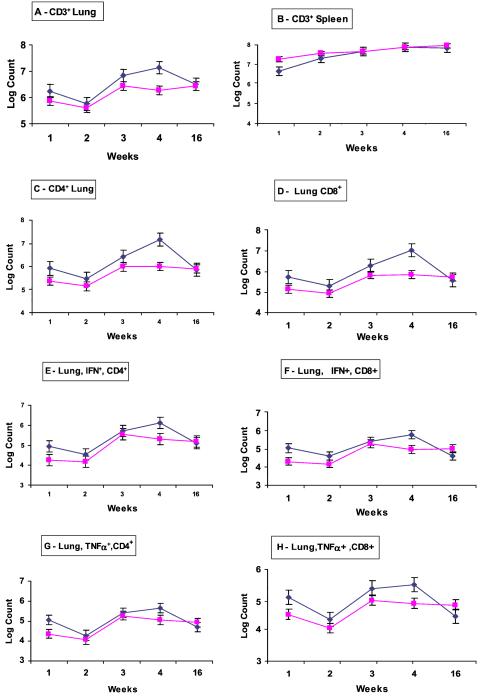
We postulated that the despite the normal bacterial survival in lung tissue, the ΔsigH mutant lacked virulence factors that induce tissue-damaging cell-mediated immune responses by the host (31, 32). Therefore we evaluated T cell responses in C57BL/6 mice after infection by both the wild type and the ΔsigH mutant by using flow cytometry of cellular suspensions obtained from mouse tissues at various times after infection. As may be seen (Fig. 3 A and B), between 3 and 4 weeks, the recruitment of CD3+ cells to the lung increased in wild-type-infected lungs, but not in mutant-infected mice. The reduced T cell recruitment to the lungs at 4 weeks by the ΔsigH mutant was observed in both total CD4+ and CD8+ cells with approximately one-log10 differences in both cell types (Fig. 3 C and D). By 16 weeks, levels of both CD4+ and CD8+ cells declined in the lungs of wild-type-infected mice but remained constant in mutant-infected mice, resulting in similar numbers of both T cell types. Intracellular cytokine staining for IFN-γ and TNF-α synthesis within pulmonary CD4+ and CD8+ cells showed that the cell-associated cytokine synthesis paralleled the T cell population curves (reduced IFN-γ and TNF-α positivity in mutant infection at 4 weeks) indicating similar rates of expression of these cytokines per cell in infection with both strains of bacteria (Fig. 3 E–H). By 16 weeks the levels of cytokine-producing CD4+ and CD8+ cells in the two types of infection had essentially re-equalized. Thus the recruitment of cytokine-producing CD4+ and CD8+ cells to the lungs (but not the spleens) of ΔsigH mutant-infected mice was significantly blunted beginning at 4 weeks after infection. Thus the pathologic disease delay phenotype seen in lungs of C57BL/6 mice after ΔsigH mutant infection correlates with diminished T cell recruitment to the lung despite a bacterial burden equivalent to that of wild type.
Figure 3.
Flow cytometry of cell populations from the tissues of C57BL/6 mice infected with wild-type M. tuberculosis (♦) and the ΔsigH mutant (■) by the aerosol route. Each time point represents the mean cell count from a group of four mice. Absolute numbers of CD3+ cells from lungs (A) and spleens (B), and absolute numbers of lung CD4+ (C) and CD8+ (D) cells are shown. Intracellular cytokine staining for IFN-γ-positive CD4+ (E) and CD8+ (F) cells and for TNF-α-positive CD4+ (G) and CD8+ (H) cells from the lungs is also shown. Error bars represent one standard error.
Time-to-Death Analyses.
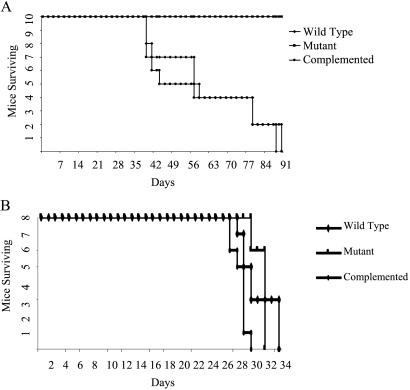
We performed time-to-death experiments to determine whether the milder immunopathology observed in ΔsigH mutant infection might manifest as a prolongation of survival. When C57BL/6 mice infected with each of the three strains were held long term for time-to-death analysis, no deaths were observed in any of the three groups for 15 months in keeping with the known innate resistance of this mouse strain to tuberculosis. However, in C3H mice the ΔsigH mutant displayed a pronounced attenuation in time-to-death with the median time-to-death (MTD) for the ΔsigH mutant exceeding 170 days (0/12 mice dead at day 170) as compared with 52 and 51.5 days for infection by the wild-type and complemented strains, respectively (Fig. 4A, P < 0.001 for mutant compared with both other strains). The differences in MTD between wild-type and complemented strains in C3H mice were not statistically significant. SCID mice infected with the ΔsigH mutant, the wild type, and the complemented strain showed no significant difference in mortality with MTD of 32, 30, and 29 days, respectively (Fig. 4B, P > 0.05 for all pairwise comparisons).
Figure 4.
Time-to-death analysis in C3H:HeJ (A) and SCID (B) mice upon intravenous. infection with M. tuberculosis wild-type strain (♦), the ΔsigH mutant (■), and the strain with complemented sigH gene (●) shows a significant mortality delay. The inoculum was 106 cfu for C3H mice (n = 10) and 104 cfu for SCID mice (n = 8).
Microarray Identification of SigH-Regulated Genes.
We studied the global expression patterns of the M. tuberculosis ΔsigH mutant by using complete genomic microarrays (33–36). Total RNA from the M. tuberculosis ΔsigH mutant and wild-type CDC1551 was prepared from cultures grown in 7H9 medium to OD600 values of 0.3, 0.6, 1.2, and >2.0. In late exponential phase of in vitro growth (OD600 = 1.2) approximately 120 genes were underexpressed and 60 genes were overexpressed in the M. tuberculosis ΔsigH mutant relative to the wild type (Table 1 and Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). Genes encoding hypothetical unknown proteins accounted for 30% of differentially expressed transcripts.
Table 1.
A summary of the genes significantly* up- or down-regulated by mutation in M. tuberculosis sigH at four different phases of growth in vitro
|
M. tuberculosis genes in the sigH mutant
|
||
|---|---|---|
| OD600 | Down-regulated by 2.0-fold or more | Up-regulated by 2.0-fold or more |
| 0.3 | 15 genes including MT4032 (thioredoxin reductase); MT4033, MT1516, and MT1517 (thioredoxins); MT2063 (ferredoxin); MT2468 and MT2469 (sulfate ABC transporters); MT1163 (citrate synthase); MT2541 (conserved hypothetical protein containing a glutaredoxin active site); MT1872 (small basic protein); MT1377 (cysteine synthase); MT0040 (AMP-binding family protein); MT1924 (conserved hypothetical protein); MT1378 (rhomboid family protein); and MT3320 (sigH) | 3 genes including MT2310 (transcriptional regulator); MT0941 (PE family protein); and MT3850 (hypothetical protein) |
| 0.6 | 23 genes including MT3320 (sigH); MT1259 (sigE); MT2783 (sigB); MT4032 (thioredoxin reductase); MT4033 (thioredoxin); MT3938; MT1297 (multidrug resistance efflux protein); and MT2541 (conserved hypothetical protein containing a glutaredoxin active site) | 9 genes including MT2310 (transcriptional regulator); MT0941 (PE family protein); MT2438 (hypothetical protein); MT0292 (PPE family protein); MT2050 (transcriptional regulator, arsR family); MT2027 (DNA-binding protein, copG family); MT2448 (peptide synthetase family); MT0146 (hypothetical protein); and MT0340 (P450 heme thiolate protein) |
| 1.2 | 121 genes including MT3320 (sigH); MT1259 (sigE); MT2783 (sigB); MT4032 (thioredoxin reductase); MT4033 (thioredoxin); MT3938; MT1297 (multidrug resistance efflux protein); MT2541 (conserved hypothetical protein containing a glutaredoxin active site); and MT0397 (clpB) | 60 genes including MT2310 (transcriptional regulator); MT0941 (PE family protein); MT2438 (hypothetical protein); MT0292 (PPE family protein); MT2050 (transcriptional regulator, arsR family); MT2027 (DNA-binding protein, copG family); MT2448 (peptide synthetase family); MT0146 (hypothetical protein); MT0340 (P450 heme thiolate protein); and MT2446 (hypothetical protein) |
| >2.0 | 164 genes including sigH; sigE; sigB; MT4033; MT4032; MT3938 (tetR transcriptional regulator); MT3555 (serine protease); MT2394 (malate oxidoreductase); MT2303 (malonyl-CoA acyl carrier protein); MT2150 (Xaa-Pro dipeptidase); MT2063 (ferredoxin); MT1896 and 1897 (urease subunits); MT1604–1606 (fumarate reductase operon); MT1516–1517 (thioredoxins); MT1297 (multidrug resistance efflux protein); MT1377 (cysteine synthase); MT1163 (citrate synthase); MT0631 (tcrA response regulator); MT0397 (clpB, ATP-binding subunit of clp protease); and MT2541 (conserved hypothetical protein containing a glutaredoxin active site) | 71 genes including MT2310 (transcriptional regulator); MT0941 (PE family protein); MT2438 (hypothetical protein); MT0292 (PPE family protein); MT2050 (transcriptional regulator, arsR family), MT2027 (DNA-binding protein, copG family), MT2448 (peptide synthetase family); MT0146 (hypothetical protein); MT0340 (P450 heme thiolate protein); and MT2446 (hypothetical protein) |
The complete list of genes significantly down-regulated by M. tuberculosis sigH mutation at OD600 of 1.2 is available as supporting information on the PNAS web site, www.pnas.org.
Significant modulation was defined as a wild-type to mutant intensity ratio of ≥2.0 or ≤0.5 in at least 75% of the evaluable spots.
Several components of the thioredoxin–thioredoxin reductase system demonstrated a relative decrease in expression in the ΔsigH mutant by microarray analysis; these include MT4032–4033 (thioredoxin reductase and thioredoxin, repressed 3.9-fold), MT1516–1517 (thioredoxins, repressed 3.8-fold), and MT0838 (a putative thioredoxin, repressed 3.2-fold). In addition, related oxidative stress response proteins MT2541 (glutaredoxin-like, repressed 3.4-fold) and MT2063 (ferredoxin, repressed 3.1-fold) showed significantly decreased relative expression in the ΔsigH mutant. Several other types of stress-response genes, including MT0265 (hsp20 family member, 3.5-fold repressed), the heat-shock gene dnaK (MT0365, hsp70 family members, 2.9-fold repressed), MT2052, MT2061, MT2698, and MT2699 (each universal stress protein family members, repressed 2.1-, 2.2-, 4.5-, and 2.5-fold, respectively), were relatively underexpressed in the ΔsigH mutant. Since thioredoxins and heat shock proteins reduce and refold proteins damaged by oxidation, their σ factor dependent expression may represent a stress-management strategy by the pathogen aimed at maintaining activity of essential proteins (37). Microarray analysis of N,N,N′,N′-tetramethylazodicarboxamide (diamide)-treated strains for the most part intensified gene expression differences between mutant and wild type (data not shown), as would be expected because diamide is a known thiol-oxidizing agent.
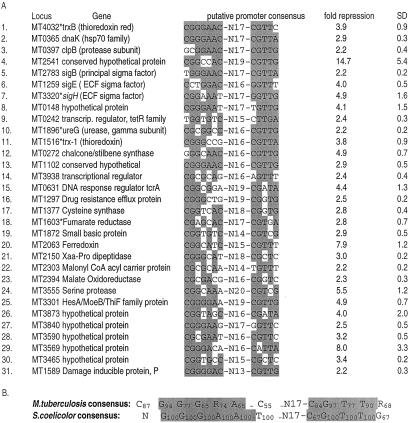
Because there is a 68-bp overlap between the sigH gene remnant in the M. tuberculosis ΔsigH mutant and the sigH amplicon spotted on our microarray, it is possible to detect the level of ineffective sigH transcription in the mutant by microarray analysis. Rather than producing a compensatory increase of ineffective sigH transcription, lack of functional SigH protein resulted in a relative decrease in sigH (MT3320) transcription in the mutant by 4.9-fold. This finding suggests that the M. tuberculosis sigH gene is self-regulated, a hypothesis strengthened by the presence of a consensus sigH-binding sequence that was found in the upstream untranslated region of the sigH gene (Fig. 5).
Figure 5.
(A) Putative sigH promoter consensus elements identified in 5′ untranslated regions of 31 genes with reduced expression in the ΔsigH mutant, and the corresponding fold-repression values and standard deviation (SD) from bacteria grown to OD600 = 1.2. Asterisks indicate that genes immediately downstream from these were also repressed and are probably part of an operon. Bases matching the consensus are shaded. (B) A consensus promoter recognition sequence for M. tuberculosis SigH derived from the 31 putative promoters, and the reported Streptomyces coelicolor SigR consensus (based on an analysis of three promoters) shown for comparison (40). Subscripts denote the percent frequency of occurrence of each base among the 31 genes shown. R = A or G.
The relative expression of gene MT3318 (Rv3222Ac), which is located distal to sigH in the same transcriptional unit and exhibits homology to the anti-σ factor gene class, was also diminished in the mutant strain (4.2-fold). The arrangement of genes at the M. tuberculosis sigH locus resembles that of the Streptomyces coelicolor A3(2) sigR locus (38, 39), and it has been shown that the gene adjacent to sigR (rsrA) inhibits SigR activity by functioning as SigR-sequestering anti-σ factor (40). Because MT3318 transcriptional levels parallel those of sigH, it appears likely that the SigH and the MT3318 gene product are produced in equimolar quantities. The expression of σ factor genes sigE (MT1259, 4.0-fold repressed) and sigB (MT2783, 3.4-fold repressed) was relatively repressed by ΔsigH mutation, suggesting that sigH may occupy a proximal position in the hierarchy of mycobacterial regulatory mechanisms.
SigH Promoter Recognition Consensus Sequence.
Analysis of the untranslated regions of genes underexpressed in the sigH mutant revealed a consensus sequence (Fig. 5) among at least 31 of these genes. These genes harboring a consensus element are likely to be under direct SigH control, whereas those genes that did not exhibit a consensus sequence may be indirectly controlled by SigH. The consensus sequence comprises two modules, CGGGRAC at the −35 region and CGTTR at the −10 region (R = A or G), both modules being separated by about 14–18 bp. This SigH consensus is in strong agreement with the consensus derived for six mycobacterial genes in a recent study by Raman et al. (41) and is similar to that observed for the sequence recognized by the S. coelicolor SigR protein (40).
Discussion
These data define a single regulatory gene and its dependent genes as effectors of virulence in M. tuberculosis. The SigH regulon comprises a global response system to manage oxidative and other denaturing stresses by inducing a family of thioredoxin and heat shock genes. Surprisingly, expression of sigH and SigH-mediated oxidative defenses appear dispensable for bacterial growth and survival in mouse tissues, yet they are required for disease induction and lethality in mice.
The virulence defect observed in the M. tuberculosis ΔsigH mutant falls in a newly appreciated phenotypic class in which there is reduced immunopathology despite normal bacterial replication and survival in host tissues. Steyn et al. (42) recently observed that mutation in M. tuberculosis whiB3 produced a mutant strain with an infection phenotype (normal tissue cfu counts, but prolonged time-to-death) similar to that of the M. tuberculosis ΔsigH mutant. Interestingly, WhiB3 appears to bind to the M. tuberculosis principal σ factor SigA and is likely to be a posttranslational regulator of σ factor activity. The common thread of defective σ factor activity in both the whiB3 and sigH mutants suggests that the ability to induce immunopathology and lethal infection requires the deployment of a regulated gene set and underscores the fact that M. tuberculosis possesses specific mechanisms that trigger deleterious host responses leading to accelerated and more extensive granulomatous inflammation (43).
Our data demonstrate that the reduced immunopathology phenotype in the ΔsigH mutant is related to reduced recruitment of both CD4+ and CD8+ T cells to the lungs at a time associated with the onset of acquired immune responses and control of bacterial replication. Concomitant with the reductions in CD4+ and CD8+ cells in the lung there were diminished populations of IFN-γ- and TNF-α-expressing T cells at the 4-week time point as well. The recruitment of T cells to the spleen was identical in the wild type and ΔsigH mutant, suggesting that the immunopathology defect of the mutant may be lung specific. It is interesting to note that whereas the entry of T cells to the lung at 3–4 weeks after infection is associated with control of bacterial growth (as in Fig. 1), replication of the mutant, which recruited 10-fold fewer CD4+ and CD8+ cells at 4 weeks, was inhibited to the same degree as wild type despite the influx of fewer T cells. This observation suggests that even modest cellular immune responses are sufficient to contain bacterial replication in a steady state at 4–6 weeks in the lungs and that the 10-fold greater numbers of CD4+ and CD8+ cells recruited by wild-type bacilli may constitute an overly exuberant cellular response that results in accelerated tissue damage and mortality.
While microarray analysis shows a role for SigH in the induction of an intracellular system of redox and stress response proteins to repair oxidative damage, SigH does not control the transcription of genes for well-characterized cell surface antigens or secreted proteins, which might explain in part its failure to elicit the typical cellular immune responses in the lung. One explanation for disease delay phenotype is that lack of SigH-mediated redox cycling may generate a higher proportion of oxidized or denatured bacterial antigens from the mutant that do not elicit tissue-damaging immune responses in the lung. Alternatively, the delay in disease progression may be independent of the mutant's impaired stress responses, but instead may be explained by diminished synthesis or release of pro-inflammatory surface antigens or secreted molecules that are SigH-dependent. In support of this notion is the fact that 30% of SigH-dependent genes are hypothetical proteins of unknown function, some of which may represent such antigens; additionally several transport proteins (MT2468–2469 sulfate ABC transporters and MT1297 efflux protein) and are controlled by SigH, suggesting diminished secretion of potentially pro-inflammatory molecules.
The unusual phenotype of the M. tuberculosis ΔsigH mutant emphasizes the importance of testing virulence by measures such as pathologic and time-to-death analysis in addition to bacterial cfu counts. The reduced immunopathology phenotype elicited by bacilli lacking a nonessential σ factor indicates that some M. tuberculosis pathogenesis mechanisms are independent of those required for survival within the host.
Supplementary Material
Acknowledgments
We thank Naomi Gauchet for helpful assistance. This work was supported by grants from the National Institutes of Health (AI 36973, AI 37856, AI 43843, and D43-TW00010), the National Vaccine Program Office, the Potts Foundation, and the Becton Dickinson Corp.
Abbreviations
- cfu
colony-forming unit(s)
- SCID
severe combined immunodeficient
- TNF-α
tumor necrosis factor α
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Dye C, Scheele S, Dolin P, Pathania V, Raviglione M C. J Am Med Assoc. 1999;282:677–686. doi: 10.1001/jama.282.7.677. [DOI] [PubMed] [Google Scholar]
- 2.Mitsos L M, Cardon L R, Fortin A, Ryan L, LaCourse R, North R J, Gros P. Genes Immun. 2000;1:467–477. doi: 10.1038/sj.gene.6363712. [DOI] [PubMed] [Google Scholar]
- 3.Turner J, Frank A A, Brooks J V, Marietta P M, Orme I M. Immunology. 2001;102:248–253. doi: 10.1046/j.1365-2567.2001.01161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kramnik I, Dietrich W F, Demant P, Bloom B R. Proc Natl Acad Sci USA. 2000;97:8560–8565. doi: 10.1073/pnas.150227197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherman D R, Sabo P J, Hickey M J, Arain T M, Mahairas G G, Yuan Y, Barry C E, 3rd, Stover C K. Proc Natl Acad Sci USA. 1995;92:6625–6629. doi: 10.1073/pnas.92.14.6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.St John G, Brot N, Ruan J, Erdjument-Bromage H, Tempst P, Weissbach H, Nathan C. Proc Natl Acad Sci USA. 2001;98:9901–9906. doi: 10.1073/pnas.161295398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Storz G, Tartaglia L A, Ames B N. Science. 1990;248:189–194. doi: 10.1126/science.2183352. [DOI] [PubMed] [Google Scholar]
- 8.Deretic V, Song J, Pagan-Ramos E. Trends Microbiol. 1997;5:367–372. doi: 10.1016/S0966-842X(97)01112-8. [DOI] [PubMed] [Google Scholar]
- 9.Manca C, Paul S, Barry C E, 3rd, Freedman V H, Kaplan G. Infect Immun. 1999;67:74–79. doi: 10.1128/iai.67.1.74-79.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piddington D L, Kashkouli A, Buchmeier N A. Infect Immun. 2000;68:4518–4522. doi: 10.1128/iai.68.8.4518-4522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sherman D R, Mdluli K, Hickey M J, Arain T M, Morris S L, Barry C E, 3rd, Stover C K. Science. 1996;272:1641–1643. doi: 10.1126/science.272.5268.1641. [DOI] [PubMed] [Google Scholar]
- 12.Holmgren A. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 13.Holmgren A. J Biol Chem. 1989;264:13963–13966. [PubMed] [Google Scholar]
- 14.Mitsui A, Hirakawa T, Yodoi J. Biochem Biophys Res Commun. 1992;186:1220–1226. doi: 10.1016/s0006-291x(05)81536-0. [DOI] [PubMed] [Google Scholar]
- 15.Carmel-Harel O, Storz G. Annu Rev Microbiol. 2000;54:439–461. doi: 10.1146/annurev.micro.54.1.439. [DOI] [PubMed] [Google Scholar]
- 16.Hondalus M K, Bardarov S, Russell R, Chan J, Jacobs W R, Jr, Bloom B R. Infect Immun. 2000;68:2888–2898. doi: 10.1128/iai.68.5.2888-2898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKinney J D, Honer zu Bentrup K, Munoz-Elias E J, Miczak A, Chen B, Chan W T, Swenson D, Sacchettini J C, Jacobs W R, Jr, Russell D G. Nature (London) 2000;406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 18.Glickman M S, Cox J S, Jacobs W R., Jr Mol Cell. 2000;5:717–727. doi: 10.1016/s1097-2765(00)80250-6. [DOI] [PubMed] [Google Scholar]
- 19.Dunn P L, North R J. J Med Microbiol. 1996;45:103–109. doi: 10.1099/00222615-45-2-103. [DOI] [PubMed] [Google Scholar]
- 20.Manganelli R, Dubnau E, Tyagi S, Kramer F R, Smith I. Mol Microbiol. 1999;31:715–724. doi: 10.1046/j.1365-2958.1999.01212.x. [DOI] [PubMed] [Google Scholar]
- 21.Graham J E, Clark-Curtiss J E. Proc Natl Acad Sci USA. 1999;96:11554–11559. doi: 10.1073/pnas.96.20.11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lonetto M A, Brown K L, Rudd K E, Buttner M J. Proc Natl Acad Sci USA. 1994;91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandes N D, Wu Q L, Kong D, Puyang X, Garg S, Husson R N. J Bacteriol. 1999;181:4266–4274. doi: 10.1128/jb.181.14.4266-4274.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee M H, Pascopella L, Jacobs W R, Jr, Hatfull G F. Proc Natl Acad Sci USA. 1991;88:3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhoades E R, Frank A A, Orme I M. Tuber Lung Dis. 1997;78:57–66. doi: 10.1016/s0962-8479(97)90016-2. [DOI] [PubMed] [Google Scholar]
- 26.Prussin C, Metcalfe D D. J Immunol Methods. 1995;188:117–128. doi: 10.1016/0022-1759(95)00209-x. [DOI] [PubMed] [Google Scholar]
- 27.Serbina N V, Flynn J L. Infect Immun. 1999;67:3980–3988. doi: 10.1128/iai.67.8.3980-3988.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caruso A M, Serbina N, Klein E, Triebold K, Bloom B R, Flynn J L. J Immunol. 1999;162:5407–5416. [PubMed] [Google Scholar]
- 29.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, 3rd, et al. Nature (London) 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 30.Valway S E, Sanchez M P, Shinnick T F, Orme I, Agerton T, Hoy D, Jones J S, Westmoreland H, Onorato I M. N Engl J Med. 1998;338:633–639. doi: 10.1056/NEJM199803053381001. [DOI] [PubMed] [Google Scholar]
- 31.Orme I M, Miller E S, Roberts A D, Furney S K, Griffin J P, Dobos K M, Chi D, Rivoire B, Brennan P J. J Immunol. 1992;148:189–196. [PubMed] [Google Scholar]
- 32.Flynn J L, Goldstein M M, Triebold K J, Koller B, Bloom B R. Proc Natl Acad Sci USA. 1992;89:12013–12017. doi: 10.1073/pnas.89.24.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Behr M A, Wilson M A, Gill W P, Salamon H, Schoolnik G K, Rane S, Small P M. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- 34.Wilson M, DeRisi J, Kristensen H H, Imboden P, Rane S, Brown P O, Schoolnik G K. Proc Natl Acad Sci USA. 1999;96:12833–12838. doi: 10.1073/pnas.96.22.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sherman D R, Voskuil M, Schnappinger D, Liao R, Harrell M I, Schoolnik G K. Proc Natl Acad Sci USA. 2001;98:7534–7539. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manganelli R, Voskuil M I, Schoolnik G K, Smith I. Mol Microbiol. 2001;41:423–437. doi: 10.1046/j.1365-2958.2001.02525.x. [DOI] [PubMed] [Google Scholar]
- 37.Motohashi K, Watanabe Y, Yohda M, Yoshida M. Proc Natl Acad Sci USA. 1999;96:7184–7189. doi: 10.1073/pnas.96.13.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paget M S, Kang J G, Roe J H, Buttner M J. EMBO J. 1998;17:5776–5782. doi: 10.1093/emboj/17.19.5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang J G, Paget M S, Seok Y J, Hahn M Y, Bae J B, Hahn J S, Kleanthous C, Buttner M J, Roe J H. EMBO J. 1999;18:4292–4298. doi: 10.1093/emboj/18.15.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paget M S, Bae J B, Hahn M Y, Li W, Kleanthous C, Roe J H, Buttner M J. Mol Microbiol. 2001;39:1036–1047. doi: 10.1046/j.1365-2958.2001.02298.x. [DOI] [PubMed] [Google Scholar]
- 41.Raman S, Song T, Puyang X, Bardarov S, Jacobs W R, Jr, Husson R N. J Bacteriol. 2001;183:6119–6125. doi: 10.1128/JB.183.20.6119-6125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steyn A J C, Collins D M, Hondalus M K, Jacobs W R, Kawakami R P, Bloom B R. Proc Natl Acad Sci USA. 2002;99:3147–3152. doi: 10.1073/pnas.052705399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dannenberg A M., Jr Am Rev Respir Dis. 1982;125:25–29. doi: 10.1164/arrd.1982.125.3P2.25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.