Abstract
Electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry coupled with capillary reverse-phase liquid chromatography was used to characterize intact proteins from the large subunit of the yeast ribosome. High mass measurement accuracy, achieved by “mass locking” with an internal standard from a dual electrospray ionization source, allowed identification of ribosomal proteins. Analyses of the intact proteins revealed information on cotranslational and posttranslational modifications of the ribosomal proteins that included loss of the initiating methionine, acetylation, methylation, and proteolytic maturation. High-resolution separations permitted differentiation of protein isoforms having high structural similarity as well as proteins from their modified forms, facilitating unequivocal assignments. The study identified 42 of the 43 core large ribosomal subunit proteins and 58 (of 64 possible) core large subunit protein isoforms having unique masses in a single analysis. These results demonstrate the basis for the high-throughput analyses of complex mixtures of intact proteins, which we believe will be an important complement to other approaches for defining protein modifications and their changes resulting from physiological processes or environmental perturbations.
Mass spectrometry has evolved into a powerful tool for analyzing biomolecules because of the development of matrix-assisted laser desorption ionization (1, 2) and electrospray ionization (ESI) (3) and advances in both mass analyzers and data processing capabilities. With these ionization methods, which are amenable to megadalton-size molecules, the broadly useful detection and characterization of biopolymers by mass spectrometric methods are feasible. For example, MS has become a preferred analytical tool for proteome analyses, in which dynamic populations of proteins are identified and quantified. Proteins are now routinely identified by mass spectrometric and tandem mass spectrometric analysis of proteolytic digests of individual protein spots on two-dimensional (2D) PAGE (4). However, the intact protein level analyses of complex protein mixtures, while potentially providing complementary and direct information for protein identification, have been a much greater experimental challenge and only rarely attempted (5, 6).
Many pivotal cellular processes are not carried out by individual proteins, but rather by large protein–protein complexes and protein–nucleic acid complexes. The analyses of such complexes have been greatly expanded by improvements in both biological separations and MS. One of the more complicated protein complexes yet studied by MS is the cytosolic ribosome complex. Efforts to define the protein composition (7, 8), modifications (9), and subunit interactions (10) of ribosomes generally have mirrored the development of biomolecular analysis techniques and experimental approaches for the study of noncovalent protein complexes.
Ribosomes are the canonical example of ribonucleoprotein complexes in both prokaryotes and eukaryotes and are responsible for cellular protein and polypeptide synthesis (translation) (11). This key biological process requires a vast investment of cellular resources, including the production of ribosomal RNA, ribosomal proteins, and accessory factors, with the result that the ribosomes are among the most abundant structures found in the cell (12). Recent structural studies of ribosomes indicate that nascent peptide bond formation is accomplished by a ribozyme core (13) and that the associated ribosomal proteins serve to maintain ribosomal structure, regulate ribosomal activity, and cotranslationally modify the nascent peptide as it exits the ribosome complex (10, 14–16). Cytoplasmic ribosomes are composed of the large and small subunits that are synthesized separately in a multistage process (17). The mature large and small subunits are assembled on cytosolic messenger RNAs and interact with many additional factors to undertake protein synthesis. Although considerably larger and more complex than prokaryotic ribosomes, the overall structure and functional regions of yeast cytosolic ribosomes are highly conserved (18). The yeast ribosome is also useful as a model system for the study of translation in higher eukaryotes, as yeast and mammalian ribosomes have high homology in gross structure, active site structure, and subunit interactions (19) and share a complete set of homologous proteins, except for rat L28 that has no apparent yeast equivalent (20). There are currently 137 established ribosomal protein genes in Saccharomyces cerevisiae, which encode 79 unique ribosomal proteins (46 large subunit and 33 small subunit) (8). The physical and biochemical characterization of yeast ribosomes has been complicated not only by the large number of proteins involved, and their highly basic amino acid content, but also by the presence of modifications, often in combination, including N-terminal processing and acetylation (9), methylation (21, 22), and phosphorylation (23, 24).
In previous work, we have demonstrated the high-throughput mass spectrometric analysis of both digested and intact proteins, using capillary reverse-phase liquid chromatography (RPLC) and capillary isoelectric focusing, respectively, coupled online with Fourier transform ion cyclotron resonance (FTICR) (25, 26). A single dimension high-efficiency capillary RPLC with long capillary columns packed with small particles can provide a peak capacity of ≈1,000 for polypeptides at pressures approaching 10,000 psi (25). FTICR provides ultra-high sensitivity, mass resolution, and mass measurement accuracy (MMA). The combined technology of capillary RPLC and FTICR provides a basis for study of complex protein mixtures with high sensitivity and a broad dynamic range.
In this study, we demonstrate the utility and potential for high-throughput mass spectrometric analysis of intact ribosomal proteins by using a capillary RPLC/FTICR technique. We show the utilization of a dual electrospray source with online LC separation to achieve high MMA for intact proteins. The high MMA of FTICR coupled with a high-resolution capillary RPLC separation facilitated unambiguous assignments of ribosomal proteins, including closely related and modified proteins.
Methods
Preparation of Large Subunit Ribosomal Proteins.
Yeast large ribosomal subunits were prepared by using a substantially modified approach based on the ribosomal purification described by Warner and Gorenstein (27). Haploid yeast BY4702 (ATCC no. 200867) was grown at 30°C in yeast extract/peptone/dextrose broth (Difco) to midlog phase (OD600 = 6.0). A cell pellet was obtained by centrifugation (Beckman JLA 10.5 rotor, 3,000 g, 15 min, 4°C) and was washed once with cold distilled water. The washed pellet was resuspended in 3 vol lysis buffer (50 mM Tris-acetate, pH 7.0/50 mMNH4Cl/12 mM MgCl2), and cells were disrupted by three cycles of French press [SLM–Aminco, Urbana, IL; 18,000 psi (1 psi = 6.89 kPa)]. The lysate was centrifuged twice (JA-20 rotor, 30,000 g, 30 and 20 min, 4°C) to remove cell debris, and the spun lysate was quick-frozen with liquid nitrogen and stored at −80°C. The substitution of French press lysis for bead-beating permitted us to obtain the large subunit from the pelleted material after ultracentrifugation of the cleared yeast lysate through a salt-sucrose cushion. Our result is consistent with observations that glass bead lysis produced a sample rich in polysomes that were effectively disrupted in preparations produced by French press (28) and with studies where the effects of hydrostatic pressure on the complex stability has been directly applied to the study of subunit dissociation/reassociation in ribosomes from Escherichia coli (29, 30).
Thawed lysate was diluted with 3 vol lysis buffer + 5 mM DTT, then centrifuged to remove aggregated material (JA-20 rotor, 18,000 g, 15 min, 4°C). In individual ultracentrifuge tubes (SW-41 Ultraclear, Beckman), the diluted spun lysate (5 ml) was placed over a high-salt sucrose cushion [7.2 ml; 20 mM Tris⋅HCl, pH 7.5/5 mM MgCl2/500 mM KCl/10% (wt/wt) sucrose/1 mM DTT], and the ribosomal large subunits were pelleted by centrifugation (SW-41Ti rotor, 200,000 g, 330 min, 4°C). The pelleted subunits were resuspended in 0.5 ml lysis buffer, and insoluble material was removed by centrifugation (Eppendorf model 5814, 17,000 g, 5 min, 4°C). The resuspended subunits were quick-frozen and stored at −80°C until analyzed. A final protein concentration of 4.1 mg/ml was determined by using a modified Bradford protein assay (BioRad) and bovine IgG as a standard.
Acid extraction (31) was performed to remove nucleic acid contaminants before each analysis, and the samples were stored on ice until injected for LC separation. The resuspended subunits were combined with 0.1 vol of 1 M MgCl2, then 2 vol of glacial acetic acid (Fisher Scientific), and mixed by inversion (Barnstead/Thermolyne Labquake rotary mixer, Dubuque, IA, 30–120 min, 4°C). The insoluble fraction was removed by centrifugation (Eppendorf model 5814, 17,000 g, 30 min, 4°C). Acid extracts were prepared freshly before each analysis and stored on ice before injection.
Capillary RPLC.
The high pressure capillary RPLC system has been described in detail (25). Briefly, mobile phases (solvent A: 35% acetonitrile, 0.1% trifluoroacetic acid in water; solvent B: 90% acetonitrile, 0.1% trifluoroacetic acid in water) were delivered by two ISCO pumps (model 100DM) at 8,000 psi. The solvents were mixed in a stainless steel mixer (vol 2.5 ml) with a magnetic stirrer before a flow splitter and the packed capillary column (150 μm i.d. × 360 μm o.d., Polymicro Technologies, Phoenix). The column was prepared by using C-5 bonded particles (5-μm particle with 300-Å pore size, Phenomenex, Terrence, CA), as described (25). Data acquisition started at sample injection (20 μl) and the gradient was started 20 min after the sample injection. Because solvent A had high content of acetonitrile (35%) to prevent protein precipitation and maximize protein recovery, several proteins were observed to elute in the breakthrough/flow-through fraction before the gradient. Gradient speed was set by a fused silica capillary flow restrictor (30 μm i.d. × 360 μm o.d.) and measured with a UV detector (Spectra 100 UV/VIS detector, Spectra-Physics) at the wavelength of 215 nm. The gradients measured were as follows: 0% B for 20 min, from 0% to 73% B over 110 min, and 73% to 100% B over 20 min.
MS.
All experiments were performed by using a previously described external ion source 7 tesla FTICR mass spectrometer (32). Briefly, the instrument is equipped with three serial quadrupoles, which are separated by two conductance limits and are operated synchronously, to guide ions produced by the ESI source to a rectangular closed cell through four stages of differential pumping. The final pumping stage is efficiently pumped by a cryopump with two concentric cryopannels extending to ICR cell, providing a pressure in the cell below 10−9 Torr. The electrospray interface was equipped with an electrodynamic ion funnel to achieve high ion transmission efficiency from the atmospheric ESI source (33).
In FTICR, mass measurements are carried out by measuring cyclotron frequencies of trapped ions and converting to m/z values by using an appropriate calibration function (34). Although a recent report on external calibration, which uses the calibration equation with matching total ion currents, has shown to provide low ppm MMA (35), its use requires accurate control of trapped ion population. This approach is especially problematic for LC/MS analysis where the population of trapped ions varies significantly and nonsystematically from spectrum to spectrum. Internal calibration should thus provide a more straightforward accounting for the frequency shifts and more accurate mass measurements. Hannis and Muddiman (36) have recently demonstrated the use of an alternating dual ESI with an external accumulation that provided high MMA, achieving 1.1 ppm for 15-mer oligonucleotide (Mr = 4548.796 Da) and 8.9 ppm for a PCR product with molecular mass of 50,849.20 Da. We have taken a similar approach by use of an internal calibrant simultaneously delivered from one emitter of a dual ESI source to achieve high MMA. The dual electrospray source consists of two spray emitters operating simultaneously, with the emitters located in front of a single heated (170°C) desolvation capillary (0.047-in i.d.). The relative positions of both emitters to the desolvation capillary were precisely and independently adjusted by two xyz-translators. The sample emitter is directly connected to HPLC by a stainless steel union and sprays effluents from the HPLC. The calibrant emitter is positioned at an angle of about 45° to the sample emitter and connected to a syringe pump that continuously injects the standard peptide solution at the flow rate of 20 μl/hr. The relative ion signals of proteins from HPLC to the standard peptide were readily adjustable by the relative positions of the two emitters to the desolvation capillary (data not shown). The ESI emitter containing “locking” peptide (γ-endorphin, Mr = 1857.9181) predominantly produced doubly charged ions (m/z = 929.9664) that typically appeared in the middle of charge state envelopes of most proteins investigated.
Achieving high MMAs for high mass species is subject to mass resolution limitations of the measured ion spectra. Larger errors in MMA of ions with higher molecular masses may occur because of the uncertainty in determination of the most abundant isotopic peak. We have estimated the MMA for two standard proteins: bovine myoglobin [monoisotopic mass 16940.9642 atomic mass units (amu); average mass 16951.6116 amu] and carbonic anhydrase II (monoisotopic mass 29138.8909 amu; average mass 29157.1198 amu). MMA measured with myoglobin, where isotopically resolved distributions were obtained, was 1.0 ppm (SD of 0.8 ppm) for the 10 spectra with various relative intensities of the standard peptide to the myoglobin. We obtained 10.3 ppm (SD of 12.1 ppm) for carbonic anhydrase II, for which imperfect isotopic resolution was obtained, and where the average mass was used.
Data Processing.
Data processing was semiautomated by using the ICR-2LS software developed at our laboratory (the ICR-2LS suite of data analysis software is publicly available for noncommercial use; requests and inquiries should be directed to: Gordon.Anderson@pnl.gov). Briefly, ICR-2LS converted the raw data into m/z spectra that were subsequently transformed to generate a table of neutral masses by using an implementation of the thrash algorithm originally developed by Horn et al. (37). The frequency shift, obtained from the comparison of the theoretical and experimental mass of the internal standard, was then applied to all neutral masses and saved to a separate data file (pek). ICR-2LS data processing was performed on the complete capillary RPLC/FTICR data set in a batch run to generate a pek file containing neutral masses of all sample components observed in the run.
The output data file (pek file) can be visualized in a 2D display of neutral mass vs. scan number, where a “spot” size represents component intensity, using software developed at our laboratory (Figs. 1B and 2). The neutral masses (monoisotopic masses for proteins with Mr <21,000 Da and average masses for Mr >21,000 Da) from the pek file were also imported into the protein search module of the ICR-2LS and were searched for matches against whole yeast protein database (38) using criteria of 10 ppm MMA for monoisotopic masses and 30 ppm for average masses. For the database search, common modifications in multiples and combinations were permitted, including methylations, acetylations, phosphorylations, oxidations, losses of leading methionine, and losses of water and ammonia.
Figure 1.
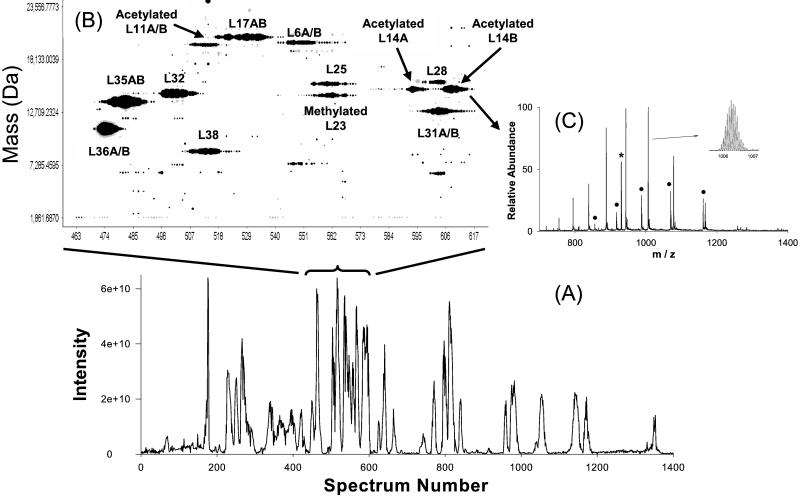
(A) Total ion current chromatogram reconstructed from FTICR mass spectra acquired during a whole capillary RPLC/FTICR experiment. (B) A 2D display of proteins in spectra 460–620. The size of the black solid circle is scaled to the intensity of a protein ion signal. Neutral masses were obtained as described in Methods. The designators AB and A/B indicate protein isoforms with identical mass and ones with different masses that eluted simultaneously, respectively. (C) Mass spectrum of proteins from spectrum number 609. The major peaks correspond to different charge states of acetylated rpL14B. rpL14B coeluted with rpL31A and rpL31B, which are denoted by ●. * indicates γ-endorphin internal standard. (Inset) An expanded mass spectrum of +15 charge state of acetylated rpL14B.
Figure 2.
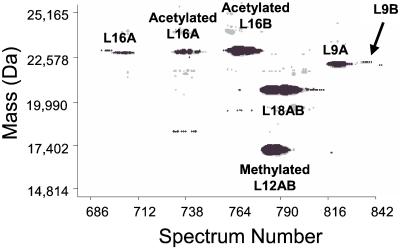
A 2D display of proteins observed in spectra 680–850 during the capillary RPLC/FTICR analysis of the yeast large ribosomal subunit (see Fig. 1 and text).
Results and Discussion
The characterization of yeast large ribosomal subunit proteins is challenging because of the number of proteins to be characterized, the simple and complex patterns of protein modification, and the existence of protein isoforms with as much as 99% sequence identity. Fig. 1A shows a capillary RPLC/FTICR chromatogram reconstructed from the mass spectra. Single peaks on the chromatogram can contain more than one protein, as illustrated by a 2D display in Fig. 1B. For example, spectrum 609 reveals two proteins (rpL31AB and acetylated rpL14B) along with the γ-endorphin internal calibrant (Fig. 1C). The long column (80 cm) provided a 1–1.5 μl/min mobile-phase linear velocity at 8000 psi that permitted a significantly improved separation efficiency compared with commercial LC instruments operating normally under 5,000 psi (39), allowing proteins with significant structural similarity to be separated. For example, rpL14A and rpL14B, isoforms with 99% sequence homology, were completely resolved as shown in Fig. 1B. The high efficiency separation also aided in the determination of protein identity, which is illustrated in Fig. 2. The 2D display clearly shows that three proteins corresponding to rpL16A, acetylated rpL16A, and acetylated rpL16B eluted in temporal proximity while exhibiting baseline separation. As predicted, acetylation increased overall protein hydrophobicity, and consequently the acetylated rpL16A elutes subsequent to the corresponding nonacetylated protein. The 2D display also provided the information on protein expression ratio between the two protein isoforms, within the limitation that small differences in ionization efficiencies (intrinsic and/or temporal) can occur for different proteins by ESI. The large subunit protein rpL9A, present in greater abundance than rpL9B, is one particularly interesting example that will be discussed later.
Observed Large Ribosomal Subunits and Subunit Modifications.
We have detected 71 sample components having a mass range of 3335.103 to 43640.510 amu, including several species not assigned by our searches (sample components with masses of 8575.975, 11299.171, 11313.364, 13384.986, 13500.528, 14446.731, 22153.002, 22170.289, and 27418.337 could not be assigned to known large subunit proteins, other ribosomal proteins, or ribosome interacting factors) (Table 1). In most cases, we observed protein masses different from those directly predicted from the coding sequence, indicating the presence of protein modifications. From this set of accurately determined masses, we have searched for a limited set of combinations of common protein modifications (methionine loss, acetylations, methylations, phosphorylations, oxidations, and loss of water and ammonia), and identified 42 of the 43 (98%) core large ribosomal subunit proteins and 58 of 64 (91%) core large subunit isoforms (Table 1).
Table 1.
Identified large ribosomal subunit proteins
| Subunit† | Calculated mass‡ | Observed mass§ | Δm (ppm)¶ | Met loss‖ | Notes** |
|---|---|---|---|---|---|
| rpL1AB | 24,410.825* | 24,410.862* | 1.5 | Y | Ac & Me(1) |
| rpL2AB | 27,277.449* | 27,277.347* | 3.7 | Y | |
| rpL3 | 43,640.806* | 43,640.510* | 6.8 | Y | Me(1) |
| rpL4A | 39,003.231* | 39,002.440* | 20.3 | Y | Ac |
| rpL4B | 38,973.205* | 38,972.675* | 13.6 | Y | Ac |
| rpL5 | 33,583.879* | 33,583.991* | 3.3 | Y | |
| rpL6A | 19,818.107 | 19,818.198 | 4.6 | Y | |
| rpL6B | 19,843.145 | 19,843.170 | 1.3 | Y | |
| rpL7A | 27,507.248* | 27,506.764* | 17.6 | Y | |
| rpL7B | — | — | Not observed | ||
| rpL8A | 27,993.763* | 27,993.634* | 4.6 | Y | |
| rpL8B | 27,980.805* | 27,980.521* | 10.2 | Y | |
| rpL9A | 21,555.648 | 21,555.702 | 2.5 | N | |
| rpL9B | 21,643.646 | 21,643.666 | 0.9 | N | |
| rpL10 | 25,230.365* | 25,230.526* | 6.4 | Y | |
| rpL11A | 19,618.365 | 19,618.379 | 0.7 | Y | Ac |
| rpL11B | 19,648.375 | 19,648.442 | 3.4 | Y | Ac |
| rpL12AB | 17,764.703 | 17,764.712 | 0.5 | Y | Me(6) |
| 17,764.666 | 17,764.712 | 2.6 | Y | Ac & Me(3) | |
| rpL13A | 22,423.013* | 22,422.896* | 5.3 | Y | |
| rpL13B | 22,393.972* | 22,393.828* | 6.4 | Y | |
| rpL14A | 15,068.558 | 15,068.575 | 1.1 | Y | Ac |
| rpL14B | 15,054.543 | 15,054.550 | 0.5 | Y | Ac |
| rpL15AB | 24,291.162* | 24,290.975* | 7.7 | Y | |
| rpL16A | 22,056.348 | 22,056.523 | 8.0 | Y | |
| rpL16A(2) | 22,098.358 | 22,098.516 | 7.1 | Y | Ac |
| rpL16B | 22,146.354 | 22,146.367 | 0.6 | Y | Ac |
| rpL17A | 20,406.023 | 20,405.841 | 8.9 | Y | |
| rpL17B | 20,408.002 | 20,407.942 | 2.9 | Y | |
| rpL18AB | 20,419.467 | 20,419.431 | 1.7 | Y | |
| rpL19AB | 21,573.078* | 21,573.159* | 3.8 | Y | |
| rpL20A | — | — | Not observed | ||
| rpL20B | — | — | Not observed | ||
| rpL21A | 18,099.734 | 18,099.778 | 2.4 | Y | |
| rpL21B | 18,131.706 | 18,131.771 | 3.6 | Y | |
| rpL22A | 13,553.958 | 13,553.964 | 0.5 | Y | |
| rpL22B | — | — | Not observed | ||
| rpL23AB | 14,430.709 | 14,430.702 | 0.5 | Y | Me(7) |
| 14,430.673 | 14,430.702 | 2.0 | Y | Ac & Me(4) | |
| rpL24A | 17,602.902 | 17,602.877 | 1.4 | N | |
| rpL24B | 17,536.903 | 17,536.881 | 1.2 | N | |
| rpL25 | 15,747.750 | 15,747.679 | 4.5 | Y | |
| rpL26A | 14,094.031 | 14,094.001‡‡ | 2.2 | Y | |
| rpL26B | 14,095.015 | 14,095.021‡‡ | 0.4 | Y | |
| rpL27A | 15,390.605 | 15,390.617 | 0.8 | Y | |
| rpL27B | — | — | Not observed | ||
| rpL28 | 16,580.021 | 16,580.038 | 1.0 | Y | |
| rpL29 | 6,533.671 | 6,533.664 | 1.1 | Y | |
| rpL30 | 11,277.230 | 11,277.256 | 2.3 | Y | |
| rpL31A | 12,814.078 | 12,814.163 | 6.6 | Y | |
| rpL31B | 12,828.094 | 12,828.081 | 1.0 | Y | |
| rpL32 | 14,631.192 | 14,631.196 | 0.3 | Y | |
| rpL33A | 12,015.526 | 12,015.514 | 1.0 | Y | |
| rpL33B | 12,029.541 | 12,029.581 | 3.3 | Y | |
| rpL34A | — | — | Not observed | ||
| rpL34B | 13,501.511 | 13,501.592 | 6.0 | Y | |
| rpL35AB | 13,769.968 | 13,769.991 | 1.7 | Y | |
| rpL36A | 10,986.257 | 10,986.274 | 1.5 | Y | |
| rpL36B | 10,997.273 | 10,997.304 | 2.8 | Y | |
| rpL37A | 9,712.957 | 9,712.943 | 1.5 | Y | |
| rpL37B | 9,730.979 | 9,730.957 | 2.2 | Y | |
| rpL38 | 8,690.060 | 8,690.073 | 1.6 | Y | |
| rpL39 | 6,206.514 | 6,206.520 | 1.1 | Y | |
| rpL40AB | 6,011.284 | 6,011.297 | 2.2 | N/A | Fragment, 77–128 |
| rpL41AB | 3,335.112 | 3,335.103 | 2.9 | N | |
| rpL42AB | 12,100.702 | 12,100.729 | 2.2 | Y | Me(2) |
| rpL43AB | 9,953.301 | 9,953.311 | 1.0 | Y | |
| rpL43AB(2) | 9,967.317 | 9,967.282 | 3.5 | Y | Me(1) |
Ribosomal subunit proteins are labeled based on a revised systematic nomenclature (20) and are consistent with the primary gene name in the yeast proteome database (YPD) (38). The combined designator AB is used for duplicated genes (e.g. rpL1AB = rpL1A and rpL1B) that yield proteins with identical sequence. Numbers in parenthesis indicate that more than one form of a protein was observed.
The mass of each ribosomal subunit, allowing for the noted modifications, was calculated by using the protein sequence provided in YPD (38), except for rpL26B and rpL42AB, which were obtained from SWISS-PROT (accession nos. P53221 and P02405, respectively) due to errors in the YPD files. Monoisotopic mass or average mass (indicated by
) was calculated by using icr-2ls.
Masses of the large ribosomal subunit components were determined as described in Methods. Data were analyzed by icr-2ls software and the observed monoisotopic or average mass (indicated by a ∗) is reported.
Values shown represent the absolute difference between predicted and observed masses in ppm units.
Indicates cleavage (Y) or retention (N) of the initiating methionine residue.
Observed modifications (Ac, acetylation; Me, methylation) are noted with multiple modifications indicated in parentheses. In two situations where potential sets of modifications cannot be resolved from our data (i.e. RPL12AB and RPL23AB), both possibilities are presented separately.
rpL26A and rpL26B differ by 0.984 Da, resulting in overlapping of two isotopic distributions. With the resolution currently available with this experiment, we were not able to resolve the two proteins, presumably coeluting from LC. Unlike other protein spectra, however, the spectra that contain rpL26A and rpL26B result in two monoisotopic masses (14094.001 and 14095.021) upon mass transformation.
We observed only a single isoform for four subunits (rpL7A, rpL22A, rpL27A, and rpL34B), and an analysis of the transcriptome level data from Holstege and coworkers (40) reveals that in each instance, the observed subunit corresponds to more than 65% of the total mRNA for that subunit (65%, 84%, 69%, and 67% respectively). Interestingly, the transcriptome data also agrees with our observation where both isoforms were observed. As mentioned earlier, rpL9A (70% of subunit mRNA) is found in large excess over rpL9B (Fig. 2), and rpL4A (64%) and rpL31A (65%) are also observed to be the dominant isoforms of their respective subunits (not shown). These results confirm an earlier finding, indicating that expression of yeast ribosomal proteins is uniformly regulated at the level of transcription (41). The failure to observe either isoform of L20 (rpL20A or rpL20B) is puzzling, as a recent analysis of ribosomal proteolytic digests by the direct analysis of large protein complexes (DALPC) technique (8) confirmed association of this protein with the large subunit. Additionally, we did not observe the three acidic proteins (five isoforms) rpP0, rpP1A, rpP1B, rpP2A, rpP2B that form a single (P1)2(P2)2P0 complex that exchanges between the cytosol and the large ribosomal subunit in a dynamic manner in the translationally active 80S ribosomal complex (42).
N-Terminal Modifications.
The most common modification to yeast proteins is the loss of the initiator methionine. Systematic studies using proteins engineered to have differing antepenultimate amino acids has revealed that in almost all cases proteins having antepenultimate amino acid residues with small uncharged side chains having a radius of gyration below 1.43 Å will cotranslationally lose the initiator methionine (14, 43, 44). Consistent with these model protein studies, our identifications (Table 1) yielded ribosomal protein N termini of MA (n = 33), MG (n = 10), MP (n = 3), MS (n = 15), MT (n = 2), and MV (n = 2) as processed N termini, whereas N termini of MK (n = 4) and MR (n = 2) retained the initiator methionine, where n is the number of proteins. The processing of the two rpL40 isoforms (N termini MQ) could not be analyzed because of proteolytic maturation (discussed later).
In eukaryotes, N-terminal acetylation, a stable modification of processed (−Met) and unprocessed (+Met) protein N termini by addition of an acetyl moiety, is a widely observed modification that occurs cotranslationally, subsequent to the opportunity for methionine processing (45). Unlike methionine processing, a simple relationship between N-terminal sequence and modification state has not been found (14, 44, 45).
The assignment of acetylation is complicated by the small mass difference between acetylation (Δm = 42.037 Da) and tri-methylation (Δm = 42.081 Da). However, based on the previous studies of ribosomal proteins (9, 46), we can confidently assign N-terminal acetylation of five large subunit proteins (10 isoforms) containing a serine residue at the N terminus: rpL1A and rpL1B (N terminus SKITSSQ), rpL4A and rpL4B (SRPQVTV), rpL11A (SAKAQNP), rpL11B (STKAQNP), rpL14A and rpL14B (STDSIVK), rpL16A (SVEPVVV), and rpL16B (SQPVVVI), while not observing acetylation of rpL3 (SHRKYEA) or rpL6A (SAQKAPK). Acetylation was not observed for other subunit isoforms containing N termini compatible with the substrate specificity (processed N termini of Ser, Ala, Gly, and Thr) of the Ard1p acetyltransferase (13). To our knowledge, the observations of acetylation in rpL4A and rpL14A are novel, but not entirely unexpected based on the conservation of N-terminal sequence with their respective isoforms. We do not observe acetylation on rp6A (N terminus SAQKAPK), rpL33A (AESHR) or rpL36A (TVKTG), in contrast to previous studies (9). In our data, only rpL16A appears partially acetylated and was observed in both the modified and unmodified forms (Fig. 2 and Table 1). Of the two large subunit proteins (four isoforms) that are not definitively assigned because of multiple methylation modifications (see below), rpL23A and rpL23B (N terminus SGNGAQG) are consistent with Ard1p modification, whereas rpL12A and rpL12B (PPKFDP) would be noncanonical for all known N-acetyltransferases.
Methylation and Phosphorylation.
Methylation and phosphorylation are the two most common posttranslational protein modifications observed on ribosomal proteins. Methylation has been previously observed on lysine and/or arginine residues in a number of yeast large ribosomal proteins (9, 21, 22); however, the low reported stoichiometry of methylation (22), and differential modification by multiple methylation enzymes (47), may have led to different subsets of methylated proteins having been identified previously. Consistent with Lhoest and coworkers (21), we observe sample components consistent with mono-methylation of rpL3 and multiple methylations of rpL12AB and rpL23AB. For rpL12AB we find a mass difference consistent with either three or six methyl groups per protein (Table 1) depending on possible occurrence of N-acetylation, and for rpL23AB we find either four or seven methylations per protein, again depending on presence/absence of N-acetylation. Given the low probability of acetylation of rpL12A based on the antepenultimate proline residue, we believe there are six methyl groups per protein, in contrast to Lhoest and coworkers who observed tri- and di-methyllysine modifications. Similarly, the high likelihood of N-acetylation of rpL23AB (based on the serine penultimate residue) would be more consistent with four methyl groups per protein, again one greater than the mono- and di-methyllysine reported by Lhoest and coworkers. Our data (Table 1) also indicate that rpL42AB is modified by di-methylation, consistent with two earlier studies (9, 22), and supports the presence of previously undetected mono-methylated rpL1AB, and mono-methylated rpL43AB, which was observed in both the methylated and nonmethylated state.
We did not observe any modifications caused by phosphorylation. It has been well established that the acidic ribosomal proteins (rpP0, rpP1A, rpP1B, rpP2A, and rpP2B) are phosphorylated in yeast (24, 48), but only a single report has identified up to two serine phosphorylation sites in a core large subunit protein (rpL5) (49). As mentioned earlier, the yeast acidic ribosome subcomplex dynamically associates with the large subunit (16, 24, 42), and the apparent absence of the acidic ribosomal proteins is likely caused by their absence in the original sample, rather than difficulties encountered during the reversed-phase separation or ESI. However, the significant difference in the chemical nature of the acidic proteins versus the overwhelmingly basic nature of other large ribosomal proteins could potentially influence both processes. Our results (Table 1) permitted us to unambiguously identify the methionine-processed form of RPL5, but sample components consistent with the mono- or di-phosphorylated forms of the protein were not observed.
Additional Ribosomal Modifications.
The genes RPL40A and RPL40B encode identical ribosomal subunit isoforms rpL40A and rpL40B as precursors coupled to the C terminus of the intracellular degradation protein ubiquitin (50). Searches of our data sets for a component with a mass consistent with the intact protein were unsuccessful, but the mature large ribosomal subunit protein (residues 77–128) was observed (Table 1) without additional modifications. This finding is consistent with the observation that the ubiquitin tail can serve an escort or chaperone function but is not required for ribosomal function (50).
It has recently been reported that rpL28 can be modified by the addition of one or more ubiquitin proteins in a manner that up-regulates ribosomal function without targeting for protein destruction (15). A search of our data set does not identify sample components containing rpL28 modified in this manner. Our observations are consistent with previous observations that L28-ubiquitination is low in the G1 phase and abundant when yeast were blocked in the S phase of the cell cycle, as the log phase population of yeast from which we harvested ribosomes would contain only a minute portion of cells undergoing S phase.
Conclusions
ESI/FTICR/MS coupled with reverse-phase capillary LC was used to characterize intact proteins from the large subunit of yeast ribosome. The high MMA coupled with high-resolution online reverse-phase capillary LC permitted us to identify 42 of the 43 core large ribosomal subunit proteins and 58 of the 64 possible core large subunit isoforms with unique masses in a single experiment. Unobserved isoforms were well correlated with data from transcriptome studies and appear to validate the model of transcriptional regulation of yeast ribosomal proteins. The ability to identify multiple intact proteins from a single experiment holds promises for high-throughput analysis of proteomes and complements conventional mass spectrometric approaches (based on protein digestion) by providing identification of the intact protein and its state of modification.
Acknowledgments
We thank Nikola Tolić for assistance in data processing. This work was supported by the U.S. Department of Energy office of Biological and Environmental Research. Pacific Northwest National Laboratory is operated for the U.S. Department of Energy by the Battelle Memorial Institute under Contract DE-AC06-76RLO 1830.
Abbreviations
- ESI
electrospray ionization
- 2D
two-dimensional
- FTICR
Fourier transform ion cyclotron resonance
- MMA
mass measurement accuracy
- amu
atomic mass unit
- RPLC
reverse-phase liquid chromatography
References
- 1.Karas M, Bachmann D, Bahr U, Hillenkamp F. Int J Mass Spectrom Ion Proc. 1987;78:53–68. [Google Scholar]
- 2.Hillenkamp F, Karas M, Beavis R C, Chait B T. Anal Chem. 1991;63:1193A–1203A. doi: 10.1021/ac00024a002. [DOI] [PubMed] [Google Scholar]
- 3.Fenn J B, Mann M, Meng C K, Wong S F, Whitehouse C M. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 4.Klose J. Electrophoresis. 1999;20:643–652. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<643::AID-ELPS643>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 5.Jensen P K, Paša-Tolić L, Peden K K, Martinović S, Lipton M S, Anderson G A, Tolić N, Wong K-K, Smith R D. Electrophoresis. 2000;21:1372–1380. doi: 10.1002/(SICI)1522-2683(20000401)21:7<1372::AID-ELPS1372>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 6.Li W, Hendrickson C L, Emmett M R, Marshall A G. Anal Chem. 1999;71:4397–4402. doi: 10.1021/ac990011e. [DOI] [PubMed] [Google Scholar]
- 7.Louie D F, Resing K A, Lewis T S, Ahn N G. J Biol Chem. 1996;271:28189–28198. doi: 10.1074/jbc.271.45.28189. [DOI] [PubMed] [Google Scholar]
- 8.Link A J, Eng J, Schieltz D M, Carmack E, Mize G J, Morris D R, Garvik B M, Yates J R., III Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 9.Arnold R J, Polevoda B, Reilly J P, Sherman F. J Biol Chem. 1999;274:37035–37040. doi: 10.1074/jbc.274.52.37035. [DOI] [PubMed] [Google Scholar]
- 10.Benjamin D R, Robindson C V, Hendrick J P, Hartl F U, Dobson C M. Proc Natl Acad Sci USA. 1998;95:7391–7395. doi: 10.1073/pnas.95.13.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank J. Am Sci. 1998;86:428–439. [Google Scholar]
- 12.Warner J R. Trends Cell Biol. 1999;24:437–440. [Google Scholar]
- 13.Nissen P, Hansen J, Ban N, Moore P B, Steitz T A. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 14.Huang S, Elliot R C, Liu P-S, Koduri R K, Weickmann J L, Lee J-H, Blair L C, Ghosh-Dastidar P, Bradshaw R A, Bryan K M, et al. Biochemistry. 1987;26:8242–8246. doi: 10.1021/bi00399a033. [DOI] [PubMed] [Google Scholar]
- 15.Spence J, Gali R R, Dittmar G, Sherman F, Karin M, Finley D. Cell. 2000;102:67–76. doi: 10.1016/s0092-8674(00)00011-8. [DOI] [PubMed] [Google Scholar]
- 16.Briones E, Briones C, Remacha M, Ballesta J P G. J Biol Chem. 1998;273:37956–37953. doi: 10.1074/jbc.273.48.31956. [DOI] [PubMed] [Google Scholar]
- 17.Kruiswijk T, Planta R J, Krop J. Biochim Biophys Acta. 1978;517:378–389. doi: 10.1016/0005-2787(78)90204-6. [DOI] [PubMed] [Google Scholar]
- 18.Verschoor A, Warner J R, Srivastava S, Grassucci R A, Frank J. Nucleic Acid Res. 1998;26:655–661. doi: 10.1093/nar/26.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgan D G, Menetret J-F, Radermacher M, Neuhof A, Akey I V, Rapoport T A, Akey C W. J Mol Biol. 2000;301:301–321. doi: 10.1006/jmbi.2000.3947. [DOI] [PubMed] [Google Scholar]
- 20.Mager W H, Planta R J, Ballesta J-P, Lee J C, Mizuta K, Suzuki K, Warner J R, Woolford J. Nucleic Acid Res. 1997;25:4872–4875. doi: 10.1093/nar/25.24.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lhoest J, Lobet Y, Costers E, Colson C. Eur J Biochem. 1984;141:585–590. doi: 10.1111/j.1432-1033.1984.tb08233.x. [DOI] [PubMed] [Google Scholar]
- 22.Kruiswijk T, Kunst A, Planta R J, Mager W H. Biochem J. 1978;175:221–225. doi: 10.1042/bj1750221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becker-Ursic D, Davies J. Biochemistry. 1976;15:2289–2296. doi: 10.1021/bi00656a007. [DOI] [PubMed] [Google Scholar]
- 24.Naranda T, Ballesta J P G. Proc Natl Acad Sci USA. 1991;88:10563–10567. doi: 10.1073/pnas.88.23.10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen Y, Tolić N, Zhao R, Paša-Tolić L, Li L, Berger S J, Harkewicz R, Anderson G A, Belov M E, Smith R D. Anal Chem. 2001;73:3011–3021. doi: 10.1021/ac001393n. [DOI] [PubMed] [Google Scholar]
- 26.Martinović S, Berger S J, Paša-Tolić L, Smith R D. Anal Chem. 2000;72:5356–5360. doi: 10.1021/ac0004557. [DOI] [PubMed] [Google Scholar]
- 27.Warner J R, Gorenstein C. In: Methods in Cell Biology. Prescott D M, editor. XX. New York: Academic; 1978. pp. 45–60. [DOI] [PubMed] [Google Scholar]
- 28.Marcus L, Ris H, Halvorson H O, Bretthauer R K, Bock R M. J Cell Biol. 1967;34:505–512. doi: 10.1083/jcb.34.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cocito C. Mol Gen Genet. 1978;162:43–50. doi: 10.1007/BF00333849. [DOI] [PubMed] [Google Scholar]
- 30.Pande C, Wishnia A. J Biol Chem. 1986;261:6272–6278. [PubMed] [Google Scholar]
- 31.Hardy S J S, Kurland C G, Voynow P, Mora G. Biochemistry. 1969;8:2897–2905. doi: 10.1021/bi00835a031. [DOI] [PubMed] [Google Scholar]
- 32.Winger B E, Hofstadler S A, Bruce J E, Udseth H R, Smith R D. J Am Soc Mass Spectrom. 1993;4:566–577. doi: 10.1016/1044-0305(93)85018-S. [DOI] [PubMed] [Google Scholar]
- 33.Kim T, Tolmachev A V, Harkewicz R, Prior D C, Anderson G, Udseth H R, Smith R D, Bailey T H, Rakov S, Futrell J H. Anal Chem. 2000;72:2247–2255. doi: 10.1021/ac991412x. [DOI] [PubMed] [Google Scholar]
- 34.Ledford E B, Rempel D L, Gross M L. Anal Chem. 1984;56:2744–2748. doi: 10.1021/ac00278a027. [DOI] [PubMed] [Google Scholar]
- 35.Eastering M L, Mize T H, Amster I J. Anal Chem. 1999;71:624–632. doi: 10.1021/ac980690d. [DOI] [PubMed] [Google Scholar]
- 36.Hannis J C, Muddiman D C. J Am Soc Mass Spectrom. 2000;11:876–883. doi: 10.1016/S1044-0305(00)00160-4. [DOI] [PubMed] [Google Scholar]
- 37.Horn D M, Zubarev R A, McLafferty F W. J Am Soc Mass Spectrom. 2000;11:320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 38.Costanzo M C, Hogan J D, Cusick M E, Davis B P, Fancher A M, Hodges P E, Kondu P, Lengieza C, Lew-Smith J E, Lingner C, et al. Nucleic Acids Res. 2000;28:73–76. doi: 10.1093/nar/28.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacNair J E, Patel K D, Jorgenson J W. Anal Chem. 1999;71:700–708. doi: 10.1021/ac9807013. [DOI] [PubMed] [Google Scholar]
- 40.Holstege F C, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 41.Li B, Nierras C R, Warner J R. Mol Cell Biol. 1999;19:5393–5404. doi: 10.1128/mcb.19.8.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saenz-Robles M T, Vilella M D, Pucciarelli G, Polo F, Remacha M, Ortiz B L, Vidales F J, Ballesta J P G. Eur J Biochem. 1988;177:531–537. doi: 10.1111/j.1432-1033.1988.tb14405.x. [DOI] [PubMed] [Google Scholar]
- 43.Tsunasawa S, Stewart J W, Sherman F. J Biol Chem. 1985;260:5382–5391. [PubMed] [Google Scholar]
- 44.Moershell R P, Hosokawa Y, Tsunasawa S, Sherman F. J Biol Chem. 1990;265:19638–19643. [PubMed] [Google Scholar]
- 45.Driessen H P C, de Jong W W, Tesser G I, Bloemendal H. CRC Crit Rev Biochem. 1985;18:281–325. doi: 10.3109/10409238509086784. [DOI] [PubMed] [Google Scholar]
- 46.Takakura H, Tsunasawa S, Miyagi M, Warner J R. J Biol Chem. 1992;267:5442–5445. [PubMed] [Google Scholar]
- 47.Lobet Y, Lhoest J, Colson C. Biochim Biophys Acta. 1989;997:224–231. doi: 10.1016/0167-4838(89)90191-x. [DOI] [PubMed] [Google Scholar]
- 48.Sanchez-Madrid F, Vidales F, Ballesta J P G. Eur J Biochem. 1981;114:609–613. doi: 10.1111/j.1432-1033.1981.tb05187.x. [DOI] [PubMed] [Google Scholar]
- 49.Campos F, Corona-Reyes M, Zinker S. Biochim Biophys Acta. 1990;1087:142–146. doi: 10.1016/0167-4781(90)90198-b. [DOI] [PubMed] [Google Scholar]
- 50.Finley D, Bartel B, Varshavsky A. Nature (London) 1989;338:394–401. doi: 10.1038/338394a0. [DOI] [PubMed] [Google Scholar]