Abstract
Age-related maculopathy (ARM), or age-related macular degeneration, is one of the most common causes of visual impairment in the elderly population of developed nations. In a combined analysis of two previous genomewide scans that included 391 families, containing up to 452 affected sib pairs, we found linkage evidence in four regions: 1q31, 9p13, 10q26, and 17q25. We now have added a third set of families and have performed an integrated analysis incorporating 530 families and up to 736 affected sib pairs. Under three diagnostic models, we have conducted linkage analyses using parametric (heterogeneity LOD [HLOD] scores under an autosomal dominant model) and nonparametric (Sall statistic) methods. There is ongoing evidence of susceptibility loci within the 1q31, 10q26, and 17q25 regions. If we treat the third set of families as a replication set, then two regions (10q26 and 17q25) are replicated, with LOD scores >1.0. If we pool all our data together, then four regions (1q31, 2q14.3, 10q26, and 17q25) show HLOD or Sall scores ⩾2.0. Within the 1q31 region, we observed an HLOD of 2.72 (genomewide P=.061) under our least stringent diagnostic model, whereas the 17q25 region contained a maximal HLOD of 3.53 (genomewide P=.007) under our intermediate diagnostic model. We have evaluated our results with respect to the findings from several new independent genomewide linkage studies and also have completed ordered subset analyses (OSAs) with apolipoprotein E alleles, smoking history, and age at onset as stratifying covariates. The OSAs generate the interesting hypothesis that the effect of smoking on the risk of ARM is accentuated by a gene in the 10q26 region—a region implicated by four other studies.
Introduction
Age-related maculopathy (ARM), or age-related macular degeneration (ARMD1 [MIM 603075]), is one of the most common causes of visual impairment in the elderly within developed nations. The condition affects nearly 10% of those >65 years of age and affects >25% of those >75 years of age (for a review, see Gorin [1998]). The medical costs for the evaluation and treatment of ARM complications are considerable and increase with more severe disease (Bonastre et al. 2003). Treatment has focused primarily on the choroidal neovascularization that is a complication of the condition, though only a relatively small percentage of patients currently benefit from existing therapies (Mandal and Chisholm 2002; Hooper and Guymer 2003). Strategies to reduce the progression of the condition, such as the intake of vitamin, mineral, and antioxidant supplements, have been shown to be effective (Age-Related Eye Disease Study Research Group 2001) but still are limited predominantly to patients with clinical evidence of the disease. There is compelling evidence of a genetic risk of ARM from epidemiologic studies, as well as from twin studies and segregation analyses (for a review, see Gorin [1998]). There is a growing impetus for these genetic studies not only to identify the biological bases for this complex disorder but to improve preventive approaches by providing a means of identifying, prior to the onset of clinical findings, individuals at high risk of developing the condition.
Several genetic approaches have been undertaken to study ARM. One strategy has been to screen those genes which are known to be causative for the monogenic juvenile macular dystrophies, such as Best disease (VMD [MIM 153700], VMD2 [MIM 607854]); Stargardt disease (STGD1 [MIM 248200], ABCA4 [MIM 601691]); malattia leventinese (MLVT [MIM 126600], EFEMPI [MIM 601548]); autosomal dominant Stargardt-like dystrophy (STGD3 [MIM 600110], ELOVL4 [MIM 605512]); cone dystrophy (CORDX1 [MIM 304020], RPGR [MIM 312610]); adult vitelliform dystrophy (AVMD [MIM 608161], RDS [MIM 179605]); and Sorsby fundus dystrophy (SFD [MIM 136900], TIMP3 [MIM 188826]). At this time, none of these genes appear to be responsible for a significant percentage of ARM cases (Weeks et al. 2000, 2001; Stone et al. 2001), though certainly they may contribute to a fraction of the ARM population. A number of candidate genes also have been investigated on the basis of their role in retinal physiology and/or retinal pigment epithelium physiology, and, yet again, there has been little evidence of a major role for any of these genes in the etiology of ARM (Stone et al. 1998, 2001; Weeks et al. 2000, 2001). Apolipoprotein E (APOE [MIM 107741]) alleles have been shown to contribute significantly to the risk of several medical conditions, including Alzheimer disease (AD2 [MIM 104310]) (Corder et al. 1994; Breitner et al. 1998; Meyer et al. 1998). A number of studies have provided fairly consistent evidence that the APOE-4 allele appears to be partially protective against ARM, whereas the APOE-2 allele may increase risk (but this has been shown less consistently) (Klaver et al. 1998; Souied et al. 1998; Pang et al. 2000; Schmidt et al. 2002; Schultz et al. 2003b).
Another genetic approach to investigating a complex genetic disorder, such as ARM, has been to use small families with the disorder to look for genomic regions that are shared in common, among two or more affected relatives, more frequently than is predicted by chance. Such an approach is relatively robust in dealing with the genetic heterogeneity of ARM and is independent from the biological hypotheses that have guided pure candidate gene studies. We have performed two genomewide scans the results of which are published elsewhere (Weeks et al. 2000, 2001). When we analyzed these together, in a combined analysis of 391 families containing up to 452 affected sib pairs, we found evidence of linkage in four regions: 1q31, 9p13, 10q26, and 17q25 (table 1). The locus on chromosome 1q31 independently confirmed a report by Klein et al. (1998), which mapped an ARM susceptibility gene to this region.
Table 1.
Peaks Seen in Genome Scans of ARM Published Elsewhere
| Chromosomeand Locus | Band | Position(cM) | Position(Mb) | HLOD | −log10 (P) | Sall | Citation |
| 1: | |||||||
| D1S1589 | 1q24 | 192.1 | 171.5 | 2.41 | … | … | Seddon et al. 2003 |
| HEMICENTIN-1 | 1q25.3–1q31.1 | … | 182.9 | … | … | … | Schultz et al. 2003a, 2003c |
| D1S202 | 1q31 | 201.6 | 184.2 | … | 2.89 | … | Iyengar et al. 2004 |
| D1S518 | 1q31 | 202.2 | 184.8 | 2.07 | … | … | Majewski et al. 2003 |
| D1S1660 | 1q31 | 212.4 | 195.9 | … | 1.00 | … | Seddon et al. 2003 |
| D1S1660 | 1q31 | 212.4 | 195.9 | 2.46 | … | … | Weeks et al. 2001 |
| D1S413 | 1q31 | 212.4 | 195.9 | 3.00a | … | … | Klein et al. 1998 |
| D1S549 | 1q41 | 239.7 | 216.7 | … | … | 2.13 | Abecasis et al. 2004 |
| 2: | |||||||
| D2S1780 | 2p25.3 | 10.0 | 3.3 | … | … | 1.96 | Abecasis et al. 2004 |
| D2S1356 | 2p21 | 64.3 | 43.3 | … | 2.39 | … | Iyengar et al. 2004 |
| D2S1384 | 2q33 | 200.4 | 205.4 | 2.37 | 1.40 | … | Seddon et al. 2003 |
| 3: | |||||||
| D3S1304 | 3p13 | 22.3 | 6.9 | 2.19 | … | … | Majewski et al. 2003 |
| D3S1763 | 3q26.1 | 176.5 | 168.6 | … | 2.09 | … | Schick et al. 2003 |
| 4: | |||||||
| D4S3360 | 4p16 | .0 | .1 | … | 2.06 | … | Iyengar et al. 2004 |
| D4S2368 | 4q32 | 167.6 | 169.4 | 2.66 | … | … | Majewski et al. 2003 |
| 5: | |||||||
| D5S814 | 5p14.1 | 39.5 | 25.9 | … | … | 1.72 | Abecasis et al. 2004 |
| D5S1506 | 5p13.3 | 49.5 | 33.9 | … | … | 2.55 | Abecasis et al. 2004 |
| D5S2500 | 5q12–5q13 | 69.2 | 58.7 | … | 1.42 | … | Schick et al. 2003 |
| D5S1456 | 5q34 | 174.8 | 169.0 | … | 2.31 | … | Iyengar et al. 2004 |
| 6: | |||||||
| D6S1031 | 6q14–6q21 | 88.6 | 77.5 | … | 1.38 | … | Schick et al. 2003 |
| D6S2436 | 6q23–6q24 | 154.6 | 154.2 | … | 1.54 | … | Schick et al. 2003 |
| 9: | |||||||
| D9S1871 | 9p24 | 8.4 | 3.8 | … | 2.32 | … | Iyengar et al. 2004 |
| D9S1118 | 9p13 | 58.3 | 31.9 | 1.79 | … | … | Weeks et al. 2001 |
| D9S938 | 9q31 | 110.9 | 101.4 | … | 2.10 | … | Iyengar et al. 2004 |
| D9S938 | 9q31.1 | 110.9 | 101.4 | … | … | 1.65 | Abecasis et al. 2004 |
| D9S934 | 9q33 | 128.0 | 116.5 | … | … | 2.01 | Majewski et al. 2003 |
| 10: | |||||||
| D10S1230 | 10q26 | 142.8 | 122.4 | 3.06 | … | … | Majewski et al. 2003 |
| D10S1230 | 10q26 | 142.8 | 122.4 | 2.00 | … | … | Weeks et al. 2001 |
| D10S1230 | 10q26 | 142.8 | 122.4 | 1.52 | … | … | Kenealy et al. 2004 |
| D10S1222 | 10q26 | 156.3 | 128.7 | 1.90 | 1.30 | … | Seddon et al. 2003 |
| D10S1248 | 10q26 | 165.3 | 130.6 | … | 2.43 | … | Iyengar et al. 2004 |
| 12: | |||||||
| D12S297 | 12q13 | 65.0 | 50.9 | … | 2.18 | … | Iyengar et al. 2004 |
| D12S1300 | 12q23–12q24 | … | 97.0 | … | 2.20 | … | Schick et al. 2003 |
| PAH | 12q22–12q23 | … | 101.7 | … | 1.45 | … | Schick et al. 2003 |
| D12S2078 | 12q23 | … | 126.3 | … | 2.70 | … | Iyengar et al. 2004 |
| 15: | |||||||
| GATA50C03 | 15q14 | 34.8 | 34.7 | … | 5.00 | … | Iyengar et al. 2004 |
| D15S659 | 15q11.1–15q14 | 43.5 | 44.1 | … | 1.32 | … | Schick et al. 2003 |
| D15S816 | 15q25–15q26 | 100.6 | 92.7 | … | 1.35 | … | Schick et al. 2003 |
| 16: | |||||||
| D16S769 | 16p12.1 | 50.6 | 26.1 | … | 1.92 | … | Iyengar et al. 2004 |
| D16S769 | 16p12.1 | 50.6 | 26.1 | … | 2.07 | … | Schick et al. 2003 |
| 17: | |||||||
| D17S928 | 17q25 | 126.5 | 80.9 | 3.16 | … | … | Weeks et al. 2001 |
| 18: | |||||||
| GATA178F11 | 18p11 | … | 2.1 | … | 2.48 | … | Iyengar et al. 2004 |
| 20: | |||||||
| D20S451 | 20q13 | … | 57.3 | … | 2.28 | … | Iyengar et al. 2004 |
| 22: | |||||||
| AGAT055Z | 22q12.1 | … | 25.8 | … | … | 2.03 | Abecasis et al. 2004 |
| D22S683 | 22q12–13 | 36.2 | 34.8 | .94 | 2.00 | … | Seddon et al. 2003 |
Parametric LOD score in a single large family.
Using a quantitative measure of ARM severity as the phenotype, Schick et al. (2003) performed a genomewide scan of 102 families from the Beaver Dam eye study, with model-free methods. This method most strongly implicated a region on chromosome 12q23–24 (near D12S346) and also implicated regions on chromosomes 5q12–13, 6q14–21, 15q11–14, and 15q25–26 (table 1).
Using a qualitative ARM phenotype on 70 large families, Majewski et al. (2003) used parametric and allele-sharing models to perform a genomewide scan. Their study found evidence for linkage in five regions—1q31, 3p13, 4q32, 9q33, and 10q26 (table 1)—in addition to finding another large family that, by itself, maps to 1q25–31 with a LOD score of 2.59.
Seddon et al. (2003) performed a genomewide scan of 158 families, using a qualitative ARM phenotype. In their study, there were 490 affected individuals, and 101 individuals were coded as “unaffected” because they were ⩾60 years old and had no evidence of AMD or minimal maculopathy. Using parametric and model-free methods, Seddon et al. found evidence for linkage on 10 different chromosomes, including the regions 1q24, 2q33, 10q26, and 22q12–13 (table 1).
Using a quantitative phenotype based on ARM severity, Iyengar et al. (2004) performed a genomewide scan of 34 extended pedigrees with model-free methods. Their strongest linkage signal was seen in the 15q14 region, and they also observed 13 regions on 11 chromosomes containing nominal evidence of linkage (table 1).
Abecasis et al. (2004) performed a 5-cM genomewide scan of 412 affected relative pairs, using nonparametric linkage analyses with two different diagnostic criteria. They found linkage in 5 regions: 1q41, 2p25, 5p13–14, 9q31, and 22q12 (table 1).
The studies to date are quite encouraging in their consistency—there are two regions (1q25–31 and 10q26) that have been implicated in ARM by multiple studies (table 1). The 1q25–31 region has been implicated by the results of five studies to date: a LOD score of 3 in one large family (Klein et al. 1998); our confirmation, with linkage to exactly the same position (Weeks et al. 2001); and further evidence of linkage in an expanded set of large families (Majewski et al. 2003), as well as in two other independent sets of families (Seddon et al. 2003; Iyengar et al. 2004). Abecasis et al. (2004) also found a linkage signal nearby in 1q41, which may be interpreted as implicating the same general region. Furthermore, a mutation in the HEMICENTIN gene (also known as “FIBULIN 6” [FIBL6 (MIM 608548)]) has been found that segregates with the disease in the initial large family (Schultz et al. 2003a). Additional evidence supporting this HEMICENTIN mutation is the identification of the mutation in a few other independent families with ARM, all of whom seem to share a common haplotype (Schultz et al. 2003c). HEMICENTIN is structurally related to the EFEMP1 gene, which has been implicated in malattia leventinese, a monogenic macular dystrophy (Stone et al. 1999). Confirmation of the role of the HEMICENTIN-1 gene by other ARM research groups is still pending at this time.
The 10q26 region has been implicated in ARM studies multiple times (table 1). Our first genomewide scan had its maximum multipoint Sall score of 1.42 near D10S1236 (Weeks et al. 2000); in our second paper,an Sall peak was seen consistently across all three diagnostic models, with a maximum Sall of 2.10 between D10S1237 and D10S1230 under model A (our strictest model) (Weeks et al. 2001). Similarly, Majewski et al. (2003) found their strongest evidence of linkage at D10S1230. Also, Seddon et al. (2003) observed a maximized heterogeneity LOD score (HLOD) of 1.90 in the 10q26 region at D10S1222, ∼13 cM from D10S1230. Iyengar et al. (2004) obtained a −log10(P) of 2.43 in the 10q26 region. Kenealy et al. (2004) obtained an HLOD of 1.52 and an MLS score of 1.52 at D10S1230 in 10q26, using 70 multiplex families containing 133 affected sib pairs.
Here, we present the results of a genome scan on a third set of families, extending our previous work (Weeks et al. 2000, 2001). We examine the implications of treating this third set as a replication data set, as well as examining the results when we simply pool all three of our genome scans together in one large data set—this combined data set is the largest number of families with ARM analyzed together to date. We also perform exploratory data analyses, using the ordered subset analysis (OSA) approach (Hauser and Boehnke 1998; Ghosh et al. 1999, 2000; Watanabe et al. 1999; Hauser et al. 2001, 2004), which attempts to improve genetic homogeneity by examining subsets of the data where families are included on the basis of relevant covariates.
Material and Methods
Families
The families in the University of Pittsburgh cohort were ascertained from a pool of 35,000 individuals, presumed to be affected by ARM, by a combination of approaches, including recruitment of families from within the retina service of the Department of Ophthalmology at the University of Pittsburgh and mailings sent to patients with ARM (on the basis of billing ICD-9 codes) from multiple retina and ophthalmic practices across the United States (Weeks et al. 2000, 2001). Patients who were interested in participating in the study forwarded a self-addressed postcard to the investigators so that they could be contacted by phone and mail. Through the initial family members, other family members were informed of the study and encouraged to participate. The informed consent process was approved by the institutional review board of the University of Pittsburgh. The families with ARM in the Duke University and Vanderbilt University cohorts were selected for the disease in a similar fashion to that used by Weeks et al. (2000, 2001). Probands were identified from the retina services of each institution, and family members were brought to the examination centers for standardized clinical evaluations and fundus photographs. Families were genotyped in three different sets (see below for details); table 2 shows the number of families included at each of the three genotyping stages.
Table 2.
Group Counts under Three Different Diagnostic Models (A, B, and C)
|
No. under Model |
||||||||||||
| A |
B |
C |
||||||||||
| Group | MGS | CIDR1 | CIDR2 | All | MGS | CIDR1 | CIDR2 | All | MGS | CIDR1 | CIDR2 | All |
| Families with ⩾2 genotyped affected members | 132 | 132 | 152 | 416 | 136 | 136 | 156 | 428 | 163 | 165 | 202 | 530 |
| Affected individuals | 376 | 328 | 395 | 1,099 | 399 | 357 | 415 | 1,171 | 457 | 415 | 501 | 1,373 |
| Genotyped affected individuals | 347 | 322 | 386 | 1,055 | 351 | 341 | 397 | 1,089 | 401 | 392 | 481 | 1,274 |
| Genotyped affected pairs: | ||||||||||||
| Sibling | 135 | 151 | 174 | 460 | 157 | 177 | 197 | 531 | 210 | 229 | 297 | 736 |
| Half-sibling | 5 | 0 | 0 | 5 | 6 | 0 | 1 | 7 | 6 | 4 | 2 | 12 |
| Avuncular | 18 | 9 | 9 | 36 | 19 | 11 | 10 | 40 | 29 | 22 | 19 | 70 |
| Cousin | 29 | 15 | 12 | 56 | 29 | 21 | 13 | 63 | 37 | 26 | 20 | 83 |
| Half-avuncular | 0 | 0 | 1 | 1 | 2 | 0 | 1 | 3 | 2 | 0 | 2 | 4 |
| Great-avuncular | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 3 | 0 | 0 | 3 |
| Parental | 1 | 2 | 4 | 7 | 5 | 3 | 4 | 12 | 13 | 8 | 10 | 31 |
| Grandparental | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 |
Documentation of the retinal features and other ocular pathology were obtained from direct clinical examinations (of local subjects when possible), eye care records (from all eye care providers), and fundus photographs. Every effort was made to document the presence or absence of drusen; the type, size, number, and confluence of drusen; the presence of pigment epithelial figures; choroidal neovascular membranes; geographic atrophy; pigment epithelial detachments (both serous and drusenoid); pigment epithelial tears; and laser treatments (for choroidal neovascular membranes). Otherocular pathology and surgery, including potential confounding diagnoses, were also recorded.
Evaluations
Clinical records were evaluated separately from fundus photographs by a single investigator (M.B.G.), and evaluations were done in separate sessions. Prior to review, each fundus photograph was masked with a unique series of letters and numbers by the clinical coordinating staff to conceal the identity of the individual. As original eye records, which often cannot be masked, were obtained, these were graded separately and then were compared with the photographs to reconcile discrepancies and determine a final grade. If the documentation was insufficient to clearly characterize the individual’s condition, additional records and/or photographs were sought. In the cases from Vanderbilt and Duke, the clinical records and fundus photographs were reviewed by a single investigator (M.B.G.) on separate days and then were reconciled. With only a few exceptions, the individual grading classifications (affected, probably affected, and possibly affected or unknown) were the same for clinical investigators from Duke (E.A.P.) and Vanderbilt (A.A.) and for M.B.G. Among the clinical investigators, there were no discrepancies in the diagnoses of ARM. Of the total 1,616 individuals submitted for genotyping in any or all of the three genomewide scans, 1,177 individuals had photographs and 1,560 individuals had records. There were 415 individuals who had only records available and 46 who had only photographs available.
Genotyping Methods
Genomic DNA was extracted from leukocytes obtained from whole blood collected into EDTA vacutainer tubes. The samples from the University of Pittsburgh were extracted with the use of a simple salting-out procedure, as described elsewhere (Miller et al. 1988), and the samples from our collaborators at Duke and Vanderbilt Universities were extracted with the use of the Puregene DNA purification method (Gentra Systems). The samples were diluted in 1 times TE buffer and sent as coded aliquots either to the National Heart, Lung, and Blood Institute (NHLBI) Mammalian Genotyping Service (MGS) or to the Center for Inherited Disease Research (CIDR) (supported by the National Institutes of Health [NIH], including the National Eye Institute). Both genotyping centers use primarily tri- and tetranucleotide repeat polymorphisms, but each institution uses their own genotyping technology, customized primers, and controls (see NHLBI MGS and CIDR Web sites). Genotyping was done without any knowledge of diagnostic status, but family structure information was used to help detect and resolve genotyping errors.
Classification of Subjects for Linkage Analyses
There are two fundamental questions with regard to the status of each participant in a genetic study of ARM: (1) How confident can one be that an individual isaffected with a macular degeneration? (i.e., diseaseseverity status) and (2) Given an affected individual, how confident can one be of the diagnosis? (i.e., diagnosis status). Three classifications were used for the disease-severity status: definitely affected, probably affected, and possibly affected or unaffected. For the diagnosis status, the following four classifications were used: ARM, ARM or another condition, another condition that is not ARM, and no evidence of ARM (from either photographs or records). Each subject was classified for both disease-severity status and diagnosis status. After all members of a given family had been evaluated, the entire family was reviewed for consistency of the diagnostic criteria and was assessed as to final eligibility for the genomewide genotyping efforts.
We established three clinical grades of ARM for genetic analyses. Under the most stringent model (model A), individuals were classified as “affected” only if they were clearly affected with ARM on the basis of extensive/coalescent drusen, pigmentary changes (including pigment epithelial detachments), and/or the presence of endstage disease (geographic atrophy and/or choroidal neovascular membranes). Model B analyses classified as “affected” all those considered affected under model A and those who were considered to be probably affected with ARM, on the basis of moderate to extensive soft drusen, extensive hard drusen with pigmentary changes, and suspected pigment epithelial detachments. Model C classified as “affected” all of those considered affected under models A and B and those whose diagnosis of ARM was less clear. Under model C, individuals were considered affected if they were definitely and probably affected with ARM or with a related maculopathy (having insufficient evidence to rule out another type ofmacular disease). Model C also included individuals with endstage disease (choroidal neovascular membranes, disciform scarring, and/or geographic atrophy), in the absence of any other documentation of macular pathology. These individuals were considered to be definitely affected, but the determination of whether the etiology was ARM or another maculopathy was considered ambiguous. Our diagnostic models were nested; for example, individuals considered affected under modes B and C, but not under model A, were coded as “unknowns” in the linkage analyses performed with model A diagnoses.
Families eligible for genomewide scans were required to have at least two affected individuals. Efforts were also made to enroll an unaffected informative family member for comparison. The average size of the families (including affected, unknown, or unaffected individuals) ascertained at the Pittsburgh, Duke, and Vanderbilt sites was 2.58 people. Of 628 families (totaling 1,616 individuals) submitted for analysis, 1,159 individuals were definitely affected with ARM and 261 were probably affected with ARM. Thus, 75.69% of affected individuals were included within model A, and 92.17% of all individuals were treated as affected in any of the models (A, B, or C).
Statistical Analyses: Error Checking
Our genotyping data were subjected to several quality checks in order to help ensure accurate genotyping. Genotypes at each locus were checked for Mendelian inconsistencies with the use of our program, PedCheck (O’Connell and Weeks 1998), which uses genotype elimination to identify the more subtle inconsistencies. We compared the number of homozygotes observed with the number of homozygotes expected on the basis of the estimated allele frequencies; a disparity in these numbers may indicate the misnaming of heterozygous genotypes as homozygous. We also examined the genotypes of several putative MZ twin pairs to assay the quality of our genotyping data.
We used the relationship-estimation program PREST (McPeek and Sun 2000) to check the accuracy of our assumed relationships between individuals against the realized level of allele sharing across the whole genome. This led to the omission of four small families, in which relationship errors could not be resolved, from our current set of families.
Allele Frequency Estimation
We estimated allele frequencies at each locus by simple allele counting in all individuals (disregarding relationships), within each data set (MGS, CIDR1, and CIDR2) separately. This approach generates unbiased estimates when applied to randomly ascertained families (Broman 2001). However, our allele-frequency estimates can be biased by the fact that the vast majority of our genotypes occur in affected individuals. Thus, if there were a strong association of a particular allele occurring more often in affected individuals than in the general population, the frequency of that allele would be overestimated. This potential effect is conservative, in that it would bias us away from linkage (Ott 1992; Göring and Terwilliger 2000). We found elsewhere (Weeks et al. 2000) that the use of more precise allele-frequency estimates that take relationships into account (obtaining these estimates is computationally time consuming) generated essentially identical results to those obtained by our simpler (and much faster) estimation approach.
We used data set–specific marker-allele frequencies in all our analyses, because the genotyping centers used different technologies that resulted in slightly different allele labels at many loci. Additionally, the technology used by CIDR to genotype the CIDR2 data set had changed since they first performed genotyping for the CIDR1 data set. Rather than laboriously attempting to establish the exact correspondence between every allele in each of the three data sets, we took a simplerapproach, employing data set–specific marker-allelefrequencies in all our analyses, especially because the capabilities of Allegro (Gudbjartsson et al. 2000) had been extended graciously (at our request) to handle correctly the data-set–specific allele frequencies.
Linkage Analysis
We performed the following analyses:
-
1.
Single-point and multipoint LOD scores under heterogeneity: Since many of our pedigrees show inheritance patterns that seem to be consistent with dominant inheritance, we chose, a priori, to compute LOD scores under a single simple dominant model (disease allele frequency = 0.0001; penetrance vector = [0.01 0.90 0.90]) and to allow for heterogeneity under an admixture model (Hodge et al. 1983). Only two disease phenotypes were used: “affected” and “unknown”; no one was given a “normal” phenotype, owing to the complexities of the ARM phenotype. The single-point and multipoint LOD scores were computed by Allegro(Gudbjartsson et al. 2000) with data set–specific marker allele frequencies (as described above). Note that the majority of our families are so small that the family-specific LOD score maximizes at either θ=0 or θ=.50. With a desire to keep the number of models used in the analyses as low as possible, we elected not to employ age-dependent penetrance models in our study.
-
2.
Single-marker and multipoint “model-free” methods: We computed single-point and multipoint “model free” LOD scores, using the Sall statistic under the linear model with Allegro (Gudbjartsson et al. 2000). Data set–specific marker-allele frequencies were used.
-
3.
Power to detect linkage: Although power estimates for complex diseases may not be very helpful, we presented simulations elsewhere (Weeks et al. 2000) that indicated that, if the genetic effect is large enough, we will have high power to detect linkage. However, as might be expected, the power depends strongly on the percentage of families linked to the region of interest.
-
4.
False-positive rates: We simulated genomewide autosomal marker data, using data set–specific allele frequencies, on all the pedigrees for each of the diagnostic models. To do this, we used Allegro to simulate marker genotypes for exactly the same people genotyped and the same disease phenotypes as in the original real data, under the assumption that the set of markers was segregating independently of ARM disease status. We then analyzed each simulated replicate, using Allegro in exactly the same manner in which we had analyzed the real data, and recorded the results. We used 1,000 replicates to compute the empirical probability of observing a LOD score greater than or equal to a given threshold anywhere in the genome.
-
5.
Ordered subsets analysis (OSA): To explore the relationship of relevant covariates to genetic heterogeneity, we applied OSAs, which use quantitative covariates to identify potentially more homogeneous subgroups from the overall sample (Hauser and Boehnke 1998; Watanabe et al. 1999; Ghosh et al. 2000; Hauser et al. 2001, 2004). In the OSA approach, families are first ranked by an appropriate family-level covariate. Then, families are added into the calculations one at a time in rank order, from one extreme to the other. After each family is added, a maximum LOD score (MLS) is computed for the current contiguous subset of the families; here we use the nonparametric LOD score based on the Sall sharing measure, with data set–specific allele frequencies. The OSA-LOD statistic is the largest MLS of all ordered subsets of the families. Three different OSA-LOD statistics are obtained—by adding families from lowest to highest rank, by adding families from highest to lowest rank, and by finding the best contiguous subset (“optimal slice”) with the largest MLS.
We tested whether the best covariate-defined subset yields a significant increase in the LOD score, as compared with what one would obtain with randomly ordered families. In other words, the P value measures how often an OSA-LOD greater than or equal to the observed OSA-LOD would occur in a subset of the randomly ranked families, on a chromosomewide basis, given our family-level LOD scores. Other simulation studies have shown that this approach has an appropriate type I error rate and can lead to increased power in the presence of heterogeneity (Hauser et al. 2001).
The family-level covariates we used were (1) mean age at onset, (2) mean pack-years, (3) proportion of affecteds with at least one APOE-2 allele, and (4) proportion of affecteds with at least one APOE-4 allele. Age at onset is the earliest age, as reported by the participant, either at which symptoms were attributed to ARM or at diagnosis. If this is not available, then it is the age at the earliest records/photos. Pack-years are self reported and are defined as the product of years and packs, where “packs” is the number of cigarette packs smoked per day, with 20 cigarettes per pack. Nonsmokers were assigned a pack-years value of zero, and cigar and pipe smokers were assigned missing values. We computed the family-specific values, using only the covariate values of the affected members in each pedigree. Therefore, it is possible for a family to have different covariate values under the different diagnostic models if who is affected differs across diagnostic models. Also, note that the OSA program only includes families with known covariate values, so the number of families used varies across covariates and diagnostic models.
Results
We have now completed a genome scan on our third set of families, which, under diagnostic model C, contains 202 families with two or more genotyped affected individuals and 297 affected sib pairs (table 2). These numbers differ slightly from the methodology section, as some samples failed to be genotyped by the genotyping centers (primarily owing to inadequate amounts of DNA or to quality control issues). Our collaborators at Vanderbilt University contributed 14 of the families containing 27 affected sib pairs (under model C), and our collaborators at Duke University contributed 41 families containing 80 affected sib pairs. One could interpret our new data in two different ways. The first approach would be to treat our current genome scan as an independent confirmation set. Our current genome-scan set is larger than those of our previous genomewide scans (table 2). The second approach is simply to pool all three genome scans together. We discuss both of these approaches below.
Third Scan as Independent Confirmation
If one treats the third genome scan as an independent replication set, then we can directly address the question of whether a peak implicated in our earlier work is replicated in our third genome scan. However, because some of the families used in our earlier studies were expanded and regenotyped for the third genome scan, this required that we recompute our results on the first two genome scans after removing these families. This did not alter our earlier conclusions, based on the combined analyses of the first two genome scans (Weeks et al. 2001), that four regions (1q31, 9p13, 10q26, and 17q25) all show multipoint HLOD or Sall scores of ⩾2.0 (under at least one model). However, when we examine the results on the third genomewide screen, only two (10q26 and 17q25) of these four regions appear to have been replicated with LOD scores >1.0. In the 10q26 region, we obtain an Sall of 1.50 at D10S212 under model B (and an Sall of 1.20 and 1.21 under models A and C, respectively, at or near D10S212). In the 17q25 region, we obtained an HLOD of 1.20 at D17S784 under model B (whereas the HLOD reaches a maximum of 0.85 at D17S784 under model A). When interpreting these results, it is important to remember that genome scans on small data sets can show remarkable variability in both evidence of linkage and location estimates (Cordell 2001).
Pooled Analyses
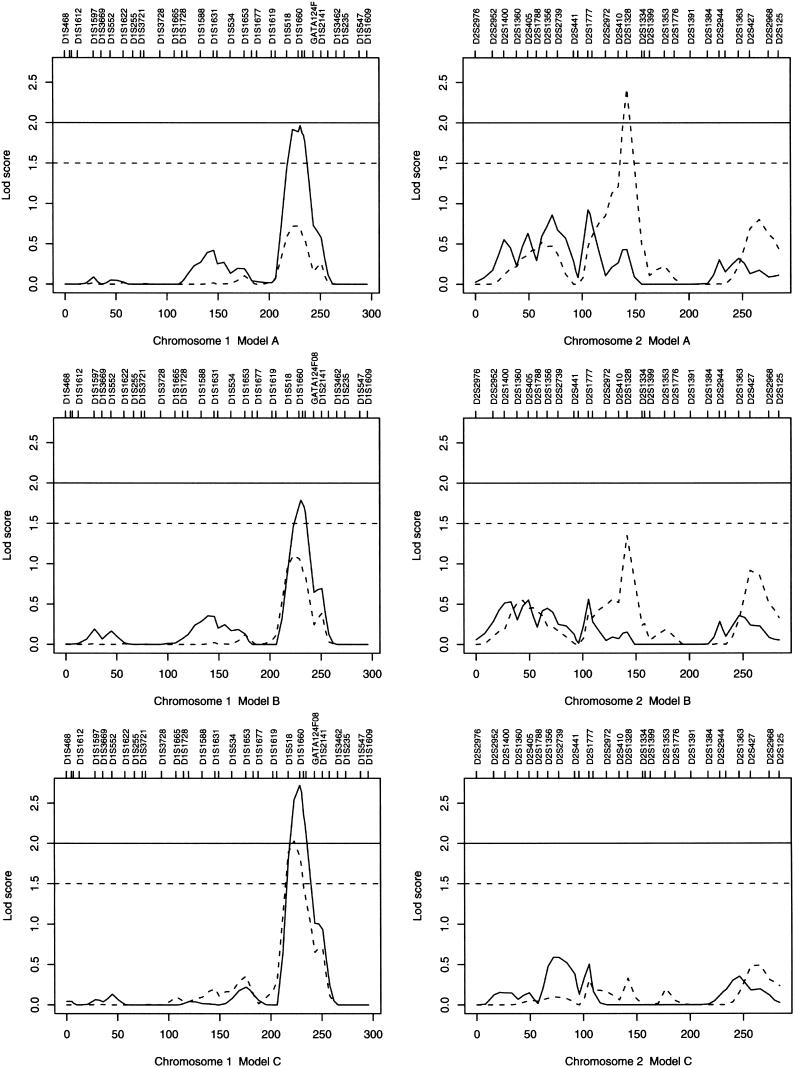
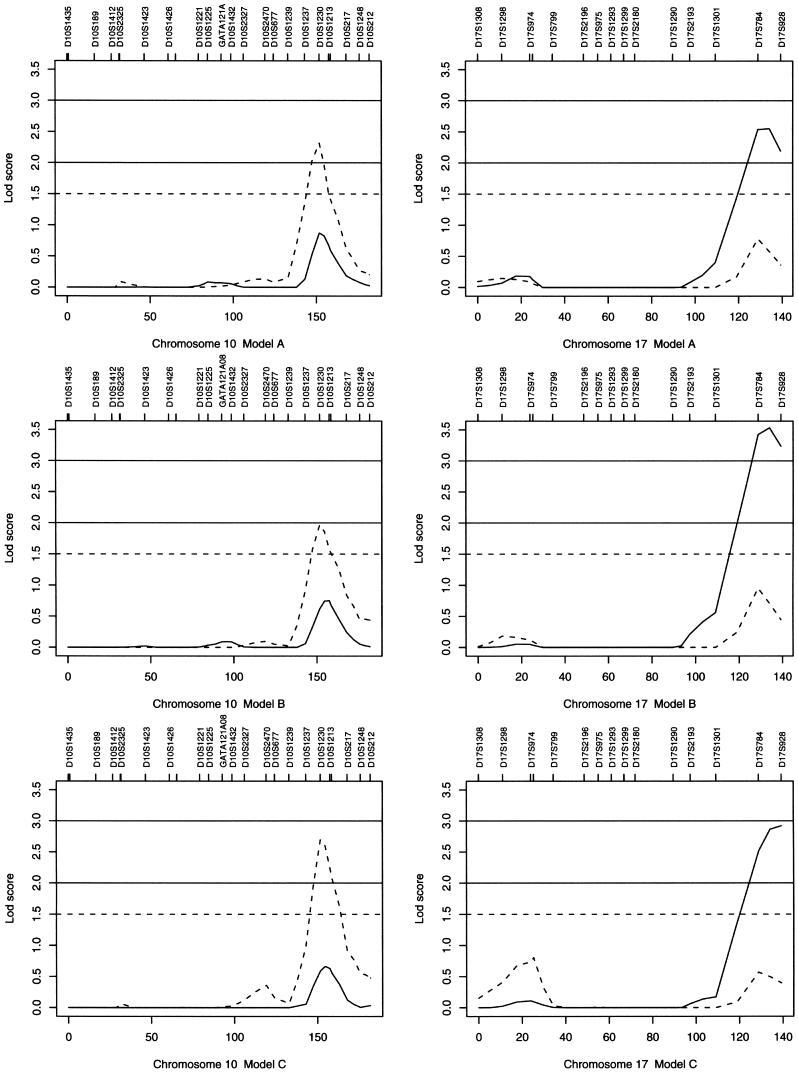
We computed both single-point and multipoint linkage statistics (HLOD and Sall) on our combined set of families; summary descriptions of the numbers of families and numbers of affected relative pairs are provided in table 2. Table 3 displays those markers that had elevated single-point or multipoint statistics ⩾1.5, and figure 1 presents the multipoint curves for chromosomes 1, 2, 10, and 17. On chromosome 1, the 1q31 region had a maximum multipoint HLOD of 2.72 at D1S1660 under model C; the corresponding genomewide empirical P=.061. Elevated HLOD peaks were also seen under models A and B (fig. 1). On chromosome 2q14.3, under model A, there is a significant single-point Sallof 3.23 (marginal single-point empirical P=.0) at D2S1328 (table 2) and a multipoint Sall of 2.42 (genomewide empirical P=.12) at the same location (fig. 1). Under model B, the single-point Sall at D2S1328 is 2.18, but the multipoint curve reaches a maximum of only 1.35, whereas there is no linkage signal under model C. On chromosome 10q26, a multipoint Sall peak is seen consistently across all three models, with a maximum Sall of 2.69 (genomewide empirical P=.06) at D10S1230 under model C. The 17q25 region contains our highest LOD score, with elevated HLODs across all three models and a maximum multipoint HLOD of 3.53 between D17S784 and D17S928 under model B (fig. 1). The genomewide empirical P value for an HLOD of 3.53 is .007.
Table 3.
Single-Point and Multipoint Parametric HLOD Scores and Nonparametric Linkage Statistics[Note]
| HLOD |
Sall |
||||||||||
| Single-Point |
Single-Point |
||||||||||
| Model,Chromosome,and Locus | Position(cM) | Combined | MGS | CIDR1 | CIDR2 | MultipointCombined | Combined | MGS | CIDR1 | CIDR2 | MultipointCombined |
| A: | |||||||||||
| 1: | |||||||||||
| D1S1660a | 228.7 | .35 | .03 | .90 | .00 | 1.89 |
.45 | .08 | .85 | .00 | .72 |
| D1S1647a | 232.3 | 1.07 | NA | .95 | .25 | 1.87 |
.20 | NA | .45 | .00 | .61 |
| D1S1678a | 234.9 | .16 | .16 | NA | NA | 1.78 |
.05 | .05 | NA | NA | .56 |
| 2: | |||||||||||
| D2S1328a | 141.7 | .71 | .01 | 1.44 | .02 | .43 | 3.23 |
.96 | 2.71 |
.25 | 2.42 |
| 3: | |||||||||||
| D3S4523 | 145.3 | 1.44 | .25 | .16 | 1.26 | .85 | 2.14 |
.27 | .28 | 2.06 |
1.78 |
| 4: | |||||||||||
| D4S2367 | 71.9 | .48 | .68 | .26 | .00 | 1.28 | 1.11 | .03 | .72 | .70 | 1.57 |
| D4S1644 | 144.4 | .16 | .35 | .00 | .01 | .21 | 1.63 |
.12 | .66 | 1.14 | 1.70 |
| 7: | |||||||||||
| D7S1808 | 37.5 | 2.00 |
.57 | .04 | 1.97 |
1.86 |
.92 | .07 | .00 | 1.85 |
1.24 |
| 10: | |||||||||||
| D10S1230a | 151.8 | 1.58 |
1.15 | .39 | .20 | .87 | 2.45 |
1.55 |
.82 | .38 | 2.32 |
| 14: | |||||||||||
| D14S599a | 31.6 | 1.21 | .12 | 1.02 | .36 | .81 | 2.58 |
.66 | 1.02 | .98 | 1.82 |
| D14S306 | 35.1 | .19 | .00 | .44 | .23 | .87 | .84 | .00 | .64 | .59 | 1.86 |
| 16: | |||||||||||
| ATA41E04a | .0 | 1.50 |
.17 | 1.52 |
.32 | .73 | 1.69 |
1.59 |
.43 | .15 | .82 |
| 17: | |||||||||||
| D17S784a | 128.8 | 1.41 | .01 | 1.45 | .62 | 2.54 |
.57 | .03 | .94 | .06 | .78 |
| D17S928a | 139.3 | .83 | .20 | .78 | .04 | 2.19 |
.03 | .00 | .42 | .00 | .36 |
| B: | |||||||||||
| 1: | |||||||||||
| D1S1660a | 228.7 | .68 | .14 | .95 | .00 | 1.71 |
.85 | .29 | .86 | .00 | 1.06 |
| D1S1647a | 232.3 | .73 | NA | .51 | .25 | 1.74 |
.23 | NA | .21 | .06 | .91 |
| D1S1678a | 234.9 | .70 | .33 | NA | NA | 1.64 |
.28 | .28 | NA | NA | .85 |
| 2: | |||||||||||
| D2S1328a | 141.7 | .48 | .03 | .88 | .00 | .15 | 2.18 |
.58 | 2.11 |
.13 | 1.35 |
| 3: | |||||||||||
| D3S2460 | 141.8 | .50 | .22 | .00 | .88 | .76 | 1.07 | .09 | .00 | 1.78 |
1.53 |
| D3S4523 | 145.3 | 1.50 |
.18 | .18 | 1.53 |
.57 | 1.88 |
.23 | .07 | 2.39 |
1.52 |
| 4: | |||||||||||
| D4S2367a | 71.9 | 1.02 | 1.58 |
.32 | .00 | 1.12 | 1.07 | .10 | .75 | .45 | 1.55 |
| D4S2394 | 129.2 | 1.13 | .00 | .00 | .45 | .20 | 1.04 | .03 | .48 | .71 | 1.50 |
| D4S1644 | 144.4 | .07 | .41 | .00 | .02 | .31 | 1.14 | .15 | .23 | .94 | 1.61 |
| 6: | |||||||||||
| D6S2427 | 49.9 | .74 | .05 | .92 | .09 | 1.26 | .69 | .01 | 1.06 | .17 | 1.76 |
| 7: | |||||||||||
| D7S1808 | 37.5 | 1.47 | .85 | .00 | 1.36 | 1.93 |
.46 | .03 | .00 | 1.32 | .83 |
| 10: | |||||||||||
| D10S1230a | 151.8 | 1.07 | 1.20 | .09 | .11 | .62 | 1.84 |
1.56 |
.16 | .46 | 1.97 |
| D10S1213 | 157.4 | .57 | .24 | .36 | NA | .75 | .54 | .32 | .22 | NA | 1.58 |
| D10S1656 | 158.5 | .00 | NA | NA | .00 | .68 | .28 | NA | NA | .28 | 1.51 |
| 14: | |||||||||||
| D14S599a | 31.6 | 1.01 | .26 | 1.02 | .06 | .40 | 2.52 |
1.08 | .74 | .71 | 1.65 |
| 17: | |||||||||||
| D17S784a | 128.8 | 2.05 |
.16 | 1.15 | 1.02 | 3.42 |
.71 | .22 | .36 | .16 | .71 |
| D17S928a | 139.3 | 1.52 |
.58 | 1.11 | .06 | 3.23 |
.07 | .09 | .13 | .00 | .45 |
| C: | |||||||||||
| 1: | |||||||||||
| D1S518a | 217.4 | 1.95 |
.42 | 1.27 | .47 | 1.80 |
1.65 |
.53 | 1.06 | .25 | 1.89 |
| D1S1660a | 228.7 | .91 | .02 | .75 | .50 | 2.72 |
1.28 | .31 | .89 | .19 | 1.83 |
| D1S1647a | 232.3 | 1.72 |
NA | .49 | 1.34 | 2.36 |
.65 | NA | .10 | .63 | 1.44 |
| D1S1678a | 234.9 | .01 | .01 | NA | NA | 2.11 |
.01 | .01 | NA | NA | 1.28 |
| D1S2141 | 250.9 | 1.59 |
.66 | .21 | .77 | .93 | .70 | .08 | .01 | 1.10 | .69 |
| 3: | |||||||||||
| D3S2432 | 57.9 | .00 | .00 | .00 | .04 | .00 | 1.45 | .56 | .11 | .98 | 1.67 |
| D3S1768 | 61.6 | 1.26 | .56 | .48 | .24 | .06 | 1.42 | .07 | .32 | 1.51 |
1.56 |
| D3S1766 | 80.2 | .11 | .00 | .00 | .59 | .06 | 1.69 |
.32 | .01 | 2.17 |
1.41 |
| 4: | |||||||||||
| D4S2367a | 71.9 | 1.00 | 1.51 |
.36 | .00 | .93 | 1.34 | .16 | 1.37 | .28 | 1.61 |
| D4S1644 | 144.4 | .56 | 1.15 | .00 | .02 | .79 | .99 | .45 | .33 | .24 | 1.56 |
| 10: | |||||||||||
| D10S1230a | 151.8 | 1.91 |
.52 | .77 | .65 | .59 | 2.74 |
1.11 | .61 | 1.03 | 2.69 |
| D10S1213 | 157.4 | .79 | .34 | .47 | NA | .63 | 1.12 | .92 | .28 | NA | 2.24 |
| D10S1656 | 158.5 | .00 | NA | NA | .00 | .56 | .30 | NA | NA | .30 | 2.09 |
| 12: | |||||||||||
| D12S2070a | 131.5 | .07 | .46 | .00 | .00 | .48 | .70 | .63 | .19 | .05 | 1.63 |
| 14: | |||||||||||
| D14S599a | 31.6 | 2.38 |
.82 | .57 | 1.01 | .52 | 3.15 |
1.70 |
.53 | 1.04 | 1.78 |
| 16: | |||||||||||
| ATA41E04 | .00 | .96 | .06 | .72 | .39 | .29 | 1.58 |
.62 | .42 | .55 | .88 |
| 17: | |||||||||||
| D17S784a | 128.8 | 1.33 | .25 | .56 | .56 | 2.52 |
.29 | .25 | .17 | .00 | .49 |
| D17S928a | 139.3 | 1.56 |
.84 | 1.63 |
.00 | 2.92 |
.04 | .30 | .21 | .00 | .40 |
Note.— Markers are displayed if they have a single-point Sall or HLOD score ⩾1.5 on the combined data set or have a multipoint Sall or HLOD score ⩾1.5 at the marker (underlined values). The cM positions do not agree exactly with those in table 1, because this table uses Haldane cM, whereas table 1 uses Kosambi cM; however, the proper recombination fractions were used in all analyses.
Marker that appeared in the comparable table in our report published elsewhere (Weeks et al. 2001).
Figure 1.
The multipoint HLOD (solid line) and the multipoint nonparametric Sall (dashed line) for chromosomes 1, 2, 10, and 17. Note that some of the marker names are not displayed because of resolution limitations.
On the basis of our simulation study, the genomewide empirical P value for a multipoint LOD score (either Sall or HLOD) of 3.0 is in the range of .03 to .04. For a multipoint LOD score of 2.5, the genomewide empirical P value is in the range of .09 to .11. Note that HLOD and Sall have approximately the same null distribution, since both involve estimating two free parameters—for HLOD, one maximizes across location and α (where α is the fraction of linked families); for Sall, one maximizes across location and δ (see Kong and Cox [1997] for a definition of δ). The genomewide empirical P value measures the chance of seeing a LOD score greater than or equal to the threshold anywhere in the genome and, thus, is larger than the marginal P value evaluated at a single point in the genome.
Exploration of Covariate Effects
The formation of subsets on the basis of relevant covariates with the OSA approach, although exploratory in nature, may result in more genetically homogenous subsets. Tables 4, 5, 6, and 7 present our results after we formed subsets on the basis of (1) mean age at onset, (2) mean pack-years, (3) proportion of affecteds with at least one APOE-2 allele, and (4) proportion of affecteds with at least one APOE-4 allele. With regard to the regions on chromosomes 1, 2, 10, and 17 implicated by the conventional linkage analyses described above, only the 10q26 region has interesting OSA-LODs (table 5). Under both models A and B, the OSAs reveal a subgroup of ∼160 highest smoking (mean pack-years ⩾26) families, with elevated OSA-LODs of 3.87 and 4.25, respectively.
Table 4.
Results of Ordered Subset Analyses with Mean Age at Onset as the Family-Specific Covariate
| Chromosome | Position(cM) | AdjacentMarker | Trait | Unconditional LOD Scores | OSA-LODScores | No. of Families Used/No. of Total Families | Cut Point | EmpiricalP | RankOrdera |
| 7 | 37.5 | D7S1808 | A | 1.216 | 3.179 | 370/562 | ⩽73.00 | .0110 | L→H |
| 8 | 21.9 | D8S1130 | A | .014 | 3.455 | 61/563 | [59.00, 63.50] | .0429 | S |
| 8 | 23.3 | D8S1130 | B | .001 | 4.086 | 60/567 | [59.00, 63.50] | .0058 | S |
| 8 | 13.3 | D8S1469 | C | .108 | 2.828 | 137/573 | ⩽65.75 | .0112 | L→H |
| 8 | 13.3 | D8S1469 | C | .108 | 4.411 | 101/573 | [59.00, 65.75] | .0040 | S |
| 12 | 12.6 | D12S2395 | A | .639 | 3.393 | 237/564 | ⩽69.00 | .0243 | L→H |
| 12 | 12.6 | D12S2395 | B | .672 | 3.449 | 240/568 | ⩽69.00 | .0345 | L→H |
| 19 | 46.5 | D19S433 | C | .098 | 1.851 | 209/571 | ⩾72.67 | .0419 | H→L |
L = low; H = high; S = optimal slice method.
Table 5.
Results of Ordered Subset Analyses with Mean Pack-Years as the Family-Specific Covariate
| Chromosome | Position(cM) | AdjacentMarker | Trait | Unconditional LOD Scores | OSA-LODScores | No. of Families Used/No. of Total Families | Cut Point | EmpiricalP | RankOrdera |
| 5 | 198.2 | D5S211 | B | .000 | 1.801 | 35/492 | ⩾59.75 | .05 | H→L |
| 8 | 39.2 | D8S1145 | A | .000 | 3.794 | 35/482 | [22.5, 27.38] | .0055 | S |
| 10 | 154.0 | D10S1230 | A | 1.824 | 3.868 | 165/482 | ⩾26.05 | .0419 | H→L |
| 10 | 154.0 | D10S1230 | B | 1.832 | 4.252 | 161/488 | ⩾26.05 | .0474 | H→L |
| 13 | 29.8 | D13S325 | A | .050 | 3.905 | 98/480 | [26.50, 49.16] | .0092 | S |
| 20 | 56.7 | D20S478 | A | .001 | 4.633 | 66/480 | [27.75, 43] | .0005 | S |
| 20 | 56.7 | D20S478 | B | .017 | 5.232 | 64/486 | [27.75, 43] | <.0001 | S |
| 20 | 56.7 | D20S478 | C | .007 | 5.655 | 44/498 | [31.50, 43] | <.0001 | S |
L = low; H = high; S = optimal slice method.
Table 6.
Results of Ordered Subset Analyses with Proportion of Affecteds Carrying at Least One APOE-2 Allele as the Family-Specific Covariate
| Chromosome | Position(cM) | AdjacentMarker | Trait | Unconditional LOD Scores | OSA-LODScores | No. of Families Used/No. of Total Families | Cut Point | EmpiricalP | RankOrdera |
| 5 | 101.1 | D5S641 | C | .294 | 1.386 | 59/410 | [.125, .667] | .0295 | S |
| 9 | 8.2 | D9S168/D9S921 | C | .259 | 2.775 | 59/406 | [.333, .750] | .0303 | S |
| 18 | 95.1 | D18S1357 | A | .402 | 1.178 | 305/396 | ⩽.333 | .0223 | L→H |
| 19 | 53.7 | D19S245 | B | .258 | .972 | 45/399 | ⩾.750 | .0240 | H→L |
L = low; H = high; S = optimal slice method.
Table 7.
Results of Ordered Subset Analyses with Proportion of Affecteds Carrying at Least One APOE-4 Allele as the Family-Specific Covariate
| Chromosome | Position(cM) | AdjacentMarker | Trait | Unconditional LOD Scores | OSA-LODScores | No. of Families Used/No. of Total Families | Cut Point | EmpiricalP | RankOrdera |
| 6 | 121.1 | D6S474 | A | .000 | 1.501 | 38/398 | ⩾.667 | .0120 | H→L |
| 7 | 191.6 | D7S559 | A | .000 | 1.442 | 38/396 | ⩾.667 | .0136 | H→L |
| 7 | 186.2 | D7S3058 | B | .000 | 1.191 | 37/399 | ⩾.667 | .0145 | H→L |
| 7 | 182.6 | D7S3058 | C | .000 | 1.505 | 31/407 | ⩾.750 | .0119 | H→L |
| 11 | 40.3 | D11S1392 | C | .089 | .744 | 87/406 | ⩾.500 | .0261 | H→L |
| 15 | 8.6 | D15S165 | C | .214 | 1.339 | 37/407 | ⩾.667 | .0327 | H→L |
L = low; H = high; S = optimal slice method.
With the use of mean pack-years, the optimal slice method generates large OSA-LODs between 4.63 and 5.66, under all three models, on chromosome 20 near D20S478 (table 5). Under models B and C, the permutation-based P value is quite small, and the optimal slice includes families with ∼27–43 mean pack-years.
When we stratify on the proportion of affecteds with at least one APOE-2 allele or on the proportion with at least one APOE-4 allele, only modest OSA-LODs are observed (tables 6 and 7). For APOE-4, however, we do observe, across all three diagnostic models, elevated OSA-LODs in the same region on chromosome 7 (table 7).
Discussion
It is difficult to study the genetics of ARM because the disease has a late age at onset, which implies that the parents are usually not available for genotyping. Furthermore, the incidence of ARM increases rapidly with advancing age, suggesting that the probability that an affected individual is a nongenetic phenocopy also increases rapidly with age. Given these potential difficulties, we find it heartening and encouraging that some regions of the genome have been implicated by multiple studies.
First, if we treat our current genome scan as an independent replication set, then two of our four previously implicated regions, 10q26 and 17q25, are replicated. Second, if we pool all three of our genome scans together, then the regions of interest are 1q31, 2q14.3, 10q26, and 17q25. As detailed in the introduction, both the 1q31 and 10q26 regions have been found in other studies (see table 1). In addition to being implicated by our studies, the 1q25–31 region has been implicated by five other studies (Klein et al. 1998; Majewski et al. 2003; Seddon et al. 2003; Abecasis et al. 2004; Iyengar et al. 2004). Furthermore, a mutation in the HEMICENTIN-1 gene has been found that segregates with the disease in the initial large family (Schultz et al. 2003a, 2003c). Similarly, the 10q26 peak has been supported consistently across all three of our studies (even when the third scan is considered by itself alone), and Majewski et al. (2003) found their strongest evidence of linkage in this region. Furthermore, as detailed in the introduction, this region has been supported by three other studies (Seddon et al. 2003; Iyengar et al. 2004; Kenealy et al. 2004).
Our identified regions of interest, based on an affecteds-only analysis, do not appear to overlap with those of Schick et al. (2003), who analyzed a quantitative measure of ARM. Their best peak was in the 12q23–24 region, which is ∼20 cM from a minor Sall peak of 1.63 at D12S2070; since their reported peak is wide, this result could constitute an overlap.
The OSA approach represents exploratory data analysis, and, as such, serves mainly as a way to use our data to generate new hypotheses that must be tested in independent data sets. One interesting hypothesis, generated by our OSA results (tables 4, 5, 6, and 7), is that the effect of smoking on risk of ARM is accentuated by a gene in the chromosome 10q26 region. That is, OSAs found a stronger linkage signal in the subgroup of families with high levels of smoking among the affected individuals. This is particularly intriguing—first, because the 10q26 region has been implicated by multiple studies and, second, because there is ample evidence that smoking markedly increases the risk of ARM (Klein et al. 1993; Schwartz 1994; Seddon et al. 1996; Vingerling et al. 1996; Klaver et al. 1997; Tamakoshi et al. 1997; Delcourt et al. 1998).
Our most significant OSA result occurs on chromosome 20 (table 5) near D20S481, with the use of the “optimal slice” method. Although the permutation results indicate that to observe a subgroup with such a large OSA-LOD by chance is extremely unlikely, this result still must be viewed with caution, since it suggests the unlikely hypothesis that smoking has a U-shaped effect on disease risk; for example, families with 27–43 mean pack-years are linked, whereas families with smoking levels below and above this range are not linked.
In summary, we have performed careful linkage analyses on the largest set of families with ARM assembled to date and have found continued support for the 1q31, 10q26, and 17q25 regions. Two of these regions, 1q31 and 10q26, have also been identified in several other independent studies. This remarkable consistency between studies is very encouraging and argues in favor of these regions containing true susceptibility genes for ARM. Finally, the exploratory OSAs generate the interesting hypothesis that the effect of smoking on risk of ARM is accentuated by a gene in the 10q26 region.
Acknowledgments
Foremost, we would like to thank the participating families and their clinicians, for their generous support that made this project possible. We would also like to thank Robert Elston and colleagues for providing us with a prepublication copy of Schick et al. (2003); Jacek Majewski and Michael Klein for an advanced copy of Majewski et al. (2003); Anand Swaroop and colleagues for an advanced summary of Abecasis et al. (2004); and Susan Santangelo and Chen-Hsing Yen for extensive discussions regarding the results of Seddon et al. (2003). Because we used methodology nearly identical to what we had used previously, some of the text of this article is taken verbatim from our article published elsewhere (Weeks et al. 2001), with the permission of Elsevier Science Incorporated. This study was funded initially by the Smith Kettlewell Eye Research Foundation, San Francisco (M.B.G. and T.O.P.), and the Pennsylvanian Lions Sight Conservation and Eye Research Foundation (M.B.G.). Additional support was provided by The Eye and Ear Foundation of Pittsburgh (NEI R01-EY09859 to M.B.G.); by Research to Prevent Blindness, New York; and by The Steinbach Foundation, New York (M.B.G.). Our first genomewide scan was genotyped by the NHLBI MGS, which is led by Dr. James Weber. The MGS is funded by NHLBI under contract N01-HV-48141 to the Marshfield Medical Research Foundation. Genotyping services for our second and third genomewide scans were provided by CIDR. CIDR is fully funded through a federal contract from the NIH to The Johns Hopkins University, contract N01-HG-65403.
Electronic-Database Information
URLs for data presented herein are as follows:
- CIDR, http://www.cidr.jhmi.edu/
- NHLBI MGS, http://research.marshfieldclinic.org/genetics/Genotyping_Service/mgsver2.htm
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
References
- Abecasis GR, Yashar BM, Zhao Y, Ghiasvand N, Zareparsi S, Branham KEH, Reddick AC, Trager EH, Yoshida S, Bahling J, Filippova E, Elner S, Johnson MW, Vine AK, Sieving PA, Jacobson SG, Richards JE, Swaroop A (2004) Agerelated macular degeneration: a high resolution genome scan for susceptibility loci in a population enriched for late-stage disease. Am J Hum Genet 74:482–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Age-Related Eye Disease Study Research Group (2001) A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report number 8. Arch Ophthalmol 119:1417–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonastre J, Le Pen C, Soubrane G, Quentel G (2003) The burden of age-related macular degeneration: results of a cohort study in two French referral centres. Pharmacoeconomics 21:181–190 [DOI] [PubMed] [Google Scholar]
- Breitner JC, Jarvik GP, Plassman BL, Saunders AM, Welsh KA (1998) Risk of Alzheimer disease with the epsilon4 allele for apolipoprotein E in a population-based study of men aged 62–73 years. Alzheimer Dis Assoc Disord 12:40–44 [DOI] [PubMed] [Google Scholar]
- Broman KW (2001) Estimation of allele frequencies with data on sibships. Genet Epidemiol 20:307–315 10.1002/gepi.2.abs [DOI] [PubMed] [Google Scholar]
- Cordell HJ (2001) Sample size requirements to control for stochastic variation in magnitude and location of allelesharing linkage statistics in affected sibling pairs. Ann Hum Genet 65:491–502 10.1046/j.1469-1809.2001.6550491.x [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC Jr, Rimmler JB, Locke PA, Conneally PM, Schmader KE, Small GW, Roses AD, Haines JL, and Pericak-Vance MA (1994) Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet 7:180–184 [DOI] [PubMed] [Google Scholar]
- Delcourt C, Diaz JL, Ponton SA, Papoz L (1998) Smoking and age-related macular degeneration: the POLA study (Pathologies Oculaires Liees a l’Age). Arch Ophthalmol 116:1031–1035 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Watanabe RM, Hauser ER, Valle T, Magnuson VL, Erdos MR, Langefeld CD, et al (1999) Type 2 diabetes: evidence for linkage on chromosome 20 in 716 Finnish affected sib pairs. Proc Natl Acad Sci USA 96:2198–2203 10.1073/pnas.96.5.2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Watanabe RM, Valle TT, Hauser ER, Magnuson VL, Langefeld CD, Ally DS, et al (2000) The Finland-United States investigation of non-insulin-dependent diabetes mellitus genetics (FUSION) study. I. An autosomal genome scan for genes that predispose to type 2 diabetes. Am J Hum Genet 67:1174–1185 [PMC free article] [PubMed] [Google Scholar]
- Gorin MB (1998) Genetics of age-related maculopathy. In: Traboulsi E (ed) Genetic diseases of the eye. Oxford University Press, London, pp 407–434 [Google Scholar]
- Göring HHH, Terwilliger JD (2000) Linkage analysis in the presence of errors. III. Marker loci and their map as nuisance parameters. Am J Hum Genet 66:1298–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Jonasson K, Frigge ML, Kong A (2000)Allegro, a new computer program for multipoint linkage analysis. Nat Genet 25:12–13 10.1038/75514 [DOI] [PubMed] [Google Scholar]
- Hauser ER, Bass MP, Martin ER, Watanabe RM, Duren WL, Boehnke M (2001) Power of the ordered subset method for detection and localization of genes in linkage analysis of complex traits. Am J Hum Genet Suppl 69:529 [Google Scholar]
- Hauser ER, Boehnke M (1998) Genetic linkage analysis of complex genetic traits by using affected sibling pairs. Biometrics 54:1238–1246 [PubMed] [Google Scholar]
- Hauser ER, Watanabe RM, Duren WL, Bass MP, Langefeld CD, Boehnke M (2004) Ordered subset analysis in genetic linkage mapping of complex traits. Genet Epidemiol 27 (in press; available at http://www3.interscience.wiley.com/cgi-bin/jissue/107560885) [DOI] [PubMed]
- Hodge SE, Anderson CE, Neiswanger K, Sparkes RS, Rimoin DL (1983) The search for heterogeneity in insulin-dependent diabetes mellitus (IDDM): linkage studies, two-locus models, and genetic heterogeneity. Am J Hum Genet 35:1139–1155 [PMC free article] [PubMed] [Google Scholar]
- Hooper CY, Guymer RH (2003) New treatments in age-related macular degeneration. Clin Experiment Ophthalmol 31:376–391 10.1046/j.1442-9071.2003.00683.x [DOI] [PubMed] [Google Scholar]
- Iyengar SK, Song D, Klein BE, Klein R, Schick JH, Humphrey J, Millard C, Liptak R, Russo K, Jun G, Lee KE, Fijal B, Elston RC (2004) Dissection of genomewide-scan data in extended families reveals a major locus and oligogenic susceptibility for age-related macular degeneration. Am J Hum Genet 74:20–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenealy SJ, Schmidt S, Agarwal A, Postel EA, De La Paz MA, Pericak-Vance MA, Haines JL (2004) Linkage analysis for age-related macular degeneration supports a gene on chromosome 10q26. Mol Vis 10:57–61 [PubMed] [Google Scholar]
- Klaver CC, Assink JJ, Vingerling JR, Hofman A, de Jong PT (1997) Smoking is also associated with age-related macular degeneration in persons aged 85 years and older: the Rotterdam study. Arch Ophthalmol 115:945 [DOI] [PubMed] [Google Scholar]
- Klaver CC, Kliffen M, van Duijn CM, Hofman A, Cruts M, Grobbee DE, van Broeckhoven C, de Jong PT (1998) Genetic association of apolipoprotein E with age-related macular degeneration. Am J Hum Genet 63:200–206 (erratum 63:1252) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein ML, Schultz DW, Edwards A, Matise TC, Rust K, Berselli CB, Trzupek K, Weleber RG, Ott J, Wirtz MK, Acott TS (1998) Age-related macular degeneration: clinical features in a large family and linkage to chromosome 1q. Arch Ophthalmol 116:1082–1088 [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BE, Linton KL, DeMets DL (1993) The Beaver Dam eye study: the relation of age-related maculopathy to smoking. Am J Epidemiol 137:190–200 [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J, Schultz DW, Weleber RG, Schain M, Edwards AO, Matise TC, Acott TS, Ott J, Klein ML (2003) Age-related macular degeneration: a genome scan in extended families. Am J Hum Genet 73:540–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal N, Chisholm IH (2002) Identifying the proportion of age related macular degeneration patients who would benefit from photodynamic therapy with verteporfin (Visudyne). Br J Ophthalmol 86:118–119 10.1136/bjo.86.1.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPeek MS, Sun L (2000) Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet 66:1076–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MR, Tschanz JT, Norton MC, Welsh BK, Steffens DC, Wyse BW, Breitner JC (1998) APOE genotype predicts when—not whether—one is predisposed to develop Alzheimer disease. Nat Genet 19:321–322 10.1038/1206 [DOI] [PubMed] [Google Scholar]
- Miller S, Dykes D, Polesky H (1988) A simple salting and procedure for extracting DNA from human nucleated cells. Nucl Acids Res 16:1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identifying genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J (1992) Strategies for characterizing highly polymorphic markers in human gene mapping. Am J Hum Genet 51:283–290 [PMC free article] [PubMed] [Google Scholar]
- Pang CP, Baum L, Chan WM, Lau TC, Poon PM, Lam DS (2000) The apolipoprotein E epsilon4 allele is unlikely to be a major risk factor of age-related macular degeneration in Chinese. Ophthalmologica 214:289–291 10.1159/000027506 [DOI] [PubMed] [Google Scholar]
- Schick JH, Iyengar SK, Klein BE, Klein R, Reading K, Liptak R, Millard C, Lee KE, Tomany SC, Moore EL, Fijal BA, Elston RC (2003) A whole genome screen of a quantitative trait of age-related maculopathy in sibships from the Beaver Dam eye study. Am J Hum Genet 72:1412–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, Klaver C, Saunders A, Postel E, De La Paz M, Agarwal A, Small K, Udar N, Ong J, Chalukya M, Nesburn A, Kenney C, Domurath R, Hogan M, Mah T, Conley Y, Ferrell R, Weeks D, de Jong P, van Duijn C, Haines J, Pericak-Vance M, Gorin M (2002) A pooled case-control study of the apolipoprotein E (APOE) gene in age-related maculopathy. Ophthalmic Genet 23:209–223 10.1076/opge.23.4.209.13883 [DOI] [PubMed] [Google Scholar]
- Schultz DW, Humpert AJ, Luzier CW, V Persun, M Schain, Weleber RG, Acott TS, Klein ML (2003a) Evidence that FIBL-6 is the ARMD1 gene. Paper presented at the Association for Research in Vision and Ophthalmology Meeting. Fort Lauderdale, Florida [Google Scholar]
- Schultz DW, Klein ML, Humpert A, Majewski J, Schain M, Weleber RG, Ott J, Acott TS (2003b) Lack of an association of apolipoprotein E gene polymorphisms with familial age-related macular degeneration. Arch Ophthalmol 121:679–683 10.1001/archopht.121.5.679 [DOI] [PubMed] [Google Scholar]
- Schultz DW, Klein ML, Humpert AJ, Luzier CW, Persun V, Schain M, Mahan A, Runckel C, Cassera M, Vittal V, Doyle TM, Martin TM, Weleber RG, Francis PJ, Acott TS (2003c) Analysis of the ARMD1 locus: evidence that a mutation in HEMICENTIN-1 is associated with age-related maculardegeneration in a large family. Hum Mol Genet 12:3315–3323 10.1093/hmg/ddg348 [DOI] [PubMed] [Google Scholar]
- Schwartz D (1994) The Beaver Dam eye study: the relation of age-related maculopathy to smoking. Surv Ophthalmol 39:84–85 [PubMed] [Google Scholar]
- Seddon JM, Santangelo SL, Book K, Chong S, Cote J (2003) A genomewide scan for age-related macular degeneration provides evidence for linkage to several chromosomal regions. Am J Hum Genet 73:780–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seddon JM, Willett WC, Speizer FE, Hankinson SE (1996) A prospective study of cigarette smoking and age-related macular degeneration in women. JAMA 276:1141–1146 [PubMed] [Google Scholar]
- Souied EH, Benlian P, Amouyel P, Feingold J, Lagarde JP, Munnich A, Kaplan J, Coscas G, Soubrane G (1998) The epsilon4 allele of the apolipoprotein E gene as a potential protective factor for exudative age-related macular degeneration. Am J Ophthalmol 125:353–359 10.1016/S0002-9394(99)80146-9 [DOI] [PubMed] [Google Scholar]
- Stone EM, Lotery AJ, Munier FL, Heon E, Piguet B, Guymer RH, Vandenburgh K, Cousin P, Nishimura D, Swiderski RE, Silvestri G, Mackey DA, Hageman GS, Bird AC, Sheffield VC, Schorderet DF (1999) A single EFEMP1 mutation associated with both malattia leventinese and Doyne honeycomb retinal dystrophy. Nat Genet 22:199–202 10.1038/9722 [DOI] [PubMed] [Google Scholar]
- Stone EM, Sheffield VC, Hageman GS (2001) Molecular genetics of age-related macular degeneration. Hum Mol Genet10:2285–2292 [DOI] [PubMed] [Google Scholar]
- Stone EM, Webster AR, Vandenburgh K, Streb LM, Hockey RR, Lotery AJ, Sheffield VC (1998) Allelic variation in ABCR associated with Stargardt disease but not age-related macular degeneration. Nat Genet 20:328–329 10.1038/3798 [DOI] [PubMed] [Google Scholar]
- Tamakoshi A, Yuzawa M, Matsui M, Uyama M, Fujiwara NK, Ohno Y (1997) Smoking and neovascular form of age related macular degeneration in late middle aged males: findings from a case-control study in Japan. Research committee on chorioretinal degenerations. Brit J Ophthalmol 81:901–904 (erratum 82:207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vingerling JR, Hofman A, Grobbee DE, de Jong PT (1996) Age-related macular degeneration and smoking: the Rotterdam study. Arch Ophthalmol 114:1193–1196 [DOI] [PubMed] [Google Scholar]
- Watanabe RM, Ghosh S, Birznieks G, Duren WL, Mitchell BD (1999) Application of an ordered subset analysis approach to the genetics of alcoholism. Genet Epidemiol Suppl 17:385–390 [DOI] [PubMed] [Google Scholar]
- Weeks DE, Conley YP, Mah TS, Paul TO, Morse L, Ngo-Chang J, Dailey JP, Ferrell RE, Gorin MB (2000) A full genome scan for age-related maculopathy. Hum Mol Genet 9:1329–1349 10.1093/hmg/9.9.1329 [DOI] [PubMed] [Google Scholar]
- Weeks DE, Conley YP, Tsai HJ, Mah TS, Rosenfeld PJ, Paul TO, Eller AW, Morse LS, Dailey JP, Ferrell RE, Gorin MB (2001) Age-related maculopathy: an expanded genome-wide scan with evidence of susceptibility loci within the 1q31 and 17q25 regions. Am J Ophthalmol 132:682–692 10.1016/S0002-9394(01)01214-4 [DOI] [PubMed] [Google Scholar]