Abstract
Two outbreaks of hand-foot-and-mouth disease (HFMD) occurred in Taiwan between 1998 and 2000. Enteroviruses were isolated from a total of 1,892 patients in this laboratory during this period. Of the virus isolates, enterovirus 71 (EV71) was diagnosed in 44.4% of the patients (132 of 297) in 1998, 2% (13 of 646) in 1999, and 20.5% (195 of 949) in 2000. Genetic analyses of the 5′-untranslated and VP1 regions of EV71 isolates by reverse transcription-PCR and sequencing were performed to understand the diversity of EV71 in these outbreaks of HFMD. Most EV71 isolates from the 1998 epidemic belonged to genotype C, while only one-tenth of the isolates were genotype B. Interestingly, all EV71 isolates tested from 1999 to 2000 belonged to genotype B. This study indicated that two genogroups of EV71 capable of inducing severe clinical illness have been circulating in Taiwan. Furthermore, the predominant EV71 genotypes responsible for each of the two major HFMD outbreaks within the 3-year period in Taiwan were different.
Enterovirus 71 (EV71) is known to be a causative agent of hand-foot-and-mouth disease (HFMD) and severe neurological complications (4). EV71 was first isolated and characterized from cases of neurological disease in California (11). Many EV71 outbreaks have been reported in different regions of the world (5, 8). In 1998, a large outbreak of HFMD (129,106 reports) occurred in Taiwan, involving 405 children with severe complications, including encephalitis, aseptic meningitis, pulmonary edema or hemorrhage, acute flaccid paralysis, and myocarditis, resulting in 78 deaths (4, 9). In 2000, there was another outbreak, with 80,677 reports of HFMD and 41 deaths in Taiwan (http://www.cdc.gov.tw/g/virus/virus3.htm).
EV71 is a human enterovirus belonging to the Picornaviridae family. The picornavirus positive-stranded RNA genome possesses an unusually long, conserved 5′-untranslated region (UTR) of approximately 740 nucleotides (nt). It has been demonstrated that this region has a crucial role in the viral life cycle (10). In contrast to the highly conserved 5′-UTR, the open reading frame encoding the capsid protein VP1 is more variable and confers distinct antigenic properties to the virus (2). Thus, this VP1 region is considered to be the most suitable region for sequence analysis to determine genetic diversity. Analysis of EV71 by RT-PCR and sequencing of the 5′-UTR and VP1 regions of the genome in this study allowed us to understand the genetic diversity of EV71 in outbreaks of HFMD and revealed the genotypes responsible for the 1998 and 2000 outbreaks.
MATERIALS AND METHODS
Virus isolation and identification.
A total of 14,901 specimens from inpatients or outpatients during the study period from 1998 to 2000 were investigated. Specimens were inoculated onto appropriate tissue cultures (Vero, A549, rhabdomyosarcoma [RD], green monkey kidney [GMK], and MRC-5 cells) to isolate enteroviruses (15). Enterovirus strains were typed antigenetically by neutralization tests using Lim and Benyesh-Melnick pools (6) or immunofluorescence tests using available monoclonal antibodies (Chemicon International Inc.).
Analysis of the 5′-UTR and VP1 sequences.
Viral RNA was extracted and used in reverse transcription-PCR (RT-PCR) as previously described (16). The primer sequences used were as follows: EV34A (sense), 5′-GGCCCACTGGGCGCTAGCA-3′, (nt 34 to 53 of EV71 MS genome), and EV1337B (antisense), 5′-TGTCCCAATGACATACTCT-3′ (nt 1356 to 1337), for the 5′-UTR; EV159 (sense), 5′-ACYATGAAAYTGTGCAAGG-3′ (nt 2385 to 2403), and EV174 (antisense), 5′-GCTGACCAAACTTTCCAAGGG-3′ (nt 3348 to 3328), for VP1 (3). DNA sequencing was performed with primers EV34A (sense), EV800B (antisense) (5′-AATGGTGGAGCCTTCTGTAG-3′ [nt 822 to 803]), EV583A (sense) (5′-TGGCTGCTTATGGTGACAA-3′ [nt 584 to 603]), and EV1019B (antisense) (5′-ACTATGATGTTTGCTGCTTC-3′ [nt 1052 to 1032]) for 5′-UTR and EV159 (sense), EV162 (antisense) (5′-CCRGTAGGKGTRCACGCRAC-3′ [nt 2869 to 2850]), EV161 (sense) (5′-CTGGGACATAGAYATAACWGG-3′ [nt 2766 to 2785]), and EV174 (antisense) for VP1 (3) using an automated DNA sequencer (model 373A; Applied Biosystems). Sequence analyses were performed on approximately 75, 13, and 40 EV71 isolates from 1998, 1999, and 2000, respectively, using programs in the University of Wisconsin Genetics Computer Group (GCG) package (version 10.1).
Phylogenetic analysis of 5′-UTR and VP1 sequences.
Fifty-three EV71 isolates were used for phylogenetic analysis using a random sampling from patients with diverse clinical presentations, ranging from uncomplicated HFMD to encephalitis and death (Table 1). Three different methods (neighbor joining, parsimony, and maximum likelihood) of phylogenetic analysis from the PHYLIP program package, version 3.573c, were used in order to make a more reliable inference of phylogeny. The nucleotide distances were calculated by the program DNADIST from the PHYLIP program package and by the use of the Kimura two-parameter model with a transition/transversion rate of 2.0. A phylogenetic tree was then constructed by use of the neighbor-joining program of the PHYLIP package. SEQBOOT was used for bootstrap analysis of data, in which 1,000 data sets were analyzed and CONSENSE was used to compute a consensus tree. The statistical estimation of the significance of branch lengths was also determined by the maximum-likelihood method. Pairwise nucleotide and amino acid sequence comparisons were performed by the Distances program of the GCG package.
TABLE 1.
EV71 isolates used for phylogenetic analysisa
| Isolate | Sampling date (mo/yr) | Clinical feature(s) on admission | Origin
|
Isolate genotype | |
|---|---|---|---|---|---|
| Patient age (yr) | Specimen | ||||
| BrCr-CA-70 | 1970 | Aseptic meningitis | 0.2 | NA | A |
| 7423-MS-87 | 1987 | Paralytic illness | 1.6 | NA | B |
| 236-TW-86 | 1986 | HFMD | NA | NA | B |
| 0756-MAA-97 | 1997 | NA | NA | NA | C |
| 0731-MAA-97 | 1997 | NA | NA | NA | B |
| AJ238447-97 | 1997 | NA | NA | NA | B |
| N4643-TW-98 | 4/1998 | Encephalitis (death) | 1.9 | Throat swab | C |
| N5033-TW-98 | 6/1998 | Encephalitis (death) | 1.6 | Gastric lavage fluid | C |
| N5055-TW-98 | 6/1998 | Encephalitis | 3.0 | Throat swab | C |
| N5062-TW-98 | 6/1998 | Herpangina | 0.9 | Stool | C |
| N5101-TW-98 | 6/1998 | HFMD | 0.8 | Throat swab | B |
| N5157-TW-98 | 6/1998 | HFMD | 6.8 | Throat swab | B |
| N5202-TW-98 | 6/1998 | HFMD | 3.3 | Throat swab | C |
| N5296-TW-98 | 6/1998 | HFMD | 5.1 | Throat swab | C |
| N5385-TW-98 | 7/1998 | HFMD | 1.0 | Throat swab | C |
| H0001-TW-98 | 7/1998 | HFMD, CNS symptoms | 0.7 | Stool | C |
| N5698-TW-98 | 8/1998 | Meningoencephalitis | 1.8 | Throat swab | C |
| N5761-TW-98 | 8/1998 | HFMD, encephalitis | 1.9 | Stool | C |
| N5811-TW-98 | 8/1998 | HFMD | 1.9 | Throat swab | C |
| H0139-TW-98 | 9/1998 | Meningoencephalitis | 1.6 | Throat swab | C |
| N6064-TW-98 | 10/1998 | HFMD | 2.2 | Throat swab | C |
| N6071-TW-98 | 10/1998 | HFMD | 3.6 | Throat swab | C |
| N6108-TW-98 | 10/1998 | HFMD | 0.7 | Stool | C |
| N6128-TW-98 | 10/1998 | Encephalitis | NA | Throat swab | C |
| N6174-TW-98 | 10/1998 | Meningoencephalitis | 1.0 | Throat swab | C |
| N6182-TW-98 | 10/1998 | HFMD, CNS symptoms | 1.1 | Throat swab | C |
| N6252-TW-98 | 10/1998 | HFMD | 2.5 | Throat swab | C |
| N6270-TW-98 | 10/1998 | HFMD | 2.6 | Throat swab | C |
| N6356-TW-98 | 10/1998 | HFMD, encephalitis (death) | 2.6 | Throat swab | C |
| S9086-TW-98 | 10/1998 | Meningitis | 1.6 | Stool | B |
| H0358-TW-98 | 12/1998 | HFMD, encephalitis | 1.6 | Stool | C |
| H0382-TW-98 | 12/1998 | Enterovirus infection | 1.0 | Throat swab | C |
| N7008-TW-99 | 2/1999 | HFMD, CNS symptoms (death) | 2.6 | Stool | B |
| N7129-TW-99 | 2/1999 | HFMD, CNS involvement | 1.0 | Throat swab | B |
| S0463-TW-99 | 11/1999 | Aseptic meningitis | 2.4 | Throat | B |
| S0042-TW-00 | 1/2000 | NA | 0.1 | Stool | B |
| N0118-TW-00 | 1/2000 | NA | 2.4 | Throat swab | B |
| M0167-TW-00 | 3/2000 | HFMD | 4.5 | Throat swab | B |
| M0177-TW-00 | 3/2000 | HFMD | 4.6 | Throat swab | B |
| M0200-TW-00 | 3/2000 | HFMD | 0.8 | Throat swab | B |
| M0201-TW-00 | 3/2000 | HFMD | 1.0 | Throat swab | B |
| M0225-TW-00 | 3/2000 | HFMD | 5.5 | Throat swab | B |
| M0226-TW-00 | 3/2000 | HFMD | 4.1 | Throat swab | B |
| H0148-TW-00 | 3/2000 | HFMD, acute pharyngitis, asthmatic bronchitis | 2.0 | Throat swab | B |
| N1034-TW-00 | 4/2000 | HFMD | 3.5 | Throat swab | B |
| N1059-TW-00 | 4/2000 | HFMD, CNS symptoms | 1.2 | Throat swab | B |
| M0267-TW-00 | 4/2000 | HFMD | 2.7 | Throat swab | B |
| N1554-TW-00 | 5/2000 | CNS symptoms | 1.1 | Throat swab | B |
| N1643-TW-00 | 5/2000 | HFMD, CNS involvement | 0.5 | Stool | B |
| H0218-TW-00 | 5/2000 | Meningoencephalitis | 2.9 | Stool | B |
| N1814-TW-00 | 6/2000 | HFMD, CNS symptoms (death) | 1.1 | Brain stem (autopsy) | B |
| M0485-TW-00 | 6/2000 | NA | 1.9 | Throat swab | B |
| H0444-TW-00 | 7/2000 | Meningoencephalitis (death) | 1.3 | Stool | B |
| H0493-TW-00 | 7/2000 | HFMD, acute gastritis (death) | 1.7 | Throat swab | B |
| M0553-TW-00 | 7/2000 | NA | 3.3 | Throat swab | B |
| H0575-TW-00 | 9/2000 | HFMD with CNS involvement | 1.4 | Stool | B |
| S0296-TW-00 | 9/2000 | HFMD, convulsion, CNS involvement | 1.8 | Stool | B |
| S0309-TW-00 | 9/2000 | NA | 4.7 | Throat swab | B |
| M0652-TW-00 | 9/2000 | HFMD | 2.9 | Throat swab | B |
The EV71 isolates used for phylogenetic analysis were a random sampling from patients with diverse clinical presentations, ranging from uncomplicated HFMD to encephalitis and death. Abbreviations: CNS, central nervous system; NA, not available.
Nucleotide sequence accession numbers.
Sequences reported in this study have been deposited in the GenBank database under accession no. AY055125 to AY055201.
RESULTS
Description of outbreaks.
From 1998 to 2000, a total of 14,901 specimens were received for viral culture in our laboratory, and enteroviruses were isolated from 2,132 specimens (1,892 patients [297, 646, and 949 patients in 1998, 1999, and 2000, respectively]). Two periods of increased incidence of EV71 infections in Taiwan occurred in each of the outbreaks in 1998 and 2000, one between May and June and the other in October. EV71 represented 44.4, 2.0, and 20.5% and CA16 comprised 18.2, 1.7, and 18.0% of enterovirus infections in 1998, 1999, and 2000, respectively. The age distributions and clinical presentations were similar in both the 1998 and 2000 epidemics (data not shown).
Phylogenetic analysis of 5′-UTR sequence.
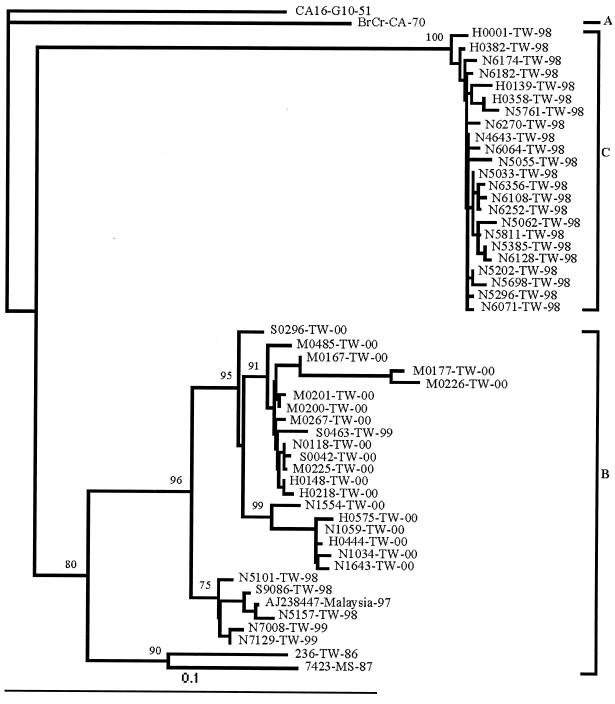
To assess the degree of heterogeneity among EV71 isolates, we performed molecular analyses by sequencing the 5′-UTR (648 nt) regions of EV71 strains obtained from 1998 to 2000. The EV71 isolates examined were from patients with diverse clinical presentations, ranging from uncomplicated HFMD to encephalitis and death (Table 1). Sequences of CA16-G10-51 were included as the outgroup, and sequences of BrCr-CA-70, 7423-MS-87 prototypes, and one Malaysian isolate, AJ238447 (440 nt), from the GenBank database were included for comparison (3). In the analysis of 5′-UTR region, a total of 26, 3, and 19 EV71 strains from 1998, 1999, and 2000, respectively, were analyzed (Fig. 1). Based on 5′-UTR sequences of 75 EV71 strains isolated from the 1998 outbreak, there was a clear segregation into two phylogenically distinct genotypes, B and C, with a ratio between genotype C and B of 10 to 1 (data not shown). In contrast, all strains from 1999 and 2000 tested belonged to genotype B, which clustered with 7423-MS-87, as did one isolate from southern Taiwan in 1986 (236-TW-86). The trees constructed by the maximum-likelihood method were very similar (data not shown).
FIG. 1.
Dendrogram of the 48 outbreak strains, four reference strains from GenBank, and one isolate from 1986, based on 648 nt (nt 98 to 745) of the 5′-UTR gene using the neighbor-joining method with the DNADIST distance measure program (PHYLIP, version 3.573c). The percentage of bootstrap frequency of each branch in tree is indicated. CA16 was included as an outgroup.
Phylogenetic analysis of VP1 sequence.
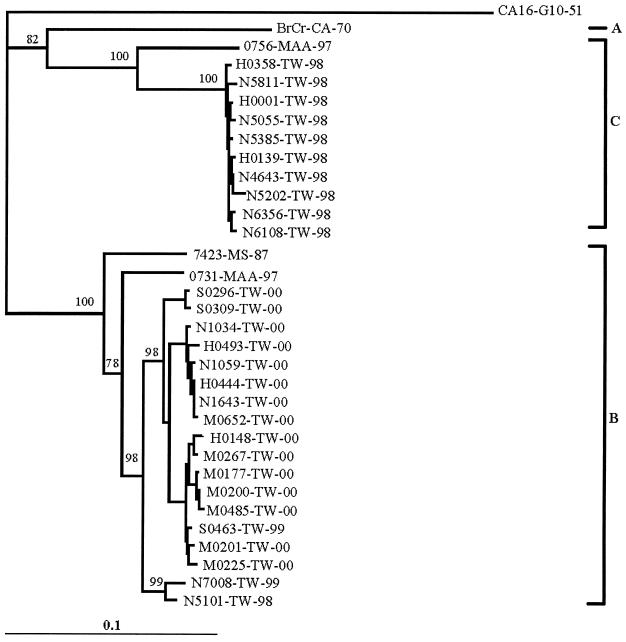
In the analysis of the VP1 region (841 nt), a total of 12, 2, and 19 EV71 strains from 1998, 1999, and 2000, respectively, were used for phylogenetic analysis (Fig. 2). The dendrogram showed only one isolate number to represent isolates with identical sequences: N6270-TW-98 and H0358-TW-98; H0575-TW-00, M0553-TW-00, N1814-TW-00, and H0444-TW-00; and H0218-TW-00 and H0148-TW-00. Sequences of CA16-G10-51, BrCr-CA-70, and 7423-MS-87 prototypes and two Malaysian isolates, 0756-MAA-97 and 0731-MAA-97, from the GenBank database were also included for comparison (3). Similar results were obtained for the phylogenetic analysis of both 5′-UTR and VP1 studies. As in the 5′-UTR analysis, VP1 sequences of 12 EV71 strains isolated from the 1998 outbreak in Taiwan were also grouped into genotypes B and C in the dendrogram, with genotype C being the most prevalent. In contrast, isolates from 1999 and 2000 grouped into the genotype B lineage.
FIG. 2.
Dendrogram of the 28 outbreak strains and five reference strains from GenBank, based on 841 nt (nt 2439 to 3280) of the VP1 gene. The branch length for the outgroup, CA16-G10-51, was reduced by 0.5 in the dendrogram.
Genetic divergence of 5′-UTR and VP1.
Pairwise nucleotide sequence comparison of the 5′-UTR showed that strains in genotype C differed by up to 1.6% within the cluster, by 17.2 to 25.9% from genotype B isolates, and by 18.4 to 19.6% from genotype A BrCr (Table 2). Genotype B isolates in Taiwan from 1998 to 2000 differed from the EV71 prototype MS strain, the 1986 Taiwan isolate (236-TW-86), and the Malaysia AJ238447 isolate by 9.2 to 15.8%, 9.7 to 16.3%, and 0.2 to 8.0%, respectively (Table 2). Pairwise nucleotide sequence comparison of the VP1 showed that strains in genotype C differed by up to 1.1% within the cluster, by 18.5 to 20.7% from genotype B isolates, and by 18.1 to 19.0% from genotype A BrCr (Table 3). The VP1 region of genotype B isolates in Taiwan from 1998 to 2000 differed from the EV71 prototype MS strain, the 1986 Taiwan isolate (236-TW-86), and the Malaysia 0731-MAA isolate by 7.4 to 9.0%, 1.3 to 4.8%, and 5.2 to 7.0%, respectively (Table 3). In contrast to the intrinsic homogeneity in genotype C (about 0 to 1.6% divergence), the genotype B isolates showed a marked divergence (about 0 to 9.5%) within the same cluster when the 5′-UTR was analyzed. More divergence (0 to 4.2%) in genotype B than genotype C (0 to 1.1%) was also seen when the VP1 region was compared (Table 3). Although the VP1 gene was considered to be the more variable region, the divergence between genotype B and C within the VP1 gene was comparable (18.5 to 20.7%) to that observed within the 5′-UTR (17.2 to 25.9%).
TABLE 2.
Pairwise nucleotide sequence comparison of 5′-UTR among EV71 isolatesa
| Isolate | % Difference in 5′-UTR sequence for comparison with:
|
|||||
|---|---|---|---|---|---|---|
| BrCr1970 | MS1987 | Taiwan1986 (236-TW-86) | Malaysia1997 (AJ238447) | Taiwan1998 (genotype C) | Taiwan1998–2000 (genotype B) | |
| BrCr1970 | 0.0 | 12.9 | 14.4 | 10.3 | 18.4–19.6 | 10.0–17.6 |
| MS1987 | 0.0 | 5.8 | 9.2 | 19.3–20.9 | 9.2–15.8 | |
| Taiwan1986 (236-TW-86) | 0.0 | 10.5 | 16.4–17.8 | 9.7–16.3 | ||
| Malaysia1997 (AJ238447) | 0.0 | 17.6–18.7 | 0.2–8.0 | |||
| Taiwan1998 (genotype C) | 0.0–1.6 | 17.2–25.9 | ||||
| Taiwan1998–2000 (genotype B) | 0.0–9.5 | |||||
The data indicate the range of sequence diversities (percent) in the pairwise comparisons among EV71 isolates. The year(s) of isolation is shown as a superscript.
TABLE 3.
Pairwise nucleotide and amino acid sequence comparisons of VP1 among EV71 isolatesa
| % Difference in VP1 sequence for comparison
| ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide
|
Amino acid
|
|||||||||||||
| BrCr1970 | MS1987 | Taiwan1986 (236-TW-86) | Malaysia1997 (0731-MAA) | Malaysia1997 (0756-MAA) | Taiwan1998 (genotype C) | Taiwan1998–2000 (genotype B) | BrCr1970 | MS1987 | Taiwan1986 (236-TW-86) | Malaysia1997 (0731-MAA) | Malaysia1997 (0756-MAA) | Taiwan1998 (genotype C) | Taiwan1998–2000 (genotype B) | |
| BrCr1970 | 0.0 | 20.7 | 21.0 | 20.2 | 20.1 | 18.1–19.0 | 19.9–21.3 | 0.0 | 6.3 | 6.7 | 6.3 | 4.8 | 4.0–5.2 | 6.0–6.7 |
| MS1987 | 0.0 | 8.0 | 6.8 | 19.6 | 19.1–19.9 | 7.4–9.0 | 0.0 | 0.7 | 0.0 | 2.9 | 2.9–3.3 | 0.4–0.7 | ||
| Taiwan1986 (236-TW-86) | 0.0 | 5.3 | 20.4 | 20.2–20.8 | 1.3–4.8 | 0.0 | 0.7 | 3.7 | 3.7–4.0 | 0.4–1.1 | ||||
| Malaysia1997 (0731-MAA) | 0.0 | 19.9 | 19.3–20.4 | 5.2–7.0 | 0.0 | 2.9 | 2.9–3.3 | 0.4–0.7 | ||||||
| Malaysia1997 (0756-MAA) | 0.0 | 8.8–9.4 | 18.8–20.5 | 0.0 | 1.1–2.2 | 2.5–3.7 | ||||||||
| Taiwan1998–2000 (genotype C) | 0.0–1.1 | 18.5–20.7 | 0.0–1.1 | 2.9–4.0 | ||||||||||
| Taiwan1998 (genotype B) | 0.0–4.2 | 0.4–1.1 | ||||||||||||
The data indicate the range of sequence diversities (percent) in the pairwise comparisons among EV71 isolates. The year(s) of isolation is shown as a superscript.
Variability of VP1 amino acid sequences.
The amino acid sequences among isolates from the 1998 and 2000 outbreaks had 96.0 to 97.1% similarity in the VP1 region. Within the same genotype, VP1 amino acid sequences of genotype B or C strains were at least 98.9% identical to one another (Table 3).
DISCUSSION
A recent report by the World Health Organization indicated that during 1997 to 1999, of 1,672 nonpoliomyelitis enterovirus isolations in the United States, EV71 had the lowest incidence, only 2.1% (1). EV71 was occasionally isolated in Taiwan in 1980 and 1986 (4); however, 132 (44.4% of enterovirus isolates in 1998) and 195 (20.5% of enterovirus isolates in 2000) strains of EV71 were isolated in 1998 and 2000, respectively. This result clearly indicated that two outbreaks occurred in Taiwan within the 3-year period. In 1999, EV71 accounted for only 2% of the isolates; however, an outbreak of coxsackievirus A10 contributed to the large number of enteroviruses isolated that year (15).
The apparent genetic distinction of EV71 isolates appeared not to correlate with the severity of the disease. EV71 from total cases were in both B and C genogroups in this study. Both genetic clusters consisted of fatal and mild HFMD cases. Similar results were obtained from studies of EV71 strains from fatal and nonfatal cases of a 1998 outbreak in Taiwan (12). In addition, the Malaysian isolates obtained from patients with uncomplicated HFMD and fatal encephalitis in 1997 were virtually identical in their VP1 regions to those in a report by Brown et al. (3). Thus, a final conclusion cannot be made from existing genetic analyses of the 5′-UTR and VP1 regions in relation to clinical manifestations.
Genetic analyses of the 5′-UTR and VP1 in this study showed consistent grouping of EV71 isolates from the 1998 outbreak into two clusters, genotypes B and C. The results suggest that recombination did not occur between these outbreak strains, an event which can lead to the evolution of highly virulent strains (12). Several studies on EV71 strains from 1998 in Taiwan also support the existence of two cocirculating genotypes (12, 13, 14). Furthermore, this study showed that EV71 isolates in the 2000 outbreak only grouped to one genogroup, genogroup B, a result of a genotype switch which occurred between two major HFMD epidemics within the 3-year study period.
The 5′-UTR of enteroviruses is generally highly conserved; in contrast, VP1 contributes to the most variable region on the surface of virion (7). However, the nucleotide sequence diversities of the 5′-UTR and VP1 regions were similar in this study. Overall these two EV71 genotypes showed 79.3 to 81.5% nucleotide identity and 96.0 to 97.1% amino acid identity in the VP1 region and 74.1 to 82.8% nucleotide identity in the 5′-UTR in these two outbreaks. Despite the wide variation in clinical presentations, this study demonstrates the narrow range of EV71 genetic diversity between two genotypes of EV71 from two large outbreaks that occurred during a 3-year period in Taiwan.
Acknowledgments
This study was supported by National Health Research Institutes grants NHRI-CN-CR8801S and NHRI-CN-CR8804P and by Department of Health grant DOH89-TD-1148.
We thank the members of the National Cheng Kung University Virology Laboratory for their efforts in isolating and typing the viruses analyzed in this study. We thank Po-Fu Chen and Ju-Shan Yu for excellent technical help in this study and David Kiang for critically reviewing the manuscript.
REFERENCES
- 1.Anonymous. 2000. Enterovirus surveillance—United States, 1997–1999. Morb. Mortal. Wkly. Rep. 49:913–916. [PubMed] [Google Scholar]
- 2.Brown, B. A., and M. A. Pallansch. 1995. Complete nucleotide sequence of enterovirus 71 is distinct from poliovirus. Virus Res. 39:195–205. [DOI] [PubMed] [Google Scholar]
- 3.Brown, B. A., M. S. Oberste, J. P. Alexander, Jr., M. L. Kennett, and M. A. Pallansch. 1999. Molecular epidemiology and evolution of enterovirus 71 strains isolated from 1970 to 1998. J. Virol. 73:9969–9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho, M., E. R. Chen, K. H. Hsu, S. J. Twu, K. T. Chen, S. F. Tsai, J. R. Wang, and S. R. Shih. 1999. An epidemic of enterovirus 71 infection in Taiwan. N. Engl. J. Med. 341:929–935. [DOI] [PubMed] [Google Scholar]
- 5.Ho, M. 2000. Enterovirus 71: the virus, its infections and outbreaks. J. Microbiol. Immunol. Infect. 33:205–216. [PubMed] [Google Scholar]
- 6.Hsiung, G. D. 1994. Picornaviridae, p.119–140. In G. D. Hsiung, C. K. Y. Fong, and M. L. Landry (ed.), Hsiung’s diagnostic virology, 4th ed. Yale University Press, New Haven, Conn.
- 7.Kunkel, U., and E. Schreier. 2000. Genetic variability within the VP1 coding region of echovirus type 30 isolates. Arch. Virol. 145:1455–1464. [DOI] [PubMed] [Google Scholar]
- 8.Landry, M. L., S. N. S. Fonseca, S. Cohen, and C. W. Bogue. 1995. Fatal enterovirus type 71 infection: rapid detection and diagnostic pitfalls. Pediatr. Infect. Dis. J. 14:1095–1100. [DOI] [PubMed] [Google Scholar]
- 9.Liu, C. C., H. W. Tseng, S. M. Wang, J. R. Wang, and I. J. Su. 2000. An outbreak of enterovirus 71 in Taiwan, 1998. I. Epidemiologic and clinical manifestations. J. Clin. Virol. 17:23–30. [DOI] [PubMed] [Google Scholar]
- 10.Muir, P., U. Kammerer, K. Korn, M. N. Mulders, T. Poyry, B. Weissbrich, R. Kandolf, G. M. Cleator, and A. M. van Loon. 1998. Molecular typing of enteroviruses: current status and future requirements. Clin. Microbiol. Rev. 11:202–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt, N. J., E. H. Lennette, and H. H. Ho. 1974. An apparently new enterovirus isolated from patients with disease of the central nervous system. J. Infect. Dis. 129:304–309. [DOI] [PubMed] [Google Scholar]
- 12.Shih, S. R., M. S. Ho, K. H. Lin, S. L. Wu, Y. T. Chen, C. N. Wu, T. Y. Lin, L. Y. Chang, K. C. Tsao, H. C. Ning, P. Y. Chang, S. M. Jung, C. Hsueh, and K. S. Chang. 2000. Genetic analysis of enterovirus 71 isolated from fatal and non-fatal cases of hand, foot and mouth disease during an epidemic in Taiwan, 1998. Virus Res. 68:127–136. [DOI] [PubMed] [Google Scholar]
- 13.Shimizu, H., A. Utama, K. Yoshii, H. Yoshida, T. Yoneyama, M. Shinniah, M. A. B. Yusof, Y. Okuno, N. Okabe, S. R. Shih, H. Y. Chen, G. R. Wang, C. L. Kao, K. S. Chang, T. Miyamura, and A. Hagiwara. 1999. Enterovirus 71 from fatal and nonfatal cases of hand, foot and mouth disease epidemics in Malaysia, Japan and Taiwan in 1997-1998. Jpn. J. Infect. Dis. 52:12–15. [PubMed] [Google Scholar]
- 14.Singh, S., V. T. K. Chow, K. P. Chan, A. E. Ling, and C. L. Poh. 2000. RT-PCR, nucleotide, amino acid and phylogenetic analyses of enterovirus type 71 strains from Asia. J. Virol. Methods 88:193–204. [DOI] [PubMed] [Google Scholar]
- 15.Tsai, H. P., P. H. Kuo, C. C. Liu, and J. R. Wang. 2001. Respiratory viral infections among pediatric inpatients and outpatients in Taiwan from 1997 to 1999. J. Clin. Microbiol. 39:111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, J. R., H. P. Tsai, P. F. Chen, Y. J. Lai, J. J. Yan, D. Kiang, K. H. Lin, C. C. Liu, and I. J. Su. 2000. An outbreak of enterovirus 71 infection in Taiwan, 1998. II. Laboratory diagnosis and genetic analysis. J. Clin. Virol. 17:91–99. [DOI] [PubMed] [Google Scholar]