Abstract
Rheumatoid arthritis (RA) is the most common systemic autoimmune disease, affecting ∼1% of the adult population worldwide, with an estimated heritability of 60%. To identify genes involved in RA susceptibility, we investigated the association between putative functional single-nucleotide polymorphisms (SNPs) and RA among white individuals by use of a case-control study design; a second sample was tested for replication. Here we report the association of RA susceptibility with the minor allele of a missense SNP in PTPN22 (discovery-study allelic P=6.6×10-4; replication-study allelic P=5.6×10-8), which encodes a hematopoietic-specific protein tyrosine phosphatase also known as “Lyp.” We show that the risk allele, which is present in ∼17% of white individuals from the general population and in ∼28% of white individuals with RA, disrupts the P1 proline-rich motif that is important for interaction with Csk, potentially altering these proteins' normal function as negative regulators of T-cell activation. The minor allele of this SNP recently was implicated in type 1 diabetes, suggesting that the variant phosphatase may increase overall reactivity of the immune system and may heighten an individual carrier’s risk for autoimmune disease.
Rheumatoid arthritis (RA [MIM 180300]) is characterized by immune cell–mediated destruction of the joint architecture and is two to three times more common in women than in men (Firestein 2003). A strong genetic component is indicated (Seldin et al. 1999; MacGregor et al. 2000), and genome scans have identified multiple regions linked to disease (Cornelis et al. 1998; Shiozawa et al. 1998; Jawaheer et al. 2001, 2003; MacKay et al. 2002; Fisher et al. 2003). Although interesting associations have been reported (Suzuki et al. 2003; Tokuhiro et al. 2003), only alleles at the HLA-DRB1 locus have consistently demonstrated both linkage and association (Seldin et al 1999).
To identify genes involved in genetic predisposition to RA, we performed a case-control association study (called “discovery study”) with assays for 87 putative functional SNPs (Botstein and Risch 2003) in RA candidate genes and/or linkage regions. The discovery study, consisting of 475 individuals with RA and 475 individually matched controls, was obtained by Genomics Collaborative, Inc. (GCI). Case samples were collected from throughout the United States, and they met the 1987 American College of Rheumatology (ACR) diagnostic criteria for RA. All case samples were from white individuals with an age at onset of RA of between 18 and 68 years and a positive rheumatoid factor of ⩾20 IU. Individuals with psoriasis, systemic lupus erythematosus (SLE), ankylosing spondylitis, or Reiter syndrome were excluded. Control samples were taken from a pool of healthy white individuals from the United States with no medical history of RA or of any of the autoimmune disorders listed above. A single control was matched to each case on the basis of sex, age (±5 years), and ethnicity (grandparental country/region of origin). All protocols and recruitment sites have been approved by national and/or local institutional review boards, and all subjects were enrolled with informed written consent.
We found association with the minor allele (T) of a missense SNP (R620W [rs2476601, 1858C→T]) in the protein tyrosine phosphatase non-receptor type 22 gene (PTPN22) (allele frequency 13.8% in cases, 8.8% in controls) (P=6.6×10-4, allelic odds ratio [OR] 1.65, 95% CI 1.23–2.20).
Replication in a second study (called the “replication study”) confirmed association. Cases in the replication study were obtained by the North American Rheumatoid Arthritis Consortium (NARAC) and consisted of members of white multiplex families. For this study, DNA was available for 840 individuals with RA from 463 families. These families were recruited from throughout the United States through the 12 participating recruitment centers of NARAC (NARAC Web site). Informed written consent was obtained from every subject, including all participating family members, and the local institutional review board’s approval was secured at each recruitment site. The enrollment criteria for family participation are described in detail elsewhere (Jawaheer et al. 2001). In brief, at least two siblings must satisfy the 1987 ACR criteria for RA, at least one sibling must have documented erosions on hand radiographs, and at least one sibling must have disease onset between the ages of 18 and 60 years. The presence of psoriasis, inflammatory bowel disease, or SLE was an exclusionary criterion for the sib pair. Controls were selected from 20,000 individuals who are part of the New York Cancer Project (NYCP), a population-based prospective study of the genetic and environmental factors that cause disease (New York Cancer Project Web site). Two control individuals were matched to a single randomly chosen affected sib on the basis of sex, age (birth decade), and ethnicity (grandparental country/region of origin). These 463 independent cases (referred to as “single sibs”) and their 926 matched controls were used for all analyses reported in this study, except where noted.
The association for single sibs was as follows: allele frequency 15.8% in cases and 8.7% in controls (P=5.6×10-8, allelic OR 1.97, 95% CI 1.55–2.50). An increase of the risk allele frequency was apparent when all affected siblings (n=840) were analyzed (allele frequency 16.8%). Genotypic analyses produced similar results, showing increased frequencies of both TT and TC genotypes in the cases, compared with the controls (table 1).
Table 1.
Frequency of PTPN22 Genotypes in Individuals with RA and Matched Controls in Two Independent Sample Sets[Note]
|
Frequency of (No. of Individuals with) PTPN22 Genotype in |
|||||
| Discovery Study |
Replication Study |
||||
| Genotype | Controls(n=475) | Cases(n=475) | Controls(n=926) | Single Sibsa(n=463) | All Sibsb(n=840) |
| TT | .006 (3) | .013 (6) | .010 (9) | .021 (10) | .025 (21) |
| TC | .164 (78) | .251 (119) | .154 (143) | .273 (126) | .287 (241) |
| CC | .830 (394) | .737 (350) | .836 (774) | .706 (327) | .688 (578) |
Note.— Genotypes were generated by use of kinetic, allele-specific PCR (Germer et al. 2000): 0.3 ng of DNA in a 15-μl reaction containing the allele-specific primers 5′-CCTCCACTTCCTGTAT/C and a common primer, 5′-CCCATCCCACACTTTATTTTATAC. Genotyping accuracy was 100%, as determined by internal comparisons by use of a different assay (Iannone et al. 2000) for the same marker on ∼6% of the samples. All case and control populations were in Hardy-Weinberg equilibrium.
Independent cases comprising one randomly selected sib per NARAC family.
All affected individuals in NARAC.
We used contingency tables and conditional-logistic regression (CLR) (Breslow and Day 1980) to assess the association of the PTPN22 R620W genotype with RA (table 2). In the discovery study, both the TC (ORCLR 1.69, P=.0012) and TT (ORCLR 2.26, P value not significant) genotypes conferred an increased risk for disease compared with the CC genotype. Further adjustment for the HLA-DRB1 genotype, the strongest known genetic risk factor (Seldin et al. 1999), had little impact on risk estimates. Similar results were observed in the replication study. The susceptible TT and TC genotypes were strongly associated with rheumatoid factor–positive (RF+) disease, even after adjustment for the HLA genotype (P=.0197 in discovery study, P=.0005 in replication study) (table 3); however, there was no evidence for association with rheumatoid factor–negative (RF−) disease. This interesting observation has been replicated in an additional cohort of patients with recent-onset RA (A. T. Lee and P. K. Gregersen, unpublished data). There was no consistent sex difference in association of the risk allele with RA (data not shown).
Table 2.
Association of PTPN22 with RA
|
Results for |
|||||||||||
| Crude Analysisa |
CLRb |
CLR, Adjusted for HLAc |
|||||||||
| Data SetandGenotype | No. ofCases | No. ofControls | ORd | 95% CI | P | ORd | 95% CI | P | ORd | 95% CI | P |
| Discovery: | |||||||||||
| TT | 6 | 3 | 2.25 | .56–9.07 | .32e | 2.26 | .56–9.14 | .25 | 2.39 | .35–16.43 | .38 |
| TC | 119 | 78 | 1.72 | 1.25–2.36 | .001 | 1.69 | 1.23–2.32 | .001 | 1.53 | 1.05–2.23 | .03 |
| TT+TCf | 125 | 81 | 1.74 | 1.27–2.38 | .0005 | 1.71 | 1.25–2.34 | .0008 | 1.55 | 1.07–2.25 | .02 |
| CC | 350 | 394 | … | … | … | … | … | … | … | … | … |
| Replication: | |||||||||||
| TT | 10 | 9 | 2.63 | 1.06–6.53 | .03 | 2.55 | 1.03–6.31 | .04 | 3.26 | 1.03–10.28 | .04 |
| TC | 126 | 143 | 2.09 | 1.59–2.74 | <.0001 | 2.08 | 1.58–2.74 | <.0001 | 1.7 | 1.19–2.42 | .004 |
| TT+TCf | 136 | 152 | 2.12 | 1.62–2.76 | <.0001 | 2.11 | 1.62–2.76 | <.0001 | 1.78 | 1.26–2.51 | .001 |
| CC | 327 | 774 | … | … | … | … | … | … | … | … | … |
Values were calculated by use of standard contingency tables (or two-tailed Fisher’s Exact Test).
CLR was performed as described elsewhere (Breslow and Day 1980) to account for individual matching of controls to cases.
To assess whether the observed associations were independent of the HLA-DRB1 genotype, we further adjusted for this known risk factor. HLA-DRB1 genotyping was performed as described elsewhere (Jawaheer et al. 2002), and, for this analysis, samples were binned according to their HLA-DRB1 genotype: high risk, two shared epitopes (HR-2SE); high risk, one shared epitope (HR-1SE; 4K,X; or 4R,X); low risk, one shared epitope (LR-1SE; 1R,X; or 10,X); and low risk, zero shared epitopes (LR-0SE; X,X) (Fries et al. 2002).
ORs were calculated relative to the major allele homozygote (CC).
P value calculated by use of Fisher’s Exact Test.
Because of the infrequency of the TT genotype, we combined it with the TC genotype and repeated the analysis.
Table 3.
Association of PTPN22 with RF+ and RF− Disease
|
Results for |
||||||||||||
| Crude Analysisa |
CLRb |
CLR, Adjusted for HLAc |
||||||||||
| Data SetandGenotype | RF Status | No. ofCases | No. ofControls | ORd | 95% CI | P | ORd | 95% CI | P | ORd | 95% CI | P |
| Discovery:e | ||||||||||||
| TT+TCf | RF+ | 125 | 81 | 1.74 | 1.27–2.38 | .0005 | 1.71 | 1.25–2.34 | .0008 | 1.55 | 1.07–2.25 | .02 |
| CC | RF+ | 350 | 394 | … | … | … | … | … | … | … | … | … |
| Replication: | ||||||||||||
| TT+TCf | RF+ | 119 | 122 | 2.38 | 1.78–3.18 | <.0001 | 2.36 | 1.76–3.16 | <.0001 | 2 | 1.35–2.97 | .0005 |
| CC | RF+ | 266 | 648 | … | … | … | … | … | … | … | … | … |
| TT+TCf | RF− | 17 | 30 | 1.17 | .60–2.29 | .64 | 1.17 | .60–2.31 | .64 | 1.09 | .51–2.36 | .82 |
| CC | RF− | 61 | 126 | … | … | … | … | … | … | … | … | … |
Values were calculated by use of standard contingency tables.
CLR was performed as described elsewhere (Breslow and Day 1980) to account for individual matching of controls to cases.
To assess whether the observed associations were independent of the HLA-DRB1 genotype, we further adjusted for this known risk factor. For this analysis, samples were binned according to their HLA-DRB1 genotype: high risk, two shared epitopes (HR-2SE); high risk, one shared epitope (HR-1SE; 4K,X; or 4R,X); low risk, one shared epitope (LR-1SE; 1R,X; or 10,X); and low risk, zero shared epitopes (LR-0SE; X,X) (Fries et al. 2002).
ORs were calculated relative to the major allele homozygote (CC).
All patients in the discovery cohort are RF+.
Because of the infrequency of the TT genotype, we combined it with the TC genotype for these analyses.
To evaluate putative modes of inheritance, we combined the two data sets and used the likelihood-ratio test (Breslow and Day 1980) on the basis of CLR models. Although we could exclude a recessive mode of inheritance (P<.0001), the data are consistent with either additive or dominant modes of inheritance. We also generated estimates of the population-attributable fraction (0.11 for discovery study [95% CI 0.05–0.17]; 0.16 for replication study [95% CI 0.10–0.21]) (Schlesselman 1982; Yang et al. 2003). However, these estimates should be interpreted with caution, since our cases were selected to have relatively severe disease and thus are not representative of the full clinical spectrum of RA.
PTPN22 is located on chromosome 1p13, ∼9 Mb centromeric to a microsatellite marker, D1S1631, that shows linkage to RA in the NARAC sib pairs (SIBPAL P=.0011) (Jawaheer et al. 2003) from which our replication study was drawn. Linkage analysis using Allegro (Gudbjartsson et al. 2000) yielded an insignificant LOD score of 0.15 for PTPN22 R620W in these sib pairs, indicating that R620W is not solely responsible for the linkage seen at D1S1631. We then conducted stratified analyses based on PTPN22 R620W genotypes of the probands. Among 7, 76, and 193 affected sib pairs for which the proband had the TT, TC, or CC genotype, respectively, the respective mean identity-by-descent–sharing values were 0.598, 0.505, and 0.486, and the respective nonparametric linkage (NPL) scores were 0.94, 0.35, and −0.46 (Allegro) (Gudbjartsson et al. 2000). These results suggested that R620W has a weak effect on the 1p13 linkage signal. Li et al. (2004) have recently shown that there is large variability in NPL scores when families are stratified by the genotype of a single, randomly selected sib, and they developed a program (Genotype-IBD Sharing Test [GIST]) in which families are weighted on the basis of the genotypes of all family members. GIST analysis that was conditional on the T allele indicated significant evidence for linkage (P<.0001). The apparent difference between the significance of the stratified analysis and the GIST results may reflect the increased power of the GIST algorithm, or, alternatively, the rarity of the TT genotype may lead to poor convergence of the GIST test statistic to the asymptotic distribution. Taken together, all these data suggest that, whereas the SNP may account for a small part of the linkage signal seen on 1p, it is clearly not responsible for the entire signal.
We also genotyped this SNP in several additional control populations (table 4). The observed frequency of the risk allele in 560 additional white subjects (8.4%) was consistent with results in our two RA control populations. When all three control data sets of whites (n=1,961) were combined, the T allele was present in 16.7% of individuals at an allele frequency of 8.7%. The T allele frequency was lower in 99 Mexican Americans (3.5%) and 409 African Americans (2.4%). This allele was not detected in 100 Han Chinese or 21 Africans. A Fisher's Exact Test showed that these differences in allele frequencies were highly significant (P<1×10-10). It will be important to expand these studies to determine whether presence of the T allele in the African American and Mexican American populations is due to admixture with whites.
Table 4.
Frequency of the PTPN22 Risk Allele in Discovery and Replication Control Samples and Other Populations
| Population | No. ofIndividuals | Risk AlleleFrequency |
| Discovery controls (white) | 475 | .088 |
| Replication controls (white) | 926 | .087 |
| Additional whitesa | 560 | .084 |
| African Americansb | 409 | .024 |
| Africansc | 21 | 0 |
| Mexican Americansd | 99 | .035 |
| Han Chinesee | 100 | 0 |
Including samples from 360 whites from throughout the United States (obtained by GCI), 147 North American whites (obtained from the National Institute of General Medical Sciences [NIGMS] Human Variation Panels [HVP]), 10 northern Europeans (NIGMS HVP), 9 Italians (NIGMS HVP), 8 Greeks (NIGMS HVP), 20 independent CEPH individuals from Utah (NIGMS), 1 CEPH individual from France (NIGMS), and 5 Adygei (an indigenous Circassian people; NIGMS). GCI obtained informed written consent from all participants and local institutional review board approval at all recruitment sites.
Including samples from 369 African Americans from throughout the United States obtained by GCI (see footnote a for comments on consent/IRB approval) and 40 African Americans (NIGMS HVP).
Including samples from 9 sub-Saharan Africans (NIGMS HVP), 7 northern Africans (north of the Sahara) (NIGMS HVP), and 5 Pygmies (NIGMS).
Including samples from 99 Mexican Americans from Los Angeles (NIGMS HVP).
Including samples from 100 Han Chinese from Los Angeles (NIGMS HVP).
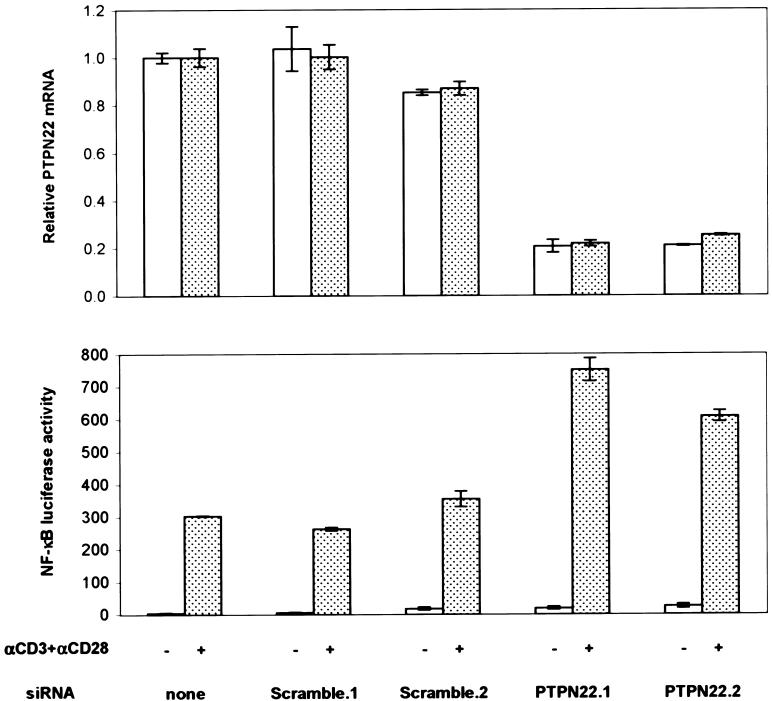
PTPN22 encodes a 110-kD cytoplasmic protein tyrosine phosphatase that consists of an N-terminal phosphatase domain and a long C-terminal region containing several proline-rich motifs (Cohen et al. 1999). The mouse ortholog, PEP (encoded by the murine gene Ptpn8), has been shown to be a potent down-regulator of T-cell receptor–dependent responses through its association with the SH3 domain of Csk (Cloutier and Veillette 1999). Although PEP and PTPN22 are only 70% identical at the amino acid level (Cohen et al. 1999), overexpression data suggest PTPN22 may play a role similar to that of PEP (Hill et al. 2002). To confirm that PTPN22 functions as a negative regulator of T-cell activation, we used RNA interference (RNAi) to decrease expression in Jurkat cells and then measured the response to antigen-receptor stimulation. mRNA knockdown by two independent siRNAs increased T-cell receptor–dependent activation by approximately two-fold, as measured by an NF-κB-reporter response (fig. 1). This is consistent with recent results in PEP-deficient mice (Hasegawa et al. 2004). These animals have rather subtle phenotypic alterations in T-cell function, with enhanced activation of Lck and expansion of memory T cells, but apparently normal naive T-cell function.
Figure 1.
PTPN22 knockdown by RNAi increases antigen-receptor signaling in Jurkat T-cell line. Knockdown and NF-κB transcriptional response after T-cell receptor stimulation for cells transfected with two control siRNAs (Scramble) and two PTPN22 siRNAs. First, 4.5 μg siRNA (Dharmacon) and 1 μg pNF-κB-luc plasmid (Stratagene) were electroporated into 2×106 Jurkat cells (ATCC, clone E6-1, homozygous for 620R) in Cell Line Nucleofector Solution V, according to manufacturer’s protocol #S18 (amaxa). After 24 h, 5.5×104 cells were stimulated with anti-CD3 (2 μg/ml [BD Pharmingen, clone UCHT1]), anti-CD28 (2 μg/ml [BD Pharmingen, clone CD28.2]), and anti-mouse IgG1 (2 μg/ml [BD Pharmingen, clone A85-1]). Cells were lysed and assayed for luminescence 6 h after stimulation by use of Bright-Glo (Promega) and a 96-well MicroBeta plate reader (Wallac). PTPN22 siRNAs that were effective for knockdown were found by screening seven candidate siRNA sequences that were designed by use of Web-based tools from Ambion and Dharmacon. siRNA sequences used in this study were PTPN22.1 (5′-AAGGCAGACAAAACCTATCCT), PTPN22.2 (5′-GAGGATTCCAGCTACATCAAT), Scramble.1 (5′-AAGAACGGCATCAAGGTGAAC), and Scramble.2 (5′-AATTCTCCGAACGTGTCACGT). RNA was harvested according to the manufacturer’s protocol (RNeasy 96 [Qiagen]), and TaqMan real-time quantitative RT-PCR (SDS-7900 [Applied Biosystems]) was used to measure PTPN22 expression levels. PTPN22 mRNA levels in each sample were normalized to total RNA amounts (RiboGreen [Molecular Probes]) and then expressed relative to PTPN22 levels in cells that were untransfected with siRNA. The PTPN22 TaqMan sequences were 5′-GGCCCAAAGCAAGAAAATTACTAAA (forward), 5′-TGTCTGCCTTGTACTTGGTAGATTG (reverse), and 5′-TTCAGCTTCAGAAATT (MGB probe).
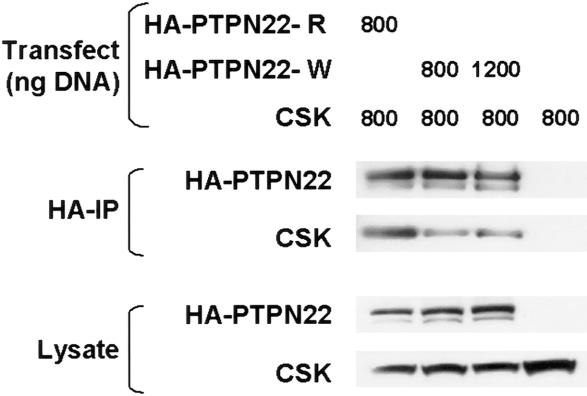
The first proline-rich domain (P1) of PEP binds the SH3 domain of the negative regulatory kinase Csk (Cloutier and Veillette 1996; Gregorieff et al. 1998). Bottini and colleagues (2004) have recently shown that the R620W SNP (which is located in P1) affects the binding of PTPN22 to Csk. We confirmed this finding by cotransfection of full-length cDNA clones for PTPN22 (R620 or W620) and Csk into 293T cells, followed by immunoprecipitation of PTPN22. Although both PTPN22 constructs were expressed at similar levels, 2.5–3-fold less Csk was coimmunoprecipitated by the W620 protein, relative to the R620 protein (fig. 2). These data suggest that the association of this missense SNP with RA may be due to the inability of the variant phosphatase to bind Csk and down-regulate T-cell activation.
Figure 2.
Western blot showing Csk coimmunoprecipitation by the two HA-tagged PTPN22 proteins (R620 and W620) expressed in 293T cells. W620 decreases the affinity of PTPN22 for Csk. Increasing the amount of PTPN22 W620 expressed does not increase the amount of Csk that coimmunoprecipitates. Quantitation of Csk band intensity indicates a 2.9-fold difference between R620 and W620. Similar results (2.6-fold) were seen in a second experiment. PTPN22 sequences were cloned by PCR and verified to encode a PTPN22 protein sequence corresponding to Swiss-Prot Q9Y2R2. The R620W variant was introduced (QuikChange [Stratagene]), and both PTPN22 variants with an N-terminal HA tag were cloned into the pCMV5 expression vector (Qbiogene). Csk was cloned by PCR, the sequence was verified, and it was introduced into the pcDNA-DEST40 expression vector (Invitrogen). The 293T (GenHunter) cells (3×105 cells/well) were seeded in 12-well plates and were transfected 24 h later by use of 3.5 μl/well of Lipofectamine 2000 (Invitrogen). Cells were washed and resuspended in lysis buffer (50 mM Tris [pH 8.0], 2 mM EDTA, 1% NP-40, 50 mM NaF, 1 mM sodium orthovanadate, and 1× protease inhibitor cocktail [Sigma]) 48 h after transfection. HA-PTPN22 immunoprecipitations were performed as described elsewhere (Cloutier and Veillette 1999), with the following modifications. Before immunoprecipatation, detergent-insoluble material was removed by centrifugation (100,000×g) and lysates were pre-cleared by use of mouse IgG agarose beads (Sigma). The lysate (100 μg) was incubated with 15 μl of anti-HA conjugated beads (Sigma) for 2 h at 4°C. Beads were washed three times in wash buffer (50 mM Tris [pH 8.0], 100 mM NaCl, 2 mM EDTA, 1% NP-40, 50 mM NaF, and 1 mM sodium orthovanadate). Precipitated proteins and lysates were analyzed by western blot, by use of anti-HA (Covance) and anti-Csk (Upstate 06-566) antibodies, and were detected by use of HRP-conjugated secondary antibodies (Pierce 31430 and Biosource ALI3404) and chemiluminescence (Pierce).
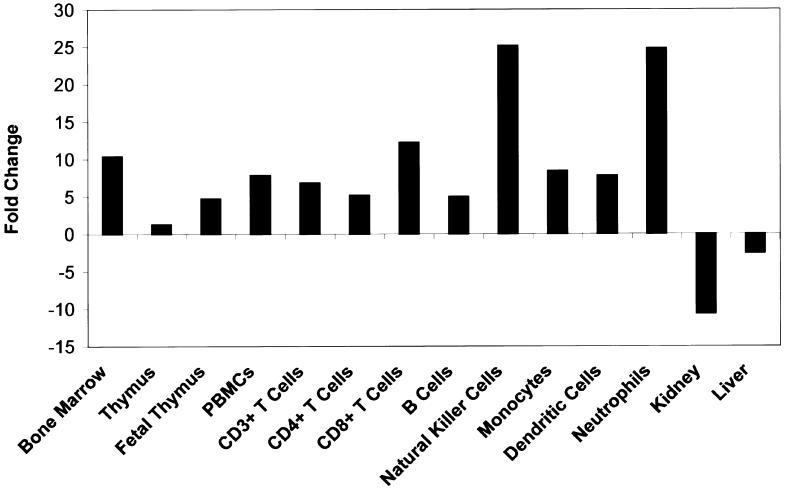
Prior studies have suggested that PTPN22 is expressed primarily in hematopoietic tissues (Cohen et al. 1999: Hill et al. 2002; Chien et al. 2003), such as thymus, spleen, bone marrow, and peripheral blood mononuclear cells (PBMCs). We confirmed and extended these findings by examining numerous tissues and specific PBMC subsets by use of semiquantitative kinetic RT-PCR (fig. 3 and table A1 [online only]). Expression appeared to be largely confined to hematopoietic tissues and was present in all subtypes of normal human PBMCs tested, including resting CD3+ T cells, CD4+ T cells, CD8+ T cells, B cells, monocytes, neutrophils, dendritic cells, and natural killer (NK) cells (fig. 3). However, there appears to be a hierarchy, with NK cells and neutrophils expressing the most PTPN22 message and B cells and CD4+ T cells expressing the least. These data raise the possibility that the association of PTPN22 with RA could also be a result of functional changes in these other cell types. In particular, monocytosis and monocyte activation are characteristic features of RA, and NK and NK-T cells may also be involved in the pathogenesis of RA (Kojo et al. 2001; Dalbeth and Callan 2002). As a consequence, a complete analysis of PTPN22 function in these cell populations will be required to fully elucidate the role of this protein in autoimmunity.
Figure 3.
RNA expression profile of PTPN22. Expression of the PTPN22 major splice variant (GenBank accession number NM_015967) was determined by kinetic RT-PCR analysis by use of total RNA from sorted hematopoeitic cells (AllCells) and selected tissues (Clontech), as described elsewhere (Rogge et al. 2000). Data for additional tissues are provided online (table A1 [online only]). To eliminate genomic DNA amplification, amplification primers (5′-GGTTGAGGAAGCTGGAGAAT and 5′-GGGAGAAGAACGATCTTGATGTA) were selected from different exons. Each RNA sample was also amplified with seven housekeeping genes, as described elsewhere (Rogge et al. 2000). The level of expression of these housekeeping genes was used to normalize the amount of message in all samples. The normalized expression levels of PTPN22 in all normal tissues (excluding tumor cell lines and purified hematopoietic cells) were averaged, and the results from each individual sample were expressed as a fold change relative to this average. A positive number indicates more PTPN22 message in the indicated sample relative to the average of all samples, whereas a negative number means there is less PTPN22 message relative to the overall average.
Familial clustering of multiple autoimmune disorders in the same family is well documented, and first-degree relatives of RA probands have a significantly higher prevalence of type 1 diabetes (T1D [MIM 222100]) than the general population (Lin et al. 1998). In addition, genomewide scans for autoimmune diseases, in both mouse and human, show colocalization of susceptibility loci (Becker et al. 1998; Griffiths et al. 1999), suggesting a possible shared genetic basis for autoimmunity. The finding that the minor allele of the PTPN22 SNP reported here is also associated with T1D (Bottini et al. 2004) supports the hypothesis that there are common genetic variants that contribute to general immune dysregulation and susceptibility to autoimmunity (Marrack et al. 2001; Wandstrat and Wakeland 2001). It will be important to examine PTPN22 associations in a wide variety of autoimmune diseases and to determine its functional role in each disease. Although a common underlying mechanism for autoimmunity is tempting to postulate, given the expression of this molecule in many immunologically relevant cell types, the possibility remains that PTPN22 may act in different ways in different autoimmune diseases.
Acknowledgments
We are grateful to the patients with RA, the controls, and the collaborating clinicians, for participation in this study; members of the Celera Diagnostics High Throughput, Biomarker, and Computational Biology groups, for invaluable help; T. L. Bugawan and E. Trachtenberg, for HLA typing; R. Lundsten, M. Kern, H. Khalili, and A. Rodrigues-Brown, for database and sample management of the NARAC collection; J. Lemaire and S. Mahan, for database and sample management at GCI; and T. White, S. Broder, R. Zamoyska, X. Hu, S. Dalrymple, J. Buggy, K. Van Orden, M. Kale, and P. Young, for discussions and insightful comments on this manuscript. Collection of the NARAC cohort has been funded by a National Arthritis Foundation grant and the National Institutes of Health, acting through the National Institute of Arthritis and Musculoskeletal and Skin Diseases and the National Institute of Allergy and Infectious Diseases (contracts N01-AR-7-2232 and R01-AR44222). These studies were performed in part in the General Clinical Research Center, Moffitt Hospital, University of California, San Francisco, with funds provided by the National Center for Research Resources (5 M01 RR-00079, U.S. Public Health Service). Support was also provided by the National Institutes of Health through grant HG02275.
Appendix A
Table A1.
RNA Expression Levels of PTPN22 in Purified Normal Cell Populations, Tissues, and Transformed Cell Lines, Expressed as a Fold Change Relative to the Average of All Normal Tissues (Excluding Tumor Cell Lines and Purified Hematopoietic Cells)[Note]
| Cell/Tissue Type | Source | Fold Changea |
| Normal cells: | ||
| NK cells | AllCells | 25.14 |
| Neutrophils | AllCells | 24.82 |
| CD8+ T cells | AllCells | 12.26 |
| Bone marrow | Clontech | 10.41 |
| Monocytes | AllCells | 8.42 |
| PBMCs | AllCells | 7.86 |
| Dendritic cells | AllCells | 7.81 |
| Pan T cells | AllCells | 6.85 |
| CD4+ T cells | AllCells | 5.21 |
| B cells | AllCells | 5.01 |
| Tissues: | ||
| Adipose | BioChain | −2.21 |
| Adrenal gland | Clontech | −4.98 |
| Bladder | Clontech | −2.64 |
| Whole brain | Clontech | −14.27 |
| Brain | Clontech | −37.66 |
| Brain (fetal) | Clontech | −12.84 |
| Breast | Stratagene | −1.68 |
| Breast | BioChain | −1.98 |
| Breast | Clontech | −5.35 |
| Breast (mammary gland) | Clontech | −1.82 |
| Cervix | Ambion | ND |
| Esophagus | BioChain | −3.61 |
| Heart | Clontech | −6.73 |
| Heart (aorta) | Clontech | −1.63 |
| Heart (diseased) | Clontech | −17.65 |
| Heart (diseased, post-infarction) | Clontech | −8.67 |
| Heart (pericardium) | BioChain | −1.56 |
| Heart (fetal) | Clontech | −22.25 |
| Intestine (large) | Stratagene | 2.99 |
| Intestine (large, tumor) | Stratagene | −1.89 |
| Intestine (small) | Clontech | −1.62 |
| Intestine (small, ileum diseased) | Stratagene | 2.54 |
| Kidney | Clontech | −10.69 |
| Kidney (fetal) | Clontech | −15.79 |
| Liver | Clontech | −2.59 |
| Liver (fetal) | Clontech | −4.96 |
| Lung | Stratagene | 4.18 |
| Lung (tumor) | Clontech | −1.07 |
| Muscle (skeletal) | Clontech | −17.45 |
| Muscle (skeletal) | Ambion | −23.81 |
| Ovary | Stratagene | −1.15 |
| Ovary (tumor) | Clontech | −8.28 |
| Pancreas | BioChain | −3.34 |
| Placenta | Clontech | −5.43 |
| Prostate | Stratagene | −1.61 |
| Prostate | Clontech | −132.45 |
| Salivary gland | Clontech | −17.23 |
| Spinal cord | Clontech | −4.75 |
| Spleen (fetal) | Clontech | −1.13 |
| Stomach | BioChain | −2.16 |
| Stomach (tumor) | Clontech | −1.87 |
| Testis | Clontech | −3.08 |
| Thymus | Clontech | 1.29 |
| Thymus (fetal) | Clontech | 4.75 |
| Thyroid (female) | Stratagene | 3.61 |
| Thyroid | Stratagene | 3.41 |
| Thyroid | Clontech | −4.13 |
| Tonsil | Clontech | −1.17 |
| Trachae | Clontech | −2.19 |
| Umbilical cord (fetal) | BioChain | −1.37 |
| Uterus | Clontech | −8.95 |
| Transformed cell lines: | ||
| Jurkat | Ambion | 9.45 |
| HL60 | Ambion | 8.66 |
| K562 | Ambion | 1.28 |
| K562 (PMA treated) | Stratagene | −1.28 |
| K562 | Stratagene | −4.33 |
| PC3 (prostate carcinoma) | Ambion | −15.1 |
| HeLa S3 | Clontech | −24.81 |
| A431 (epidermal carcinoma) | Ambion | ND |
| HeLa S3 | Ambion | ND |
Note.— ND = not detected.
Expression of the PTPN22 major splice variant was determined by kinetic RT-PCR analysis. Each RNA sample was also amplified with seven housekeeping genes, as described elsewhere (Rogge et al. 2000). The level of expression of these housekeeping genes was used to normalize the amount of message in all samples. The normalized expression levels of PTPN22 in all normal tissues (excluding tumor cell lines and purified hematopoietic cells) were averaged, and the results from each individual sample were expressed as a fold change relative to this average. A positive number indicates more PTPN22 message in the indicated sample relative to the average of all samples, whereas a negative number means there is less PTPN22 message relative to the overall average.
Electronic-Database Information
The accession number and URLs for data presented herein are as follows:
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for PTPN22 major splice variant [accession number NM_015967])
- Genotype-IBD Sharing Test (GIST), http://phg.mc.vanderbilt.edu/GIST.shtml [DOI] [PMC free article] [PubMed]
- New York Cancer Project, http://www.amdec.org
- NARAC, http://www.naracdata.org/index.asp
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for RA and T1D) [PubMed]
References
- Becker KG, Simon RM, Bailey-Wilson JE, Freidlin B, Biddison WE, McFarland HF, Trent JM (1998) Clustering of non-major histocompatibility complex susceptibility candidate loci in human autoimmune diseases. Proc Natl Acad Sci USA 95:9979–9984 10.1073/pnas.95.17.9979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, Risch N (2003) Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet Suppl 33:228–237 10.1038/ng1090 [DOI] [PubMed] [Google Scholar]
- Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, MacMurray J, Meloni GF, Lucarelli P, Pellecchia M, Eisenbarth GS, Comings D, Mustelin T (2004) A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet 36:337–338 10.1038/ng1323 [DOI] [PubMed] [Google Scholar]
- Breslow NE, Day NE (1980) Statistical methods in cancer research, volume I: the analysis of case-control studies. IARC Sci Publ 32:5–338 [PubMed] [Google Scholar]
- Chien W, Tidow N, Williamson EA, Shih LY, Krug U, Kettenbach A, Fermin AC, Roifman CM, Koeffler HP (2003) Characterization of a myeloid tyrosine phosphatase, Lyp, and its role in the Bcr-Abl signal transduction pathway. J Biol Chem 278:27413–27420 10.1074/jbc.M304575200 [DOI] [PubMed] [Google Scholar]
- Cloutier JF, Veillette A (1996) Association of inhibitory tyrosine protein kinase p50csk with protein tyrosine phosphatase PEP in T cells and other hemopoietic cells. EMBO J 15:4909–4918 [PMC free article] [PubMed] [Google Scholar]
- Cloutier JF, Veillette A (1999) Cooperative inhibition of T-cell antigen receptor signaling by a complex between a kinase and a phosphatase. J Exp Med 189:111–121 10.1084/jem.189.1.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Dadi H, Shaoul E, Sharfe N, Roifman CM (1999) Cloning and characterization of a lymphoid-specific, inducible human protein tyrosine phosphatase, Lyp. Blood 93:2013–2024 [PubMed] [Google Scholar]
- Cornelis F, Faure S, Martinez M, Prud’homme JF, Fritz P, Dib C, Alves H, et al (1998) New susceptibility locus for rheumatoid arthritis suggested by a genome-wide linkage study. Proc Natl Acad Sci USA 95:10746–10750 10.1073/pnas.95.18.10746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalbeth N, Callan MF (2002) A subset of natural killer cells is greatly expanded within inflamed joints. Arthritis Rheum 46:1763–1772 10.1002/art.10410 [DOI] [PubMed] [Google Scholar]
- Firestein GS (2003) Evolving concepts of rheumatoid arthritis. Nature 423:356–361 10.1038/nature01661 [DOI] [PubMed] [Google Scholar]
- Fisher SA, Lanchbury JS, Lewis CM (2003) Meta-analysis of four rheumatoid arthritis genome-wide linkage studies: confirmation of a susceptibility locus on chromosome 16. Arthritis Rheum 48:1200–1206 10.1002/art.10945 [DOI] [PubMed] [Google Scholar]
- Fries JF, Wolfe F, Apple R, Erlich H, Bugawan T, Holmes T, Bruce B (2002) HLA-DRB1 genotype associations in 793 white patients from a rheumatoid arthritis inception cohort: frequency, severity, and treatment bias. Arthritis Rheum 46:2320–2329 10.1002/art.10485 [DOI] [PubMed] [Google Scholar]
- Germer S, Holland MJ, Higuchi R (2000) High-throughput SNP allele-frequency determination in pooled DNA samples by kinetic PCR. Genome Res 10:258–266 10.1101/gr.10.2.258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorieff A, Cloutier JF, Veillette A (1998) Sequence requirements for association of protein-tyrosine phosphatase PEP with the Src homology 3 domain of inhibitory tyrosine protein kinase p50(csk). J Biol Chem 273:13217–13222 10.1074/jbc.273.21.13217 [DOI] [PubMed] [Google Scholar]
- Griffiths MM, Encinas JA, Remmers EF, Kuchroo VK, Wilder RL (1999) Mapping autoimmunity genes. Curr Opin Immunol 11:689–700 10.1016/S0952-7915(99)00038-2 [DOI] [PubMed] [Google Scholar]
- Gudbjartsson DF, Jonasson K, Frigge ML, Kong A (2000) Allegro, a new computer program for multipoint linkage analysis. Nat Genet 25:12–13 10.1038/75514 [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Martin F, Huang G, Tumas D, Diehl L, Chan AC (2004) PEST domain-enriched tyrosine phosphatase (PEP) regulation of effector/memory T cells. Science 303:685–689 10.1126/science.1092138 [DOI] [PubMed] [Google Scholar]
- Hill RJ, Zozulya S, Lu YL, Ward K, Gishizky M, Jallal B (2002) The lymphoid protein tyrosine phosphatase Lyp interacts with the adaptor molecule Grb2 and functions as a negative regulator of T-cell activation. Exp Hematol 30:237–244 10.1016/S0301-472X(01)00794-9 [DOI] [PubMed] [Google Scholar]
- Iannone MA, Taylor JD, Chen J, Li MS, Rivers P, Slentz-Kesler KA, Weiner MP (2000) Multiplexed single nucleotide polymorphism genotyping by oligonucleotide ligation and flow cytometry. Cytometry 39:131–140 [DOI] [PubMed] [Google Scholar]
- Jawaheer D, Li W, Graham RR, Chen W, Damle A, Xiao X, Monteiro J, Khalili H, Lee A, Lundsten R, Begovich A, Bugawan T, Erlich H, Elder JT, Criswell LA, Seldin MF, Amos CI, Behrens TW, Gregersen PK (2002) Dissecting the genetic complexity of the association between human leukocyte antigens and rheumatoid arthritis. Am J Hum Genet 71:585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawaheer D, Seldin MF, Amos CI, Chen WV, Shigeta R, Etzel C, Damle A, et al (2003) Screening the genome for rheumatoid arthritis susceptibility genes: a replication study and combined analysis of 512 multicase families. Arthritis Rheum 48:906–916 10.1002/art.10989 [DOI] [PubMed] [Google Scholar]
- Jawaheer D, Seldin MF, Amos CI, Chen WV, Shigeta R, Monteiro J, Kern M, Criswell LA, Albani S, Nelson JL, Clegg DO, Pope R, Schroeder HW Jr, Bridges SL Jr, Pisetsky DS, Ward R, Kastner DL, Wilder RL, Pincus T, Callahan LF, Flemming D, Wener MH, Gregersen PK (2001) A genomewide screen in multiplex rheumatoid arthritis families suggests genetic overlap with other autoimmune diseases. Am J Hum Genet 68:927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojo S, Adachi Y, Keino H, Taniguchi M, Sumida T (2001) Dysfunction of T cell receptor AV24AJ18+, BV11+ double-negative regulatory natural killer T cells in autoimmune diseases. Arthritis Rheum 44:1127–1138 [DOI] [PubMed] [Google Scholar]
- Li C, Scott LJ, Boehnke M (2004) Assessing whether an allele can account in part for a linkage signal: the Genotype-IBD Sharing Test (GIST). Am J Hum Genet 74:418–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JP, Cash JM, Doyle SZ, Peden S, Kanik K, Amos CI, Bale SJ, Wilder RL (1998) Familial clustering of rheumatoid arthritis with other autoimmune diseases. Hum Genet 103:475–482 10.1007/s004390050853 [DOI] [PubMed] [Google Scholar]
- MacGregor AJ, Snieder H, Rigby AS, Koskenvuo M, Kaprio J, Aho K, Silman AJ (2000) Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum 43:30–37 [DOI] [PubMed] [Google Scholar]
- MacKay K, Eyre S, Myerscough A, Milicic A, Barton A, Laval S, Barrett J, Lee D, White S, John S, Brown MA, Bell J, Silman A, Ollier W, Wordsworth P, Worthington J (2002) Whole-genome linkage analysis of rheumatoid arthritis susceptibility loci in 252 affected sibling pairs in the United Kingdom. Arthritis Rheum 46:632–639 10.1002/art.10147 [DOI] [PubMed] [Google Scholar]
- Marrack P, Kappler J, Kotzin BL (2001) Autoimmune disease: why and where it occurs. Nat Med 7:899–905 10.1038/90935 [DOI] [PubMed] [Google Scholar]
- Rogge L, Bianchi E, Biffi M, Bono E, Chang SY, Alexander H, Santini C, Ferrari G, Sinigaglia L, Seiler M, Neeb M, Mous J, Sinigaglia F, Certa U (2000) Transcript imaging of the development of human T helper cells using oligonucleotide arrays. Nat Genet 25:96–101 10.1038/75671 [DOI] [PubMed] [Google Scholar]
- Schlesselman JJ (1982) Case-control studies: design, conduct, analysis. Oxford University Press, New York [Google Scholar]
- Seldin MF, Amos CI, Ward R, Gregersen PK (1999) The genetics revolution and the assault on rheumatoid arthritis. Arthritis Rheum 42:1071–1079 [DOI] [PubMed] [Google Scholar]
- Shiozawa S, Hayashi S, Tsukamoto Y, Goko H, Kawasaki H, Wada T, Shimizu K, Yasuda N, Kamatani N, Takasugi K, Tanaka Y, Shiozawa K, Imura S (1998) Identification of the gene loci that predispose to rheumatoid arthritis. Int Immunol 10:1891–1895 10.1093/intimm/10.12.1891 [DOI] [PubMed] [Google Scholar]
- Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M, Nagasaki M, Nakayama-Hamada M, Kawaida R, Ono M, Ohtsuki M, Furukawa H, Yoshino S, Yukioka M, Tohma S, Matsubara T, Wakitani S, Teshima R, Nishioka Y, Sekine A, Iida A, Takahashi A, Tsunoda T, Nakamura Y, Yamamoto K (2003) Functional haplotypes of PADI4 encoding citrullinating enzyme peptidylarginine deaminase 4, are associated with rheumatoid arthritis. Nat Genet 34:395–402 10.1038/ng1206 [DOI] [PubMed] [Google Scholar]
- Tokuhiro S, Yamada R, Chang X, Suzuki A, Kochi Y, Sawada T, Suzuki M, Nagasaki M, Ohtsuki M, Ono M, Furukawa H, Nagashima M, Yoshino S, Mabuchi A, Sekine A, Saito S, Takahashi A, Tsunoda T, Nakamura Y, Yamamoto K (2003) An intronic SNP in a RUNX1 binding site of SLC22A4 encoding an organic cation transporter is associated with rheumatoid arthritis. Nat Genet 35:341–348 10.1038/ng1267 [DOI] [PubMed] [Google Scholar]
- Wandstrat A, Wakeland E (2001) The genetics of complex autoimmune diseases: non-MHC susceptibility genes. Nat Immunol 2:802–809 10.1038/ni0901-802 [DOI] [PubMed] [Google Scholar]
- Yang Q, Khoury MJ, Friedman JM, Flanders WD (2003) On the use of population attributable fraction to determine sample size for case-control studies of gene-environment interaction. Epidemiology 14:161–167 10.1097/00001648-200303000-00009 [DOI] [PubMed] [Google Scholar]