Abstract
Using a large sample of cases and controls from a single center, we show that a T→C substitution in exon 9 (Y402H) of the complement factor H gene is strongly associated with susceptibility to age-related macular degeneration, the most common cause of blindness in the elderly. Frequency of the C allele was 0.61 in cases, versus 0.34 in age-matched controls (P<1×10-24). Genotype frequencies also differ markedly between cases and controls (χ2=112.68 [2 degrees of freedom]; P<1×10-24). A multiplicative model fits the data well, and we estimate the population frequency of the high-risk C allele to be 0.39 (95% confidence interval 0.36–0.42) and the genotype relative risk to be 2.44 (95% confidence interval 2.08–2.83) for TC heterozygotes and 5.93 (95% confidence interval 4.33–8.02) for CC homozygotes.
Age-related macular degeneration (AMD) (ARMD1 [MIM 603075]), the leading cause of untreatable blindness among the elderly in Western populations, is a clinically heterogeneous and genetically complex disease with multiple genetic and environmental risk factors (Age-Related Eye Disease Study Research Group 2000). Mutations in several genes (e.g., ABCA4 [MIM 601691], TIMP3 [MIM 188826], RDS/peripherin [MIM 179605], and ELOVL4 [MIM 605512]) can cause early-onset macular diseases, but they do not appear to contribute significantly to AMD susceptibility (Stone et al. 2001). Particularly interesting are the fibulin-3 gene (MIM 601548) and related genes. Fibulin-3 mutations underlie drusen formation in Doyne honeycomb retinal dystrophy (MIM 126600), a disease that is phenotypically similar to AMD (Stone et al. 1999; Marmorstein et al. 2002). A mutation screen of five other fibulin genes detected missense mutations in fibulin-5 (MIM 604580) in 1.7% of patients with AMD (Stone et al. 2004). In addition, a fibulin-6 (MIM 608548) variant has been reported to cosegregate with the ARMD1 locus in one large pedigree (Schultz et al. 2003). However, this change does not appear to play a significant role in AMD (Abecasis et al. 2004; Hayashi et al. 2004; Iyengar et al. 2004).
Linkage studies have suggested several chromosomal regions that may harbor AMD susceptibility genes. Klein and colleagues (1998) were the first to map a susceptibility locus (ARMD1) to chromosome 1q25-q31 in a large pedigree with AMD. Since then, many studies have been performed, and, overall, their results provide support for susceptibility loci on several chromosomes, including chromosomes 1q, 9q, 10q, and 22q (Weeks et al. 2000, 2004; Majewski et al. 2003; Seddon et al. 2003; Abecasis et al. 2004; Iyengar et al. 2004; Schmidt et al. 2004). Association studies have also been performed, and some of the identified loci appear to contribute to disease susceptibility. For example, an association between AMD and allelic variants of apolipoprotein E (APOE [MIM 107741]) has been widely documented, with the APOE-ɛ4 allele linked to lower risk of disease and the APOE-ɛ2 allele linked to higher risk (Klaver et al. 1998; Schmidt et al. 2002; Baird et al. 2004; Zareparsi et al. 2004). Recently, we reported an association between increased risk of AMD and the D299G variation in toll-like receptor 4 (TLR4 [MIM 603030]), a protein involved in innate immunity and phagocytosis by retinal pigment epithelium (Zareparsi et al. 2005). Despite these advances, the alleles that account for most of the genetic susceptibility to AMD remain undiscovered.
Recently, three independent studies have suggested that a polymorphism (Y402H) in the complement factor H gene (CFH [MIM 134370]) makes a substantial contribution to AMD susceptibility (Y402H has a T→C substitution at nucleotide 1277 in exon 9, which results in a tyrosine→histidine change). All three studies relied on linkage disequilibrium in the region and advances in SNP genotyping for gene identification (Abecasis et al. 2005). CFH maps to a region of chromosome 1q where several genome scans showed substantial evidence for linkage. One study (Klein et al. 2005), a genomewide association scan of 96 AMD cases and 50 controls by use of an Affymetrix 100K chip, identified two neighboring SNPs that were significantly associated with AMD. Another study (Edwards et al. 2005) examined noncoding SNPs across 14 Mb of the ARMD1 locus on chromosome 1q in a sample of 224 cases and 134 controls; genotyping of 14 SNPs spanning CFH was performed in a larger sample. The third study (Haines et al. 2005) also focused on examination of SNPs distributed across the ARMD1 locus, but it included a sample of 495 unrelated cases, 185 controls, and 182 families. In each study, initial associations were followed by additional genotyping that eventually led to the identification of a peak of association, which suggested the CFH-Y402H (C/T) variant as the susceptibility allele. These studies reported odds ratios (ORs) for AMD ranging between 2.4 and 4.6 for carriers of the C allele and between 3.3 and 7.4 for CC homozygotes (Edwards et al. 2005; Haines et al. 2005; Klein et al. 2005).
Independent replication studies are required to accurately assess the contribution of the associated alleles to disease susceptibility, because initial reports of association are vulnerable to a “winner’s curse” effect, which can produce overestimates of the effect size (Goring et al. 2001; Lohmueller et al. 2003). We used a large sample of cases (N=616) and controls (N=275), collected at the Kellogg Eye Center in Ann Arbor, MI, to provide independent estimates of the effect of this common variant (CFH-Y402H) on AMD susceptibility. A majority of patients with AMD had late-stage AMD that presented as choroidal neovascularization (CNV [N=238]), geographic atrophy (GA [N=143]), or both CNV and GA (N=133). The remaining patients had large macular drusen (LMD) in both eyes (N=102) (Abecasis et al. 2004; Zareparsi et al. 2004). Control individuals were at least 68 years old and did not present any evidence of AMD in either eye after ophthalmic examination. All patients and controls reported their ethnicity as “white, not of Hispanic origin” and were recruited after informed consent. The human-genetics investigations described here were approved by the University of Michigan institutional review board. Genotyping was performed without knowledge of disease status. As shown in table 1, we detected a significantly higher frequency of the C allele in patients with AMD than in controls (0.61 vs. 0.34; χ2 = 110.96 [1 df]; P<1×10-24). Genotype frequencies are also significantly different in affected and unaffected individuals (χ2=112.68 [2 df]; P<1×10-24). OR calculations show that individuals carrying at least one copy of the C allele have a 4.36-fold increase in the risk of AMD (95% CI 3.13–6.08), whereas homozygous CC individuals exhibit a 5.52-fold increase in the risk of developing AMD (95% CI 3.54–8.59). Our results are within the range reported by the original studies (Edwards et al. 2005; Haines et al. 2005; Klein et al. 2005), and they validate the gene-dosage effect reported in two of the studies (Haines et al. 2005; Klein et al. 2005). Inclusion of age and sex as covariates in the logistic regression analysis did not affect the conclusions. Homozygous individuals for the putative risk allele were more common in all subtypes of AMD than in controls; specifically, those homozygous for the C allele included 38% of patients with GA, 34% of those with CNV, and 33% of those with LMD in both eyes, as compared with only 9% of control individuals. Interestingly, the homozygous genotype (CC) was more common in patients with a family history of AMD (38% of 261 cases) than in those without a family history (33% of 308 cases), a difference that was not statistically significant (P>.05). Finally, we note that the diagnosis of AMD occurred earlier for patients with at least one copy of the risk allele (mean in years [± SD] 70.9 ± 8.6) than for those without the risk allele (73.7 ± 9.2; P=.01).
Table 1.
Genotypic and Allele Frequencies for the T→C (Y402H) Variation in CFH Exon 9
|
No. (Frequency) of Genotypeor Allele in |
||
| Genotypeor Allele | Individuals with AMD[N=616] | Controls[N=275)] |
| TT | 86 (.14) | 114 (.415) |
| TC | 311 (.50) | 136 (.495) |
| CC | 219 (.36) | 25 (.09) |
| T allele | 483 (.39) | 364 (.66) |
| C allele | 749 (.61) | 186 (.34) |
To characterize the contribution of the Y402H polymorphism to AMD susceptibility, we fitted a series of genetic models to the data by maximum-likelihood estimation (results summarized in table 2). Model fitting was performed using the software and methods of Li et al. (2005). We performed our analysis with the assumption of a disease prevalence (K) of 0.20 (table 2), which is compatible with published estimates for elderly populations >70 years of age (Friedman et al. 2004), and without constraints on disease prevalence (table 2). With the use of a model with constrained prevalence (K=0.20), the data suggest that the population frequency of the C allele is 0.39 (95% CI 0.36–0.42) and that the relative risk is 2.81 (95% CI 2.17–3.65) for TC heterozygotes and 6.30 (95% CI 4.53–8.72) for CC homozygotes. We also used the Akaike Information Criterium (AIC) to compare all models, including those with constrained and unconstrained prevalence, and to select the best-fitting genetic model. Our results suggest a multiplicative model—with K=0.20, a disease-allele frequency of 0.39 (95% CI 0.36–0.42), and genotype relative risks of 2.44 (95% CI 2.08–2.83) for TC heterozygotes and 5.93 (95% CI 4.33–8.02) for CC homozygotes—as the most parsimonious model. The fitted log likelihood was 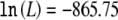 for the multiplicative model with K=0.20 (two parameters, one for the genotype relative risk and another for the disease-allele frequency), versus
for the multiplicative model with K=0.20 (two parameters, one for the genotype relative risk and another for the disease-allele frequency), versus 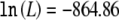 for an unconstrained model (four parameters, corresponding to three penetrances and one disease-allele frequency) and
for an unconstrained model (four parameters, corresponding to three penetrances and one disease-allele frequency) and 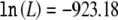 for a model under the assumption of no contribution to disease susceptibility (one parameter, corresponding to the marker-allele frequency). Using this multiplicative model, we estimate the contribution of this allele to sibling-specific recurrence risk of AMD to be λs=1.21 and the population-attributable fraction to be 0.62. This is the locus-specific λs, which can be calculated as a function of penetrances and disease-allele frequencies by use of the formulas of Risch (1990). The model predicts frequencies of 0.43, 0.47, and 0.10 among controls and 0.15, 0.48, 0.37 among cases for the TT, TC, and CC genotypes, respectively; the predicted values closely match the observed frequencies (see table 1) (goodness-of-fit χ2=2.61 [2 df], not significant). An additive model also fits the data well, but dominant or recessive models are excluded. The agreement between our fitted model and observed genotype counts suggests that population stratification is not a major concern (Wittke-Thompson et al. 2005), a conclusion consistent with our analysis of previous genome-scan data.
for a model under the assumption of no contribution to disease susceptibility (one parameter, corresponding to the marker-allele frequency). Using this multiplicative model, we estimate the contribution of this allele to sibling-specific recurrence risk of AMD to be λs=1.21 and the population-attributable fraction to be 0.62. This is the locus-specific λs, which can be calculated as a function of penetrances and disease-allele frequencies by use of the formulas of Risch (1990). The model predicts frequencies of 0.43, 0.47, and 0.10 among controls and 0.15, 0.48, 0.37 among cases for the TT, TC, and CC genotypes, respectively; the predicted values closely match the observed frequencies (see table 1) (goodness-of-fit χ2=2.61 [2 df], not significant). An additive model also fits the data well, but dominant or recessive models are excluded. The agreement between our fitted model and observed genotype counts suggests that population stratification is not a major concern (Wittke-Thompson et al. 2005), a conclusion consistent with our analysis of previous genome-scan data.
Table 2.
Comparison of Fitted Genetic Models[Note]
| Prevalence and Model(No. of Parameters) | ln(L) | P(C)a | f(TT)b | f(TC)b | f(CC)b | GRR1c | GRR2d | K | AttributableFraction | λs |
| Constrained to K=.20: | ||||||||||
| No effect (1) | −923.18 | .52 | .20 | .20 | .20 | 1 | 1 | .20 | … | … |
| General (3) | −864.86 | .39 | .08 | .21 | .47 | 2.81 | 6.30 | .20 | .62 | 1.21 |
| Multiplicativee (2) | −865.75f | .39 | .08 | .20 | .49 | 2.44 | 5.93 | .20 | .59 | 1.21 |
| Additiveg (2) | −869.14 | .41 | .07 | .23 | .38 | 3.05 | 5.10 | .20 | .63 | 1.14 |
| Dominanth (2) | −893.28 | .46 | .09 | .24 | .24 | 2.62 | 2.62 | .20 | .53 | 1.05 |
| Recessivei (2) | −897.67 | .46 | .16 | .16 | .36 | 1.00 | 2.24 | .20 | .21 | 1.07 |
| Unconstrained: | ||||||||||
| No effect (1) | −923.18 | .52 | NA | NA | NA | 1 | 1 | NA | … | … |
| General (4) | −864.46 | .49 | .30 | .57 | .83 | 1.88 | 2.77 | .56 | .46 | 1.06 |
| Multiplicativee (3) | −865.54 | .43 | .15 | .31 | .66 | 2.10 | 4.40 | .32 | .53 | 1.14 |
| Additiveg (3) | −864.46 | .49 | .30 | .57 | .83 | 1.88 | 2.76 | .56 | .46 | 1.06 |
| Dominanth (3) | −884.30 | .52 | .41 | .75 | .75 | 1.83 | 1.83 | .68 | .39 | 1.02 |
| Recessivei (3) | −884.77 | .53 | .63 | .63 | .91 | 1.00 | 1.43 | .71 | .11 | 1.01 |
Note.— NA = not applicable.
Estimated frequency of allele C.
Estimated probability of disease for genotype.
GRR1=f(TC)/f(TT).
GRR2=f(CC)/f(TT).
Constraint: GRR2=GRR12.
The best-fitting model, selected using the AIC.
Constraint: GRR1-1=GRR2-GRR1.
Constraint: GRR1=GRR2.
Contraint: GRR1=1.0.
CFH plays an essential role in regulation of complement activation, a major component of innate immunity against microbial infection. This regulation is achieved because CFH can bind to C3b (generated by cleavage of the α-chain of complement 3 [C3]), leading to the production of terminal C5b-9 complex (Giannakis et al. 2003). Many proteins of the complement system, including C5b-9 complex, have been detected in drusen from the eyes of patients with AMD (Mullins et al. 2000; Hageman et al. 2001; Johnson et al. 2001). CFH has three binding sites for C3b and additional binding sites for heparin and C-reactive protein (CRP). The Y402H change is expected to alter these interactions, since it is located within the cluster of positively charged amino acids implicated in the binding of CRP and heparin (Giannakis et al. 2003). Notably, associations have been reported between AMD and increased levels of CRP (Seddon et al. 2004) and between AMD and a polymorphism in TLR4, a key gene involved in innate immunity (Zareparsi et al. 2005). Hence, it is possible that certain microbial infections may be environmental triggers for AMD pathogenesis.
In summary, our data provide strong evidence for the Y402H variant being a common susceptibility allele for AMD. It is possible that another allele in strong disequilibrium with Y402H may cause disease susceptibility; this hypothesis can be tested only by evaluation of all polymorphisms and/or mechanistic functional evidence to explain the role of the Y402H variant. Our resequencing of CFH exon 9 (∼500 bp) in all cases and controls did not identify any additional nearby coding variants. It will also be important to determine if the CFH-Y402H variant can explain the chromosome 1q linkage signal that is observed in independent genome scans (Klein et al. 1998; Majewski et al. 2003; Seddon et al. 2003; Abecasis et al. 2004; Iyengar et al. 2004; Weeks et al. 2004). Although CFH and TLR4 are the first major AMD susceptibility genes to be identified, comprehensive studies of genetic variations are expected to lead to the identification of additional variants that contribute to this complex and debilitating disease.
Acknowledgments
We thank all the individuals who participated in this study, and we are grateful to numerous ophthalmologists and clinical staff for insights and assistance. This research was supported by grants from the National Institutes of Health (F32-EY014085, R01-EY015771, P30-EY007003, and K25-HG000060), the Foundation Fighting Blindness, the Macula Vision Research Foundation, the Elmer and Sylvia Sramek Foundation, Research to Prevent Blindness, and the Harold F. Falls Collegiate Professorship.
Web Resources
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ARMD1, ABCA4, TIMP3, RDS/peripherin, ELOVL4, fibulin-3, Doyne honeycomb retinal dystrophy, fibulin-5, fibulin-6, APOE, TLR4, and CFH)
References
- Abecasis GR, Ghosh D, Nichols TE (2005) Linkage disequilibrium: ancient history drives the new genetics. Hum Hered 59:118–124 [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Yashar BM, Zhao Y, Ghiasvand NM, Zareparsi S, Branham KE, Reddick AC, Trager EH, Yoshida S, Bahling J, Filippova E, Elner S, Johnson MW, Vine AK, Sieving PA, Jacobson SG, Richards JE, Swaroop A (2004) Age-related macular degeneration: a high-resolution genome scan for susceptibility loci in a population enriched for late-stage disease. Am J Hum Genet 74:482–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Age-Related Eye Disease Study Research Group (2000) Risk factors associated with age-related macular degeneration. A case-control study in the age-related eye disease study: Age-Related Eye Disease Study Report Number 3. Ophthalmology 107:2224–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird PN, Guida E, Chu DT, Vu HT, Guymer RH (2004) The epsilon2 and epsilon4 alleles of the apolipoprotein gene are associated with age-related macular degeneration. Invest Ophthalmol Vis Sci 45:1311–1315 [DOI] [PubMed] [Google Scholar]
- Edwards AO, Ritter R 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA (2005) Complement factor H polymorphism and age-related macular degeneration. Science 308:421–424 [DOI] [PubMed] [Google Scholar]
- Friedman DS, O’Colmain BJ, Munoz B, Tomany SC, McCarty C, de Jong PT, Nemesure B, Mitchell P, Kempen J (2004) Prevalence of age-related macular degeneration in the United States. Arch Ophthalmol 122:564–572 [DOI] [PubMed] [Google Scholar]
- Giannakis E, Jokiranta TS, Male DA, Ranganathan S, Ormsby RJ, Fischetti VA, Mold C, Gordon DL (2003) A common site within factor H SCR 7 responsible for binding heparin, C-reactive protein and streptococcal M protein. Eur J Immunol 33:962–969 [DOI] [PubMed] [Google Scholar]
- Goring HH, Terwilliger JD, Blangero J (2001) Large upward bias in estimation of locus-specific effects from genomewide scans. Am J Hum Genet 69:1357–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman GS, Luthert PJ, Victor Chong NH, Johnson LV, Anderson DH, Mullins RF (2001) An integrated hypothesis that considers drusen as biomarkers of immune-mediated processes at the RPE-Bruch’s membrane interface in aging and age-related macular degeneration. Prog Retin Eye Res 20:705–732 [DOI] [PubMed] [Google Scholar]
- Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, Schnetz-Boutaud N, Agarwal A, Postel EA, Pericak-Vance MA (2005) Complement factor H variant increases the risk of age-related macular degeneration. Science 308:419–421 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Merriam JE, Klaver CC, Zernant J, Bergen AA, Smith RT, Chang S, Merriam JC, Allikmets R (2004) Evaluation of the ARMD1 locus on 1q25-31 in patients with age-related maculopathy: genetic variation in laminin genes and in exon 104 of HEMICENTIN-1. Ophthalmic Genet 25:111–119 [DOI] [PubMed] [Google Scholar]
- Iyengar SK, Song D, Klein BE, Klein R, Schick JH, Humphrey J, Millard C, Liptak R, Russo K, Jun G, Lee KE, Fijal B, Elston RC (2004) Dissection of genomewide-scan data in extended families reveals a major locus and oligogenic susceptibility for age-related macular degeneration. Am J Hum Genet 74:20–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LV, Leitner WP, Staples MK, Anderson DH (2001) Complement activation and inflammatory processes in drusen formation and age related macular degeneration. Exp Eye Res 73:887–896 [DOI] [PubMed] [Google Scholar]
- Klaver CC, Kliffen M, van Duijn CM, Hofman A, Cruts M, Grobbee DE, van Broeckhoven C, de Jong PT (1998) Genetic association of apolipoprotein E with age-related macular degeneration. Am J Hum Genet 63:200–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein ML, Schultz DW, Edwards A, Matise TC, Rust K, Berselli CB, Trzupek K, Weleber RG, Ott J, Wirtz MK, Acott TS (1998) Age-related macular degeneration: clinical features in a large family and linkage to chromosome 1q. Arch Ophthalmol 116:1082–1088 [DOI] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, Sangiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J (2005) Complement factor H polymorphism in age-related macular degeneration. Science 308:385–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Boehnke M, Abecasis GR (2005) Joint modeling of linkage and association: identifying SNPs responsible for a linkage signal. Am J Hum Genet 76:934–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN (2003) Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet 33:177–182 [DOI] [PubMed] [Google Scholar]
- Majewski J, Schultz DW, Weleber RG, Schain MB, Edwards AO, Matise TC, Acott TS, Ott J, Klein ML (2003) Age-related macular degeneration: a genome scan in extended families. Am J Hum Genet 73:540–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein LY, Munier FL, Arsenijevic Y, Schorderet DF, McLaughlin PJ, Chung D, Traboulsi E, Marmorstein AD (2002) Aberrant accumulation of EFEMP1 underlies drusen formation in malattia leventinese and age-related macular degeneration. Proc Natl Acad Sci USA 99:13067–13072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RF, Russell SR, Anderson DH, Hageman GS (2000) Drusen associated with aging and age-related macular degeneration contain proteins common to extracellular deposits associated with atherosclerosis, elastosis, amyloidosis, and dense deposit disease. FASEB J 14:835–846 [PubMed] [Google Scholar]
- Risch N (1990) Linkage strategies for genetically complex traits. I. Multilocus models. Am J Hum Genet 46:222–228 [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, Klaver C, Saunders A, Postel E, De La Paz M, Agarwal A, Small K, Udar N, Ong J, Chalukya M, Nesburn A, Kenney C, Domurath R, Hogan M, Mah T, Conley Y, Ferrell R, Weeks D, de Jong PT, van Duijn C, Haines J, Pericak-Vance M, Gorin M (2002) A pooled case-control study of the apolipoprotein E (APOE) gene in age-related maculopathy. Ophthalmic Genet 23:209–223 [DOI] [PubMed] [Google Scholar]
- Schmidt S, Scott WK, Postel EA, Agarwal A, Hauser ER, De La Paz MA, Gilbert JR, Weeks DE, Gorin MB, Haines JL, Pericak-Vance MA (2004) Ordered subset linkage analysis supports a susceptibility locus for age-related macular degeneration on chromosome 16p12. BMC Genet 5:18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DW, Klein ML, Humpert AJ, Luzier CW, Persun V, Schain M, Mahan A, Runckel C, Cassera M, Vittal V, Doyle TM, Martin TM, Weleber RG, Francis PJ, Acott TS (2003) Analysis of the ARMD1 locus: evidence that a mutation in HEMICENTIN-1 is associated with age-related macular degeneration in a large family. Hum Mol Genet 12:3315–3323 [DOI] [PubMed] [Google Scholar]
- Seddon JM, Gensler G, Milton RC, Klein ML, Rifai N (2004) Association between C-reactive protein and age-related macular degeneration. JAMA 291:704–710 [DOI] [PubMed] [Google Scholar]
- Seddon JM, Santangelo SL, Book K, Chong S, Cote J (2003) A genomewide scan for age-related macular degeneration provides evidence for linkage to several chromosomal regions. Am J Hum Genet 73:780–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EM, Braun TA, Russell SR, Kuehn MH, Lotery AJ, Moore PA, Eastman CG, Casavant TL, Sheffield VC (2004) Missense variations in the fibulin 5 gene and age-related macular degeneration. N Engl J Med 351:346–353 [DOI] [PubMed] [Google Scholar]
- Stone EM, Lotery AJ, Munier FL, Heon E, Piguet B, Guymer RH, Vandenburgh K, Cousin P, Nishimura D, Swiderski RE, Silvestri G, Mackey DA, Hageman GS, Bird AC, Sheffield VC, Schorderet DF (1999) A single EFEMP1 mutation associated with both Malattia Leventinese and Doyne honeycomb retinal dystrophy. Nat Genet 22:199–202 [DOI] [PubMed] [Google Scholar]
- Stone EM, Sheffield VC, Hageman GS (2001) Molecular genetics of age-related macular degeneration. Hum Mol Genet 10:2285–2292 [DOI] [PubMed] [Google Scholar]
- Weeks DE, Conley YP, Mah TS, Paul TO, Morse L, Ngo-Chang J, Dailey JP, Ferrell RE, Gorin MB (2000) A full genome scan for age-related maculopathy. Hum Mol Genet 9:1329–1349 [DOI] [PubMed] [Google Scholar]
- Weeks DE, Conley YP, Tsai HJ, Mah TS, Schmidt S, Postel EA, Agarwal A, Haines JL, Pericak-Vance MA, Rosenfeld PJ, Paul TO, Eller AW, Morse LS, Dailey JP, Ferrell RE, Gorin MB (2004) Age-related maculopathy: a genomewide scan with continued evidence of susceptibility loci within the 1q31, 10q26, and 17q25 regions. Am J Hum Genet 75:174–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittke-Thompson JK, Pluzhnikov A, Cox NJ (2005) Rational inferences about departures from Hardy-Weinberg equilibrium. Am J Hum Genet 76:967–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zareparsi S, Buraczynska M, Branham KE, Shah S, Eng D, Li M, Pawar H, Yashar BM, Moroi SE, Lichter PR, Petty HR, Richards JE, Abecasis GR, Elner VM, Swaroop A (2005) Toll-like receptor 4 variant D299G is associated with susceptibility to age-related macular degeneration. Hum Mol Genet (http://hmg.oupjournals.org/cgi/reprint/ddi154v1) (electronically published April 13, 2005; accessed May 9, 2005) [DOI] [PubMed] [Google Scholar]
- Zareparsi S, Reddick AC, Branham KE, Moore KB, Jessup L, Thoms S, Smith-Wheelock M, Yashar BM, Swaroop A (2004) Association of apolipoprotein E alleles with susceptibility to age-related macular degeneration in a large cohort from a single center. Invest Ophthalmol Vis Sci 45:1306–1310 [DOI] [PubMed] [Google Scholar]


