Abstract
Human immunodeficiency virus type 1 (HIV-1) is a difficult target for vaccine development, in part because of its ever-expanding genetic diversity and attendant capacity to escape immunologic recognition. Vaccine efficacy might be improved by maximizing immunogen antigenic similarity to viruses likely to be encountered by vaccinees. To this end, we designed a prototype HIV-1 envelope vaccine using a deduced ancestral state for the env gene. The ancestral state reconstruction method was shown to be >95% accurate by computer simulation and 99.8% accurate when estimating the known inoculum used in an experimental infection study in rhesus macaques. Furthermore, the deduced ancestor gene differed from the set of sequences used to derive the ancestor by an average of 12.3%, while these latter sequences were an average of 17.3% different from each other. A full-length ancestral subtype B HIV-1 env gene was constructed and shown to produce a glycoprotein of 160 kDa that bound and fused with cells expressing the HIV-1 coreceptor CCR5. This Env was also functional in a virus pseudotype assay. When either gp160- or gp140-expressing plasmids and recombinant gp120 were used to immunize rabbits in a DNA prime-protein boost regimen, the artificial gene induced immunoglobulin G antibodies capable of weakly neutralizing heterologous primary HIV-1 strains. The results were similar for rabbits immunized in parallel with a natural isolate, HIV-1 SF162. Further design efforts to better present conserved neutralization determinants are warranted.
Human immunodeficiency virus type 1 (HIV-1) has high replication and mutation rates that permit rapid generation of viruses that can escape immune recognition. Within an infected host, the HIV-1 population diversifies over time, producing mostly defective viruses but nonetheless persisting and accumulating mutations at a rate of up to 1% per year in its env gene (57). HIV sequences sampled from a population of infected individuals recapitulate a star-like phylogeny (1), i.e., most of the variants sampled at the same time are positioned on long branches roughly equidistant from the center of the tree. Thus, any given variant should be approximately twice this distance from any other circulating strain. A primary concern in designing protective AIDS vaccines, then, is the choice of strains likely to best provide protection against the expanding population of HIV-1 variants (46, 47). It is further assumed that key epitopes and conformational determinants must be conserved. A vaccine that is genetically conserved would thus be advantageous.
Several methods for choosing a vaccine candidate on the basis of genetic or protein sequence data have been put forth recently. First, and the approach followed in current clinical trials, is to choose one or a small number of laboratory-grown or primary viral isolates, typically chosen to approximate a “circulating” strain or to simply match the HIV-1 subtype(s) in the targeted population (16, 18, 24, 52). An advantage of this approach is that it typically employs viral genes derived from a viable virus and thus produces antigens likely to adopt “native” conformations. However, as a result of HIV-1 mutational radiation, any given “circulating” strain will be genetically, and presumably antigenically, maximally dissimilar to other non-epidemiologically linked strains likely to be encountered by the vaccinee, with the degree of dissimilarity proportional to the length of time the virus has been circulating within the population. Thus, unless key epitopes are conserved, vaccines based on specific viral isolates are unlikely to be effective against a broad range of circulating viruses. The results of the first phase III AIDS vaccine trial suggest that monomeric envelope proteins that are derived from such isolates are insufficient to provide protective immunity (51), although it remains an open question whether more native presentations of these Env proteins might be effective vaccine components. To enhance the breadth of the elicited immune response, a second approach is to include components from as many diverse HIV-1 isolates as possible in the vaccine, with the intention of inducing multiple responses against divergent viral proteins (18, 29).
A third approach to vaccine strain choice is to build a consensus sequence based on either circulating strains or strains in the HIV database (23). This approach was recently tested using a group M consensus immunogen and shown to elicit broad T-cell responses and weak neutralizing antibody in small animal models (21). A consensus sequence will be genetically closer to circulating strains than any given natural virus isolate, but its sequence may be biased by sampling and may link polymorphisms in combinations not found in any natural or viable virus, thus potentially resulting in inappropriate structural conformations. Consequently, there is a need for new, effective methods of identifying candidate sequences for vaccine development to treat and/or prevent HIV infection (47). To this end, we and others have proposed the use of an HIV population ancestral sequence as a vaccine candidate (20, 23, 36, 37, 45, 49). Such a vaccine might correspond to an ancestor of all known HIV strains, an HIV sequence subtype, or viruses circulating in a given geographic region or risk group. The ancestral viral sequence is reconstructed from a phylogenetic tree describing the historical relationships of sequences sampled from the population of interest and is thus expected to correspond to the most recent common ancestor (MRCA) of the viral strains sampled from the targeted population. It is also likely that such an ancestral sequence will encompass elements conserved within the sampled virus population.
Here we show that a predicted ancestral env sequence for subtype B HIV-1 encodes a functional, CCR5 coreceptor-specific glycoprotein capable of complementing env-defective HIV-1 genomes. We employed DNA vaccines expressing full-length gp160 or the soluble form of gp140 to immunize rabbits and found that these elicited neutralizing antibodies to primary isolates at a low level, comparable to antibodies elicited by a codon-optimized natural isolate (HIV-1 SF162). Boosting with homologous gp120 protein did not significantly enhance the neutralization capacity of the sera.
MATERIALS AND METHODS
Ancestral state simulations.
To assess the accuracy of the ancestral state reconstruction method, we performed the following in silico experiment. Using the program Seq-Gen (55), the evolution of HIV-1B env sequences was simulated, given an ancestral sequence and following the phylogenetic histories depicted in the trees in Fig. 1a, below, and the model of evolution provided in Table S1 in the supplemental material. One hundred replicate data sets were generated for both tree shapes. We then used PAUP* (63) to generate a maximum-likelihood tree and an ancestral sequence for all 200 data sets (100 simulations on both the star- and caterpillar-shaped phylogenies). To be conservative, the simulations were performed on trees with very long external branches (having a genetic distance of 20%), and the caterpillar trees had internal branch lengths of 1% genetic distance.
FIG. 1.
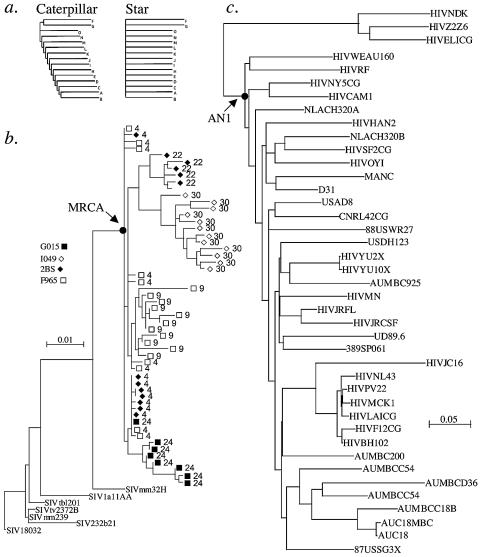
Phylogenetic relationships. (a) Idealized phylogenetic trees with caterpillar (left) and star (right) shapes. (b) Sequences taken from macaques inoculated with molecularly cloned virus SIVmac251(BK28). The legend within the figure indicates the animal identifier, and at each tip of the tree the identifier is followed by the time, in months, from the time of virus inoculation to sampling. (c) Thirty-eight HIV-1 subtype B env sequences and three subtype D (outgroup) sequences used to root the subtype B sequences (see Table 2). The subtype B sequences were from the following nine countries, representing a broad sample of subtype B diversity: Australia, n = 8; China, 1; France, 6; Gabon, 1; Germany, 2; Great Britain, 2; The Netherlands, 2; Spain, 1; United States, 15. The node corresponding to the position of the ancestral, most recent common ancestor is indicated with an arrow in panels b and c.
Ancestral state reconstruction of the SIVmacBK28 env sequence.
SIVmac sequences obtained from a series of experimental infections of rhesus macaques (15) were used to reconstruct an ancestral sequence to compare to the plasmid-derived simian immunodeficiency virus (SIV) clone used to inoculate these monkeys. A region of env from position 8265 to 8827 of the SIVmacBK28 genome was amplified by nested PCR from uncultured peripheral blood mononuclear cell (PBMC) DNA from the SIV-infected macaques. First-round primers were UP-3, positions (SIVMM239 coordinates [8]) 8117 to 8130, AGACTGCAGATGTGAAGAGGTACAC, and PEXTM6 (8977 to 8953), GGATCTGGTATGCTCATAGCAA. Second-round primers were PEXTM7 (8265 to 8286), GATACTGCAGCAACAGCAACAGCTG, and UP-5 (8827 to 8810), GCAAAGCTTCTCTGGTTGGCAGTG. Amplified products were then cloned and sequenced as described previously (14). The GenBank accession numbers for these sequences are AY169007 to AY169163. Methods for SIVmacBK28 ancestor reconstruction were identical to those outlined below. The model of evolution (Table S1) and the reconstructed sequence (Fig. S1) are provided in material available at the website http://ubik.mullins.microbiol.washington.edu/HIV/Doria-Rose2005/.
Ancestral state reconstruction of an HIV-1 B env sequence.
Thirty-eight HIV-1 subtype B env genes were selected from GenBank, with three additional sequences from clade D used as an outgroup for rooting (35) (see Fig. 1c, below, and also Table S2 in the supplemental material). Sequences were aligned using CLUSTALW (64), and the alignment was refined using GDE (58). Inferred amino acid sequences were used to guide the introduction of alignment gaps such that they were inserted between codons. This alignment was then modified for phylogenetic analysis such that regions that could not be unambiguously aligned were removed (38).
An appropriate evolutionary model for phylogeny and ancestral state reconstructions was selected using the Akaike information criterion as implemented in Modeltest 3.0 (53) (Table S1). Evolutionary trees were inferred using maximum-likelihood estimation methods as implemented in PAUP* version 4.0b10 (63). Ten different subtree-pruning-regrafting heuristic searches were performed using a different random-addition order each time and using a neighbor-joining tree of the subtype B sequences as a backbone constraint. The ancestral nucleotide sequence was inferred as the sequence at the basal node of the clade using the inferred phylogenies. The nucleotide at each site of the ancestral sequence had the highest likelihood, integrated over all state assignments at other nodes. The nucleotide sequence is provided in material posted on the website http://ubik.mullins.microbiol.washington.edu/HIV/Doria-Rose2005/ (Fig. S2).
To predict the amino acid sequences for the complete gp160, the inferred ancestral sequences were visually aligned to the complete alignment prior to gap stripping and then translated using GDE (58). Since the highly variable regions were deleted as complete codon triplets, the translations into amino acids were in the correct reading frame and codons were properly maintained. The ancestral amino acid sequences for the regions deleted from the gap-stripped alignment were predicted visually and refined using parsimony-based sequence reconstruction for these sites using the program MacClade (42). The complete gp160 amino acid sequences were converted to DNA sequences optimized for expression in human cells using the BACKTRANSLATE program of the Wisconsin Sequence Analysis Package (GCG), version 10, and a human gene codon usage table from the Codon Usage database (http://www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species = Homo+sapiens+[gbpri]) (48).
The AN1-envB gene was chemically synthesized commercially by Midland Certified Reagent Company (Midland, TX). It was sequence verified and inserted into a mammalian expression vector, pVR1012 (27), to produce the plasmid pVR1012-AN1env. To generate a gp140 expression plasmid, pVR1012-AN1env was subjected to site-directed mutagenesis (QuikChange kit; Stratagene) to introduce two stop codons after the codon for amino acid lysine 711 (nucleotides 2134 to 2139). Mutagenesis was verified by sequencing.
Fusion assay.
GHOST-CCR5 or GHOST-CXCR4 cells (gift of S. Zolla-Pazner) (10) were seeded in two-chamber culture slides (Falcon, Franklin Lakes, NJ) at 5 × 104 cells/well and transfected the next day with 0.5 μg env plasmid using Fugene-6 (Roche, Indianapolis, IN). Transfected cells were allowed to fuse for 2 days and then were fixed and stained for immunofluorescence using the monoclonal antibody b12 (gift of D. Burton) and 1:2,500 fluorescein isothiocyanate-conjugated goat anti-human immunoglobulin G (IgG; Cappel). The presence of large multinucleated cells indicated that cell-cell fusion had occurred.
Pseudotype assay.
293T cells were transfected in 75-cm flasks with 4 μg env plasmid, 4 μg pNL43lucR-E- (28), and 12 μl Fugene-6. Two days later, medium was collected and spun at 3,000 × g, and the resulting cell-free pseudovirus was stored at −80°C. Pseudovirus was used to infect TZM-bl cells (12) in the presence of 7.5 μg/ml DEAE-dextran. Two days later, infection was measured by luciferase expression using a commercial kit (Bright-Glo; Promega, Madison, WI) and an EG&G Berthold MicroLumat Plus luminometer. Alternatively, 293T cells were transfected with env plasmids, and Q23Δenv (40) (gift of J. Overbaugh) and pseudovirus were used to infect cMAGI cells (11) (gift of J. Overbaugh) in the presence of 10 μg/ml DEAE-dextran. After 2 days, cells were fixed and stained for β-galactosidase expression (indicating infection).
Western blot analysis.
293T cells were transfected in six-well dishes with 1 μg env plasmid and 6 μl Fugene-6, with medium and cell lysates collected 48 h later. Twenty-one microliters of each sample was used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a polyvinylidene difluoride membrane, probed with heat-inactivated pooled human HIV+ serum (gift of L. M. Frenkel), and horseradish peroxidase-conjugated goat anti-human IgG (Sigma, St. Louis, MO) and then developed with the color substrate 3-amino-9-ethylcarbazole.
Recombinant protein production.
Recombinant gp120 from HIV-1 SF162 was prepared as described elsewhere (3). AN1-EnvB rgp120 was derived from the synthetic gp160 gene by site-directed mutagenesis to place double termination codons immediately after the gp120 reading frame. The BstEII-NotI fragment of the gene, containing most of the gp120 and all of the gp41 reading frame, was excised and cloned into the cognate region of plasmid pSF128, which consists of the pcDNA3.1+/Zeo expression vector (Invitrogen, Carlsbad, CA) and a synthetic gene for HIV (Ba-L) gp160, which in HIV-1 signal peptide was replaced with that for human CD5. Expression of this plasmid in eukaryotic cells is regulated by the cytomegalovirus early promoter and the bovine growth hormone polyadenylation site. The resultant clone, pSF182, encodes a protein with the CD5 signal peptide, which was removed during expression, and a gp120 in which the first 10 amino acid residues are derived from HIV (Ba-L) and the remainder are from AN1-Env B.
pSF128 was purified using an Endofree pDNA kit (QIAGEN) and transfected into A293 cells using Fugene-6 (Roche) at a ratio of 6 μl of Fugene to 2 μg DNA. Cells were selected with zeocin at 200 μg/ml and 10% fetal bovine serum for 5 days and then transferred to T-75 flasks. When foci appeared, weaning to serum-free medium was initiated using an increasing proportion of 293-SFM II medium from Invitrogen and gradually reducing the zeocin concentration. Cells were considered weaned when they were able to grow for three passages in 100% 293-SFM II-20 μg/ml zeocin. Large quantities of 293 cells expressing AN1-Env B gp120 were then grown in roller bottles in 100% 293-SFM II-20 μg/ml of zeocin. The culture supernatant was subjected to lectin affinity chromatography using a plant-derived lectin isolated from snow drop (Galanthus nivalis) bulb. The affinity matrix, in which the lectin was covalently linked to agarose beads, was washed thoroughly to remove nonspecifically bound proteins. The bound protein was then eluted with alpha-methyl-d-mannopyranoside and dialyzed against phosphate-buffered saline (PBS). Production of gp120 was monitored by enzyme-linked immunosorbent assay (ELISA) at all stages of production, and purity was verified by SDS-PAGE. The SF162 protein was prepared as previously described (60).
Rabbit immunizations.
New Zealand White rabbits were vaccinated by Gene Gun (Bio-Rad, Hercules, CA) (41). Eighteen immunizations of 2 μg DNA each were given for each dose in clusters of three nonoverlapping positions at six sites (back, legs, and abdomen); rabbits were shaved in these areas prior to immunization. Animals were DNA vaccinated at weeks 1, 5, 10, 19, 23, and 46. Protein boosts, consisting of 44 μg recombinant gp120 mixed with an equal volume of incomplete Freund's adjuvant, were injected intramuscularly at weeks 68 and 78. Blood was collected the day before and 2 weeks after each immunization. Animals were housed at R+R Rabbitry, Marysville, WA, and procedures followed IACUC-approved protocols.
IgG purification.
Individual samples (800 μl) of rabbit sera, collected at weeks 5 and 80 of vaccination, were heat inactivated at 56°C for 1 hour prior to purification. The samples were each mixed with 800 μl of 4 Fast Flow Protein A Sepharose (Pharmacia) slurry and incubated for 1 hour at room temperature. Resin was collected and washed three times in PBS using two Handee spin cups (Pierce, Rockford, IL). IgG was eluted in 400 μl 0.1 M glycine-HCl, pH 2.7, for 5 minutes; the eluate was collected immediately by centrifugation and neutralized with 40 μl of 3 M Tris, pH 9.5. Samples were then equilibrated to PBS by dialysis using Slide-a-lyzer cassettes, with a molecular mass cutoff of 10,000 Da (Pierce, Rockford, IL). Thirty-five to 60% of the binding activity was recovered as determined by ELISA. Purity was ∼90%, as estimated by Coomassie staining of SDS-PAGE gels.
Antibody assays.
Binding antibody responses to HIV-1 envelope antigens were measured by ELISA as described previously (13). Briefly, Immunosorp plates (Nalge Nunc, Rochester, NY) were coated with 2 μg/ml HIV-1 SF162 rgp120. Diluted plasma was incubated for 1 hour on the plates and detected with horseradish peroxidase-conjugated anti-rabbit IgG (Kirkegaard and Perry, Gaithersburg, MD). The endpoint titer was defined as the reciprocal of the highest dilution that gave an absorbance value more than twice that of preimmune serum at the corresponding dilution. Values were standardized to a positive control rabbit serum sample (kind gift of L. Stamatatos) that was included with every assay.
Neutralization assays: cMAGI assay.
Assays were performed as described in reference 13. Briefly, serial dilutions of sera were incubated with virus for 1 hour and then added to duplicate wells of cMAGI cells (11). After 2 days, cells were fixed and stained for β-galactosidase expression (indicating infection). The percent neutralization at a given titer was calculated by using the equation [(Vo − Vn)/Vo] × 100, where Vn is the number of infected cells in the virus plus antibody wells and Vo is the number of positive cells in virus-alone wells. We graphed percent neutralization versus dilution for each sample and matched prebleed sera. Sera that failed to achieve 80% neutralization or were not distinguishable from prebleed samples were scored as negative. Subtraction of prebleed activity at each dilution resulted in the same value assignments for titers. Titers were normalized to that of a standard HIV+ human serum pool (gift of L. M. Frenkel) that was included on each assay plate. Viruses HIV-1 SF162, 92US657 (also called 301657), 92TH014, 92HT593, 91US056, and 93IN101 were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health, and were propagated in anonymous donor human PBMCs.
Neutralization assays: luciferase reduction assay (M7-luc assay).
Neutralization was measured as a function of the reduction in luciferase reporter gene expression after multiple rounds of virus replication in 5.25.EGFP.Luc.M7 cells (43). Briefly, 5,000 50% tissue culture infective doses of virus was mixed with serial dilutions of serum in triplicate and incubated for 1 h at 37°C in 96-well microculture plates. A total of 5 × 104 5.25.EGFP.Luc.M7 cells (gift of N. Landau), suspended in RPMI with 12% heat-inactivated fetal bovine serum and 10 μg/ml DEAE-dextran, were added to each well and incubated for 3 to 4 days. These cells possess Tat-responsive reporter genes for luciferase (Luc) and green fluorescence protein (GFP). Luciferase was measured using a commercial kit (Bright-Glo; Promega, Madison, WI) and a Victor-2 luminometer.
Neutralization titers were calculated as the dilution at which the relative luminescence units (RLU) were 50% that of virus-only wells. For the large panel of viruses, sera from week 80 (post-eighth vaccination) were assayed at a 1:10 dilution in triplicate, and the percent reduction in RLU was calculated relative to the corresponding week-5 serum sample (shown to be equivalent to preimmunization sera in these assays) for each rabbit. A reduction of 50% is considered significant in this assay. Virus stocks for both assays were generated in phytohemagglutinin-stimulated normal human PBMCs and made cell free by 0.45-μm filtration prior to storage at −80°C.
RESULTS
Phylogenetic method validation.
We first evaluated the ability of likelihood-based phylogenetic methods to accurately predict ancestral sequences using both a simulation and an experimental approach. HIV-1 is widely considered to have a star-like phylogeny, with all external branches radiating from the same central point on the tree (1). However, there is at least some level of substructure in the phylogeny (i.e., some short internal branches at the base of the tree). In terms of tree space, the realized HIV-1 tree (Fig. 1) is between a true star and what can be called a caterpillar tree (i.e., long external branches with short internal branches). To estimate the accuracy of ancestral state reconstruction, we performed 100 simulations with a known ancestor at each of the two extremes of the star-caterpillar continuum. For the caterpillar tree, the MRCA was estimated with 95.4% accuracy. For the star phylogeny, the MRCA was estimated with 98.2% accuracy. Since the true HIV phylogeny falls somewhere between the caterpillar and star structures, we estimate our MRCA reconstruction accuracy to be in the range of 95 to 98%.
We also evaluated viral gene sequences taken from four rhesus macaques infected with a molecularly defined strain of SIV, SIVmac251-BK28 (34), in an effort to assess the reconstruction of the infecting viral sequence. From a phylogenetic tree (Fig. 1b) of 1.1-kb env gene fragments taken between 1 and 3 years postinfection (15), 99.8% of the infecting viral sequence (373 of 376 amino acids), including 98.2% of the 170 variable sites, were accurately predicted (see Fig. S1 at http://ubik.mullins.microbiol.washington.edu/HIV/Doria-Rose2005/).
Derivation of subtype B ancestor.
Using the same procedures, we derived an ancestral nucleotide sequence of the envelope gene of HIV-1 subtype B, the most extensively evaluated HIV-1 subtype to date. We used the sequences listed in Table S2 in the supplemental material and the phylogeny shown in Fig. 1c. This ancestral sequence produced an open reading frame encoding a complete, 884-amino-acid gp160 gene product (AN1-EnvB). The nucleotide sequence is available as Fig. S2 in the supplemental material. The amino acid sequence is shown in Fig. 2, along with potential N-linked glycosylation sites (NXS/T sequons). The amino acid distances between the ancestral sequence and the natural subtype B strains used to estimate it were 12.3% on average (range, 8.0 to 21.0%), while these sequences were 17.3% different from each other (range, 13.3 to 23.2%). At 884 amino acids, the encoded protein is long relative to most sequences (average, 858 amino acids); much of the additional length is in the variable loops. Also unusual is the number of NXS/T sequons: the median for gp120 is 25 (33, 67), while AN1-EnvB has 31 in gp120. Based on V3 sequence features, AN1-EnvB is also predicted to use CCR5 as a coreceptor (30).
FIG. 2.
Amino acid sequence of An1-EnvB. Potential N-linked glycosylation sites are shown in bold.
In vitro validation of ancestor protein expression.
We next sought to evaluate the functional characteristics of the ancestral env sequence. We reengineered the deduced sequence to reflect the dominant patterns of human codon usage (48) without changing any of the encoded amino acids (codon optimized) and then chemically synthesized the gene and cloned it into a mammalian expression vector, pVR1012 (27). Transfection of this plasmid into COS-7 or 293T cells resulted in the production of high levels of viral protein gp160 and its cleavage products, gp120 and gp41. A truncated gp140 form, which is expected to be secreted but retain the oligomeric properties of gp160, was also generated, expressed, and shown to bind soluble CD4 in an ELISA, as did gp160 (data not shown). Western blot analysis of 293T cells transfected with either the gp160 or gp140 form showed that the expressed proteins were of the predicted sizes and were recognized by anti-HIV antibodies (Fig. 3a). Cell lysates contained both gp140 and gp160, while only the gp140 form appeared in the medium, showing, as expected, that it was secreted. Lysates of cells transfected with gp160 showed bands for gp41, suggesting that Env was cleaved to its mature form intracellularly. The cleavage was not complete, as full-length gp160 was also visible. Little, if any cleavage to gp120 was noted in the gp140 samples. The apparent molecular weight and somewhat diffuse nature of the gp140 and gp160 bands suggest that the AN1-EnvB proteins were highly glycosylated. Western blot and quantitative ELISA showed that levels of the gp160 form of AN1-EnvB gp160 were comparable to levels detected following transfection of the widely employed, humanized Env-gp160 gene of SHIV89.6P (4) and higher than that of nonoptimized HIV-1 89.6 Env (Fig. 3a). Expression of the gp140 form was similar to that of a codon-optimized gp140 from HIV-1 SF162 (3) (data not shown).
FIG. 3.
Analysis of AN1-Env B expression. (a) 293T cells were transfected with the indicated plasmid. After 48 h, medium and cell lysates were collected. Samples were analyzed by Western blotting and probed with human HIV+ sera. Lanes: 1, culture medium and AN1-EnvB gp160; 2, culture medium and AN1-EnvB gp140; 3, culture medium and HIV-1 89.6 gp160; 4, culture medium and empty vector; 5, lysate and AN1-EnvB gp160; 6, lysate and AN1-EnvB gp140; 7, lysate and HIV-1 89.6 gp160; 8, lysate and empty vector. (b and c) GHOST-CCR5 (b) or GHOST-CXCR4 cells (c) (10) were transfected with AN1-EnvB gp160. After 2 days they were fixed and stained for immunofluorescence using anti-HIV monoclonal antibody b12. Large multinucleated cells indicate cell-cell fusion mediated by Env.
Expression of gp160 in transfected cells was confirmed by an immunofluorescence assay (Fig. 3b and c). Cell-cell fusion occurred when GHOST-CCR5 cells (10) expressing the HIV-1 receptors CD4 and CCR5 were transfected with AN1-EnvB gp160 (Fig. 3b). In contrast, no fusion occurred when GHOST-CXCR4 cells (10) expressing CD4 plus CXCR4 were transfected (Fig. 3c), although Env was expressed in these cells. These results indicate that AN1-EnvB used only CCR5, as predicted by its sequence, and that it was folded properly and fully functional for directing membrane fusion. To confirm the functionality of AN1-EnvB, we used pVR1012AN1env (encoding full-length gp160) to complement the env-defective genome Q23Δenv (40). The resulting pseudotyped virus particles were able to infect permissive cMAGI cells (data not shown). We also used the same plasmid to complement the env-defective genome encoded by pNL4-3lucR-E- (28), and the resulting pseudovirus was infectious on TZM-bl cells (data not shown).
Immunogenicity of AN1-EnvB.
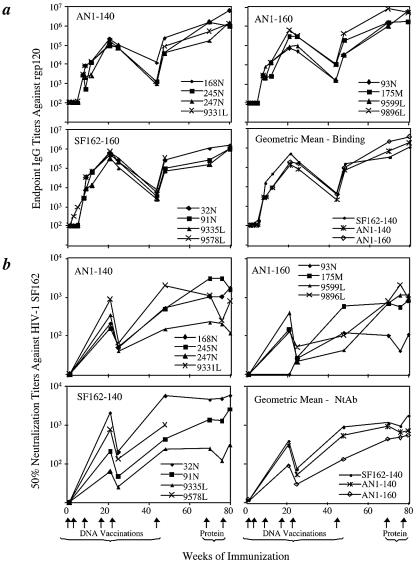
To assess the humoral immune response elicited by An1-EnvB, we immunized mice with pVR1012AN1env using a Gene Gun and found binding of antibody to HIV-1 gp120 at titers up to 1:300 (data not shown). We then immunized rabbits with DNA encoding gp160 and gp140 versions of the protein via the Gene Gun (19, 41) followed by boosting with purified recombinant AN1-EnvB gp120 protein produced in CHO cells. As controls, we immunized an additional group of rabbits with DNA encoding a codon-optimized Env gp140 gene from a primary HIV-1 isolate, HIV-1 SF162, and recombinant HIV-1 SF162 gp120 (rgp120) (3), or with the empty vector and adjuvant. All envelope plasmids elicited envelope-specific binding antibody, with endpoint titers in rabbits of up to 1:400,000 (Fig. 4a). Binding antibody titers increased after protein boosting (roughly eightfold when geometric mean titers of all animals at each time point are compared).
FIG. 4.
Rabbit humoral immune responses to vaccination. (a) Reciprocal endpoint binding titers against rgp120 from HIV-1 SF162. Binding of IgG was measured by ELISA. Arrows indicate the times of vaccinations. (b) Neutralization of HIV-1 SF162 by sera from immunized rabbits. Neutralization was measured in the M7-luc assay. Titers are the reciprocal dilutions able to inhibit viral infectivity by 50%. Arrows at the bottom of the figure indicate the times of vaccinations. NtAb, neutralizing antibody.
Virus neutralization.
We next measured antibody neutralization, first using the cMAGI assay (11, 32) with five heterologous subtype B, primary R5 HIV-1 isolates, SF162, 92US657, 92TH014, 91US056, and 92HT593, and one subtype C primary isolate, 93IN101 (Table 1). As a positive control, we included HIV+ sera pooled from several HIV-infected patients from the Seattle, Wash., area. All isolates tested were neutralized by this latter pool, but with differing sensitivities. Sera from the rabbits were analyzed after four or five immunizations with DNA against the same panel of isolates and compared with the preimmunization sera. After four DNA immunizations, all four gp140-SF162-immunized rabbit sera had neutralizing activity with 80% neutralization titers of >8 against the homologous virus. AN1-immunized rabbits had neutralization detected at this time point only using lower cutoff criteria (50 to 75%) (data not shown). After five immunizations, neutralization was assessed against the full virus panel. At this time, sera from only one SF162-immunized animal neutralized homologous virus, and one other rabbit had neutralizing activity against a heterologous virus (92US657). However, sera from this time point had atypically low neutralizing activity (Fig. 4b). In contrast, all eight rabbits immunized with AN1-EnvB genes neutralized isolate 92TH014 and four of eight neutralized 92US657. No other neutralizing activity was observed. Hence, while not very broad, more heterologous neutralizing activity was evident in rabbits that received AN1-EnvB compared to those immunized with SF162 Env. No rabbit sera neutralized 92HT593, 91US056, or the clade C virus 93IN101.
TABLE 1.
Neutralization of HIV-1 by sera from immunized rabbits in the cMAGI assay
| Rabbit no. | DNA immunogen | Neutralization titera after:
|
||||||
|---|---|---|---|---|---|---|---|---|
| Fourth dose | Fifth dose
|
|||||||
| SF162 clade B | SF162 clade B | 92US657 clade B | 92TH014 clade B | 92HT593 clade B | 91US056 clade B | 93IN101 clade C | ||
| 32N | SF162-gp140 | 15b | <8 | 10 | <8 | <4 | <4 | <8 |
| 91N | SF162-gp140 | 18 | <8 | <8 | <8 | <4 | <4 | <8 |
| 9335L | SF162-gp140 | 9 | <8 | <8 | <8 | <4 | <4 | <8 |
| 9578L | SF162-gp140 | 13 | 9 | <8 | NTc | <4 | <4 | <8 |
| 168N | AN1-gp140 | <8 | <8 | 12 | 16 | <4 | <4 | <8 |
| 245N | AN1-gp140 | <8 | <8 | 16 | 48 | <4 | <4 | <8 |
| 247N | AN1-gp140 | <8 | <8 | <8 | 20 | <4 | <4 | <8 |
| 9331L | AN1-gp140 | <8 | <8 | 13 | 36 | <4 | <4 | <8 |
| 93N | AN1-gp160 | <8 | <8 | 8 | 16 | <4 | <4 | <8 |
| 175N | AN1-gp160 | <8 | <8 | <8 | 10 | <4 | <4 | <8 |
| 9599L | AN1-gp160 | <8 | <8 | <8 | 24 | <4 | <4 | <8 |
| 9896L | AN1-gp160 | <8 | <8 | <8 | 16 | <4 | <4 | <8 |
| 8848L | Control | <8 | <8 | <8 | <8 | <4 | <4 | <8 |
| 9827L | Control | <8 | <8 | <8 | <8 | <4 | <4 | <8 |
| Control | HIV+ serum pool | NT | 500 | 300 | 100 | 120 | 300 | 200 |
Titer is the reciprocal of the last dilution that gave 80% reduction in infectivity (except for 92TH014, where it was 90%). Sera were obtained 2 weeks following the immunization dose indicated, at 21 and 25 weeks, respectively.
Numbers in bold are greater than the cutoff.
NT, not tested.
Whereas the above sera had low or no neutralization of HIV-1 SF162 in the cMAGI assay, activity was usually detected by a second neutralization assay that targets the luciferase-expressing 5.25.EGFP.Luc.M7 cells (7), herein referred to as the M7-luc assay. Sera were tested for neutralization activity against HIV-1 SF162 2 weeks after each immunization with DNA or with a gp120 subunit protein boost. Titers were calculated as the dilution that gave 50% reduction of luciferase relative to wells with no antibody. Significant titers of neutralizing antibodies against HIV-1 SF162 were found in all Env-immunized rabbits (Fig. 4b). M7-luc neutralizing activity generally increased over the course of vaccination and reached a mean titer of 800 to 1,000 in the rabbits immunized with DNA expressing gp140 after six immunizations (Fig. 4b). Neutralization titers were boosted roughly twofold with gp120 protein immunization at week 67 and sustained with a second protein boost at 78 weeks. Titers of neutralizing antibody against HIV-1 SF162 were not significantly different between any of the groups at weeks 21, 25, 48, or 80 (Mann-Whitney U test).
Using the M7-luc assay, sera from all rabbits were tested after the final DNA vaccination and after each protein boost against a panel of heterologous B, C, and E primary isolates. In this assay, all sera were diluted 1:10, results were recorded as percent reduction of luciferase compared to week 5 sera, and values greater than 50% were considered significant. Serum from one control rabbit scored at the 51% level, but other samples from controls were negative. Week 5 samples were used as controls because of a scarcity of preimmunization samples; these had no or minimal Env binding activity, and background neutralization titers were very close to those of corresponding preimmunization sera for the same animals (data not shown). Data from samples taken after the last protein boost, reported as %RI, are shown in Table 2. Sera from three of three rabbits that received HIV-1 SF162 immunogens (the fourth died prior to protein boosting) could neutralize Bx08, two were active against Ba-L, and one of those was also active against QH0692. Sera from rabbits immunized with AN1-EnvB gp160 or gp140 also had activity against these three viral isolates. Three of eight rabbit sera neutralized Bx08, and two of those sera also had activity against Ba-L and QH0692. Activity against BaL and Bx08 was also detected at weeks 48 and 70 in several animals (not shown). No rabbit sera neutralized clade B isolate 6101, clade C isolate S080, or clade E isolate CM244.
TABLE 2.
Neutralization of HIV-1 by rabbit serum and purified IgG at week 80, post-eighth vaccination, in the M7-luc assay
| Animal | Envelope immunogen
|
SF162 clade B
|
Bal clade B
|
Bx08 clade B
|
QH0692 clade B
|
JR-FL clade B
|
S080 clade C
|
CM244 clade E
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA | rgp120 | Titera | IC50b | %RIc | Titer | IC50 | %RI | Titer | IC50 | %RI | Titer | IC50 | %RI | Titer | IC50 | %RI | Titer | IC50 | %RI | Titer | IC50 | |
| 32N | SF162-gp140 | SF162 | 4,176 | 1.6 | —d | 38 | — | 65 | 456 | 14 | — | — | — | — | — | — | — | — | — | — | — | — |
| 91N | SF162-gp140 | SF162 | 3,728 | 6.7 | 62 | 46 | — | 68 | 867 | 11 | 53 | 27 | — | — | — | — | — | 25 | — | — | — | — |
| 9335L | SF162-gp140 | SF162 | 494 | 61.6 | 66 | 38 | — | 66 | 390 | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 9578L | SF162-gp140 | SF162 | — | — | — | — | — | — | ||||||||||||||
| 168N | AN1-gp140 | AN1-Env B | 1,593 | 6.6 | 69 | 39 | — | 73 | 572 | 123 | 65 | — | — | — | — | — | — | — | — | — | — | — |
| 245N | AN1-gp140 | AN1-Env B | — | — | — | — | — | — | ||||||||||||||
| 247N | AN1-gp140 | AN1-Env B | — | — | — | — | — | — | ||||||||||||||
| 9331L | AN1-gp140 | AN1-Env B | — | — | — | — | — | — | ||||||||||||||
| 93N | AN1-gp160 | AN1-Env B | — | — | — | — | — | — | ||||||||||||||
| 175M | AN1-gp160 | AN1-Env B | 623 | 13.9 | 71 | 40 | — | 61 | 508 | — | 62 | — | — | — | 24 | — | — | — | — | — | 34 | — |
| 9599L | AN1-gp160 | AN1-Env B | 2,481 | 5.1 | — | 34 | — | 59 | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 9896L | AN1-gp160 | AN1-Env B | — | — | — | — | — | — | ||||||||||||||
| 8848L | Control | Adjuvant | — | — | — | — | — | — | ||||||||||||||
| 9827L | Control | Adjuvant | 51 | — | — | — | — | — | ||||||||||||||
Titer, 50% neutralization titers of selected serum samples. The limit of detection was <20, the lowest dilution tested. Titers are the reciprocal serum dilution at which RLUs were reduced 50% relative to no serum. No neutralization was detected in this assay against HIV-1 clade B strain 6101.
IC50 values are the purified IgG concentration, in μg/ml, at which RLUs were reduced 50% relative to no sample. The limit of detection was the highest IgG concentration tested and ranged from 235 to 510 μg/ml.
%RI, percent reduction in infectivity in relative luciferase units, compared to the corresponding week 5 sample. All sera were diluted 1:10. No neutralization was detected in this assay against HIV-1 clade B strains JR-FL and 6101, clade C S080, and clade E CM244.
—, no neutralization detected at 50% level. Blank cells indicate that the sample was not tested.
To confirm that the observed virus-neutralizing activity was due to antibodies and not to nonspecific inhibition by other components of the serum, we purified IgG from sera of six of the rabbits and used these in our assays. Three rabbits had received AN1-EnvB immunogens, and three had received the HIV-1 SF162 immunogens. IgG and the corresponding sera (Table 2) were tested in the M7-luc system. For this subset of samples, we obtained titers for 50% neutralization of each isolate, allowing more precise quantitation of activity in the serum. All week 80 IgG samples had strong neutralizing activity against HIV-1 SF162, with 50% inhibitory concentration (IC50) values from 1.6 μg/ml to 61 μg/ml; likewise, the sera from which they were purified had high titers against this virus, from 1:494 up to 1:4,176. The sera with the highest titers yielded the IgG samples with the lowest IC50 values, as expected. IgG from two of the six also had strong activity, and one showed weak activity, against Bx08. In this set of assays, no significant neutralization of isolate 6101 was found. Weak neutralization of BaL was noted in all serum samples, and sporadic low-level activity against three other isolates was noted in several sera, but not in the corresponding IgG; this may be due to the limits of detection of the assay. IgG samples from week 5 had no detectable activity at the highest concentrations tested. These data demonstrate that the observed neutralizing activity of the rabbit sera is attributable in large part to IgG.
Comparing data across the two assays, we found that five of the eight primary heterologous clade B viruses tested were weakly neutralized by AN1-Env B sera. The clade B viral amino acid sequences used here were, on average, 9% distant from AN1-EnvB and 13% from HIV-1 SF162. The non-B amino acid sequences were on average 24% and 22% divergent from AN1-EnvB and SF162, respectively. Within the clade B group, neutralization of individual viruses by vaccinated rabbit sera did not correlate with total sequence distance between the vaccine immunogen and the tested virus. The number of viruses neutralized by serum from each rabbit was not statistically different between the AN1-EnvB- and SF162-immunized rabbit groups (Mann-Whitney U test). Overall, the neutralizing antibodies elicited both by AN1-EnvB and by HIV-1 SF162 immunogens had modest potencies and breadth.
DISCUSSION
Artificial HIV gene sequences have the potential to be more effective vaccines and reagents than natural isolates, since they can be engineered to contain more of the conserved features and epitopes than found in any one natural isolate. We have put this concept into practice by predicting, synthesizing, and evaluating an ancestor of the HIV-1 clade B env gene. We found that the AN1-EnvB protein can be stably expressed in mammalian cell lines, is glycosylated, binds CD4, directs cell-cell fusion, and complements env-defective genomes in a virus infectivity assay. AN1-EnvB uses CCR5 but not CXCR4 as a coreceptor, as predicted by patterns in its sequence and confirmed by cell fusion assays in GHOST cell lines. This functional protein elicited high titers of Env-binding antibodies in vaccinated mice (data not shown) and rabbits (Fig. 4a). After DNA priming with or without protein boosting, immunized rabbit sera neutralized a variety of primary isolates at a low level. Only HIV-1 isolates characterized as easily neutralized by human monoclonal antibodies (5) were neutralized to any great degree by the rabbit sera. The gp140 construct gave slightly higher titers (binding and neutralizing) than gp160, but this difference was not statistically significant. Binding titers were similar to those elicited by HIV-1 SF162-based immunogens.
Neutralization of a broad range of primary HIV-1 isolates is a major goal of contemporary vaccine research (5, 59). We tested sera from rabbits for virus neutralization at various times in the course of vaccination. In an effort to examine breadth in an impartial manner, we tested sera using two different assays, performed in two different laboratories. No standard panel of HIV-1 primary isolates has been established to date for comparisons of serum neutralizing activity between laboratories; in this study, each laboratory used a distinct panel of laboratory and primary viruses with a single isolate in common, HIV-1 SF162. Results in both laboratories indicated that sera from the AN1-Env B-immunized rabbits could reproducibly neutralize heterologous HIV-1 strains to a modest degree. Purified IgG from rabbit sera was also found to have neutralizing activity. No statistically significant differences between vaccine groups were found for the titers against HIV-1 SF162 or the numbers of isolates neutralized. These data, for both the easily neutralized HIV-1 SF162 strain and the panels of primary isolates, imply that the artificial sequence was at least as immunogenic as this natural isolate. No significant increase in breadth, however, was found using the AN1-EnvB immunogen.
A recent study by Gao et al. (21) evaluated the use of a group M consensus envelope as an immunogen. An artificial gene, CON6, was derived from a consensus of multiple HIV-1 subtypes, instead of clade B, as we used. CON6 derived its hypervariable domains from a clade C env sequence, instead of the parsimony-reconstructed hypervariable regions we used. CON6 encoded a functional protein that, like AN1-EnvB, was competent in pseudotype assays. To study neutralizing antibody, guinea pigs were inoculated with CON6 protein in RIBI; this contrasts with the study reported here, in which we inoculated rabbits by DNA priming and boosting with protein in incomplete Freund’s adjuvant. However, one of the assays used to examine neutralizing antibody—the M7-luc assay—was the same in both studies and was conducted in the same laboratory, and therefore we can directly compare the results. The CON6 sera showed somewhat more breadth than sera in our study, with several HIV isolates neutralized by samples from more than one rabbit. However, the CON6 study did not include a comparison with a natural isolate immunogen (e.g., SF162, as we used), making it difficult to conclude that the somewhat-improved breadth relative to ours and that of other studies can be attributed to the immunogen sequence; vaccine delivery and choice of animal model may have also played a role. In sum, both the consensus and ancestor approaches to immunogen design yielded low-level neutralizing antibody with some breadth.
Other recent vaccine experiments have shown that DNA expressing unmodified HIV-1 Env proteins can elicit low-level antibodies that neutralize the homologous primary isolate (2). Low-titer cross-reactive neutralizing antibodies have been elicited using modified envelope DNA vaccines or recombinant proteins (2, 17, 25, 39). Because the assays used and viruses tested have differed from study to study, it is difficult to compare the results directly. In general, our data compare well to these published reports; however, to date, no vaccine candidate has elicited antibodies with a breadth or potency against primary isolates that is close to what is found in patients (9, 44). Clearly, significant advances will be required for an effective humoral immunogen.
The codon-optimized vectors used here elicited high titers of binding antibodies with DNA vaccination alone, consistent with the findings of Barnett et al. (2). Many groups, including ours, have shown that responses to DNA vaccines can be effectively boosted with recombinant viral vectors or recombinant glycoproteins (2, 13). Immunization of the DNA-primed rabbits with homologous gp120 subunit protein in adjuvant resulted in boosting of binding antibody titers against HIV-1 SF162, including in rabbits immunized with AN1-EnvB gp120. However, gp120 protein failed to boost neutralizing antibody responses significantly. We hypothesize that the gp140 and gp160 DNA vaccines presented native Env conformations in vivo that were boosted poorly or not at all by these adjuvanted, and probably at least partially denatured, gp120 monomer proteins. Although native gp120 can bind conformation-dependent broad neutralizing antibodies (61), elicit neutralizing antibodies (26), and boost DNA priming (3), there is evidence that oligomeric envelope proteins are more effective in boosting broader responses against primary HIV-1 isolates (14, 60).
AN1-Env B gp120 contains 31 potential N-linked glycosylation sites (sequons), many more than the median of 25 reported for primary isolates (33, 67). By comparison, HIV-1 SF162, for which we found equivalent immunogenicity to AN1-EnvB, has 21 sequons, and CON6 has 26 (21). This hyperglycosylation may affect the immunogenicity of AN1-EnvB, independent of the sequence-distance effects we have postulated here. However, it is difficult to predict whether the effect would be to increase or decrease the elicited neutralization titer and breadth, relative to artificial sequences with more typical numbers of sequons or to natural isolates. On the one hand, the already high level of glycosylation found on natural isolates has been shown to affect T-cell recognition (6, 62), and it has been suggested to shield neutralizing epitopes via steric hindrance of antibody binding (65, 66). Based on the hypothesis that removal of glycans will expose previously occluded epitopes, there have been several studies of mutant immunogens with fewer sequons; these have yielded conflicting results, with some (31, 56) although not all (54) studies showing increased neutralization activity in sera of animals vaccinated with hypoglycosylated mutants compared to immunization with the parental strain. Thus, we might expect that if removing glycans improves immunogenicity, then conversely adding glycans would reduce immunogenicity. On the other hand, added glycosylation sites can be used to shield nonneutralizing or poorly conserved epitopes and redirect immune responses to conserved, neutralizing epitopes (22, 50). Thus, increased breadth of immunogenicity of hyperglycosylated Env might be expected. Therefore, although it is likely that the hyperglycosylation of AN1-EnvB had an influence on neutralization, we cannot assume that it had an overall positive or negative impact. Future investigation using ancestor sequences with more typical numbers or targeted removal of sequons may address this question.
In this study, we have not explored the development of cross-reactive cellular immune responses, due to limitations in the rabbit model used. As with glycosylation sites, the AN1-EnvB protein is also highly enriched for consensus CTL epitope sequences (F. Li and J. I. Mullins, unpublished observations). Thus, it is possible that ancestral sequences will be more effective in eliciting broad cellular immunity than humoral immunity, and investigation of this hypothesis is ongoing. Expanded studies to explore the ability of these and related immunogens to elicit both broad cellular and humoral immunity are needed.
Acknowledgments
This work was supported by a grant from the Boeing Foundation and one from the U.S. Public Health Service to the University of Washington Center for AIDS Research. N.D.-R. was supported by PHS T32A107509 and T32CA0922925.
We thank Nancy Miller and Marv Reitz for producing the AN1-EnvB gene and rgp120-AN1, Lisa Frenkel for discussions and HIV+ serum, Leonidas Stamatatos for rabbit serum, Indresh Srivastava for rgp120-SF162, and W. F. Sutton for growth of virus stocks. Viruses were obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
REFERENCES
- 1.Anderson, J., G. Learn, A. Rodrigo, X. He, Y. Wang, H. Weinstock, M. Kalish, K. E. Robbins, L. Hood, and J. Mullins. 2003. Predicting demographic group structures based on DNA sequence data. Mol. Biol. Evol. 20:1168-1180. [DOI] [PubMed] [Google Scholar]
- 2.Barnett, S. W., S. Lu, I. Srivastava, S. Cherpelis, A. Gettie, J. Blanchard, S. Wang, I. Mboudjeka, L. Leung, Y. Lian, A. Fong, C. Buckner, A. Ly, S. Hilt, J. Ulmer, C. T. Wild, J. R. Mascola, and L. Stamatatos. 2001. The ability of an oligomeric human immunodeficiency virus type 1 (HIV- 1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J. Virol. 75:5526-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett, S. W., S. Rajasekar, H. Legg, B. Doe, D. H. Fuller, J. R. Haynes, C. M. Walker, and K. S. Steimer. 1997. Vaccination with HIV-1 gp120 DNA induces immune responses that are boosted by a recombinant gp120 protein subunit. Vaccine 15:869-873. [DOI] [PubMed] [Google Scholar]
- 4.Barouch, D. H., A. Craiu, M. J. Kuroda, J. E. Schmitz, X. X. Zheng, S. Santra, J. D. Frost, G. R. Krivulka, M. A. Lifton, C. L. Crabbs, G. Heidecker, H. C. Perry, M. E. Davies, H. Xie, C. E. Nickerson, T. D. Steenbeke, C. I. Lord, D. C. Montefiori, T. B. Strom, J. W. Shiver, M. G. Lewis, and N. L. Letvin. 2000. Augmentation of immune responses to HIV-1 and simian immunodeficiency virus DNA vaccines by IL-2/Ig plasmid administration in rhesus monkeys. Proc. Natl. Acad. Sci. USA 97:4192-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Botarelli, P., B. A. Houlden, N. L. Haigwood, C. Servis, D. Montagna, and S. Abrignani. 1991. N-Glycosylation of HIV-gp120 may constrain recognition by T lymphocytes. J. Immunol. 147:3128-3132. [PubMed] [Google Scholar]
- 7.Brandt, S. M., R. Mariani, A. U. Holland, T. J. Hope, and N. R. Landau. 2002. Association of chemokine-mediated block to HIV entry with coreceptor internalization. J. Biol. Chem. 277:17291-17299. [DOI] [PubMed] [Google Scholar]
- 8.Calef, C., J. Mokili, D. H. O'Connor, D. I. Watkins, and B. Korber. 2002. Numbering positions in SIV relative to SIVMM239, p. 171-181. In C. Kuiken, B. Foley, B. Hahn, P. Marx, F. McCutchan, J. W. Mellors, S. Wolinsky, and B. Korber (ed.), HIV Sequence Compendium 2001. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 9.Cao, Y., L. Qin, L. Zhang, J. Safrit, and D. D. Ho. 1995. Virologic and immunologic characterization of long-term survivors of HIV-1 infection. N. Engl. J. Med. 332:201-208. [DOI] [PubMed] [Google Scholar]
- 10.Cecilia, D., V. N. KewalRamani, J. O'Leary, B. Volsky, P. Nyambi, S. Burda, S. Xu, D. R. Littman, and S. Zolla-Pazner. 1998. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J. Virol. 72:6988-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chackerian, B., E. M. Long, P. A. Luciw, and J. Overbaugh. 1997. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J. Virol. 71:3932-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, X. Wu, W. A. O'Brien, L. Ratner, J. C. Kappes, G. M. Shaw, and E. Hunter. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74:8358-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doria-Rose, N. A., C. Ohlen, P. Polacino, C. C. Pierce, M. T. Hensel, L. Kuller, T. Mulvania, D. Anderson, P. D. Greenberg, S.-L. Hu, and N. L. Haigwood. 2003. Multigene DNA priming-boosting vaccines protect macaques from acute CD4+-T-cell depletion after simian-human immunodeficiency virus SHIV89.6P mucosal challenge. J. Virol. 77:11563-11577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Earl, P. L., W. Sugiura, D. C. Montefiori, C. C. Broder, S. A. Lee, C. Wild, J. Lifson, and B. Moss. 2001. Immunogenicity and protective efficacy of oligomeric human immunodeficiency virus type 1 gp140. J. Virol. 75:645-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edmonson, P., M. Murphey-Corb, L. N. Martin, C. Delahunty, J. Heeney, H. Kornfeld, P. R. Donahue, G. H. Learn, L. Hood, and J. I. Mullins. 1998. Evolution of a simian immunodeficiency virus pathogen. J. Virol. 72:405-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esparza, J., S. Osmanov, C. Pattou-Markovic, C. Toure, M. L. Chang, and S. Nixon. 2002. Past, present and future of HIV vaccine trials in developing countries. Vaccine 20:1897-1898. [DOI] [PubMed] [Google Scholar]
- 17.Fouts, T., K. Godfrey, K. Bobb, D. Montefiori, C. V. Hanson, V. S. Kalyanaraman, A. DeVico, and R. Pal. 2002. Crosslinked HIV-1 envelope-CD4 receptor complexes elicit broadly cross-reactive neutralizing antibodies in rhesus macaques. Proc. Natl. Acad. Sci. USA 99:11842-11847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francis, D. P., T. Gregory, M. J. McElrath, R. B. Belshe, G. J. Gorse, S. Migasena, D. Kitayaporn, P. Pitisuttitham, T. Matthews, D. H. Schwartz, and P. W. Berman. 1998. Advancing AIDSVAX to phase 3. Safety, immunogenicity, and plans for phase 3. AIDS Res. Hum Retrovir. 14(Suppl. 3):S325-S331. [PubMed] [Google Scholar]
- 19.Fynan, E. F., R. G. Webster, D. H. Fuller, J. R. Haynes, J. C. Santoro, and H. L. Robinson. 1993. DNA vaccines: protective immunizations by parenteral, mucosal, and gene-gun inoculations. Proc. Natl. Acad. Sci. USA 90:11478-11482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao, F., B. T. Korber, E. Weaver, H. X. Liao, B. H. Hahn, and B. F. Haynes. 2004. Centralized immunogens as a vaccine strategy to overcome HIV-1 diversity. Expert Rev. Vaccines 3:S161-S168. [DOI] [PubMed] [Google Scholar]
- 21.Gao, F., E. A. Weaver, Z. Lu, Y. Li, H. X. Liao, B. Ma, S. M. Alam, R. M. Scearce, L. L. Sutherland, J. S. Yu, J. M. Decker, G. M. Shaw, D. C. Montefiori, B. T. Korber, B. H. Hahn, and B. F. Haynes. 2005. Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group m consensus envelope glycoprotein. J. Virol. 79:1154-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garrity, R. R., G. Rimmelzwaan, A. Minassian, W. P. Tsai, G. Lin, J. J. de Jong, J. Goudsmit, and P. L. Nara. 1997. Refocusing neutralizing antibody response by targeted dampening of an immunodominant epitope. J. Immunol. 159:279-289. [PubMed] [Google Scholar]
- 23.Gaschen, B., J. Taylor, K. Yusim, B. Foley, F. Gao, D. Lang, V. Novitsky, B. Haynes, B. H. Hahn, T. Bhattacharya, and B. Korber. 2002. Diversity considerations in HIV-1 vaccine selection. Science 296:2354-2360. [DOI] [PubMed] [Google Scholar]
- 24.Graham, B. S. 2002. Clinical trials of HIV vaccines. Annu. Rev. Med. 53:207-221. [DOI] [PubMed] [Google Scholar]
- 25.Grundner, C., Y. Li, M. Louder, J. Mascola, X. Yang, J. Sodroski, and R. Wyatt. 2005. Analysis of the neutralizing antibody response elicited in rabbits by repeated inoculation with trimeric HIV-1 envelope glycoproteins. Virology 331:33-46. [DOI] [PubMed] [Google Scholar]
- 26.Haigwood, N. L., P. L. Nara, E. Brooks, G. A. Van Nest, G. Ott, K. W. Higgins, N. Dunlop, C. J. Scandella, J. W. Eichberg, and K. S. Steimer. 1992. Native but not denatured recombinant human immunodeficiency virus type 1 gp120 generates broad-spectrum neutralizing antibodies in baboons. J. Virol. 66:172-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartikka, J., M. Sawdey, F. Cornefert-Jensen, M. Margalith, K. Barnhart, M. Nolasco, H. L. Vahlsing, J. Meek, M. Marquet, P. Hobart, J. Norman, and M. Manthorpe. 1996. An improved plasmid DNA expression vector for direct injection into skeletal muscle. Hum. Gene Ther. 7:1205-1217. [DOI] [PubMed] [Google Scholar]
- 28.He, J., S. Choe, R. Walker, P. Di Marzio, D. O. Morgan, and N. R. Landau. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 69:6705-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heeney, J. L. 2004. Requirement of diverse T-helper responses elicited by HIV vaccines: induction of highly targeted humoral and CTL responses. Expert Rev. Vaccines 3:S53-S64. [DOI] [PubMed] [Google Scholar]
- 30.Jensen, M. A., F.-S. Li, A. B. van 't Wout, D. C. Nickle, D. Shriner, H. He, S. McLaughlin, R. Shankarappa, J. B. Margolick, and J. I. Mullins. 2003. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J. Virol. 77:13376-13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson, W. E., H. Sanford, L. Schwall, D. R. Burton, P. W. Parren, J. E. Robinson, and R. C. Desrosiers. 2003. Assorted mutations in the envelope gene of simian immunodeficiency virus lead to loss of neutralization resistance against antibodies representing a broad spectrum of specificities. J. Virol. 77:9993-10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korber, B., B. Gaschen, K. Yusim, R. Thakallapally, C. Kesmir, and V. Detours. 2001. Evolutionary and immunological implications of contemporary HIV-1 variation. Br. Med. Bull. 58:19-42. [DOI] [PubMed] [Google Scholar]
- 34.Kornfeld, H., N. Riedel, G. A. Viglianti, V. Hirsch, and J. I. Mullins. 1987. Cloning of HTLV-4 and its relation to simian and human immunodeficiency viruses. Nature 326:610-613. [DOI] [PubMed] [Google Scholar]
- 35.Kuiken, C., B. Korber, and R. W. Shafer. 2003. HIV sequence databases. AIDS Rev. 5:52-61. [PMC free article] [PubMed] [Google Scholar]
- 36.Learn, G., and J. I. Mullins. 2000. Presented at the Twelfth Joint Scientific Meeting of the AIDS Panels for the U.S.-Japan Cooperative Medical Science Program, Santa Fe, N.Mex.
- 37.Learn, G., and J. I. Mullins. 2000. Presented at the Seventh International Discussion Meeting on HIV Dynamics & Evolution, Seattle, Wash.
- 38.Learn, G. H., Jr., B. T. Korber, B. Foley, B. H. Hahn, S. M. Wolinsky, and J. I. Mullins. 1996. Maintaining the integrity of human immunodeficiency virus sequence databases. J. Virol. 70:5720-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liao, H. X., S. M. Alam, J. R. Mascola, J. Robinson, B. Ma, D. C. Montefiori, M. Rhein, L. L. Sutherland, R. Scearce, and B. F. Haynes. 2004. Immunogenicity of constrained monoclonal antibody A32-human immunodeficiency virus (HIV) Env gp120 complexes compared to that of recombinant HIV type 1 gp120 envelope glycoproteins. J. Virol. 78:5270-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Long, E. M., S. M. Rainwater, L. Lavreys, K. Mandaliya, and J. Overbaugh. 2002. HIV type 1 variants transmitted to women in Kenya require the CCR5 coreceptor for entry, regardless of the genetic complexity of the infecting virus. AIDS Res. Hum Retrovir. 18:567-576. [DOI] [PubMed] [Google Scholar]
- 41.Lu, S., R. Wyatt, J. F. Richmond, F. Mustafa, S. Wang, J. Weng, D. C. Montefiori, J. Sodroski, and H. L. Robinson. 1998. Immunogenicity of DNA vaccines expressing human immunodeficiency virus type 1 envelope glycoprotein with and without deletions in the V1/2 and V3 regions. AIDS Res. Hum. Retrovir. 14:151-155. [DOI] [PubMed] [Google Scholar]
- 42.Maddison, W. P., and D. R. Maddison. 1992. MacClade: analysis of phylogeny and character evolution, version 3. Sinauer Associates, Inc., Sunderland, Mass.
- 43.Montefiori, D. C., B. Metch, M. J. McElrath, S. Self, K. J. Weinhold, and L. Corey. 2004. Demographic factors that influence the neutralizing antibody response in recipients of recombinant HIV-1 gp120 vaccines. J Infect. Dis. 190:1962-1969. [DOI] [PubMed] [Google Scholar]
- 44.Moog, C., H. J. Fleury, I. Pellegrin, A. Kirn, and A. M. Aubertin. 1997. Autologous and heterologous neutralizing antibody responses following initial seroconversion in human immunodeficiency virus type 1-infected individuals. J. Virol. 71:3734-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mullins, J. I., D. C. Nickle, L. Heath, A. G. Rodrigo, and G. H. Learn. 2004. Immunogen sequence: the fourth tier of AIDS vaccine design. Expert Rev. Vaccines 3(Suppl. 1):S151-S159. [DOI] [PubMed] [Google Scholar]
- 46.Nabel, G., W. Makgoba, and J. Esparza. 2002. HIV-1 diversity and vaccine development. Science 296:2335. [DOI] [PubMed] [Google Scholar]
- 47.Nabel, G. J. 2002. HIV vaccine strategies. Vaccine 20:1945-1947. [DOI] [PubMed] [Google Scholar]
- 48.Nakamura, Y., T. Gojobori, and T. Ikemura. 2000. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nickle, D. C., M. A. Jensen, G. S. Gottlieb, D. Shriner, G. H. Learn, A. G. Rodrigo, and J. I. Mullins. 2003. Consensus and ancestral state HIV vaccines. Science 299:1515-1518. [DOI] [PubMed] [Google Scholar]
- 50.Pantophlet, R., I. A. Wilson, and D. R. Burton. 2003. Hyperglycosylated mutants of human immunodeficiency virus (HIV) type 1 monomeric gp120 as novel antigens for HIV vaccine design. J. Virol. 77:5889-5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pitisuttithum, P. 2005. HIV-1 prophylactic vaccine trials in Thailand. Curr. HIV Res. 3:17-30. [DOI] [PubMed] [Google Scholar]
- 52.Pitisuttithum, P., S. Nitayaphan, P. Thongcharoen, C. Khamboonruang, J. Kim, M. de Souza, T. Chuenchitra, R. P. Garner, D. Thapinta, V. Polonis, S. Ratto-Kim, P. Chanbancherd, J. Chiu, D. L. Birx, A. M. Duliege, J. G. McNeil, and A. E. Brown. 2003. Safety and immunogenicity of combinations of recombinant subtype E and B human immunodeficiency virus type 1 envelope glycoprotein 120 vaccines in healthy Thai adults. J. Infect. Dis. 188:219-227. [DOI] [PubMed] [Google Scholar]
- 53.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 54.Quinones-Kochs, M. I., L. Buonocore, and J. K. Rose. 2002. Role of N-linked glycans in a human immunodeficiency virus envelope glycoprotein: effects on protein function and the neutralizing antibody response. J. Virol. 76:4199-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rambaut, A., and N. C. Grassly. 1997. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput. Appl. Biosci. 13:235-238. [DOI] [PubMed] [Google Scholar]
- 56.Reitter, J. N., R. E. Means, and R. C. Desrosiers. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679-684. [DOI] [PubMed] [Google Scholar]
- 57.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith, S. W., R. Overbeek, C. R. Woese, W. Gilbert, and P. M. Gillevet. 1994. The Genetic Data Environment: an expandable GUI for multiple sequence analysis. Comput. Appl. Biol. Sci. 10:671-675. [DOI] [PubMed] [Google Scholar]
- 59.Spearman, P. W. 2003. HIV vaccine research: lessons from the past and promise for the future. Curr. HIV Res. 1:101-120. [DOI] [PubMed] [Google Scholar]
- 60.Srivastava, I. K., L. Stamatatos, H. Legg, E. Kan, A. Fong, S. R. Coates, L. Leung, M. Wininger, J. J. Donnelly, J. B. Ulmer, and S. W. Barnett. 2002. Purification and characterization of oligomeric envelope glycoprotein from a primary R5 subtype B human immunodeficiency virus. J. Virol. 76:2835-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steimer, K. S., C. J. Scandella, P. V. Skiles, and N. L. Haigwood. 1991. Neutralization of divergent HIV-1 isolates by conformation-dependent human antibodies to gp120. Science 254:105-108. [DOI] [PubMed] [Google Scholar]
- 62.Surman, S., T. D. Lockey, K. S. Slobod, B. Jones, J. M. Riberdy, S. W. White, P. C. Doherty, and J. L. Hurwitz. 2001. Localization of CD4+ T cell epitope hotspots to exposed strands of HIV envelope glycoprotein suggests structural influences on antigen processing. Proc. Natl. Acad. Sci. USA 98:4587-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swofford, D. L. 1999. PAUP* 4.0: phylogenetic analysis using parsimony (*and other methods), version 4.0b2a. Sinauer Associates, Inc., Sunderland, Mass.
- 64.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 66.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, M., B. Gaschen, W. Blay, B. Foley, N. Haigwood, C. Kuiken, and B. Korber. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14:1229-1246. [DOI] [PubMed] [Google Scholar]