Abstract
The hereditary spastic paraplegias (HSPs) are a complex group of neurodegenerative disorders characterized by lower-limb spasticity and weakness. Silver syndrome (SS) is a particularly disabling dominantly inherited form of HSP, complicated by amyotrophy of the hand muscles. Having excluded the multiple known HSP loci, we undertook a genomewide screen for linkage of SS in one large multigenerational family, which revealed evidence for linkage of the SS locus, which we have designated “SPG17,” to chromosome 11q12-q14. Haplotype construction and analysis of recombination events permitted the minimal interval defining SPG17 to be refined to ∼13 cM, flanked by markers D11S1765 and D11S4136. SS in a second family was not linked to SPG17, demonstrating further genetic heterogeneity in HSP, even within this clinically distinct subtype.
“Hereditary spastic paraplegia” (HSP) is a general term used to describe the group of disorders characterized by progressive lower-limb spasticity and weakness. When spastic paraplegia is the only clinical feature, the disease is described as “pure” HSP; if accompanied by other neurological features, such as deafness, dementia, and mental retardation, the disease is described as “complicated” HSP. The pathological feature common to both subtypes is axonal degeneration involving the longest fibers of the corticospinal tracts and of the dorsal columns, which may be associated with a loss of anterior horn cells (Harding 1993). Although variability in age at onset, in rate of progression, and in severity are features of HSP (Harding 1981; Polo et al. 1993; Dürr et al. 1994; De Jonghe et al. 1996; Nielsen et al. 1997), the first signs of disease typically are apparent by the 2d or 3d decade of life.
In addition to marked clinical variation, HSP demonstrates considerable genetic heterogeneity, with eight autosomal dominant, four autosomal recessive, and three X-linked loci mapped to date (Hazan et al. 1993, 1994; Hentati et al. 1994; Jouet et al. 1994; Saugier-Veber et al. 1994; Fink et al. 1995; Casari et al. 1998; Hedera et al. 1999; Martinez Murrilo et al. 1999; Reid et al. 1999, 2000; Seri et al. 1999; Claes et al. 2000; Fontaine et al. 2000; Vazza et al. 2000).
An especially disabling autosomal dominant form of complicated HSP, termed “Silver syndrome” (SS [MIM 270685]), is characterized by lower-limb spasticity associated with marked amyotrophy of and weakness of the small muscles of the hands and the feet (Silver 1966). Previously, we have excluded linkage of this form of HSP to any of the known HSP loci (Patel et al., in press). Having performed a genomewide screen for linkage, we now present evidence for linkage of SS to chromosome 11q12-q14 and for marked phenotypic variability in this form of HSP.
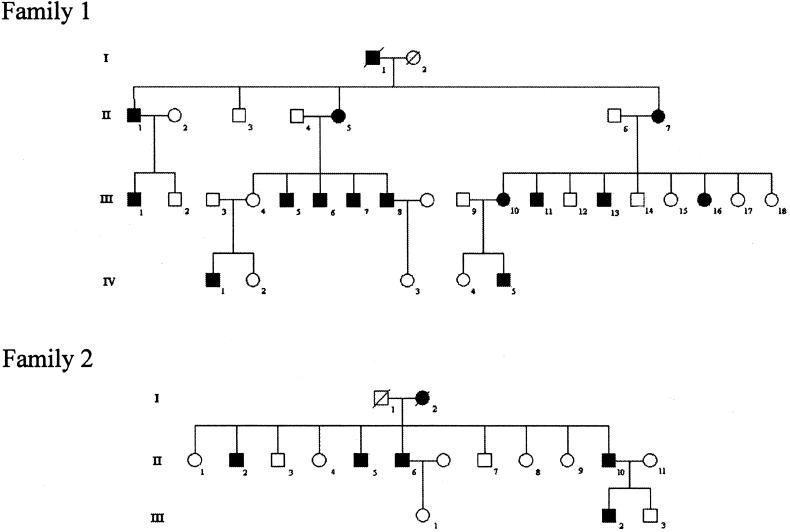
Clinical evaluation of two families was undertaken by two independent examiners (P.H. and T.T.W.), and the data are summarized in table 1. Patients in two families with SS (fig. 1) were classified either as definitely, probably, or possibly affected by HSP or as unaffected, on the basis of standard criteria (Fink et al. 1996). The presence of wasting of the small muscles of the hands and the feet also indicated affected status, but individuals were classified as definitely affected only if they presented with lower-limb hyperreflexia and extensor-plantar responses, which could occur without amyotrophy of the small muscles of the hands and the feet.
Table 1.
Clinical Features of Affected Members of Families 1 and 2[Note]
|
Age at(years) |
Status ofa |
|||||||
| Pyramidal Signs |
||||||||
| Patient | Examination | Onset | Sphincter Involvement | Pes cavus | Amyotrophy of Hand Muscles | Upper Limb | Lower Limb | Vibration Sense |
| Family 1: | ||||||||
| II.1 | 75 | 25 | … | … | + | + | ++ | N |
| II.5 | 77 | 40s | ++ | ++ | + | + | +++ | N |
| II.7 | 81 | … | … | ++ | ++ | − | ++ | N |
| III.1 | 45 | 20s | … | + | − | + | + | N |
| III.3 | 18 | 15 | … | + | − | − | + | N |
| III.5 | 49 | 20s | + | − | + | − | + | N |
| III.6 | 53 | 8 | … | + | + | − | ++ | N |
| III.8 | 55 | 28 | … | + | + | − | ++ | N |
| III.11 | 57 | … | … | − | − | − | − | … |
| III.13 | 61 | 40 | … | − | + | + | + | Impairment in ankles |
| III.16 | 42 | … | … | + | ++ | − | + | N |
| Family 2: | ||||||||
| II.6 | 64 | 35 | … | − | − | − | ++ | N |
| II.10 | 52 | … | … | − | ++ | − | + | Impairment in ankles |
| III.2 | 23 | 7 | ++ | + | ++ | − | ++ | N |
Note.— Data for affected individuals III.7 and IV.1 in family 1 and for affected individuals II.2 and II.5 in family 2 were not available.
− = Absent; + = mild; ++ = moderate; +++ = severe; N = normal.
Figure 1.
Structure of SS pedigrees 1 and 2. Blackened symbols denote affected individuals.
Age at onset in family 1 varied for gait abnormalities (8–40 years) and for hand involvement (14–60 years). Lower-limb spasticity and amyotrophy of intrinsic hand muscles was present in the majority of affected individuals. All individuals' intrinsic hand muscles were weak, with severe amyotrophy most marked in the thenar eminence. There was also impairment of vibration sense in the lower limbs of older individuals.
Assignment of affection status to family members was essentially straightforward, with the exception of individual III.11, whose medical history was complicated by previous pulmonary sarcoidosis and type II diabetes. Individual III.11 did have clinical features consistent with a diagnosis of HSP—namely, spastic gait, bilateral clawing of the toes, hypertonia of the leg muscles, brisk knee jerks, and an extensor–left-plantar response—and it was considered highly likely that he was affected. However, although there were signs compatible with an ulnar-nerve lesion at the left elbow, there were no other signs of amyotrophy.
Previously, linkage of SS in families 1 and 2 to any of the known autosomal dominant HSP loci (on chromosomes 2p21-p22, 2q24-q34, 8q24, 10q23.3-q24.1, 12q13, 14q11.2-q24.3, 15q11.1, and 19q13) has been excluded (Patel et al., in press). To map the position of the SS variant of HSP, a 10-cM–resolution, genomewide screen for linkage (ABI PRISM Linkage Mapping Set Version 2 markers; Perkin Elmer Biosystems) was undertaken in family 1. Since male-to-male transmission of the disorder has been observed in these families, linkage analysis was confined to the 22 human autosomes. PCRs were performed in either a GeneAmp PCR system 9700 (Perkin Elmer Biosystems) or a Techne Genius PCR machine. PCR products were then pooled and size fractionated on a 4% denaturing polyacrylamide gel (BioRad), by an ABI377 sequencer (Perkin Elmer Biosystems), according to recommended protocols. The data were analyzed by GeneScan Analysis Software 2.1 and GENOTYPER (version 2.1) (Perkin Elmer Biosystems), to generate genotypes for linkage analysis.
To investigate regions of uninformativity, further saturation, with an additional 223 markers, was performed with unlabeled primers (Genosys), with detection on 8% denaturing PAGE and silver staining. These PCRs were performed with 200 ng of genomic DNA, reaction buffer IV containing 1.5 mM MgCl2 (Advanced Biotechnologies), 5 mM each dNTP, 50 pmol of each primer, and 0.15 units of Red Hot DNA polymerase (Advanced Biotechnologies), in a final volume of 25 ml. Amplifications were performed with a “hot-start” cycle of 2× (96°C, +4°C above calculated annealing, and 72°C—all for 30 s each), 2× (96°C, +2°C above calculated annealing, and 72°C—all for 30 s each), and 35× (96°C, at annealing, and 72°C—all for 30 s each).
Two-point LOD scores were calculated, for each marker, by MLINK (version 5.1) of the LINKAGE package (Lathrop et al. 1984, 1985), as implemented in FASTLINK (version 4.1) (Schäffer et al. 1994). For linkage computations, the gene frequency of HSP was assumed to be 10−4, and male and female recombination rates (θ) were assumed to be equal. LOD scores were calculated with equal allele frequencies assumed for all markers. The map order and distances between markers were based on the microsatellite-marker map of the Center for Medical Genetics, Marshfield Medical Research Foundation.
In view of the variable penetrance in HSP, LOD scores were initially computed by a conservative approach, with affected family members only. In the absence of published data on age-specific population risks of spasticity and of amyotrophy, a nominal phenocopy risk of 10−4 was assumed. LOD scores were also calculated by assigning family members to liability classes. Although the penetrance of SS cannot be reliably calculated from a single family, age-specific risks were estimated by survival analysis (through symptoms specific to age at onset or through symptoms common within the family). Under this model, the disease gene confers a cumulative risk of 24% by the age of 20 years and of 70% by the age of 45 years and a lifetime risk of 85%.
Linkage of SS was initially assessed using affected family members only and by assigning individual III.11 to an “unknown” phenotype. This analysis generated pairwise LOD scores of 3.51 and 3.49 for chromosome 11q12-q14 markers D11S987 and D11S4113, respectively. Pairwise LOD scores generated under the assumption that individual III.11 is affected for all chromosome 11q12-q14 markers are shown in table 2. A second analysis was then performed by assigning family members to liability classes. The LOD scores generated in this manner were not significantly different from those determined in the analysis based on an affecteds-only model.
Table 2.
LOD Scores for Chromosome 11 Markers in Family 1
|
LOD Score at θ = |
|||||||
| Locus | .00 | .01 | .10 | .20 | .30 | .40 | .50 |
| D11S986 | −.11 | .14 | 1.01 | 1.30 | 1.24 | .86 | .35 |
| D11S4191 | −.71 | −.45 | .46 | .79 | .83 | .58 | .20 |
| D11S1765 | −.29 | −.04 | .84 | 1.13 | 1.08 | .74 | .29 |
| D11S4205 | 3.77 | 3.72 | 3.45 | 3.10 | 2.34 | 1.50 | .62 |
| D11S1883 | 3.80 | 3.75 | 3.48 | 3.13 | 2.37 | 1.53 | .64 |
| D11S4178 | 3.10 | 3.05 | 2.82 | 2.51 | 1.86 | 1.13 | .40 |
| D11S1889 | 3.80 | 3.74 | 3.47 | 3.12 | 2.36 | 1.53 | .64 |
| D11S913 | 3.77 | 3.72 | 3.45 | 3.10 | 2.34 | 1.50 | .62 |
| D11S987 | 3.88 | 3.82 | 3.55 | 3.20 | 2.44 | 1.60 | .69 |
| D11S4155 | .56 | .55 | .50 | .44 | .32 | .20 | .09 |
| D11S4113 | 3.88 | 3.82 | 3.55 | 3.20 | 2.44 | 1.60 | .69 |
| D11S4095 | 3.54 | 3.49 | 3.24 | 2.92 | 2.22 | 1.46 | .63 |
| D11S1337 | .51 | .50 | .45 | .38 | .25 | .13 | .04 |
| D11S4136 | −.80 | .07 | .61 | .73 | .63 | .39 | .14 |
| D11S4196 | 2.38 | 2.34 | 2.16 | 1.93 | 1.42 | .88 | .34 |
| D11S1314 | 1.00 | 1.84 | 2.28 | 2.25 | 1.84 | 1.23 | .52 |
| D11S4184 | 1.82 | 1.79 | 1.64 | 1.45 | 1.04 | .62 | .23 |
| D11S916 | −.50 | .36 | .89 | .98 | .83 | .53 | .22 |
| D11S4207 | −.80 | .07 | .61 | .73 | .63 | .39 | .14 |
| D11S4128 | .23 | .97 | 1.45 | 1.49 | 1.23 | .80 | .31 |
| D11S1321 | .91 | 1.75 | 2.19 | 2.16 | 1.76 | 1.16 | .48 |
| D11S4081 | .81 | .79 | .72 | .64 | .46 | .28 | .12 |
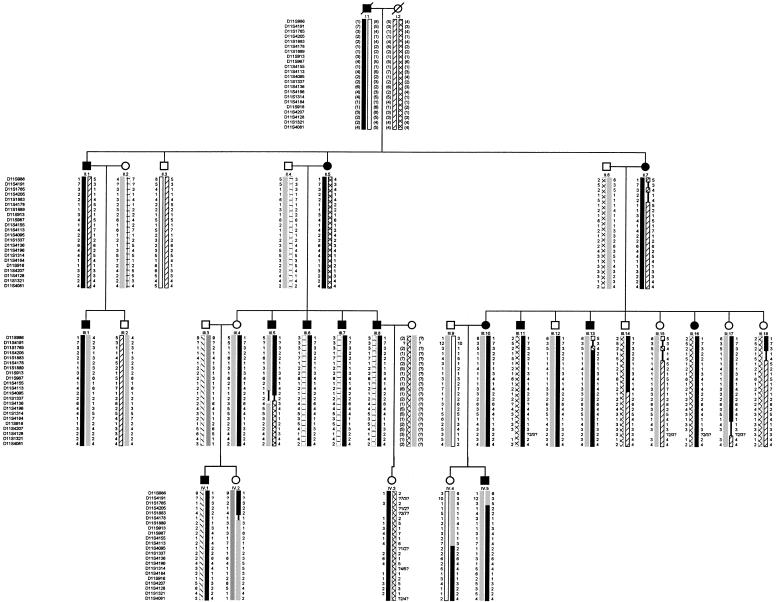
Haplotype construction revealed that each of the 14 affected family members had inherited an identical region of chromosome 11q12-q14 (fig. 2). Recombination events across this region positioned the locus, which we have designated “SPG17,” within a 13-cM interval flanked by markers D11S1765 and D11S4136. Haplotype analysis also showed that four individuals—III.4, III.12, III.17, and IV.3, clinically unaffected at the ages of 41, 55, 45, and 35 years, respectively—have inherited the whole affected haplotype (i.e., D11S1765–D11S4136). These four individuals were independently examined on two occasions, but none showed signs of disease. Of these four individuals, III.4 has passed on the complete affected haplotype to an affected daughter, age 18 years, but is herself clinically unaffected at the age of 41 years (fig. 2). Three other unaffected individuals—III.14, IV.2, and IV.4—have inherited a portion of the affected haplotype—D11S1765–D11S4095, D11S1765–D11S1889, and D11S4113–D11S4136, respectively. These data suggest that the penetrance of SPG17, as determined in family 1, is ∼.66 by the age of 40 years and .92 by the age of 55 years.
Figure 2.
Chromosome 11q12-q14 haplotype. The haplotype segregating with SS is in black. Reconstructed genotypes are in parentheses. Recombination events in affected individuals restrict the critical interval to an ∼13-cM region between D11S1765 and D11S4136. Blackened symbols denote affected individuals.
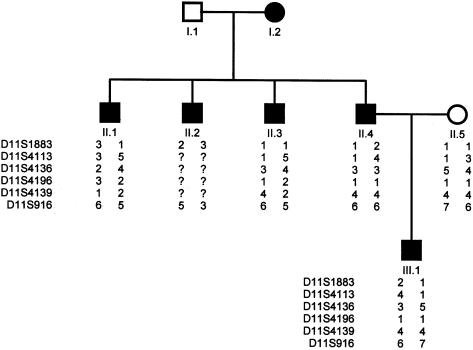
Linkage of SS to chromosome 11q12-q14 was also examined in the affected members of family 2, who display a similar pattern of amyotrophy of the hand muscles and a highly variable age at onset, for markers D11S1883, D11S4113, D114136, D114196, D11S4139, and D11S916 (fig. 3). In family 2, SS clearly does not segregate with these markers. Two-point LOD scores generated by an affecteds-only model (θ=.001), for markers D11S1883, D11S4113, D114136, D114196, D11S4139, and D11S916, were −2.88, −2.53, −2.30, −2.30, −2.30, and −2.04, respectively, which demonstrates further genetic heterogeneity of HSP, even within this rare subtype.
Figure 3.
Haplotype for pedigree 2, showing microsatellites in the region that shows linkage of SS in family 1. Blackened symbols denote affected individuals.
Despite the large number of HSP loci already established, many families do not show evidence of linkage to any of these regions (McDermott et al. 2000). Using a single large family (i.e., family 1), we have identified, on chromosome 11q12-q14, a locus containing a gene, SPG17, that causes SS, a complicated form of HSP. Variable penetrance appears to be a characteristic of SS, consistent with other studies that suggest a highly variable age at onset of other forms of the disease (Gigli et al. 1993; Byrne et al. 1998; Fontaine et al. 2000).
Recent studies have begun to determine the molecular pathogenesis of some forms of this heterogeneous disease. Mutations of the genes encoding paraplegin and spastin underlie, respectively, autosomal recessive and autosomal dominant forms of HSP. These genes encode AAA proteins (i.e., ATPases associated with diverse cellular activities). Although distantly related, paraplegin and spastin are thought to have divergent roles, sharing little homology outside their AAA motifs. Although little is known about the nuclear molecule spastin, paraplegin is thought to be a mitochondrial chaperone with homology to the yeast ATPase AFG3. In yeast, AFG3 is part of a membrane-bound complex with ATP-dependent protease activity. Similarly, AFG3 exhibits a chaperone-like function, mediating assembly of membrane-associated ATP synthase (Arlt et al. 1996).
The gene for copper chaperone for superoxide dismutase (CCS) has recently been mapped within the SS locus on chromosome 11q (i.e., SPG17) (Bartnikas et al. 2000). Although its function is clearly different from that of paraplegin, CCS donates copper to the homodimeric enzyme SOD1, the principal function of which is believed to be the detoxification of superoxide radicals. SOD1 is mutated in ∼20% of patients with familial amyotrophic lateral sclerosis (ALS) (Deng et al. 1993; Rosen et al. 1993; Casareno et al. 1998). ALS and HSP—particularly, the SS variant—share a number of features, including spasticity, muscle weakness and amyotrophy, and pathological changes in corticospinal tracts and in anterior horn cells (Harding 1993). In view of this, we are currently evaluating patients with SS, for disease-associated mutations of CCS.
Acknowledgments
This work was supported by the Birth Defects Foundation (U.K.), by St. George’s Hospital Medical School, and by The Wellcome Trust.
Electronic-Database Information
The accession number and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/ (for microsatellite-marker map)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SS [MIM 270685])
References
- Arlt H, Tauer R, Feldmann H, Neupert W, Langer T (1996) The YTA10-12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell 85:875–885 [DOI] [PubMed] [Google Scholar]
- Bartnikas TB, Waggoner DJ, Casareno RL, Gaedigk R, White RA, Gitlin JD (2000) Chromosomal localization of CCS, the copper chaperone for Cu/Zn superoxide dismutase Mamm Genome 11:409–411 [DOI] [PubMed] [Google Scholar]
- Byrne PC, Webb S, McSweeney F, Burke T, Hutchinson M, Parfrey NA (1998) Linkage of AD HSP and cognitive impairment to chromosome 2p: haplotype and phenotype analysis indicates variable expression and low or delayed penetrance. Eur J Hum Genet 6:275–282 [DOI] [PubMed] [Google Scholar]
- Casareno RL, Waggoner D, Gitlin JD (1998) The copper chaperone CCS directly interacts with copper/zinc superoxide dismutase. J Biol Chem 273:23625–23628 [DOI] [PubMed] [Google Scholar]
- Casari G, De Fusco M, Ciarmatori S, Zeviani M, Mora M, Fernandez P, De Michele G, Filla A, Cocozza S, Marconi R, Durr A, Fontaine B, Ballabio A (1998) Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell 93:973–983 [DOI] [PubMed] [Google Scholar]
- Claes S, Devriendt K, Van Goethem G, Roelen L, Meireleire J, Raeymaekers P, Cassiman JJ, Fryns JP (2000) Novel syndromic form of X-linked complicated spastic paraplegia. Am J Med Genet 94:1–4 [DOI] [PubMed] [Google Scholar]
- De Jonghe P, Krols L, Michalik A, Hazan J, Smeyers G, Lofgren A, Weissenbach J, Martin JJ, Van Broeckhoven C (1996) Pure familial spastic paraplegia: clinical and genetic analysis of nine Belgian pedigrees. Eur J Hum Genet 4:260–266 [DOI] [PubMed] [Google Scholar]
- Deng HX, Hentati A, Tainer JA, Iqbal Z, Cayabyab A, Hung WY, Getzoff ED, Hu P, Herzfeldt B, Roos RP, Warner C, Deng G, Soriano E, Smyth C, Parge HE, Ahmed A, Roses AD, Hallewell RA, Pericak-Vance MA, Siddique T (1993) Amyotrophic lateral sclerosis and structural defects in Cu,Zn superoxide dismutase. Science 261:1047–1051 [DOI] [PubMed] [Google Scholar]
- Dürr A, Brice A, Serdaru M, Rancurel G, Derouesne C, Lyon-Caen O, Agid Y, Fontaine B (1994) The phenotype of “pure” autosomal dominant spastic paraplegia. Neurology 44:1274–1277 [DOI] [PubMed] [Google Scholar]
- Fink JK, Heiman-Patterson T, Bird T, Cambi F, Dube MP, Figlewicz DA, Haines JL, Heiman-Patterson T, Hentati A, Pericak-Vance MA, Raskind W, Rouleau GA, Siddique T (1996) Hereditary spastic paraplegia: advances in genetic research. Neurology 46:1507–1514 [DOI] [PubMed] [Google Scholar]
- Fink JK, Jones SM, Sharp GB, Lange BM, Otterud B, Leppert M (1995) Autosomal dominant familial spastic paraplegia: tight linkage to chromosome 15q. Am J Hum Genet 56:188–192 [PMC free article] [PubMed] [Google Scholar]
- Fontaine B, Davoine CS, Durr A, Paternotte C, Feki I, Weissenbach J, Hazan J, Brice A (2000) A new locus for autosomal dominant pure spastic paraplegia, on chromosome 2q24-q34. Am J Hum Genet 66:702–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigli GL, Diomedi M, Bernardi G, Placidi F, Marciani MG, Calia E, Maschio MC, Neri G (1993) Spastic paraplegia, epilepsy, and mental retardation in several members of a family: a novel genetic disorder. Am J Med Genet 45:711–716 [DOI] [PubMed] [Google Scholar]
- Harding AE (1981) Hereditary “pure” spastic paraplegia: a clinical and genetic study of 22 families. J Neurol Neurosurg Psychiatry 44:871–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (1993) Hereditary spastic paraplegias. Semin Neurol 13:333–336 [DOI] [PubMed] [Google Scholar]
- Hazan J, Fontaine B, Bruyn RPM, Lamy C, van Deutekom CT, Rime CS, Durr A, Melki J, Lyon-Caen O, Agid Y, Munnich A, Padberg GW, de Recondo J, Frants RR, Brice A, Weissenbach J (1994) Linkage of a new locus for autosomal dominant familial spastic paraplegia to chromosome 2p. Hum Mol Genet 3:1569–1573 [DOI] [PubMed] [Google Scholar]
- Hazan J, Lamy C, Melki J, Munnich A, de Recondo J, Weissenbach J (1993) Autosomal dominant familial spastic paraplegia is genetically heterogeneous and one locus maps to chromosome 14q. Nat Genet 5:163–167 [DOI] [PubMed] [Google Scholar]
- Hedera P, Rainier S, Alvarado D, Zhao X, Williamson J, Otterud B, Leppert M, Fink JK (1999) Novel locus for autosomal dominant hereditary spastic paraplegia, on chromosome 8q. Am J Hum Genet 64:563–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentati A, Pericak-Vance MA, Hung WY, Belal S, Laing N, Boustany RM, Hentati F, Ben Hamida M, Siddique T (1994) Linkage of ‘pure’ autosomal recessive familial spastic paraplegia to chromosome 8 markers and evidence of genetic locus heterogeneity. Hum Mol Genet 3:1263–1267 [DOI] [PubMed] [Google Scholar]
- Jouet M, Rosenthal A, Armstrong G, MacFarlane J, Stevenson R, Paterson J, Metzenberg A, Ionasescu V, Temple K, Kenwrick S (1994) X-linked spastic paraplegia (SPG1), MASA syndrome and X-linked hydrocephalus result from mutations in the L1 gene. Nat Genet 7:402–407 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier JM, Ott J (1984) Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 81:3443–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (1985) Multilocus linkage analysis in humans: detection of linkage and estimation of recombination. Am J Hum Genet 37:482–498 [PMC free article] [PubMed] [Google Scholar]
- Martinez Murrilo F, Kobayashi H, Pegoraro E, Galluzzi G, Creel G, Mariani C, Farina E, Ricci E, Alfonso G, Pauli RM, Hoffman EP (1999) Genetic localization of a new locus for recessive familial spastic paraparesis to 15q13-q15. Neurology 53:50–56 [DOI] [PubMed] [Google Scholar]
- McDermott C, White K, Bushby K, Shaw P (2000) Hereditary spastic paraparesis: a review of new developments. J Neurol Neurosurg Psychiatry 69:150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JE, Koefoed P, Abell K, Hasholt L, Eiberg H, Fenger K, Niebuhr E, Sorensen SA (1997) CAG repeat expansion in autosomal dominant pure spastic paraplegia linked to chromosome 2p21-p24. Hum Mol Genet 6:1811–1816 [DOI] [PubMed] [Google Scholar]
- Patel H, Hart PE, Warner T, Allen I, Phillimore HE, Silver JR, Wood NW, Jeffery S, Patton MA, Crosby AH. Silver syndrome is not linked to any of the previously established autosomal dominant hereditary spastic paraplegia loci. Am J Med Genet (in press) [DOI] [PubMed] [Google Scholar]
- Polo JM, Calleja J, Combarros O, Berciano J (1993) Hereditary “pure” spastic paraplegia: a study of nine families. J Neurol Neurosurg Psychiatry 56:175–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid E, Dearlove AM, Osborn O, Rogers MT, Rubinsztein DC (2000) A locus for autosomal dominant “pure” hereditary spastic paraplegia maps to chromosome 19q13. Am J Hum Genet 66:728–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid E, Dearlove AM, Rhodes M, Rubinsztein DC (1999) A new locus for autosomal dominant “pure” hereditary spastic paraplegia mapping to chromosome 12q13 and evidence of further genetic heterogeneity. Am J Hum Genet 65:757–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, et al (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362:59–62 [DOI] [PubMed] [Google Scholar]
- Saugier-Veber P, Munnich A, Bonneau D, Rozet JM, le Merrer M, Gil R, Boespflug-Tanguy O (1994) X-linked spastic paraplegia and Pelizaeus-Merzbacher disease are allelic at the proteolipid protein locus. Nat Genet 6:257–262 [DOI] [PubMed] [Google Scholar]
- Schäffer AA, Gupta K, Shriram K, Cottingham RW (1994) Avoiding recomputation in linkage analysis. Hum Hered 44:225–237 [DOI] [PubMed] [Google Scholar]
- Seri M, Cusano R, Forabosco P, Cinti R, Caroli F, Picco P, Bini R, Morra VB, De Michele G, Lerone M, Silengo M, Pela I, Borrone C, Romeo G, Devoto M (1999) Genetic mapping to 10q23.3-q24.2, in a large Italian pedigree, of a new syndrome showing bilateral cataracts, gastroesophageal reflux, and spastic paraparesis with amyotrophy. Am J Hum Genet 64:586–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver JR (1966) Familial spastic paraplegia with amyotrophy of the hands. J Neurol Neurosurg Psychiatry 29:135–144 [Google Scholar]
- Vazza G, Zortea M, Boaretto F, Micaglio GF, Sartori V, Mostacciuolo ML (2000) A new locus for autosomal recessive spastic paraplegia associated with mental retardation and distal motor neuropathy, SPG14, maps to chromosome 3q27-q28. Am J Hum Genet 67:504–509 [DOI] [PMC free article] [PubMed] [Google Scholar]