Abstract
Infection of a human melanoma cell line by a retroviral vector resulted in transmission of a mouse VL30 (mVL30-1) retroelement RNA to some of the cells infected by the retrovirus, followed by synthesis, integration, and expression of the mVL30-1 cDNA. One vector carried a tissue factor (TF) transgene that generated high TF melanoma clones, and another vector was a control without the TF transgene that generated low TF clones. Some high TF melanoma clones contained the mVL30-1 retroelement and others did not, and some low TF melanoma clones contained the mVL30-1 retroelement and others did not. Each type of melanoma clone was tested for its metastatic potential in severe combined immunodeficient (SCID) mice, by i.v. injection of the cells to generate lung tumors. None of the low TF clones that either contained or lacked the mVL30-1 retroelement generated lung tumors, consistent with earlier results showing that high TF expression promoted metastasis. The high TF clones containing the mVL30-1 retroelement were strongly metastatic, in contrast to the high TF clones lacking the mVL30-1 retroelement, which were weakly metastatic. Southern hybridization analyses showed that the mVL30-1 cDNA integrated into different genomic sites in different melanoma clones, suggesting that the effect of the mVL30-1 retroelement on metastasis depends not on integration per se but instead on expression of the mVL30-1 RNA. A role for the mVL30-1 RNA in metastasis and possibly other cell functions is an unexpected finding, because the RNA appears to lack significant coding potential for a functional protein. The metastatic effect might be mediated directly by a noncoding mVL30-1 RNA or by a peptide or small protein encoded by one of the short ORFs in the mVL30-1 RNA.
A mouse VL30 retrotransposable element (mVL30 retroelement) was first identified as a 30S RNA molecule encapsulated in mouse leukemia virus (MLV) particles that were produced in a mouse packaging cell line (1–4). Mouse leukemia virus and other retroviruses can transmit an encapsulated mVL30 RNA to cells infected by the retrovirus, resulting in the synthesis of VL30 cDNA mediated by retrovirus-encoded reverse transcriptase, followed by integration of the mVL30 cDNA into the cell genome (5, 6). This remarkable process of retrovirus-mediated transmission of a retroelement RNA provides a mechanism for dissemination of an otherwise noninfectious retroelement, limited only by the host range of the retrovirus.
The mVL30 genome has several structural features of a retroviral genome. These features include direct 5′ and 3′ long terminal repeats (LTRs), a tRNA primer binding site, a polypurine tract, a splice donor site, a packaging signal, and an internal region containing about 3.7 kb (7). In contrast to the internal region of a retroviral genome that encodes the gag, pol, and env proteins required to form an infectious retrovirus, the corresponding region of a mVL30 genome has numerous stop codons in all reading frames. A limited homology of the mVL30 genome to the gag and pol genes of mouse leukemia virus suggests that the mVL30 genome evolved as a densely mutated noninfectious descendent of an infectious retrovirus. A mouse genome contains upwards of 200 members of the mVL30 family, varying in size from solo LTRs to a complete mVL30 genome. Some integrated mVL30 retroelements are transcribed, generating a mVL30 RNA that is potentially infectious, depending on the availability of a retroviral carrier.
Because retroviral vectors are widely used in human gene therapy protocols, and several packaging cell lines for constructing the vectors are derived from murine components, there is a significant risk that a mVL30 RNA will be transmitted to patients participating in such a gene therapy trial (8). Integration of the mVL30 cDNA could generate a somatic mutation that activates or inactivates a cellular gene, and recombination of the mVL30 cDNA with a cellular gene could lead to transmission of the cellular gene to other cells. The latter mechanism probably was involved in the formation of the oncogenic sarcoma viruses Ha-MuSV and Ki-MuSV (9–11).
In an earlier study, it was reported that the metastatic potential of a human melanoma cell line was increased by retrovirus-mediated transfection of tissue factor cDNA, resulting in high level expression of tissue factor on the cell surface (12). In the study reported here, we show that the increase in metastatic potential of the transfected melanoma cells depends not only on tissue factor expression, but also on transmission of the mVL30-1 RNA, presumably by the retroviral vector, and integration and expression of the mVL30-1 cDNA.
Materials and Methods
Cell Lines.
The human melanoma cell line YU-SIT1, the retroviral packaging cell lines PE501 and PA317, and several of the retrovirus-mediated transfected melanoma clones have been described (12). The cells were grown as attached monolayers in DMEM containing 10% FBS, penicillin, and streptomycin in a CO2 incubator at 37°C. Cultures were routinely monitored for mycoplasma contamination, which was not detected in any of the cultures.
Retrovirus-Mediated Transfection of Melanoma Cells.
The retroviral vector LXSN (12) was used for transfection of a cDNA encoding human TF into YU-SIT1 cells. The cells were plated at a density of 5 × 105 cells per 6-cm culture dish and incubated overnight. On the following day, the medium was replaced with a solution containing either the vector carrying a TF cDNA insert or a control vector without the insert and supplemented with 16 μg Polybrene. The cells were incubated for 24 h, and then were replated in culture media containing 1 mg/ml G418 and incubated until G418-resistant colonies were formed. Single clones were prepared by generating cell lines from the colonies.
TF Expression.
Cell surface expression of TF was analyzed by flow cytometry, as follows (12). Melanoma cells were detached from a culture flask by a brief incubation with PBS containing 2 mM EDTA, blocked with PBS containing 2% BSA, and suspended for 20 min at 4°C in PBS containing polyclonal rabbit anti-TF antibody or rabbit IgG as a control. After washing the cells with PBS to remove unbound antibody, the cells were incubated for 20 min at 4°C with goat anti-rabbit IgG antibody conjugated to fluorescein, washed with PBS, and analyzed on a Becton Dickinson fluorescence-activated cell sorter (FACS) Start Analyzer. All of the clones transfected with the TF cDNA showed a comparable high level of TF expression, as indicated by the displacement of the FACS curve (12).
Analysis of Differential Gene Expression in Retrovirus-Infected Melanoma Cells.
The procedure used has been described (13). Total RNA was isolated from melanoma cells grown to 60–70% confluence, and 0.2 μg of the RNA was reverse transcribed by using a preamplification kit (GIBCO/BRL) with 3′ anchor primers (E1T12M) containing the sequences CGGAATTCGGGT-12M (M = A,G,C). The resulting single-strand cDNAs were amplified by PCR using three E1T12M primers and 24 different arbitrary primers containing the sequences CGTGAATTCG-arbitrary 10mers. The PCR mixture contained 0.4 μM of each primer, 2.0 μM dNTP, 10 μCi (1 Ci = 37 GBq) [35S]dATP (1200 Ci/mmol) and 1.0 unit of AmpliTaq DNA polymerase. PCR was done under low stringency annealing conditions and differential display was done under high stringency annealing conditions. The thermal cycling parameters were as follows. For low stringency annealing of primers, one cycle at 95° for 1 min, 40°C for 4 min, and 72°C for 1 min. For high stringency annealing of primers, 35 cycles at 94°C for 45 s, 60°C for 2 min, and 72°C for 1 min. The reaction products were separated on a 6% polyacrylamide DNA sequencing gel, and the gel was autoradiographed. The experiment was done in triplicate by using three independent RNA preparations. The cDNA bands showing consistent differential expression were cloned into a TA vector (Invitrogen) and sequenced. Differential expression of each band was checked by reverse transcription (RT)-PCR.
Synthesis and Sequencing of VL30-1 cDNA Derived from RNA of Retrovirus-Infected Melanoma Cells.
RNA isolation.
Melanoma cells were grown as attached cultures to about 80% confluence in a 75-cm2 flask, washed with ice-cold PBS, lysed in 7.5 ml TRIZOL (GIBCO/BRL), and incubated for 5 min at 25°C. Chloroform was added, and the mixture was agitated vigorously for 15 s, incubated for 3 min at 25°C, and centrifuged at 12,000 × g for 15 min at 4°C. The aqueous phase was transferred to a fresh tube, 3.75 ml isopropyl alcohol was added, and the solution was incubated for 10 min at 25°C and centrifuged at 12,000 × g for 10 min at 4°C. The RNA pellet was rinsed with 75% ethanol, air-dried, and dissolved in 50 μl H2O.
cDNA synthesis.
One microliter of a 20-mM oligo(dT) primer solution was added to 1.0 μg RNA in 11.5 μl H2O. The RNA/oligo(dT) mixture was kept for 2 min at 70°C and chilled on ice, and 3.5 μl of 5× reaction buffer was added; the buffer contained 1.0 μl 100 mM DTT, μl dNTP mix (10 mM each), 0.5 μl RNase inhibitor (40 units/μl), and 1.0 μl Moloney murine leukemia virus (MMLV) reverse transcriptase (20 units/μl). The mixture was incubated for 1 h at 42°C and then for 5 min at 94°C to stop the reaction.
The PCR procedure was done in a solution containing 25.0 μl of Taq PCR master mix, 2.0 μl of cDNA, 21.0 μl of H2O, and 1.0 μl of of primers 1 and 2: primer 1, GAGCTCTGGTCTGTGTGCTTTCATGT (5′ LTR 441–466); primer 2, TACACCTTTGCACAGGGTAGAGGAGT (3′ LTR 384–359). The thermal cycling parameters were 5 min at 94°C, 45 s at 94°C, 1 min at 55°C, and 3 min at 72°C, for a total of 35 cycles, followed by 10 min at 72°C. The PCR product was electrophoresed in 1.0% agarose gel, and the 4.3-kb SVL-1 band was isolated, cloned into the pCR4-TOPO vector (CLONTECH), and sequenced by using T3 and T7 primers.
Sequencing the Complete LTR in SVL-1 cDNA.
The PCR was done in a solution containing 25.0 μl of Taq PCR master mix, 2.0 μl of cDNA, 21.0 μl of H2O, and 1.0 μl each of the following primers: primer 3, TGAAGAATAAAAAATTACTGGCC (left end of the LTR); primer 4, TGAAAGATTTTCGAATTCCCGG (right end of the LTR). The thermal cycling parameters were 5 min at 94°C, 45 s at 94°C, 1 min at 55°C, and 2 min at 72°C for 35 cycles, followed by 10 min at 72°C. The PCR product was electrophoresed in 1.0% agarose gel, and the 600-bp band was isolated, cloned into the pCR4-TOPO vector, and sequenced by using T3 primer.
PCR Screens for SVL-1 cDNA in Genomic DNA of Melanomal Clones.
Genomic DNA was isolated from melanoma cells by using a DNeasy tissue kit (Qiagen). The PCR procedure was done in a solution containing 25.0 μl of Taq PCR master mix, 1.0 μl of a 1.0 mg/ml solution of DNA, 1.0 μl of primer 4 (see above), 1.0 μl of primer 5 (see below), and 22 μl of H2O. Primer 5: CTTCATGACCAAACCCTTCA. The thermal cycling parameters were 5 min at 94°C, 45 s at 94°C, 1 min at 59°C, and 3 min at 72°C for 30 cycles, followed by 10 min at 72°C.
RT-PCR Screens for SVL-1 RNA in Melanoma Clones.
The RNA isolation and RT-PCR procedures were done as described above. For the final PCR step, the reaction mixture contained 25.0 μl Taq PCR master mix, 1.0 μl cDNA, 1.0 μl primer 5 (see above),1.0 μl Primer 6 and 22.0 μl H2O. Primer 6: GTATGAGTTTCTTCTGCCA. The thermal cycling parameters were 5 min at 94°C, 45 s at 94°C, 1 min at 58°C, and 2 min at 72°C for 30 cycles, followed by 10 min at 72°C.
SCID Mouse Model of Metastasis.
Human melanoma cells were detached from the culture flask by a brief incubation with PBS containing 2 mM EDTA, suspended in DMEM/10% FBS, washed, and resuspended in PBS. Female 6-week-old SCID CB-17 mice (Charles River Breeding Laboratories) were injected via the lateral tail vein with 6 × 105 melanoma cells. The animals were euthanized and dissected 10 to11 weeks later, and the number of tumors on the surface of the lungs was counted.
Southern Blot Analysis for SVL-1 RNA in Retrovirus-Infected Melanoma Clones.
Genomic DNA was isolated from cells with the DNeasy Tissue kit (Qiagen). DNA (10 g) was digested with PstI and electrophoresed on a 0.7% agarose gel in a Tris-acetate electrophoresis buffer. The digested DNA was transferred to a nylon membrane (Schleicher & Schuell) and fixed at 80°C for 2 h. Probe was generated by PCR with primer 7 and primer 8: primer 7, GTGTATACTGTAAAGAAA; primer 8, CTTTTAGCCACGAACAGG. Probe was labeled with 32P. Hybridization was performed with ExpressHyb Hybridization Solution (CLONTECH) as recommended by the manufacturer.
Results
The initial purpose of this study was to determine whether TF activates an intracellular signaling pathway in human melanoma cells that results in altered gene expression and enhanced metastatic potential (12). A test for differential gene expression was done with two cloned lines derived from the human melanoma line YU-SIT1, which expresses a low level of TF and is weakly metastatic in SCID mice. One clone (T2) was infected with the retroviral vector LXSN containing a TF cDNA insert; this clone expresses a high level of TF and is strongly metastatic in SCID mice. The other clone (L8) was infected with the LXSN control vector lacking the TF insert; this clone expresses a low level of TF and is weakly metastatic in SCID mice, similar to the parental line YU-SIT1. The test for differential gene expression involved screening cDNA libraries prepared from the RNA of the T2 and L8 clones. The cDNAs from each clone were fractionated by gel electrophoresis, and the banding patterns in the gels were compared. The only reproducible difference detected was an intense cDNA band present in the T2 gel but not in the L8 gel. The T2 cDNA was sequenced and found to be identical to the DNA spanning positions 1517 to 2945 in the mVL30 clone NVL-3 (7). To determine whether the T2 cDNA was derived from a complete VL30 RNA, a longer T2 cDNA was synthesized by RT-PCR using primers derived from conserved sequences in mouse VL30 LTRs (7). The longer T2 cDNA contains 4,939 bp, with the characteristic organization of a complete mVL30 retroelement (Fig. 1). Comparison of the sequence of this mVL30 clone, called mVL30-1, with the NVL-3 sequence showed 65% identity for the LTRs and 99% identity for the internal region between the LTRs. Most of the sequence divergence occurs in the U3 region of the LTR, as reported for other mVL30 clones (7).
Figure 1.
(A) Sequence and (B) map of the complete mVL30-1 cDNA synthesized by RT-PCR from the RNA of a human melanoma clone infected with a retroviral vector. The regions identified on the map are as follows. U3, 1–340 and 4321–4670; R, 341–544 and 4671–4864; U5, 545–619 and 4865–4939; tR (tRNA primer binding site), 622–639; SD (splice donor site), 676–682; and PP (polypurine tract), 4305–4322
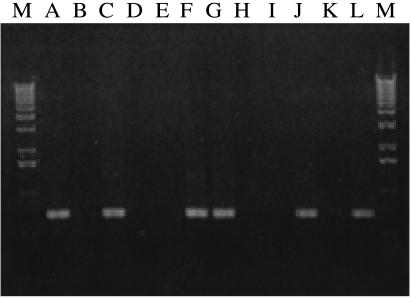
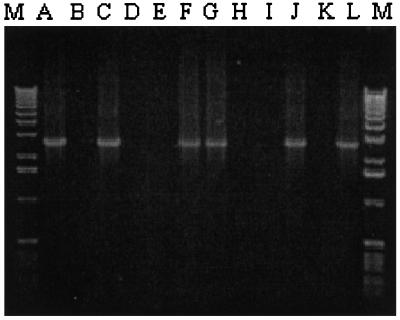
Additional human melanoma clones were generated by infection of the parental line YU-SIT1 with the retroviral vector containing a TF cDNA insert or with the control vector, and the clones were tested for mVL30-1 RNA by RT-PCR and for mVL30-1 cDNA in the genomic DNA by PCR. Both the mVL30-1 RNA and cDNA were present in four of the eight high TF clones and in two of the four low TF clones examined (Figs. 2 and 3). The other high TF and low TF clones, and also the parental line YU-SIT1, did not contain either mVL30-1 RNA or cDNA. Thus, about 50% of the melanoma cells infected by either retroviral vector integrate and express mVL30-1 RNA, which presumably was transmitted to the cells by the infecting retrovirus. It was fortuitous for this study that a mVL30-1 RNA was transmitted and integrated into the founder cell of the high TF clone but not of the low TF clone used in the screen for differential gene expression.
Figure 2.
RT-PCR screen for mVL30-1 RNA in human melanoma clones infected with a retroviral vector. The samples were electrophoresed in agarose gel and photographed. The clone tested in each lane is as follows. A, T2; B, T4; C, Td; D, Te; E, Tf; F, Th; G, Tj; H, Tk; I, L1; J, L3; K, L4; L, L8; and M, 1 kb DNA ladder. The size of the RT-PCR products is 728 bp.
Figure 3.
Screen for mVL30-1 cDNA in the genomic DNA of human melanoma clones infected with a retroviral vector. The clones tested in each lane are the same as in Fig. 2. The size of the PCR products is 2,652 bp.
The metastatic potential of the eight high TF melanoma clones and the four low TF melanoma clones was determined in a SCID mouse model, which involves injecting the melanoma cells intravenously and counting the number of lung tumors formed several weeks later (Tables 1 and 2). None of the low TF clones, either those that contain or lack mVL30-1 RNA, produced any lung tumors. The high TF clones lacking mVL30-1 RNA produced an average of 2.1 lung tumors, as compared with an average of 26.7 lung tumors produced by the high TF clones containing mVL30-1 RNA. These results confirm earlier results showing that the metastatic potential of melanoma clones derived by retrovirus-mediated transfection of the YU-SIT1 cell line depends on the level of expression of TF protein on the cell surface (12). An additional finding is that the metastatic potential of the melanoma clones also depends on integration and expression of a mVL30 cDNA.
Table 1.
Effect of high TF and the mVL30-1 retroelement on the metastatic potential of human melanoma clones in SCID mice
| Melanoma clone | Total mice per clone | mVL30-1 RNA and cDNA | Number of lung tumors per mouse |
|---|---|---|---|
| T2 | 9 | + | 18, 0, 1, 0, 39, 0, 30, 4, 13 |
| Td | 3 | + | 3, 21, 22 |
| Th | 4 | + | 46, 71, 20, 71 |
| Tj | 2 | + | 114, 25 |
| Total: 480 | |||
| Average per mouse: 26.7 | |||
| T4 | 14 | − | 0, 1, 0, 0, 0, 1, 0, 1, 0, 0, 1, 0, 2, 0 |
| Te | 2 | − | 2, 0 |
| Tf | 3 | − | 23, 0, 0 |
| Tk | 3 | − | 7, 8, 0 |
| Total: 46 | |||
| Average per mouse: 2.1 |
Table 2.
Effect of low TF and the mVL30-1 retroelement on the metastatic potential of human melanoma clones in SCID mice
| Melanoma clone | Total mice per clone | mVL30-1 | Number of lung tumors per mouse |
|---|---|---|---|
| L3 | 4 | + | 0, 0, 0, 0 |
| L8 | 3 | + | 0, 0, 0 |
| Total: 0 | |||
| Average per mouse: 0 | |||
| L1 | 3 | − | 0, 0, 0 |
| L4 | 3 | − | 0, 0, 0 |
| Total: 0 | |||
| Average per mouse: 0 |
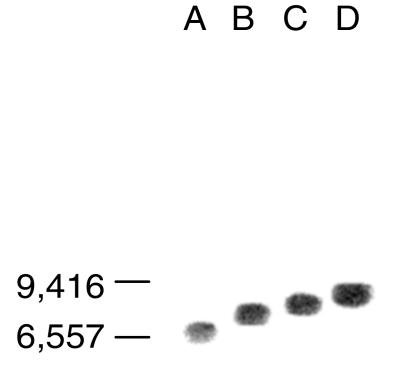
To determine whether the metastatic effect of the mVL30-1 retroelement depends on the integration site of the mVL30-1 cDNA, the integration site in four melanoma clones was compared by Southern hybridization (Fig. 4). The results show that the mVL30-1 cDNA integrated at a different site in each clone, suggesting that the metastatic effect does not depend on the integration site.
Figure 4.
Comparison of the genomic insertion sites of mVL30-1 cDNA in human melanoma clones infected with a retroviral vector. Genomic DNA was digested with HindIII and analyzed by Southern hybridization. The clone tested in each lane is as follows: A, T2; B, Td; C, Th; and D, Tj
Discussion
Retroviruses prepared in a mouse packaging cell line can encapsulate and transmit a mouse VL30 (mVL30) RNA to retrovirus-infected cells, which synthesize, integrate, and express the mVL30 cDNA (reviewed in ref. 7). Earlier experiments showed that the metastatic potential of a human melanoma cell line was increased by retrovirus-mediated transfection of TF cDNA, resulting in high level expression of TF on the tumor cell surface (12). The experiments reported here show that some of the retrovirally infected melanoma cells contain a mVL30 cDNA and RNA. The retroviral vector presumably transmitted to the melanoma cells the mVL30 RNA that was reverse-transcribed by the retroviral enzyme, and the resulting cDNA was integrated into the melanoma cell genome and transcribed.
The genome of the mVL30 retroelement transmitted to human melanoma cells, called mVL30-1, has a retrovirus-like organization, with 5′ and 3′ LTRs that flank a 4-kb internal region, similar to other full-length mVL30 genomes (Fig. 1). The internal region has multiple stop triplets in each reading frame, in contrast to the corresponding region of an infectious retrovirus that has three long ORFs encoding the gag, pol, and env genes, which are essential for the formation of mature infectious particles. Nevertheless, this region has retained about 99% sequence identity to the internal regions of at least one other mVL30 genome, whereas the U3 region of the LTR shows significant divergence within the mVL30 family (7). The strong sequence conservation of the internal region suggests that the sequence is required for an indispensable physiologic function.
The metastatic potential of clones of retrovirally infected human melanoma cells was tested in a SCID mouse model of metastasis, in which tumor cells are injected intravenously and the number of metastatic lung tumors formed by the bloodborne melanoma cells is determined (12). The melanoma cells were first infected either with a retroviral vector encoding TF cDNA to generate high TF clones, or with a control retroviral vector to generate low TF clones. None of the low TF melanoma clones had lung tumors, regardless of the presence or absence of mVL30-1 RNA and cDNA. The high TF melanoma clones containing mVL30-1 RNA and cDNA produced an average of 26.7 lung tumors, in contrast to the high TF clones lacking mVL30-1 RNA and cDNA, which produced an average of 2.1 lung tumors. These findings indicate that the metastatic potential of the melanoma clones depends on a high level expression of TF protein on the tumor cell surface, as reported previously (12), and also on the integration of mVL30-1 cDNA and expression of mVL30-1 RNA. One mechanism for the metastatic effect of a retroelement is the integration of the cDNA into a site that causes disruption of the regulation or function of a key gene involved in oncogenesis. The finding that the mVL30-1 cDNA integrated into different genomic sites in several of the metastatic melanoma clones suggests that metastasis depends not on integration per se but instead on expression of the mVL30-1 RNA. A role for the mVL30-1 RNA in metastasis and possibly other cell functions is an unexpected finding, because the RNA appears to lack significant coding potential for a functional protein. The metastatic effect might be mediated directly by noncoding mVL30-1 RNA or by a polypeptide encoded by one of the short ORFs in the RNA. Because a mVL30 retroelement can be transmitted to human cells in the course of a retrovirus-mediated gene therapy procedure, there is a risk that the retroelement could promote an oncogenic response in an infected cell.
Acknowledgments
The research reported here was supported in part by a Program Project Grant HL29019 from the National Institutes of Health and by generous gifts from private donors.
Abbreviations
- LTR
long terminal repeat
- TF
tissue factor
- RT-PCR
reverse transcription–PCR
- SCID
severe combined immunodeficient
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF486451).
References
- 1.Howk R S, Troxler D H, Lowy D, Duesberg P H, Scolnick E M. J Virol. 1978;25:115–123. doi: 10.1128/jvi.25.1.115-123.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sherwin S A, Rapp U R, Benveniste R E, Sen A, Todaro G J. J Virol. 1978;26:257–264. doi: 10.1128/jvi.26.2.257-264.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besmer P, Olshevsky U, Baltimore D, Dolberg D, Fan H. J Virol. 1979;29:1168–1176. doi: 10.1128/jvi.29.3.1168-1176.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams S E, Rathjen P D, Stanway C A, Fulton S M, Malim M H, Wilson W, Ogden J, King L, Kingsman S M, Kingsman A J. Mol Cell Biol. 1988;8:2989–2998. doi: 10.1128/mcb.8.8.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scolnick E M, Vass W C, Howk R S, Duesberg P H. J Virol. 1979;29:964–972. doi: 10.1128/jvi.29.3.964-972.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter A T, Norton J D, Avery R J. Nucleic Acids Res. 1983;11:6243–6254. doi: 10.1093/nar/11.18.6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.French N S, Norton J D. Biochim Biophys Acta. 1997;1352:33–47. doi: 10.1016/s0167-4781(97)00009-2. [DOI] [PubMed] [Google Scholar]
- 8.Chakraborty A K, Zink M A, Hodgson C P. Cancer Gene Ther. 1994;1:113–118. [PubMed] [Google Scholar]
- 9.Giri C P, Hodgson C P, Elder P K, Courtney M G, Getz M J. Nucleic Acids Res. 1983;11:305–319. doi: 10.1093/nar/11.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roop D R, Lowy D R, Tambourin P E, Strickland J, Harper J R, Balaschak M, Spangler E F, Yuspa S H. Nature (London) 1986;323:822–824. doi: 10.1038/323822a0. [DOI] [PubMed] [Google Scholar]
- 11.Velu T J, Vass W C, Lowy D R, Tambourin P E. J Virol. 1989;63:1384–1392. doi: 10.1128/jvi.63.3.1384-1392.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bromberg M E, Konigsberg W H, Madison J F, Pawashe A, Garen A. Proc Natl Acad Sci USA. 1995;92:8205–8209. doi: 10.1073/pnas.92.18.8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao S, Ooi S, Pardee B A. BioTechniques. 1995;18:842–850. [PubMed] [Google Scholar]