Abstract
Iron serves as a signal in Pseudomonas aeruginosa biofilm development. We examined the influence of mutations in known and putative iron acquisition-signaling genes on biofilm morphology. In iron-sufficient medium, mutants that cannot obtain iron through the high-affinity pyoverdine iron acquisition system form thin biofilms similar to those formed by the parent under low iron conditions. If an iron source for a different iron acquisition system is provided to a pyoverdine mutant, normal biofilm development occurs. This enabled us to identify iron uptake gene clusters that likely serve in transport of ferric citrate and ferrioxamine. We suggest that the functional iron signal for P. aeruginosa biofilm development is active transport of chelated iron or the level of internal iron. If the signal is internal iron levels, then a factor likely to be involved in iron signaling is the cytoplasmic ferric uptake regulator protein, Fur, which controls expression of iron-responsive genes. In support of a Fur involvement, we found that with low iron a Fur mutant was able to organize into more mature biofilms than was the parent. The two known Fur-controlled small regulatory RNAs (PrrF1 and F2) do not appear to mediate iron control of biofilm development. This information establishes a mechanistic basis for iron control of P. aeruginosa biofilm formation.
Keywords: pyoverdine
Biofilms consist of groups of bacteria attached to surfaces and encased in a hydrated polymeric matrix. Pseudomonas aeruginosa biofilms cause persistent infections in individuals with underlying health problems. For example, most people with the genetic disease cystic fibrosis are plagued by chronic P. aeruginosa biofilm infections of their lungs (reviewed in refs. 1-3). Iron starvation can prevent bacterial growth. Recent work shows that with sufficient iron for growth the levels of this metal serve as a signal for biofilm development (4, 5). By sequestering iron, subgrowth inhibitory concentrations of the mammalian iron chelator lactoferrin block the ability of P. aeruginosa biofilms to mature from thin layers of cells attached to a surface into large multicellular biofilm structures (4). The influence of lactoferrin on biofilm development is thought to be related to the fact that at low iron concentrations P. aeruginosa exhibits incessant twitching motility on surfaces, and cells do not form sessile structures (4, 5). The mechanism for iron control of twitching motility is unknown, as is the mechanism of iron signaling in biofilm development.
Iron is essential for, yet toxic to, bacteria. For most pathogens, including P. aeruginosa, there is intense competition for iron with the host (6-8). P. aeruginosa has multiple systems to sense and sequester iron in its environment, and it is able to regulate cellular iron acquisition and storage through an assortment of positive and negative regulatory factors (reviewed in refs. 6, 9, and 10). The two best-studied P. aeruginosa iron acquisition systems are the high-affinity pyoverdine system and the lower-affinity pyochelin system. Pyoverdine and pyochelin bind extracellular iron (Fe3+), which is then transported into the cell together with these siderophores. Pyoverdine synthesis and secretion are regulated by means of the extracytoplasmic function (ECF) σ factor PvdS. Expression of PvdS is regulated by iron and the ferric uptake regulator (Fur). In this acquisition system, pyoverdine binds to an outer membrane-associated receptor, which results in transmission of a signal to a membrane-spanning anti-σ factor that governs the activity of PvdS, thereby controlling expression of pyoverdine synthesis. The signal also regulates expression of other genes, including extracellular virulence factors such as exotoxin A (9, 11, 12). There are at least 10 other gene clusters in the P. aeruginosa genome that may encode iron-responsive ECF σ factor-regulated systems (12). For example, there is one gene cluster encoding proteins with extensive sequence similarity to the FecIR, ferric citrate system in Escherichia coli (12, 13).
As in many other bacteria, P. aeruginosa has a Fur protein that functions as a global regulator of iron-responsive genes (14, 15). Fur has both negative and positive regulatory effects on gene expression. It represses gene expression by direct binding to the operators of iron starvation-inducible genes. It activates gene expression indirectly through control of a pair of small regulatory RNAs (sRNAs), PrrF1 and PrrF2 (16). In P. aeruginosa, fur appears to be an essential gene, and the only fur mutants available either produce reduced levels of wild-type Fur or have missense mutations that exhibit high reversion rates (17, 18).
We sought to better understand the iron-signaling cascade critical for normal biofilm development. We asked whether any of the known or putative iron-uptake and iron-starvation ECF σ factors are involved in regulating biofilm development. Our data indicate that any iron-uptake system that can provide sufficient levels of internal iron to P. aeruginosa can function in the iron-signaling pathway. This finding suggests that a critical level of intracellular iron serves as the signal for biofilm development. Our data also indicate that intracellular iron signaling is mediated by Fur but not by means of its regulation of PrrF1 and PrrF2.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions. We used P. aeruginosa PAO1 as the wild-type strain. Strain PAO1 and most of the P. aeruginosa transposon insertion mutants used were from the Comprehensive P. aeruginosa transposon mutant library at the University of Washington Genome Center (19). We used the following mutants with transposons in iron starvation ECF σ-factor homologs: PTL4103 (PA0149), PTL52421 (PA0472), PTL9556 (PA0675), PTL16978 (PA1300), PTL37473 (PA1363), P TL16034 (PA1912), P TL18118 (PA 2050), PTL51829 (PA3410), PTL7851 (PA3899), and PTL49853 (PA4896). We also used mutants with sequence similarity to various iron receptors: PTL40529 and PTL14626 (both PA0151); PTL20210 (PA0470) with similarity to a hydroxamate type receptor; PTL45373 (PA0674), a ferripyoverdine receptor homolog; PTL18189 (hxuC), a heme receptor homolog; PTL5195 (PA1365), a ferribactin receptor homolog; PTL4009 (PA1910); PTL11509 (PA2466), a ferrioxamine receptor homolog; PTL9628 (hasR), a heme uptake homolog; and PTL14263 (fecA). The pyochelin mutant (pchA) was PTL5032. All of the mutants contained either a ISlacZ/hah or an ISphoA/hah transposon insertion. Further information on these mutants is at www.genome.washington.edu/UWGC/Pseudomonas/index.cfm. The locations of all of the mutations were confirmed by PCR with primers flanking the insertion sites. We also used PAO1ΔpvdS (20); PAO1ΔpvdA (14); PAO1ΔfpvA (11), a strain with a fur point mutation; PAO1furC6Tc (17, 18), a PAO1ΔprrF1-F2 mutant (16); PAO1ΔpvdD ΔpchEF (provided by Urs Ochsner, Replidyne Inc., Louisville, CO); and several strains with mutations generated as described below.
For growth curve and flow cell biofilm experiments we used 1% Bacto Tryptic Soy Broth (TSB) (Becton Dickinson). Incubation was at room temperature (23-25°C). Where indicated, 20 μg/ml human lactoferrin (Sigma) was added to the medium. Also as indicated, 50 or 100 μM ferric chloride, 1 μM ferric dicitrate, 1.5 μM desferrioxamine mesylate (Sigma), or pyoverdine-conditioned medium was added. Pyoverdine-conditioned medium was prepared by growing P. aeruginosa PAO1 in Kings B medium (21) for 12 h at 37°C. The cells were removed by centrifugation and filtration through a 0.2-μm filter. Pyoverdine-conditioned medium was added at 1% as indicated. As a control for the pyoverdine experiments, we used medium conditioned by the PvdA mutant PAO1ΔpvdA. Escherichia coli was used for recombinant manipulations and was grown in Luria-Bertani (LB) broth. We used Pseudomonas Isolation Agar (Becton Dickinson) for selection of transconjugants. Antibiotics were used at the following concentrations: 100 μg/ml gentamicin (Gm) for E. coli and P. aeruginosa,10 μg/ml tetracycline (Tc) for E. coli, and 100 μg/ml Tc for P. aeruginosa.
Construction of Plasmids and P. aeruginosa Mutants. Although most of the P. aeruginosa mutants were from a preexisting library, some were constructed as described here. For construction of the PA2387 mutant (EB101), a 1.4-kb HindIII-NotI DNA fragment upstream of PA2387 (from bp 2638897 to bp 2640299 on the P. aeruginosa chromosome) and a 1.5-kb NotI-XbaI DNA fragment downstream of PA2387 (from bp 2640893 to bp 2674792 on the P. aeruginosa chromosome) were amplified and cloned into a HindIII-XbaI-digested pEX18Tc gene replacement vector (22). The resulting plasmid was digested with NotI and ligated with a NotI-digested aacC1 cassette to create pEB2. This plasmid was mobilized from E. coli SM10 (23) into P. aeruginosa PAO1 by conjugation. Selection for GmR colonies followed by screening for TcS colonies yielded a PA2387 mutant, P. aeruginosa EB101. The mutation was confirmed by PCR analysis. The PA2467-2468 mutant (EB102) was constructed in a similar fashion. A 1.4-kb HindIII-NotI DNA fragment corresponding to a region upstream of PA2467 (from bp 2783773 to bp 2785221 on the P. aeruginosa chromosome) and a NotI-XbaI 1.5-kb fragment downstream of PA2468 (from bp 2786704 to bp 2788240 on the P. aeruginosa chromosome) were amplified and cloned into HindIII-XbaI-digested pEX18Tc. The resulting plasmid was then digested with NotI and ligated with a NotI-digested aacC1 cassette to create pEB3. A transconjugant was isolated, and the mutation was confirmed as described above.
We constructed the following double mutants using a method described in ref. 24: EB104 (PAO1 ΔpvdA,ISlacZ/hah PA3901), EB107 (PAO1 ΔpvdA,ISlacZ/hah pchA), EB110 (PAO1 ΔpvdA, ISlacZ/hah PA0470), and EB111 (PAO1 ΔpvdA, ISlacZ/hah PA2466). In all cases we transformed PAO1ΔpvdA with genomic DNA (40-80 μg) from PTL14263 to construct EB104, PTL5032 to construct strain EB107, PTL20210 to construct strain EB110, or PTL11509 to construct strain EB111. After transformation, colonies were selected on LB agar plates containing Tc (100 μg/ml). The mutation in each strain was confirmed by PCR.
The triple mutant EB112 (PAO1ΔpvdA, 63-bp insertion in PA0470, ISlacZ/hah PA2466) was constructed by using the cre-recombinase system (25). Strain EB110 was mated with E. coli carrying pCre1. Transconjugants were selected on Pseudomonas isolation agar. A TcS strain was isolated, and the 63-bp insertion was confirmed by PCR. This strain was transformed with PTL11509 genomic DNA, a transformant was selected, and all of the mutations were confirmed by PCR.
To construct the pvdS-gfp fusion strain, a 250-bp EcoRI-HindIII fragment containing the pvdS promoter was amplified (from bp 2721925 to bp 2722175) and cloned into an EcoRI- HindIII-digested promoterless gfp pPROBE AT (26). The plasmid was transferred to PAO1ΔpvdD ΔpchEF by conjugation. The resulting strain, EB125, contains the pvdS-gfp fusion as a single chromosomal copy.
For complementation of the pvdA mutation, a 2-kb fragment containing pvdA was amplified (from bp 2638500 to bp 2640501) and cloned in EcoRI-digested pJN105 (27). The resulting plasmid pEB4 was used to transform PAO1ΔpvdA. For complementation of the pvdS mutation we used pPVD27 (28).
Biofilm Experiments. We used flow cells to follow biofilm growth on a glass surface (24). The flow cells were inoculated with a 1:50 dilution (in 1% TSB) of a P. aeruginosa stationary phase culture, and flow was initiated after 1 h. To image biofilms, we used confocal scanning laser microscopy (CSLM). The CSLM was a Radiance 2100 system (Bio-Rad) with a Nikon Eclipse E600 microscope. Generally, we imaged GFP in P. aeruginosa containing pMRP9-1, which has a constitutively expressed gfp (29). Where indicated, we counterstained the biofilm with propidium iodide (4 μM) and imaged the propidium iodide fluorescence by CSLM (30). The image acquisition software was lasersharp 2000 (Bio-Rad). Images were processed with confocal assistant or volocity (Improvision, Lexington, MA) software.
Twitching Motility Assays. For twitching motility assays we used Petri plates with 1% TSB plus 1% Noble agar (Becton Dickinson) with or without 2 mM desferrioxamine to chelate iron as indicated. Plates were dried overnight at room temperature, and cells from a colony grown overnight on a LB-agar plate were point inoculated at the bottom of the agar plate. After 3 days, the twitching motility distance along the plastic-agar interface (at the bottom of the agar plate) was measured.
Results
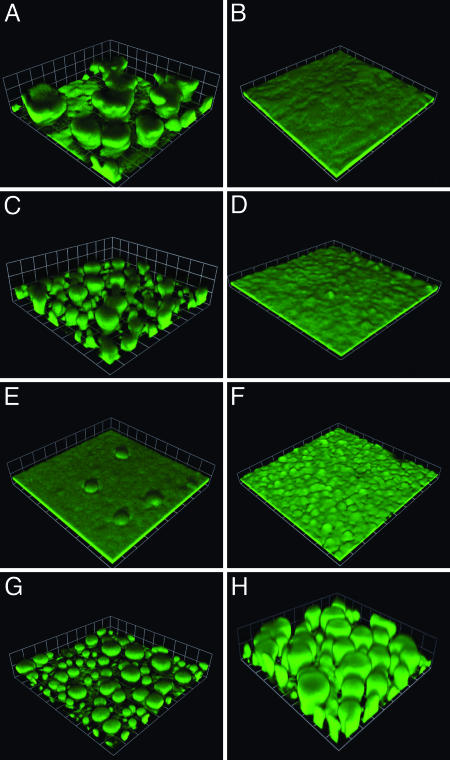
Biofilm Development in Pyoverdine and Pyochelin Synthesis Mutants. We first followed biofilm development in P. aeruginosa mutants incapable of synthesizing the two well known siderophores pyoverdine and pyochelin, PAO1ΔpvdA and PAO1pchA::TcR, respectively. Planktonic growth in 1% TSB and twitching motility (data not shown) of these mutants were indistinguishable from the parent. Biofilm formation of the pyochelin mutant under a flow of 1% TSB is similar to that of the parent. Both strains form mushroom-like structures on the glass surface. The pyoverdine mutant, however, forms a thin uniform layer of growth on the glass surface that is similar to the biofilms formed by the parent and pyochelin mutant in the presence of lactoferrin (Fig. 1). PvdA is required for pyoverdine synthesis, and PvdS codes for the σ factor required to activate pyoverdine synthesis and to regulate expression of other genes. A pvdS mutant, PAOΔpvdS, shows a biofilm phenotype similar to that of the pvdA mutant (Fig. 1). Complementation of the pvdA and pvdS mutations restores the normal biofilm architecture (Fig. 1). Furthermore, addition of pyoverdine-conditioned medium (see Materials and Methods) allows biofilms of the PvdA mutant to form normally (Fig. 1).
Fig. 1.
Biofilm formation of P. aeruginosa pyoverdine and pyochelin mutants. The parent without (A) and with (B) lactoferrin (20 μg/ml), the pyochelin mutant PAO1pchA::TcR without (C) and with (D) lactoferrin (20 μg/ml), the pyoverdine synthesis mutant PAO1ΔpvdA (E), the pyoverdine ECF σ-factor PvdS mutant PAOΔpvdS without lactoferrin (F), the pyoverdine mutant carrying the pvdA expression vector pEB4 without lactoferrin (G), and the pyoverdine mutant grown in pyoverdine-conditioned medium without lactoferrin (H). The P. aeruginosa cells contained the gfp plasmid pMRP9-1. Images are from 6-day biofilms (the squares are 61 μm on a side).
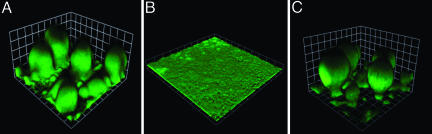
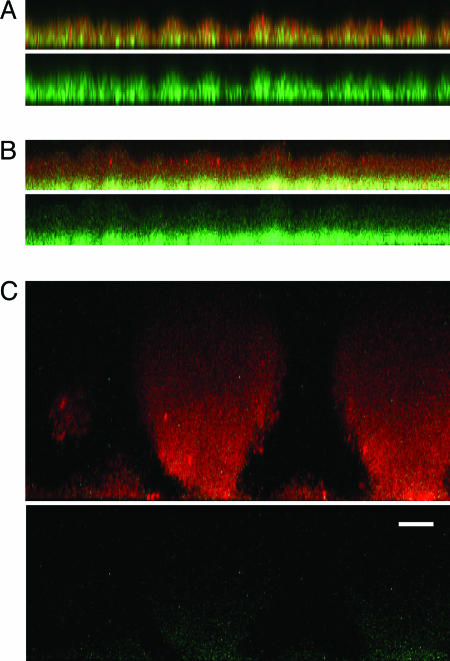
Biofilm Formation in Flow Cells Requires Active Iron Transport. Compared to pyoverdine, pyochelin has a low affinity for iron (9). To ask whether the pyoverdine mutant can acquire iron for signaling biofilm development through pyochelin in the presence of sufficiently high environmental iron concentrations, we supplemented the medium with 50 μM FeCl3. In this iron-rich medium the pyoverdine mutant produces biofilms that are similar to the parent. A pvdA,pchA double mutant, EB107, forms thin, flat biofilms in high-iron medium, even at 100 μM ferric chloride (Fig. 2). We obtained similar results with a pvdD,pchEF double mutant (data not shown). Addition of desferrioxamine or ferric dicitrate, both of which P. aeruginosa can use as an iron source (31, 32), restores biofilm formation by the double mutant (Fig. 2). These experiments suggest that an iron uptake system is required for normal biofilm development on glass and that passive diffusion of iron does not provide sufficient intracellular iron for biofilm development. To test the hypothesis that P. aeruginosa biofilms cannot obtain sufficient iron for signaling by passive diffusion even at high external iron concentrations, we examined the expression of an iron-repressed, Fur-regulated promoter (pvdS)-driven gfp in our pyoverdine, pyochelin double mutant (EB125). With or without added iron (100 μM FeCl3), biofilms are flat (<15 μm), and the cells express GFP (Fig. 3). In the presence of added iron, the cells on the biofilm surface express less GFP than those at the base of the biofilm (Fig. 3). This finding suggests that there is sufficient iron for Fur to serve as a repressor in cells near the biofilm surface, but without an iron uptake system passive diffusion does not provide sufficient iron for Fur repression throughout the biofilm. When the double mutant is grown in the presence of desferrioxamine, which can be taken up by the ferrioxamine transport system, mushroom-like structures form and the cells express low levels of GFP (Fig. 3). These data are consistent with the hypothesis that an active iron transport system is needed for normal biofilm development.
Fig. 2.
Active iron transport is required for normal biofilm development in flow cells. (A) Biofilm of the pvdA mutant PAO1ΔpvdA grown with added ferric chloride (50 μM). Biofilm of a pyoverdine, pyochelin double mutant (EB107) grown with 100 μM ferric chloride (B) or 1.5 μM desferrioxamine (C) (similar results were obtained when 1 μM ferric dicitrate was added; data not shown). All strains contained the gfp plasmid pMRP9-1. Images represent 6-day biofilms (the squares are 30 μm on a side).
Fig. 3.
Expression of pvdS-gfp in biofilms of a pyoverdine, pyochelin double mutant (EB125) in the presence of FeCl3 or desferrioxamine. (A) Biofilms grown without FeCl3.(B) Biofilms grown with 100 μM FeCl3.(C) Biofilms grown with 1.5 μM desferrioxamine. Images represent 6-day biofilms counterstained with propidium iodide. (A-C Upper) A reconstructed side view of the combined red and green channels. Red shows the biofilm, and green shows regions where cells have expressed GFP. (A-C Lower)A side view of GFP expression alone. (Scale bar in Lower, 15 μm; all of the side images are the same magnification.)
Do Mutations in Other Iron-Responsive ECF σ-Factor Systems Influence Biofilm Formation? Visca et al. (12) identified 19 ORFs encoding putative ECF σ-factors in the P. aeruginosa genome. Thirteen of these show substantial sequence similarity with iron starvation σ factors, and they contain putative Fur binding sites in their promoter region. Several are in clusters that include genes predicted to encode outer-membrane receptors involved in iron transport. We wanted to determine whether any of these putative iron acquisition signaling systems is involved in biofilm development; therefore, we studied mutants with defects in the putative iron-responsive ECF σ-factor genes and the putative FecA iron receptor homologs. Mutants with insertions in the following ECF σ-factor genes were examined: PA0149, PA0472, PA0675, PA1300, PA1363, PA1912, PA2050, PA2387, PA2468, PA3410, PA3899, and PA4896. Iron receptor mutants with insertions in PA0151, PA0470, PA0674, PA1302, PA1910, PA1365, PA2398, PA2466, PA3408, and PA3901 were also examined. None of the mutations has an appreciable affect on planktonic growth or twitching motility (data not shown). With a single exception, biofilm formation of the mutants is indistinguishable from the parent in the presence (thin, flat biofilms) or absence (large, mushroom-like structures) of lactoferrin. The single exception is the PA2398 mutant, PAO1fpvA::GmR. This fpvA mutant forms thin, flat biofilms in the absence of lactoferrin that are similar in appearance to those formed by pvd mutants in the absence of lactoferrin (data not shown). FpvA is known to function in the pyoverdine signaling and uptake cascade (33).
Alternate Sources of Iron Can Signal Biofilm Development in a Pyoverdine Synthesis Mutant. Without sufficiently high levels of environmental iron, pyoverdine mutants form thin, flat biofilms (Fig. 1), and we believe that cytoplasmic iron levels or transport of chelated iron cues biofilm development. However, we know that P. aeruginosa can use a variety of chelated forms of environmental iron and that it can acquire iron chelated by other organisms (9). For example, it can use iron chelated with citrate or with desferrioxamine (31, 32, 34). If iron signaling in biofilm formation is mediated by a cytoplasmic receptor or active transport of chelated iron, then provision of ferric dicitrate or desferrioxamine should allow the pvdA mutant to form biofilms with the wild-type mushroom-like structures. In fact, biofilms of the pvdA mutant grown in the presence of 1 μM ferric dicitrate show the mushroom-like appearance of the parent strain (Fig. 4).
Fig. 4.
Ferric citrate and ferrioxamine mediate development of mushroom-like structures in the pyoverdine synthesis mutant PAO1ΔpvdA. The pvdA mutant biofilm in medium supplemented with 1 μM ferric dicitrate (A) or 1.5 μM desferrioxamine (B). (C) A biofilm of the pvdA, PA3901 double mutant EB104 in medium containing 1 μM ferric dicitrate. (D) A biofilm of EB112 (the pyoverdine, PA2466, and PA0470 triple mutant) in medium with 1.5 μM desferrioxamine. All strains contained pMRP9-1. Images represent 6-day biofilms (the squares are 62 μm on a side).
The P. aeruginosa PA3899-3901 gene cluster codes for polypeptides showing extensive sequence identity with the E. coli ferric dicitrate uptake polypeptides, the Fec polypeptides (35). Thus, we constructed P. aeruginosa EB104, which contains an insertion in PA3901 and a deletion in pvdA. This mutant shows no appreciable differences in growth and twitching motility compared with the parent; however, it forms thin, flat biofilms even in the presence of ferric dicitrate (Fig. 4). Similar to the results obtained with ferric dicitrate as an alternative iron scavenger, addition of desferrioxamine (1.5 μM) also restores mushroom formation to the pvdA mutant. In the P. aeruginosa genome there are two gene clusters (PA0470-0472 and PA2466-2468) that show extensive sequence identity with ferrioxamine uptake systems from other bacteria (15). We constructed two double mutants that contain a deletion in pvdA and an insertion in PA0470 or PA2466 (EB110 and EB111, respectively). Both mutants form mushroom-like structures in the presence of desferrioxamine (data not shown). A triple mutant (EB112) that contains deletions in pvdA and PA2466 and an insertion in PA0470 forms a sparse biofilm (Fig. 4). This triple mutant exhibits normal twitching motility, but its growth in broth containing desferrioxamine is poor (data not shown). These data are consistent with the conclusion that with sufficient levels of cytoplasmic iron P. aeruginosa biofilms form mushroom-like structures. These experiments also provide evidence suggesting that PA3899-3901 are involved in ferric dicitrate uptake and that the PA0470 and PA2466 gene clusters are involved in ferrioxamine uptake. Further work is required to critically establish these gene clusters as ferric dicitrate and desferrioxamine uptake systems.
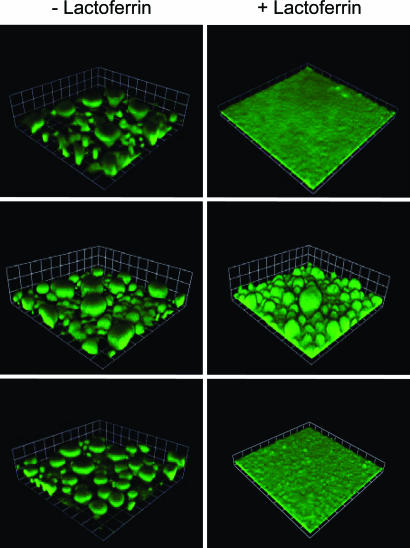
Intracellular Iron Signaling Is Mediated by Fur. The experiments described above are consistent with the hypothesis that biofilm development is regulated by intracellular iron. The major intracellular iron regulator in Proteobacteria including P. aeruginosa is Fur (6, 15). The P. aeruginosa Fur appears to be an essential gene; however, Fur missense mutants have been isolated (17, 18). Fur is an iron-dependent transcription repressor. Thus, one might predict that a P. aeruginosa Fur mutant could build structured biofilms even under lactoferrin-induced iron limitation. Accordingly, we followed biofilm development of a Fur mutant in the presence and absence of lactoferrin. Without lactoferrin the mutant forms mushroom structures that resembled parent structures without lactoferrin (Fig. 5). In the presence of lactoferrin, the mutant forms structured biofilms whereas the parent forms the expected flat, thin biofilms (Fig. 5). These observations suggest that Fur controls genes that are crucial in biofilm development.
Fig. 5.
Biofilm formation by Fur and PrrF mutants. (Top) Biofilms of the parent PAO1. (Middle) Biofilms of the Fur mutant PAO1furC6Tc. (Bottom) Biofilms of the PAO1ΔprrF1-F2 double mutant. Where indicated, biofilms were grown in the presence of 20 μg/ml lactoferrin. Strain PAO1 and PAO1ΔprrF1-F2 contained the gfp plasmid pMRP9-1. The Fur mutant was stained with propidium iodide (we could not introduce pMRP9-1 into the strain). Images in Left are of 6-day biofilms, and the squares are 60 μm on a side. Images in Right are also images of 6-day biofilms, but the squares are 30 μm on a side.
Wilderman et al. (16) recently identified two Fur-repressed sRNA molecules in P. aeruginosa. These sRNAs (PrrF1 and PrrF2) are >95% identical to each other, and expression of both is repressed by iron. However, each can also be independently regulated by other factors (e.g., heme). Deletion of both sRNAs is required to affect the iron-dependent regulation of an array of genes, including those involved in resistance to oxidative stress, iron storage, and intermediary metabolism. Unlike the Fur mutant, our PrrF1F2 double mutant formed biofilms similar to the parent mushroom-like structures in the absence of lactoferrin and thin, flat biofilms in the presence of lactoferrin (Fig. 5). Thus, we conclude that these sRNAs are not required for biofilm formation under the conditions used in this study.
Discussion
Singh et al. (4) discovered that the mammalian iron chelator lactoferrin restricts the development of P. aeruginosa biofilms. In the presence of lactoferrin, P. aeruginosa forms a thin layer of cells on glass surfaces and the individual cells exhibit incessant twitching motility. In the absence of lactoferrin, P. aeruginosa constructs thicker biofilms consisting of cells clustered in mushroom-like structures. The lactoferrin effect was correlated to its activity as an iron chelator. These experiments indicated that iron serves as an environmental signal for P. aeruginosa biofilm development. To better understand the role of iron in P. aeruginosa biofilm formation, we studied a variety of mutants with defects in known or suspected iron-uptake and regulatory functions. In the absence of a functional iron uptake system, P. aeruginosa forms thin, flat biofilms in the absence of lactoferrin (Fig. 1). We show that P. aeruginosa can acquire iron and support normal biofilm formation by using the endogenous siderophores pyoverdine (at relatively low external iron concentrations) or pyochelin (at higher iron levels). It can also use specific iron-responsive ECF σ-factor systems to acquire ferric citrate or ferrioxamine for biofilm formation (Figs. 1, 2, 3, 4). It cannot acquire sufficient amounts of iron for normal biofilm development by passive diffusion in the flow-cell system (Fig. 3). All of our data on biofilm formation by iron acquisition mutants are consistent with the conclusion that biofilm formation in flow cells requires active iron transport and sufficient intracellular iron serves as a cue for development of mushroom-like structures. Although the pyoverdine uptake system has a dual function in that it also serves to regulate expression of a number of virulence genes directly (11, 36), this does not appear to be its role in iron regulation of biofilm formation.
Strains with mutations in the other predicted iron uptake systems formed normal biofilms. This finding is inconsistent with a recent report that a specific virulence mutant had a defect in a putative iron receptor (PA0151) and had a defect in biofilm formation (37). We tested two PA0151 mutants, and both formed normal biofilms in the absence of lactoferrin. This discrepancy could result from the differences in experimental protocols or strain variation. Our results show that, under the conditions we used and with derivatives of strain PAO1, normal biofilms form as long as there is a functional pyoverdine system and there is a sufficient level of environmental iron for the pyoverdine system to function.
As discussed above, a mutant strain that could not produce the high-affinity siderophore pyoverdine or the low-affinity siderophore pyochelin is unable to form mushroom-like structures even in the presence of high levels of inorganic iron, but it can form these structures when given ferric dicitrate or desferrioxamine (Fig. 2). We know that P. aeruginosa can use ferric citrate and other chelated forms of iron (9, 31, 33), but there is no experimental evidence linking specific genes to ferric dicitrate utilization. Among the various mutants with specific defects in putative iron-responsive ECF σ-factor systems that we tested, there is one with a mutation in the PA3899-3901 gene cluster. The polypeptides coded by genes in this cluster show >50% identity to the E. coli ferric uptake gene products FecA, FecI, and FecR (35). This E. coli system is responsible for ferric dicitrate uptake. We thus constructed a mutant incapable of producing pyoverdine and this putative ferric citrate system. This mutant produced thin, flat biofilms even when ferric dicitrate was provided (Fig. 4). This finding suggests that the PA3899-3901 cluster codes for a ferric dicitrate uptake system. We also have similar data suggesting that two gene clusters PA2466-2468 and PA0470-0472 facilitate ferrioxamine uptake (Fig. 4). Further characterization of these gene clusters is required to verify their role in iron transport. We are currently employing this screening approach to correlate specific iron-responsive ECF σ-factor systems with the ability to use different chelated forms of iron for biofilm development.
If, as we conclude, cytoplasmic iron functions as the signal for development of mushroom-like structures, a next question is what is the intracellular iron response regulator? A logical candidate is Fur, the major iron-responsive transcriptional regulator in P. aeruginosa (15). If Fur controls a gene or genes required for biofilm development into mushroom-like structures, one might predict that a Fur mutant could form such clusters in the presence of lactoferrin where the parent forms thin, flat biofilms. We tested a strain with a point mutation in this essential gene. This mutation results in low Fur activity (17). The influence of lactoferrin on the biofilm development of this mutant was not as severe as the influence of lactoferrin on the parent. Whereas the parent formed flat, thin biofilms in the presence of lactoferrin, the mutant formed large clusters of cells reminiscent of, but not identical to, the parent in the absence of lactoferrin (Fig. 5). Fur appears to mediate iron signaling in biofilm development through its regulation of multiple iron acquisition systems and their regulatory genes (e.g., ECF σ factors). The fact that Fur mutants produce high amounts of pyoverdine (17) may account in part for the biofilm architecture of our Fur mutant. We do not know what other Fur-regulated factors might be involved in biofilm formation. Nevertheless, we ruled out a contribution of the Fur-repressed sRNAs in biofilm growth (Fig. 5).
Biofilm development is thought to occur through a series of discrete steps. The first step involves attachment of cells to a surface. The second step is microcolony formation. A third step involves maturation of microcolonies into mushroom-like structures. Finally, there is a detachment step. Previous work suggests that iron sensing might serve as a checkpoint in the first two steps. O'Toole and Kolter (38) reported that the defect in certain attachment mutants can be overcome if sufficient iron is added to the medium. Singh et al. (4) showed that low iron stimulates twitching motility and blocks the formation of microcolonies on glass surfaces. Our data suggest that there may be another iron checkpoint for development of microcolonies that is not related to twitching. In support of a third checkpoint, the pyoverdine mutants (PvdA, FpvA, and PvdS) did not show a twitching motility phenotype. Furthermore, time-lapse microscopy of the mutants developing as biofilms did not show the incessant twitching described by Singh et al. (data not shown). The Fur mutant appears to bypass all three hypothetical checkpoints and form mushroom-like structures even in the presence of lactoferrin.
Previous studies showed that the formation of mushroom-like structures by P. aeruginosa biofilms depended on the carbon and energy source provided in the growth medium (39, 40). For biofilms grown in minimal medium, glucose supported formation of mushroom-like structures, but citrate did not. This finding may suggest that the iron-regulatory pathway and the carbon source-regulatory pathway might converge at a point downstream of Fur involvement.
It is not surprising that multiple iron uptake systems play a role in biofilm formation. It is likely that the different environments where P. aeruginosa resides dictate which forms of iron might be available. For example, ferric citrate or ferrioxamine are not likely to be available iron sources in a mammalian host. Finally, it must be said that no one yet understands the significance of the formation of mushroom-like structures on glass surfaces to biofilm formation in natural environments. Nevertheless, understanding the pathways leading to formation of these structures informs us about regulatory networks involved in biofilm development. It seems clear that iron plays a critical role in biofilm formation. It may be that there are several iron-regulated steps in biofilm formation. We show that at least one of these depends on the ability of cells to import sufficient levels of iron for Fur activity.
Acknowledgments
We thank Michael Jacobs, Colin Manoil, and Maynard Olson for providing us with the P. aeruginosa mutants from the University of Washington Genome Center P. aeruginosa PAO1 transposon mutant library. We thank Pradeep Singh for his advice and comments. This work was supported by a grant from the W. M. Keck Foundation (to E.P.G.), National Institute of General Medical Sciences Grant GM59026 (to E.P.G.), and National Institute of Allergy and Infectious Diseases Grant AI15940 (to M.L.V.). E.B. was partially funded by the Fulbright program.
Author contributions: E.B. and E.P.G. designed research; E.B. performed research; E.B., M.L.V., and E.P.G. analyzed data; E.B., M.L.V., and E.P.G. wrote the paper; and M.L.V. contributed new reagents/analytic tools.
Abbreviations: ECF, extracytoplasmic function; Tc, tetracycline; Fur, ferric uptake regulator; TSB, Bacto Tryptic Soy Broth; sRNA, small regulatory RNA.
References
- 1.Costerton, J. W., Stewart, P. S. & Greenberg, E. P. (1999) Science 284, 1318-1322. [DOI] [PubMed] [Google Scholar]
- 2.Hall-Stoodley, L., Costerton, J. W. & Stoodley, P. (2004) Nat. Rev. Microbiol. 2, 95-108. [DOI] [PubMed] [Google Scholar]
- 3.Lyczak, J. B., Cannon, C. L. & Pier, G. B. (2000) Microbes Infect. 2, 1051-1060. [DOI] [PubMed] [Google Scholar]
- 4.Singh, P. K., Parsek, M. R., Greenberg, E. P. & Welsh, M. J. (2002) Nature 417, 552-555. [DOI] [PubMed] [Google Scholar]
- 5.Singh, P. K. (2004) Biometals 17, 267-270. [DOI] [PubMed] [Google Scholar]
- 6.Ratledge, C. & Dover, L. G. (2000) Annu. Rev. Microbiol. 54, 881-941. [DOI] [PubMed] [Google Scholar]
- 7.Braun, V. (1997) Biol. Chem. 378, 779-786. [PubMed] [Google Scholar]
- 8.Griffiths, E. (1999) in Iron and Infection: Molecular, Physiological and Clinical Aspects, eds. Bullen, J. J. & Griffiths, E. (Wiley, New York), 2nd Ed., pp. 1-26.
- 9.Poole, K. & McKay, G. A. (2003) Front. Biosci. 8, d661-d686. [DOI] [PubMed] [Google Scholar]
- 10.Wandersman, C. & Delepelaire, P. (2004) Annu. Rev. Microbiol. 58, 611-647. [DOI] [PubMed] [Google Scholar]
- 11.Lamont, I. L., Beare, P. A., Ochsner, U., Vasil, A. I. & Vasil, M. L. (2002) Proc. Natl. Acad. Sci. USA 99, 7072-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Visca, P., Leoni, L., Wilson, M. J. & Lamont, I. L. (2002) Mol. Microbiol. 45, 1177-1190. [DOI] [PubMed] [Google Scholar]
- 13.Braun, V. & Braun, M. (2002) FEBS Lett. 529, 78-85. [DOI] [PubMed] [Google Scholar]
- 14.Ochsner, U. A., Wilderman, P. J., Vasil, A. I. & Vasil, M. L. (2002) Mol. Microbiol. 45, 1277-1287. [DOI] [PubMed] [Google Scholar]
- 15.Vasil, M. L. & Ochsner, U. A. (1999) Mol. Microbiol. 34, 399-413. [DOI] [PubMed] [Google Scholar]
- 16.Wilderman, P. J., Sowa, N. A., FitzGerald, D. J., FitzGerald, P. C., Gottesman, S., Ochsner, U. A. & Vasil, M. L. (2004) Proc. Natl. Acad. Sci. USA 101, 9792-9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barton, H. A., Johnson, Z., Cox, C. D., Vasil, A. I. & Vasil, M. L. (1996) Mol. Microbiol. 21, 1001-1017. [DOI] [PubMed] [Google Scholar]
- 18.Ochsner, U. A., Vasil, A. I., Johnson, Z. & Vasil, M. L. (1999) J. Bacteriol. 181, 1099-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs, M. A., Alwood, A., Thaipisuttikul, I., Spencer, D., Haugen, E., Ernst, S., Will, O., Kaul, R., Raymond, C., Levy, R., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 14339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ochsner, U. A. & Vasil, M. L. (1996) Proc. Natl. Acad. Sci. USA 93, 4409-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King, E. O., Ward, M. K. & Raney, D. E. (1954) J. Lab. Clin. Med. 44, 301-307. [PubMed] [Google Scholar]
- 22.Hoang, T. T., Karkhoff-Schweizer, R. R., Kutchma, A. J. & Schweizer, H. P. (1998) Gene 212, 77-86. [DOI] [PubMed] [Google Scholar]
- 23.de Lorenzo, V. & Timmis, K. N. (1994) Methods Enzymol. 235, 386-405. [DOI] [PubMed] [Google Scholar]
- 24.Schuster, M., Hawkins, A. C., Harwood, C. S. & Greenberg, E. P. (2004) Mol. Microbiol. 51, 973-985. [DOI] [PubMed] [Google Scholar]
- 25.Bailey, J. & Manoil, C. (2002) Nat. Biotechnol. 20, 839-842. [DOI] [PubMed] [Google Scholar]
- 26.Miller, W. G., Leveau, J. H. & Lindow, S. E. (2000) Mol. Plant-Microbe Interact. 13, 1243-1250. [DOI] [PubMed] [Google Scholar]
- 27.Newman, J. R. & Fuqua, C. (1999) Gene 227, 197-203. [DOI] [PubMed] [Google Scholar]
- 28.Ochsner, U. A., Johnson, Z., Lamont, I. L., Cunliffe, H. E. & Vasil, M. L. (1996) Mol. Microbiol. 21, 1019-1028. [DOI] [PubMed] [Google Scholar]
- 29.Davies, D. G., Parsek, M. R., Pearson, J. P., Iglewski, B. H., Costerton, J. W. & Greenberg, E. P. (1998) Science 280, 295-298. [DOI] [PubMed] [Google Scholar]
- 30.Yarwood, J. M., Bartels, D. J., Volper, E. M. & Greenberg, E. P. (2004) J. Bacteriol. 186, 1838-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cox, C. D. (1980) J. Bacteriol. 142, 581-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cornelis, P., Moguilevsky, N., Jacques, J. F. & Masson, P. L. (1987) Antibiot. Chemother. 39, 290-306. [DOI] [PubMed] [Google Scholar]
- 33.Poole, K., Neshat, S., Krebes, K. & Heinrichs, D. E. (1993) J. Bacteriol. 175, 4597-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brock, J. H., Liceaga, J. & Kontoghiorghes, G. J. (1988) FEMS Microbiol. Immunol. 1, 55-60. [DOI] [PubMed] [Google Scholar]
- 35.Mahren, S., Enz, S. & Braun, V. (2002) J. Bacteriol. 184, 3704-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beare, P. A., For, R. J., Martin, L. W. & Lamont, I. L. (2003) Mol. Microbiol. 47, 195-207. [DOI] [PubMed] [Google Scholar]
- 37.Potvin, E., Lehoux, D. E., Kukavica-Ibrulj, I., Richard, K. L., Sanschagrin, F., Lau, G. W. & Levesque, R. C. (2003) Environ. Microbiol. 5, 1294-1308. [DOI] [PubMed] [Google Scholar]
- 38.O'Toole, G. A. & Kolter, R. (1998) Mol. Microbiol. 28, 449-461. [DOI] [PubMed] [Google Scholar]
- 39.O'Toole, G. A., Gibbs, K. A., Hager, P. W., Phibbs, P. V., Jr., & Kolter, R. (2000) J. Bacteriol. 182, 425-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klausen, M., Aaes-Jorgensen, A., Molin, S. & Tolker-Nielsen, T. (2003) Mol. Microbiol. 50, 61-68. [DOI] [PubMed] [Google Scholar]