Abstract
In contrast to many other virus infections, primary cytomegalovirus (CMV) infection does not fully protect against reinfection. Accordingly, clinical data have revealed a coexistence of multiple human CMV variants/strains in individual patients. Notably, the phenomenon of multiple infection was found to correlate with increased virus load and severity of CMV disease. Although of obvious medical relevance, the mechanism underlying this correlation is unknown. A weak immune response in an individual could be responsible for a more severe disease and for multiple infections. Alternatively, synergistic contributions of variants that differ in their biological properties can lead to qualitative changes in viral fitness by direct interactions such as genetic recombination or functional complementation within coinfected host cells. We have addressed this important question paradigmatically with the murine model by differently designed combinations of two viruses employed for experimental coinfection of mice. Specifically, a murine cytomegalovirus (MCMV) mutant expressing Cre recombinase was combined for coinfection with a mutant carrying Cre-inducible green fluorescent protein gene, and attenuated mutants were combined for coinfection with wild-type virus followed by two-color in situ hybridization studies visualizing the replication of the two viruses in infected host organs. These different approaches concurred in the conclusion that coinfection of host cells is more frequent than statistically predicted and that this coinfection alters virus fitness by functional trans-complementation rather than by genetic recombination. The reported findings make a major contribution to our molecular understanding of enhanced CMV pathogenicity in the multiply infected host.
Human cytomegalovirus (HCMV), a betaherpesvirus that is highly prevalent in the population, is currently the most frequent cause of viral congenital infection in the western world and a major cause of morbidity and fatal disease in patients undergoing bone marrow or organ transplantation, as well as in patients suffering from acquired or inborn immunodeficiency.
Apparently, primary HCMV infection offers only partial protection against reinfection by a different strain of the virus, as defined by serology and the genotype of variants of the CMV glycoprotein H (6). This implies that multiple CMV strains can accumulate in a person during a lifetime. Genomes of multiple HCMV strains have been found to coexist in the immunodeficient host (12). This important finding has been reported for immunocompetent patients as well (18, 32). Therefore, the coinfection observed in the immunocompromised patient could be caused by simultaneous reactivation of previously acquired multiple strains and not exclusively by reinfections with novel CMV strains upon immunosuppression. The presence of multiple CMV strains/variants during congenital HCMV infection strongly correlated with lethal outcome during gestation (4). Likewise, studies of transplantation recipients revealed a correlation between multiple infections and CMV load (11), delayed virus clearance (21), graft rejection (11), and disease progression (11, 43).
The mechanism underlying this correlation is not known. It is an inherent problem that clinical data cannot clarify whether the presence of multiple HCMV strains is the cause or the result of the patient's disease condition. A weak immune response in an individual could be responsible for a more severe disease and for multiple infections. In that case, the host condition defines the propensity for multiple infections. On the other hand, if different virus strains/variants complement each other to increase their “collective fitness,” multiple infections lead to enhanced pathogenicity. It is generally held that coinfection with different microorganisms leads to aggravated disease (15). Polymorphism is widely observed among HCMV genes (27, 38), which is likely to provide a molecular basis for differences in biological properties between HCMV variants.
Enhancement of pathogenicity by multiple infections can result from extracellular cooperativity of independently replicating viruses, such as by differential expression of homologs/analogs of cytokines and chemokines and their receptors, by which one variant may modulate the inflammatory environment to the benefit of other variants. Alternatively, but not mutually exclusively, variants may interact directly. Thus, genetic recombination, as well as trans-complementation within coinfected cells, bears a risk of enhancement of virus fitness and pathogenic potential.
Although the isolation of recombinant HCMVs from patients carrying multiple HCMV strains (4, 18, 32) implied coinfection of individual host cells, the likelihood of this event under in vivo conditions awaits closer investigation. However, a systematic analysis is not feasible in patients, and HCMV cannot be studied in animal models due to the species specificity of CMVs (33).
Previous studies in an animal infection model showed that herpes simplex virus (HSV) variants interact during primary infection (13, 22, 44). However, HSV differs from HCMV in many biological aspects that define the chance for virus interactions, for instance, cell tropism and dissemination routes. Therefore, it is not clear whether these findings can be extrapolated to explain the clinical findings in CMV infections. The mouse CMV (MCMV) resembles its human counterpart by sharing many homologous genes, as well as in most aspects of biology, including cell tropism or blood-borne dissemination. It thus represents the most intensively studied in vivo model of CMV infection to date (36).
Here, we show that viable, attenuated MCMV variants, as in the case of HSV coinfection, benefit from coinfection. Moreover, we extend the previous studies by showing direct evidence of functional complementation of virus variants in vivo. We also show that MCMV variants regularly meet in individual cells upon separate systemic infections. Finally, we demonstrate that attenuated CMV variants codisseminate to distant organs and coinfect tissue cells in these organs at a frequency that can be explained only if the viruses meet each other at the site of infection and comigrate during their dissemination.
MATERIALS AND METHODS
Cells and mice.
NIH 3T3 (ATCC catalog no. CRL-1658) and M2-10B4 (ATCC catalog no. CRL-1972) were grown as previously described (29). Murine embryonic fibroblasts (MEFs) were prepared and maintained as described previously (36). Female BALB/c mice were purchased from Harlan-Winkelmann, infected at 7 to 8 weeks of age, and kept under specific-pathogen-free conditions throughout. Animal experiments were approved by the responsible state office (approval no. 211-2531-38/99).
Viruses and viral mutagenesis.
Wild-type MCMV (WT-MCMV) was derived from pSM3fr (52), a molecular clone with in vivo growth properties comparable to the MCMV Smith strain. All of the viruses were propagated on M2-10B4 cells, as described previously (29). Virus quantification was done by plaque assay on MEFs, essentially as previously described (36). Tn-M36.A and ΔM36-MCMV have been described previously (29).
Generation of Cre-MCMV.
A 1.2-kbp BamHI-SalI fragment of pGS403 carrying part of the Cre open reading frame interrupted by an intron (47) was inserted into BamHI/SalI-digested pMC-Cre (17). A 2.2-kbp XhoI fragment with the Cre gene was transferred into PmeI-digested pHMM5 (2) to provide the flanking sequences required for recombination into the MCMV ie2 locus, and then a 9.3-kbp HindIII fragment from the resulting plasmid was inserted into pST-TRαI, which is a pST76K-derived shuttle plasmid (37) encoding tetracycline resistance, RecA, and the lacZ α-peptide. The Cre gene was inserted into bacterial artificial chromosome (BAC) pSM3fr (52) by employing a two-step replacement mutagenesis procedure in Escherichia coli strain DH10B as previously described (30), which resulted in the formation of BAC pSM3fr-Cre. Bacterial clones carrying resolved cointegrates were identified by their white color on agar plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside).
Generation of loxPGFP-MCMV.
pCMV-LneoL-βGal (14) was digested with HindIII and SacI. A HindIII/SacI-cut 900-bp DNA fragment carrying the enhanced green fluorescent protein (GFP) open reading frame without the ATG start codon and followed by the simian virus 40 polyadenylation signal, which was generated by PCR using primers gfplox.for (5′-GCGAAGCTTGGTGAGCAAGGGCGAGGAGCTGT-3′) and gfplox.rev (5′-GGCGAGCTCCAGACATGATAAGATACATTGATGAG-3′), was inserted, resulting in pCMV-LneoL-gfp. A 3.1-kbp fragment from pCMV-LneoL-gfp obtained by NsiI/SacI digestion and blunt ended by Klenow treatment was inserted in PmeI-digested pHMM5-Kan, leading to pHMM-GFP-STOP. pHMM5-Kan is a derivative of pHMM5 with a kanamycin resistance gene flanked by FLP recognition target sites from pCP15 (9) inserted into the NsiI site of pHMM5. An 8.2-kbp AvrII fragment was isolated from pHMM-GFP-STOP and used for red α,β,γ-mediated recombination with the MCMV BAC pSM3fr (52) as previously described (51). The kanamycin resistance marker was subsequently removed by FLP-mediated recombination as previously described (9), which resulted in BAC pSM3fr-GFP-STOP. The BACs pSM3fr-Cre and pSM3fr-GFP-STOP were transfected into murine cells to give rise to the Cre-MCMV and loxPGFP-MCMVs, respectively.
In vivo coinfection protocols. Purified virus stocks were diluted in cold sterile phosphate-buffered saline and kept separate at all time points before infection to prevent a possible aggregation of virions. Thus, we minimized a bias to the favor of coinfection of cells. To avoid channeling of viruses towards the same cell populations at the beginning of infection, mice were infected intraperitoneally (i.p.) by two consecutive injections, with an interval of 45 min between injections or (where indicated) intravenously (i.v.), using the same time interval.
Quantification of infectious virus in organs.
Infectious virus was quantitated by plaque assay according to established procedures essentially as described previously (40) with some modification. In brief, organs were homogenized in 5 ml of Dulbecco's modified Eagle medium (supplemented with 10% fetal calf serum) and diluted in 1:10 steps. Diluted homogenates were layered on MEFs and centrifuged at 1,000 × g for 30 min for enhancement of infectivity. The centrifugation was followed by 30 min of incubation at 37°C. Finally, supernatants were discarded, and cells were overlaid with carboxymethylcellulose (Sigma; catalog no. C-4888) to prevent secondary viral spread. For quantification of GFP-expressing viruses from Cre-MCMV- and loxPGFP-MCMV-coinfected organs, homogenates were titrated in triplicate at 1:3.2 steps, which ensured an optimal number of 20 to 40 plaques/well in each sample. At this concentration, the assay allowed the detection of as few as 1 GFP-positive plaque among a total of 100 plaques. The low plaque density in combination with a carboxymethylcellulose overlay prevented the merging of plaques and minimized the possibility of in vitro recombination events. To exclude such events from the counts, only plaques in which all infected cells expressed GFP were considered to have resulted from in vivo recombination events.
Quantitative PCR assays.
Whole organs were homogenized on disposable cell strainers (100-μm pore size; Falcon BD catalog no. 352360) in a total volume of 5 ml. DNA from 200 μl of submandibular salivary gland homogenates was isolated by using the High-Pure viral nucleic acid kit (Roche), according to the manufacturer's instructions. A 1/25 aliquot of DNA was used for quantitative PCR. Thus, the theoretical detection limit was 625 MCMV genome copies for the salivary glands. Due to the larger amount of genomic DNA, homogenates of lungs and spleen were diluted 1:10 prior to DNA isolation. Accordingly, the detection limit in these organs approximates 6,250 viral genomes per organ. Quantitative PCR was done in the LightCycler apparatus using the QuantiTect SYBR Green PCR kit (QIAGEN GmbH, Hilden, Germany; catalog no. 204243). ΔM36 MCMV genomes were quantified with primers specific for the kanamycin resistance gene (kan): Kan-fw (5′-GGTTTGGTTGATGCGAGTGAT-3′) and Kan-rev. (5′-GAC GACTGAATCCGGTGAGAAT-3′). Cycling conditions were 45 cycles, with each cycle consisting of 95°C for 5 s, 61°C for 15 s, and 72°C for 10 s with signal detection at 78°C at the end of each cycle. The specificity of each PCR was monitored by melting-curve analysis.
Qualitative PCR assays.
DNA was isolated as described above. The Cre-mediated GFP recombination was detected by PCR using primers GFPloxP-fw (5′-GCGTGGATAGCGGTTTGACT-3′) and GFPloxP-rev. (5′-GTCGTGCTGCTTCATGTGGT-3′). Cycling was done for 40 cycles, each cycle consisting of 94°C for 30 s, 59°C for 30s, and 72°C for 60 s with standard PCR buffer supplemented with 8% dimethyl sulfoxide. PCR for the MCMV M36 exon 1 was done using primers M36EX1-fw (5′-TCATCACACAGACCACCGTAT C-3′) and M36EX1-rev. (5′-GCAAGAGGAACAACAACCCG-3′). Cycling was done as follows: 40 cycles, each consisting of 95°C for 30 s, 59°C for 30 s, and 72°C for 45 s. PCR specific for the MCMV M54 gene was done using primers MCMV-fw (5′-ATCATCCGTTGCATCTCGTTG-3′) and MCMV-rev. (5′-CGCCATCT GTATCCGTCCAT-3′). Cycling was done for a total of 30 cycles, with each cycle consisting of 95°C for 30 s, 58°C for 30 s, and 72°C for 30 s.
Two-color in situ hybridization (2C-ISH).
2C-ISH was used for simultaneous detection of two different viruses in serial tissue sections from coinfected mice. Procedures and applications have been described previously in greater detail (16, 36). In essence, a red label is used to visualize the viral gene M36 in intranuclear inclusion bodies of cells infected with WT-MCMV, whereas a black label is assigned to the kan gene sequence present in mutant ΔM36-MCMV (29). DNA hybridization was performed with deparaffinized serial sections of liver tissue as explained previously (16, 36), except that the technique was further improved by generating 1-μm sections to increase the probability for multiple sections within individual intranuclear inclusion bodies. The hybridization probe M36-P spans 1,644 bp of gene M36 from nucleotide (nt) positions 47,624 to 49,248 of the MCMV strain Smith genome (GenBank/EMBL/DDBJ, accession no. U68299; complete sequence) (39) It was synthesized by PCR using plasmid pCR3-M36 (29) as the template and oligonucleotides 5′-ATATCCCCGTGTCATCTTAA-3′ (nt 47,624 to 47,643) and 5′-ATGTATGAGCAAGAGGAACA-3′ (nt 49,267 to 49,248) as primers. M36-P was tagged during PCR by incorporation of fluorescein-conjugated dUTP (fluorescein-12-dUTP; catalog. no. 1373242, Roche Laboratories). The hybridization probe Kan-P spans 558 bp of the kanamycin resistance gene kan (transposon Tn903; EMBL accession no. V00359). It was synthesized by PCR using pDrive Cloning Vector (from the QIAGEN PCR Cloning kit) as the template and oligonucleotides 5′-TCAGGTGCGACAATCTATCG-3′ and 5′-TCCGACTCGTCCAACATCAA-3′ as primers. Kan-P was tagged during the PCR by incorporation of digoxigenin-11-dUTP (catalog no. 1093088; Roche Laboratories). Red staining was achieved by using alkaline phosphatase-conjugated anti-fluorescein antibody (catalog. no. 1426338; Roche Laboratories) with fuchsin as the chromogenic substrate. Black staining was achieved by using peroxidase-conjugated antidigoxigenin antibody (catalog no. 1207733; Roche Laboratories) with diaminobenzidine tetrahydrochloride as the substrate, followed by color enhancement with ammonium nickel sulfate hexahydrate. Quantitative microscopic analysis and the photodocumentation were performed as described previously (16).
Statistical analysis.
The probability of random coinfection of liver cells was evaluated by using the Poisson distribution function F(n) = λn × e−λ/n! (factorial), which gives a probability for single hit of F(1) = λ × e−λ and a probability for dual hit of F(2) = F(1) × λ/2 (25).
RESULTS
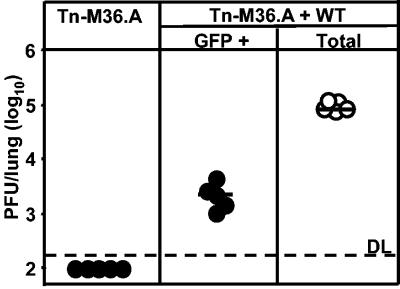
WT-MCMV promotes the in vivo growth of attenuated MCMV variants. To study whether interactions between CMV variants provide them with replication advantages, we used coinfection of mice with WT-MCMV and a growth-attenuated, GFP-expressing mutant MCMV as a first paradigmatic model. Replication of the two viruses in host organs was quantified by a plaque assay of organ homogenates, counting white plaques and fluorescent plaques, respectively. Comparison of the virus titers in the coinfected group with those in a control group that was singly infected with the same dose of mutant virus defined the influence of WT virus on the growth of the mutant. Specifically, as an example of an attenuated mutant, we used here Tn-M36.A MCMV, a previously characterized mutant in which the antiapoptotic M36 gene is disrupted by a GFP expression cassette leading to a growth deficiency phenotype in macrophages (29). To avoid a bias for coinfection through aggregation of virions in the inoculum suspension, coinfection was performed consecutively, with Tn-M36.A MCMV administered first, followed 45 min later by WT-MCMV. The assay was performed on day 5 after i.p. infection of BALB/c mice with 105 PFU of each virus. Upon single infection, mutant Tn-M36.A MCMV was undetectable in the lungs, indicating an essential role of the M36 protein for in vivo infection and virus dissemination to distant target organs. In contrast, GFP-expressing virus was found to be present in the lungs of all coinfected mice with a median frequency of ∼2% of the overall infection of the lungs (Fig. 1).
FIG. 1.
Coinfection with WT MCMV rescues an attenuated MCMV in vivo. BALB/c mice were infected i.p. with 5 × 105 PFU of Tn-M36.A virus alone or in combination with the same dose of WT-MCMV. Lungs were assayed for infectious virus at day 5 postinfection (dpi 5), using fluorescence microscopy to quantify Tn-M36.A virus (•) and light microscopy to quantify total MCMV (○). Circles represent individual mice. Horizontal bars indicate group medians. DL, detection limit.
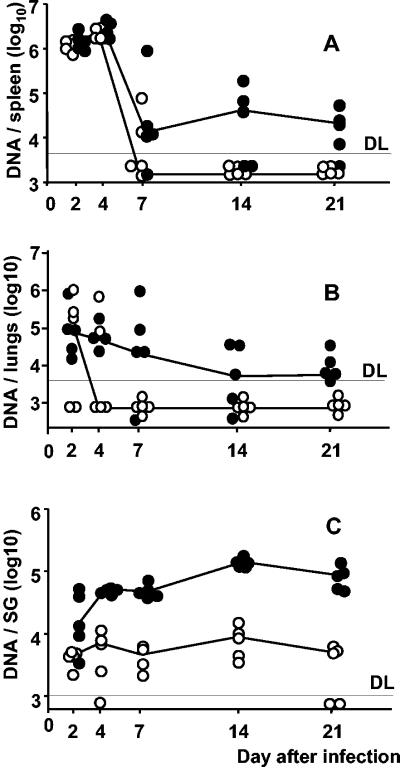
To monitor the influence of WT-MCMV on mutant virus replication over time, we tested in vivo growth kinetics in spleen, lungs, and salivary glands, again comparing mutant virus replication in a coinfected group with its replication after single infection. This experiment was performed with ΔM36-MCMV, a mutant virus in which gene M36 is replaced by the kanamycin resistance gene kan (29). A primer pair specific for kan was used to detect and quantitate ΔM36-MCMV genomes in organ homogenates by real-time quantitative PCR until day 21 after infection (Fig. 2). In essence, in the long-term course of in vivo infection, the mutant virus DNA load was consistently higher after coinfection with WT-MCMV. The most profound impact of WT presence on mutant virus growth was observed in the salivary glands, a known organ site of secondary infection and persistence (20). For the sake of clarity, it should be noted that detection of viral DNA until day 4 does not necessarily represent virus replication, since real-time PCR cannot discriminate between inoculated and replicating virus genomes. Accordingly, the number of ΔM36-MCMV genomes was found to drop between days 4 and 7 unless growth was rescued by trans-complementation with WT-MCMV. There was no significant effect of the ΔM36-MCMV on WT-MCMV replication (not shown). Thus, WT-MCMV supports not only the growth but also the dissemination of ΔM36-MCMV.
FIG. 2.
WT-mediated rescue of ΔM36 occurs at secondary sites of infection at late time points. BALB/c mice were infected i.p. with 105 PFU of ΔM36 (○). In addition, the coinfected group received an equal amount of WT-MCMV by a separate injection (•). Organs were assayed for ΔM36 genomes by real-time PCR at dpi 2, 4, 7, 14, and 21. ΔM36 genome copies per spleen (A), lungs (B) or salivary glands (C) are shown. Circles represent individual mice. Median values are connected. DL, detection limit.
To determine whether WT-MCMV supports the growth of other attenuated mutants, not just specifically of those lacking the M36 gene, we tested as an independent third example a previously described mutant that lacks the immune evasion gene m152. The m152/gp40 immune evasion protein has a dual function: by retention of peptide-loaded major histocompatibility complex class I (MHC-I) molecules in a pre-Golgi compartment, it inhibits the recognition of infected cells by CD8 T cells (24, 54); by down-modulation of ligands of the activating receptor NKG2D, it simultaneously inhibits recognition of infected cells by NK cells (23).
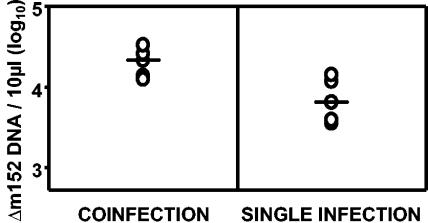
In the specific mutant used here, the m152 gene was replaced by the zeocin resistance gene zeo (51). Accordingly, mutant virus genomes were detected and quantitated by real-time quantitative PCR using mutant-specific primers directed against the zeo cassette. Viruses lacking m152 were growth attenuated in vivo by a factor of ∼10 (reference 24 and data not shown). Compared with the mutant genome copy number found in organs on day 5 after single infection with the mutant virus Δm152-MCMV, coinfection with WT-MCMV induced a threefold median increase in the replication of the mutant in the spleen (Fig. 3), as well as in the lungs (not shown), although only the change in the spleen was statistically significant (P = 0.029). Similarly, a GFP-expressing MCMV lacking the M45 antiapoptotic gene (7), a gene that is essential for viral replication in vivo (26), was able to grow in vivo if coinfected with WT-MCMV (not shown).
FIG. 3.
Coinfection with WT-MCMV rescues Δm152-MCMV in vivo. Mice were infected i.p. with 3 × 105 PFU of Δm152-MCMV. In addition, the coinfected group received an equal amount of WT-MCMV by a separate injection. DNA from organ homogenates was assayed for Δm152 genomes by real-time PCR at dpi 5. Genome copies per 10 μl of spleen homogenate are shown. Circles represent individual mice. Median values are indicated by horizontal bars.
In conclusion, these data have collectively shown that functional complementation is not restricted to the combination of WT-MCMV and a particular attenuated mutant but appears to apply more widely to different types of mutants.
The helper effect of WT-MCMV is not based on recombination.
Thus far the data indicated that WT-MCMV supports the in vivo growth and dissemination of attenuated mutants. This helper effect could be due to trans-complementation or DNA recombination. Homologous or illegitimate DNA recombinations of mutant and WT genomes can reinsert a missing gene in the mutant genome and thus restore the gene function and the virus fitness. However, homologous recombination would have reinserted the original sequence in the mutated locus, a process during which the mutants would have lost the gene expressing GFP or the mutant DNA sequence that was used for detection and quantification in PCR. Therefore, the presence of GFP-expressing MCMV and the increase in virus genomes containing the mutant-specific DNA fragment could not be explained by homologous recombinations. Illegitimate recombinations, which reinsert the WT gene in an ectopic position of the mutant genome, remained to be considered. In that case, the mutant would reacquire WT-like virulence and contain both the WT as well as the mutated sequences.
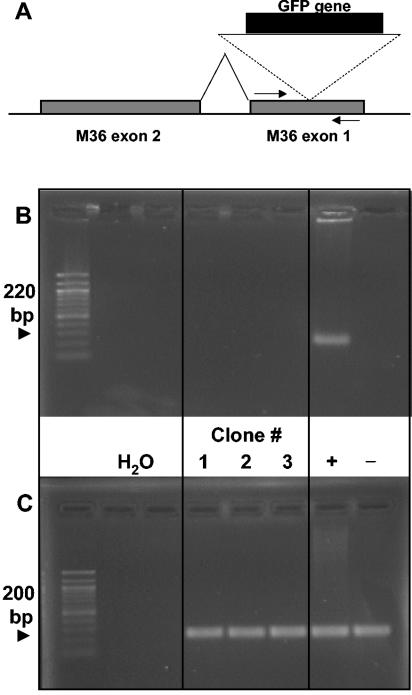
To evaluate a possible contribution of illegitimate recombination, GFP-positive plaques derived from mice coinfected with WT-MCMV and mutant Tn-M36.A MCMV were used for the cloning of progeny virus by repeated plaque purification. A PCR was designed with primers flanking the transposon insertion in gene M36 to give a 200-bp amplificate with the WT sequence but no product in the presence of the intermitting, 3-kb-long transposon (Fig. 4A). The attenuated phenotype of the transposon mutant (Fig. 1) (29) proved that this genomic locus was essential for the function of the M36 gene. As shown in Fig. 4, DNA from representative GFP-positive clones did not yield a PCR product; that is, it did not contain a functional, uninterrupted M36 gene. While this experiment cannot exclude rare events of illegitimate recombination, it was obvious from our data that the frequency of recombination was too low to account quantitatively for the observed helper effect. We therefore conclude that the viruses interact primarily by trans-complementation.
FIG. 4.
WT-mediated rescue is not due to DNA recombination. Mice were separately infected i.p. with 5 × 105 PFU of Tn-M36.A MCMV and 5 × 105 PFU of WT-MCMV. (A) Schematic representation of the transposon insertion which disrupts the exon 1 of the M36 gene, rendering M36 nonfunctional and Tn-M36.A MCMV attenuated. Tn-M36.A MCMV can be identified by GFP expression. Arrows indicate the primers used in the experiment shown in panel B. (B) Three independent GFP-expressing clones, isolated from lungs of three separate mice, were plaque purified by serial passaging on permissive cells. Supernatants of clones 1, 2, and 3 were assayed for ectopic insertion of the M36 gene by PCR, using the primer pair shown in panel A, specific for the M36 exon 1 and yielding an amplificate of 220 bp. +, supernatant from cells infected with WT-MCMV; −, supernatant from cells infected with ΔM36-MCMV. (C) The same supernatants were tested for the presence of MCMV genomes by a PCR specific for the viral DNA polymerase, yielding an amplificate of 200 bp. Ethidium bromide-stained gels are shown.
Cre-mediated functional complementation upon coinfection in vitro.
As an approach to provide direct molecular evidence for functional complementation between two CMVs, we used a reporter system in which, by design, reporter gene expression requires functional complementation between two viruses. Specifically, one mutant, referred to as loxPGFP-MCMV, was constructed to carry an inactive GFP gene introduced into ie2, a locus of the MCMV genome that is dispensable for viral growth in vitro and in vivo (8, 31). The open reading frame of the inserted GFP gene was interrupted by a 3-kbp-long stop cassette, which was flanked by loxP sites. Therefore, the GFP is not expressed unless Cre-mediated recombination removes the stop cassette. Accordingly, mutant Cre-MCMV was constructed to carry the gene encoding Cre recombinase. Thus, loxP sites in loxPGFP-MCMV can only be recombined and GFP only be expressed if Cre recombinase is present within the same cells, accomplished by coinfection of cells with both viruses.
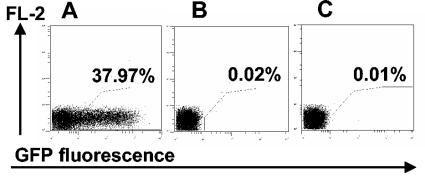
Specifically, NIH 3T3 fibroblasts were infected with Cre-MCMV at a multiplicity of infection (MOI) of 0.5. Supernatant containing released Cre-MCMV virions was harvested after 48 h, a stage at which a substantial proportion of infected cells was already lysed, and was added to cultures of NIH 3T3 cells infected with loxPGFP-MCMV at an MOI of 0.5. Cells singly infected with Cre-MCMV or loxPGFP-MCMV were used as negative controls. Cells were analyzed for GFP expression by flow cytometry at 24 h postinfection. Approximately 40% of the cells that were exposed to Cre-MCMV and loxPGFP-MCMV were positive for GFP expression (Fig. 5A), whereas no GFP expression was found in cells infected with either loxPGFP-MCMV (Fig. 5B) or Cre-MCMV (not shown) alone.
FIG. 5.
Direct evidence for CMV functional complementation. NIH 3T3 cells were infected at an MOI of 1 with loxPGFP-MCMV and analyzed by flow cytometry for GFP expression at 24 h post infection. (A) Cells were additionally treated with supernatant derived from cells infected for 48 h with Cre-MCMV. (B) Cells received loxPGFP-MCMV only. (C) Cells were additionally treated with supernatant derived from cells infected for 48 h with Cre-MCMV filtered through 100-nm-pore-size filters.
These data show that Cre recombinase was active within cells that were infected with the reporter virus loxPGFP-MCMV and exposed to the supernatant of cells infected with Cre-MCMV. Clearly, expression of Cre recombinase within coinfected cells is the straightforward idea and most reasonable mechanism. However, free Cre recombinase released into the supernatant may have been taken up by the reporter cells. To evaluate the possibility of Cre recombinase release and uptake, supernatant of cells infected with Cre-MCMV was passed through 0.1-μm filters to exclude the transfer of infectious virions to the reporter cells. Reporter cells exposed to this filtered supernatant were found not to express GFP (Fig. 5C). In conclusion, reporter gene expression required coinfection of cells.
Functional complementation and single-cell coinfection in vivo.
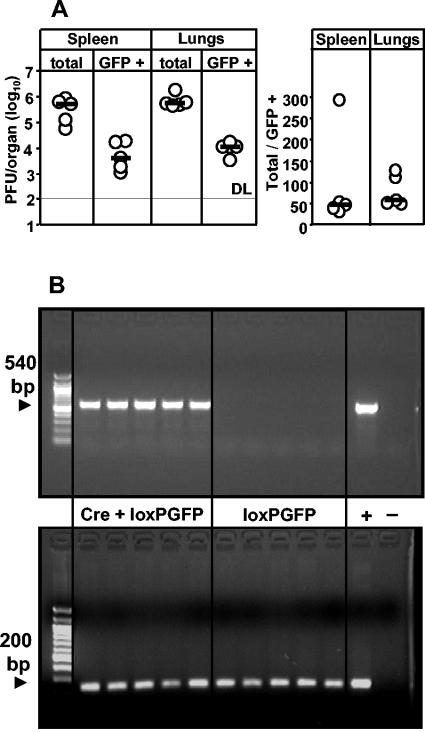
To estimate the incidence of functional complementation in vivo, mice were i.v. coinfected with Cre- and loxPGFP-MCMV. Total and GFP-expressing viruses from organs of coinfected mice were quantified by plaque assay on day 5 postinfection. GFP-expressing viruses were detected in homogenates of spleen and lungs of all mice tested, with the median frequencies being roughly 2% in both organs (Fig. 6A).
FIG. 6.
CMV functional complementation indicates single-cell coinfection events in vivo. Mice were separately infected i.v. with 2 × 105 PFU of Cre-MCMV and 2 × 105 PFU of loxPGFP-MCMV. (A) Organ homogenates were assayed for infectious virus at dpi 5, using fluorescence microscopy to quantify recombined loxPGFP-MCMV and light microscopy to quantify total virus. Circles represent individual mice. Horizontal bars mark the median values. DL, detection limit. The right panel indicates the ratio of total versus GFP-expressing plaques. (B) Organ homogenates of the same mice were assayed by PCR. Control mice (loxPGFP) were infected with loxPGFP-MCMV alone. Recombination events were detected by using a primer pair specific for recombined GFP, yielding an amplificate of 540 bp (top). The presence of MCMV genomes was detected by a PCR specific for the viral DNA polymerase, yielding an amplificate of 200 bp (bottom). +, cell supernatant derived from NIH 3T3 cells coinfected with Cre-MCMV and loxPGFP-MCMV; −, murine genomic DNA.
While this finding unequivocally demonstrated the occurrence of Cre-mediated recombination, it remained a critical question whether the recombination had indeed happened already in vivo or rather during the assay in vitro (see above). Although the probability for in vitro recombination was here technically minimized by a very low MOI in the plaque assay and by the prevention of secondary plaque formation, formal proof for the occurrence of recombination in vivo is only provided by direct analysis of viral genomes in the organ homogenates. We therefore designed a recombination-specific PCR with primers binding to sequences in the GFP gene that flank the loxP sites. Thus, a positive PCR signal can be seen only upon Cre-recombination, which excises the 3-kbp stop cassette from the virus genome. For control, a group of mice was infected with the loxPGFP virus alone to monitor putative Cre-independent recombination events. All five coinfected mice showed a positive GFP PCR signal in the spleen (Fig. 6B, top) and lungs (not shown), whereas control mice did not. Presence of MCMV DNA in all tested samples was controlled by a PCR specific for gene M54 encoding the viral DNA polymerase (Fig. 6B, bottom).
In conclusion, Cre-dependent recombination had already occurred in vivo in the coinfected host. Most importantly, this set of data has shown that functional complementation occurs not only between WT virus and attenuated variants but also between two viruses that are both fully replication competent.
Functional complementation is based on single-cell coinfection.
All data presented above demonstrated functional complementation between two CMVs and provided strong but indirect evidence for in vivo coinfection of cells as the underlying mechanism. Direct evidence for simultaneous replication of two viruses in the same cell requires the detection of the two CMV genomes at the site of DNA replication, which is in the nucleus. In the late phase of the viral replicative cycle at the beginning of virion morphogenesis, viral DNA is packaged into nucleocapsids, which accumulate in a demarcated region of the nucleus that becomes histologically visible as an intranuclear inclusion body. 2C-ISH of serial tissue sections is the method of choice to verify and visualize coinfection of single cells in vivo (16, 36).
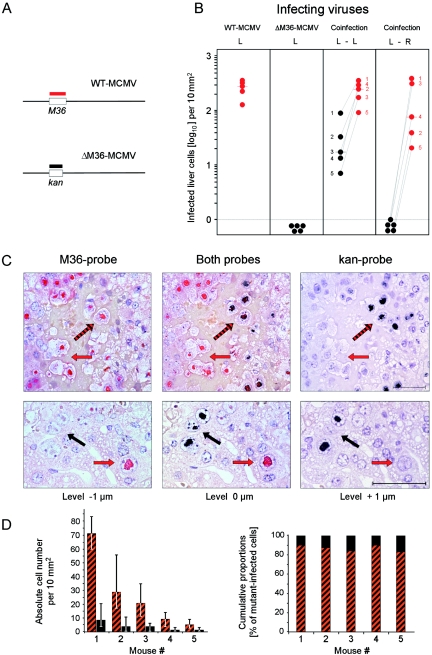
Since CMV disease develops only in the immunocompromised host and since trans-complementation between virus variants most likely occurs in an immunocompromised CMV-positive host after reactivation of one variant and superinfection with another variant or by simultaneous reactivation of variants acquired previously by sequential primary infections (see Discussion), we used the coinfection of immunocompromised mice here as a model for trans-complementation under conditions of CMV disease. The liver was chosen for two reasons: first, because hepatitis is a relevant manifestation of CMV disease; second, because hepatocyte nuclei are large enough for an analysis of viral replication in an individual nucleus by 2C-ISH of serial sections. Specifically, in the experiment shown in Fig. 7, gamma-irradiated BALB/c mice were coinfected subcutaneously by the intraplantar route with WT-MCMV and mutant ΔM36-MCMV, 105 PFU of each. For discrimination of the two viruses in the infected liver, the red-tagged and black-tagged hybridization probes M36-P and Kan-P were used to stain intranuclear inclusion bodies containing WT-MCMV DNA and ΔM36-MCMV DNA, respectively (Fig. 7A). Infected cells were detected by 2C-ISH and quantitated (Fig. 7B). In the single-infection control groups, red-tagged WT-MCMV was found to replicate in the liver, whereas black-tagged ΔM36-MCMV failed. Upon coinfection with WT-MCMV, however, black-tagged ΔM36-MCMV disseminated to the liver and replicated in liver cells. Importantly, the deficiency of ΔM36-MCMV was complemented by WT-MCMV only after ipsilateral intraplantar coinfection, that is, if the two viruses were administered to the same footpad, whereas contralateral intraplantar coinfection failed. This finding indicated that the two viruses had to meet for complementation not within the liver but at the local site of coinfection before they jointly disseminated to the liver. Once in the liver, the two viruses replicated jointly and independently in liver tissue cells, with the hepatocyte as the main target cell type (Fig. 7C). As expected, the fully dissemination-competent and replication-competent WT-MCMV disseminated with higher efficacy, which led to a ∼10-fold median excess of red-stained over black-stained cells in liver tissue sections after ipsilateral intraplantar coinfection (Fig. 7B and C). A detailed serial section-based 2C-ISH analysis of black-stained cells revealed the existence of coinfected hepatocytes (Fig. 7C, top) as well as of hepatocytes in which the mutant virus replicated independently (Fig. 7C, bottom). Notably, although infection density in liver tissue varied considerably between individual mice, which is a usual observation with this experimental system, the proportions of coinfected cells and mutant-only infected cells were found to be remarkably constant, with ∼10 to 20% of the black-stained cells containing only the mutant (Fig. 7D). It is also important to note that cells singly infected by the mutant virus were not regularly found in close proximity to double-infected cells but were usually seen as separate foci of infection (Fig. 5C, bottom). Moreover, a quantitative histometric analysis of foci revealed a higher frequency of red-stained WT-MCMV foci, but no significant size distribution difference between red-stained and black-stained foci (not shown). So, we conclude that mutant virus ΔM36-MCMV grows like WT-MCMV once it has made its way into the liver.
FIG.7.
Coinfection of individual cells in host organs. (A) Maps (not drawn to scale) illustrate the design of DNA probes for discrimination between WT-MCMV and ΔM36-MCMV in host tissues by 2C-ISH. The red-tagged hybridization probe M36-P identifies WT-MCMV DNA in intranuclear inclusion bodies of infected cells, the black-tagged hybridization probe Kan-P identifies the mutant virus DNA. (B) Functional complementation after ipsilateral local coinfection. Groups of BALB/c mice (n = 5) were immunocompromised by total-body gamma irradiation with a dose of 7 Gy and were infected subcutaneously by the intraplantar route with WT-MCMV (red symbol) and/or mutant ΔM36-MCMV (black symbol). L, single infection at the left hind footpad with 105 PFU of the respective virus; L-L, ipsilateral coinfection at the left hind footpad with both viruses, 105 PFU of each; L-R, contralateral coinfection with mutant and WT virus, 105 PFU of each, administered to the left and right hind footpads, respectively. On day 12 after local infection, infected liver cells were detected by 2C-ISH and quantitated by counting in representative 10-mm2 areas of 2-μm sections of liver tissue. Dot symbols represent data from individual mice. The median values are marked. In the coinfected groups, individual mice are numbered according to the rank defined by replication of the mutant virus. (C) 2C-ISH histological images of serial 1-μm sections of liver tissue on day 12 after ipsilateral intraplantar coinfection. Directly proximal sections, defining cut levels 0, −1, and +1 μm were hybridized with the probes indicated. Red and black arrows point to intranuclear inclusion bodies of hepatocytes infected exclusively with WT-MCMV and ΔM36-MCMV, respectively. Black-and-red-striped arrows point to inclusion bodies of coinfected cells containing both types of DNA. Counterstaining was performed with hematoxylin. Bar, 50 μm. (D) Quantitation of coinfected cells and of cells infected exclusively by the mutant. Cells were counted in 10 representative 10-mm2 areas derived from 10 independent 2-μm sections of liver tissue for each individual mouse corresponding to the data shown in panel B. Bars represent the median values with the ranges indicated. Cumulative proportions are based on the median values.
The observed frequency of coinfected foci provides an additional argument against random coinfection of liver cells by dual hits of free virions within the liver. For calculating the statistically expected frequency we need to know the actual multiplicity of infection in the liver; this is difficult to predict, as it depends on unknown parameters such as the portion of coinfection inoculum that reaches the liver passively through the circulation and the amplification by local virus replication at the site of inoculation prior to passive dissemination. However, we can empirically determine the multiplicity of infection from the ratio of the number of nucleated liver cells and the number of foci of infection actually detected in the liver. For a representative liver section area of 100 mm2, this ratio was found to be ∼770:1, which is equivalent to an empirical frequency of productive hits of F(>0) = ∼0.0013 = 1 − F(0). From F(0) = e−λ, the expected frequency of single hits, F(1), of dual hits, F(2), and of multiple hits, F(>2), can be estimated by using the Poisson distribution function. Extrapolated to 2 × 108 permissive cells in the liver, this calculation predicted ∼130,000 single-hit events with each of the two viruses and only ∼168 dual-hit events, with multiple-hit events of negligible frequency. Considering the probability for dual hit by any combination of the two viruses, which is 0.25, the number of liver cells coinfected randomly by WT-MCMV and mutant ΔM36-MCMV is just 84. This calculation predicts a ratio of coinfected foci to single-infected foci of either type as 84/130,000 = ∼1:1,550. By experiment, however, we found 486 red foci, 22 double-stained foci, and only 4 black foci in the representative 100-mm2 area. So, with an observed ratio of 1:22, the dual-infection events were much more frequent than statistically predicted, whereas the events of single infection by mutant ΔM36-MCMV were much less frequent than statistically predicted. Although foci of infection caused by ΔM36-MCMV alone were rare in the liver after intraplantar coinfection, the mere fact that they existed at all (Fig. 7C, bottom) demonstrated that mutant virus replication in hepatocytes, as well as spread between hepatocytes, does not depend upon trans-complementation. In accordance with this notion, systemic infection by the intravenous route favors the seeding of independently growing mutant ΔM36-MCMV to the liver (data not shown).
In conclusion, the quantitative 2C-ISH analysis revealed a dissemination deficiency of mutant ΔM36-MCMV and a coinfection of liver cells by WT-MCMV and the mutant in a frequency that cannot be explained by random dual hits. From the collective evidence, we must propose that WT-MCMV trail-ropes the dissemination-deficient mutant for migration to the liver and that the viruses most likely meet each other in a migratory cell type at the local site of coinoculation.
DISCUSSION
Clinical trials have revealed a correlation between infections by multiple CMV strains and virus load and delayed clearance (11, 21). However, clinical studies did not define whether CMV strains interacted to increase their collective fitness. Here, we have shown that CMV interactions indeed increase the fitness of CMV variants in terms of improved growth and dissemination in the host. Mice infected only with virus mutants showed significantly lower titers in vivo and faster mutant clearance rates than mice that received WT-MCMV in addition to the mutant. By using inbred and sex- and age-matched mice, we excluded potential differences in host susceptibility to infection as contributors to poor mutant clearance in individual mice. Thus, CMV-CMV interactions provide a replication advantage to attenuated viruses.
We and others have previously observed that coinfection with a virulent MCMV can promote the growth of attenuated or even nonviable viral mutants in vivo (36, 42), but the significance of this finding, the frequency of the coinfection events, and the underlying mechanisms were not studied in greater depth. To the best of our knowledge, the data presented here provide for the first time direct and firm evidence of in vivo trans-complementation of CMV variants. The data also allow us to conclude that cis-complementation by homologous or illegitimate recombination is rare and cannot explain the increase in mutant virus titers that was observed in the presence of WT-MCMV.
For the first set of experiments, we chose MCMV mutants whose deleted genes are known to control antiapoptotic or immune-modulating functions. Therefore, the attenuated viruses could only get benefit from the products of the WT genes when both viruses infected the same individual cell in vivo. The approaches of Cre-loxP recombination and in situ hybridization directly demonstrated and even visualized that this is what indeed happens in the tissues of coinfected mice. Moreover, Cre-loxP recombination showed that trans-complementation by coinfection is not dependent on a selective pressure caused by growth deficits of an attenuated coinfection partner. trans-Complementation also occurs regularly between fully replication-competent viruses, between loxPGFP-MCMV and Cre-MCMV, in the specific case shown here as a paradigm. Thus, trans-complementation by coinfection is not just a mechanism for rescuing attenuated variants but bears the potential to further increase fitness and pathogenicity of already highly virulent virus strains.
It is worth noting that our findings do not exclude the possibility that other viral properties, for instance, the expression of viral chemokine homologs (28, 35), may provide a functional trans-complementation even when the viruses do not infect the same cells, namely, by secretion and uptake or receptor-mediated signaling between neighboring or even distantly located cells. In that case, the chance for viral trans-complementation leading to growth advantage and pathogenicity enhancement should be even higher, as such a mechanism is not dependent on the probability of dual or multiple hits at individual target cells.
Interestingly, ΔM36-MCMV inoculated in the footpad disseminated to the liver only in the presence of WT-MCMV. The same applied to infection of the salivary glands, another secondary site of infection. These data argued that mutant MCMV codisseminates with the WT to distant sites in the organism. One explanation for this phenomenon may be the morphogenetic propensity of MCMV to form multicapsid virions in cell culture and in vivo (10, 53), that is, virions harboring several capsids and thus several genomes. So in the case of coinfection of the same cell, two or more different genotypes can be packaged into one multicapsid virion and disseminate jointly. It must be noted, however, that a formal proof for infectivity of these structures is pending.
Another conceivable explanation for codissemination is that multiple CMV mutants codisseminate through joint transport by infected cells. Dendritic cells (1) as well as monocytes-macrophages (48), have been implicated in the dissemination of MCMV, and ΔM36-MCMV has a strong growth deficit in macrophages (29). Reports from the group of E. S. Mocarski indicated that MCMV attracts CD11b− MHC-II− cells of hematopoietic origin to sites of infection and inflammation, exploiting them for dissemination (41, 42, 48). Remarkably, the dissemination of an MCMV mutant lacking a viral chemokine homolog was found to be improved when it was injected in a pool with WT virus (42).
Our work extends the existing knowledge of herpesvirus coinfections (13, 22, 44). The pioneering study had reported on lethal infection of mice by a mixture of two HSV strains, which were both shown to be avirulent in the respective single infections (22). A follow-up study by the same group showed that the majority of HSV clones, plaque purified from coinfected animals, had not gained virulence. This implicated that DNA recombination was not the mechanism underlying the increase in virulence observed in the coinfection experiment (44). Thus, although complementation through coinfection of individual cells was not documented, functional complementation was a likely explanation. Notably, the increase in virulence occurred only when the HSV strains were coinoculated at the same site in a mixture but not when injected separately, which argued for the need of direct viral interactions (22). We provide here direct evidence for single-cell coinfection and trans-complementation as an underlying mechanism of virus interaction. Moreover, alpha- and betaherpesviruses differ in their dissemination routes. The codissemination of alphaherpesviruses is facilitated by the fact that the viruses are channeled into the same axons at the site of primary infection, whereas betaherpesviruses disseminate by utilizing blood cells attracted to the site of infection. Thus, the codissemination of CMVs could not be predicted from the previous work on HSV-1 coinfection. Whereas HSV pathogenesis and biology are determined mainly by infection of epithelial cells and neurons, CMV has a wide range of potential target cells, including dendritic cells, monocyte-macrophages, endothelial cells, smooth muscle cells, epithelial cells, fibroblasts, and bone marrow stromal cells (1, 19, 34, 36, 45, 46, 48). A broad range of permissive cells to choose from should diminish the probability of coinfection of cells, and minor differences in cell tropism between virus variants may even favor a segregation of variants into different host cell types. Yet, as we have shown here, coinoculation at the same subcutaneous site was also important for trans-complementation of CMV, at least for WT-MCMV and the dissemination-deficient mutant ΔM36-MCMV. In this context, it is worth emphasizing that MCMV variants showed a high propensity to coinfect host tissue cells even upon systemic and time-spaced inoculations, as exemplified by replication-competent viruses expressing Cre and Cre-inducible GFP, respectively. Again, as discussed above, it is likely that a migratory cell type in the circulation is the primary target cell that can be hit twice while circulating before it transports the two viruses jointly into host tissues. A limited number of these primary target cells would explain why events of coinfection in tissues are observed more frequently than is statistically predicted from random dual hits in organs with high levels of cellularity.
Multiple CMV strains have been found in autopsy material derived from congenitally infected infants (4), indicating that simultaneous or sequential infection by multiple viruses occurs under conditions of primary infection. It is unknown whether simultaneous infections can also occur later in life in healthy individuals. Can reactivation from latency possibly complement a superinfecting virus or vice versa? Such clinically important questions can now be addressed with the mouse model of CMV coinfection. If reinfection of healthy adults is the principle that initially leads to an accumulation of multiple CMV strains, which can then reactivate under conditions of immunosuppression, CMV strains are likely to interact mainly upon reactivation from latency. Notably, in accordance with this idea, multiple strains were observed to emerge over time in patients who underwent an episode of immunosuppression (3), and the correlation of the presence of multiple strains with high virus load and poor prognosis was observed with immunocompromised adults (11, 21, 43). Several studies reported a high percentage of adult individuals (3, 49), but not infants (5, 50), carrying multiple viral strains. This difference between adult and pediatric patients may also indicate that multiple strains are acquired primarily through consecutive superinfections over time, rather than through a single infection with codisseminating strains, as we see no rationale that could explain why codissemination should affect pediatric patients with lower probability.
The mechanism of trans-complementation by coinfection documented here bears a risk of “fatal alliances” between variants with otherwise moderate pathogenicity and offers an explanation for the poor clinical prognosis of CMV disease in the multiply infected patient.
Acknowledgments
L.C.-S. and U.H.K. were supported by DFG, focus project “New vaccination strategies.” J.P. and M.J.R. were supported by the DFG, Collaborative Research grants 432 (project A10) and 490 (project B1), respectively.
We thank M. Wagner, W. Brune, and C. Menard for assistance with viral constructs. We thank Aysel Rojan for expert technical assistance in the histological studies. We thank P. Chambon and D. Metzger for providing plasmid pCMV-LneoL-βGal and L. Enquist for pGS403.
REFERENCES
- 1.Andrews, D. M., C. E. Andoniou, F. Granucci, P. Ricciardi-Castagnoli, and M. A. Degli-Esposti. 2001. Infection of dendritic cells by murine cytomegalovirus induces functional paralysis. Nat. Immunol. 2:1077-1084. [DOI] [PubMed] [Google Scholar]
- 2.Angulo, A., P. Ghazal, and M. Messerle. 2000. The major immediate-early gene ie3 of mouse cytomegalovirus is essential for viral growth. J. Virol. 74:11129-11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aquino, V. H., and L. T. Figueiredo. 2000. High prevalence of renal transplant recipients infected with more than one cytomegalovirus glycoprotein B genotype. J. Med. Virol. 61:138-142. [PubMed] [Google Scholar]
- 4.Arav-Boger, R., R. E. Willoughby, R. F. Pass, J. C. Zong, W. J. Jang, D. Alcendor, and G. S. Hayward. 2002. Polymorphisms of the cytomegalovirus (CMV)-encoded tumor necrosis factor-alpha and beta-chemokine receptors in congenital CMV disease. J. Infect. Dis. 186:1057-1064. [DOI] [PubMed] [Google Scholar]
- 5.Baldanti, F., A. Sarasini, M. Furione, M. Gatti, G. Comolli, M. G. Revello, and G. Gerna. 1998. Coinfection of the immunocompromised but not the immunocompetent host by multiple human cytomegalovirus strains. Arch. Virol. 143:1701-1709. [DOI] [PubMed] [Google Scholar]
- 6.Boppana, S. B., L. B. Rivera, K. B. Fowler, M. Mach, and W. J. Britt. 2001. Intrauterine transmission of cytomegalovirus to infants of women with preconceptional immunity. N. Engl. J. Med. 344:1366-1371. [DOI] [PubMed] [Google Scholar]
- 7.Brune, W., C. Menard, J. Heesemann, and U. H. Koszinowski. 2001. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Science 291:303-305. [DOI] [PubMed] [Google Scholar]
- 8.Cardin, R. D., G. B. Abenes, C. A. Stoddart, and E. S. Mocarski. 1995. Murine cytomegalovirus IE2, an activator of gene expression, is dispensable for growth and latency in mice. Virology 209:236-241. [DOI] [PubMed] [Google Scholar]
- 9.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 10.Chong, K. T., and C. A. Mims. 1981. Murine cytomegalovirus particle types in relation to sources of virus and pathogenicity. J. Gen. Virol. 57:415-419. [DOI] [PubMed] [Google Scholar]
- 11.Coaquette, A., A. Bourgeois, C. Dirand, A. Varin, W. Chen, and G. Herbein. 2004. Mixed cytomegalovirus glycoprotein B genotypes in immunocompromised patients. Clin. Infect. Dis. 39:155-161. [DOI] [PubMed] [Google Scholar]
- 12.Drew, W. L., E. S. Sweet, R. C. Miner, and E. S. Mocarski. 1984. Multiple infections by cytomegalovirus in patients with acquired immunodeficiency syndrome: documentation by Southern blot hybridization. J. Infect. Dis. 150:952-953. [DOI] [PubMed] [Google Scholar]
- 13.Efstathiou, S., S. Kemp, G. Darby, and A. C. Minson. 1989. The role of herpes simplex virus type 1 thymidine kinase in pathogenesis. J. Gen. Virol. 70:869-879. [DOI] [PubMed] [Google Scholar]
- 14.Feil, R., J. Wagner, D. Metzger, and P. Chambon. 1997. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem. Biophys. Res. Commun. 237:752-757. [DOI] [PubMed] [Google Scholar]
- 15.Griffiths, P. D. 2000. Biological synergism between infectious agents. Rev. Med. Virol. 10:351-353. [DOI] [PubMed] [Google Scholar]
- 16.Grzimek, N. K., J. Podlech, H. P. Steffens, R. Holtappels, S. Schmalz, and M. J. Reddehase. 1999. In vivo replication of recombinant murine cytomegalovirus driven by the paralogous major immediate-early promoter-enhancer of human cytomegalovirus. J. Virol. 73:5043-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu, H., J. D. Marth, P. C. Orban, H. Mossmann, and K. Rajewsky. 1994. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science 265:103-106. [DOI] [PubMed] [Google Scholar]
- 18.Haberland, M., U. Meyer-Konig, and F. T. Hufert. 1999. Variation within the glycoprotein B gene of human cytomegalovirus is due to homologous recombination. J. Gen. Virol. 80:1495-1500. [DOI] [PubMed] [Google Scholar]
- 19.Hahn, G., R. Jores, and E. S. Mocarski. 1998. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc. Natl. Acad. Sci. USA 95:3937-3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henson, D., and A. J. Strano. 1972. Mouse cytomegalovirus. Necrosis of infected and morphologically normal submaxillary gland acinar cells during termination of chronic infection. Am. J. Pathol. 68:183-202. [PMC free article] [PubMed] [Google Scholar]
- 21.Humar, A., D. Kumar, C. Gilbert, and G. Boivin. 2003. Cytomegalovirus (CMV) glycoprotein B genotypes and response to antiviral therapy, in solid-organ-transplant recipients with CMV disease. J. Infect. Dis. 188:581-584. [DOI] [PubMed] [Google Scholar]
- 22.Javier, R. T., F. Sedarati, and J. G. Stevens. 1986. Two avirulent herpes simplex viruses generate lethal recombinants in vivo. Science 234:746-748. [DOI] [PubMed] [Google Scholar]
- 23.Krmpotic, A., D. H. Busch, I. Bubic, F. Gebhardt, H. Hengel, M. Hasan, A. A. Scalzo, U. H. Koszinowski, and S. Jonjic. 2002. MCMV glycoprotein gp40 confers virus resistance to CD8+ T cells and NK cells in vivo. Nat. Immunol. 3:529-535. [DOI] [PubMed] [Google Scholar]
- 24.Krmpotic, A., M. Messerle, I. Crnkovic-Mertens, B. Polic, S. Jonjic, and U. H. Koszinowski. 1999. The immunoevasive function encoded by the mouse cytomegalovirus gene m152 protects the virus against T cell control in vivo. J. Exp. Med. 190:1285-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lefkowitz, I., and H. Waldmann (ed.). 1979. Limiting dilution analysis of cells in the immune system. Cambridge University Press, Cambridge, United Kingdom.
- 26.Lembo, D., M. Donalisio, A. Hofer, M. Cornaglia, W. Brune, U. Koszinowski, L. Thelander, and S. Landolfo. 2004. The ribonucleotide reductase R1 homolog of murine cytomegalovirus is not a functional enzyme subunit but is required for pathogenesis. J. Virol. 78:4278-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lurain, N. S., K. S. Kapell, D. D. Huang, J. A. Short, J. Paintsil, E. Winkfield, C. A. Benedict, C. F. Ware, and J. W. Bremer. 1999. Human cytomegalovirus UL144 open reading frame: sequence hypervariability in low-passage clinical isolates. J. Virol. 73:10040-10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacDonald, M. R., X. Y. Li, and H. W. Virgin IV. 1997. Late expression of a beta chemokine homolog by murine cytomegalovirus. J. Virol. 71:1671-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menard, C., M. Wagner, Z. Ruzsics, K. Holak, W. Brune, A. E. Campbell, and U. H. Koszinowski. 2003. Role of murine cytomegalovirus US22 gene family members in replication in macrophages. J. Virol. 77:5557-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Messerle, M., G. M. Keil, and U. H. Koszinowski. 1991. Structure and expression of murine cytomegalovirus immediate-early gene 2. J. Virol. 65:1638-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer-Konig, U., K. Ebert, B. Schrage, S. Pollak, and F. T. Hufert. 1998. Simultaneous infection of healthy people with multiple human cytomegalovirus strains. Lancet 352:1280-1281. [DOI] [PubMed] [Google Scholar]
- 33.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2674. In D. M. Knipe, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 34.Myerson, D., R. C. Hackman, J. A. Nelson, D. C. Ward, and J. K. McDougall. 1984. Widespread presence of histologically occult cytomegalovirus. Hum. Pathol. 15:430-439. [DOI] [PubMed] [Google Scholar]
- 35.Penfold, M. E., D. J. Dairaghi, G. M. Duke, N. Saederup, E. S. Mocarski, G. W. Kemble, and T. J. Schall. 1999. Cytomegalovirus encodes a potent alpha chemokine. Proc. Natl. Acad. Sci. USA 96:9839-9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Podlech, J., R. Holtappels, N. K. Grzimek, and M. J. Reddehase. 2002. Animal models: murine cytomegalovirus, p. 493-525. In S. H. E. Kaufmann and D. Kabelitz (ed.), Methods in microbiology, 2nd ed., vol. 32. Academic Press, London, United Kingdom.
- 37.Posfai, G., M. D. Koob, H. A. Kirkpatrick, and F. R. Blattner. 1997. Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J. Bacteriol. 179:4426-4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prichard, M. N., M. E. Penfold, G. M. Duke, R. R. Spaete, and G. W. Kemble. 2001. A review of genetic differences between limited and extensively passaged human cytomegalovirus strains. Rev. Med. Virol. 11:191-200. [DOI] [PubMed] [Google Scholar]
- 39.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reddehase, M. J., F. Weiland, K. Munch, S. Jonjic, A. Luske, and U. H. Koszinowski. 1985. Interstitial murine cytomegalovirus pneumonia after irradiation: characterization of cells that limit viral replication during established infection of the lungs. J. Virol. 55:264-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saederup, N., S. A. Aguirre, T. E. Sparer, D. M. Bouley, and E. S. Mocarski. 2001. Murine cytomegalovirus CC chemokine homolog MCK-2 (m131-129) is a determinant of dissemination that increases inflammation at initial sites of infection. J. Virol. 75:9966-9976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saederup, N., Y. C. Lin, D. J. Dairaghi, T. J. Schall, and E. S. Mocarski. 1999. Cytomegalovirus-encoded beta chemokine promotes monocyte-associated viremia in the host. Proc. Natl. Acad. Sci. USA 96:10881-10886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarcinella, L., T. Mazzulli, B. Willey, and A. Humar. 2002. Cytomegalovirus glycoprotein B genotype does not correlate with outcomes in liver transplant patients. J. Clin. Virol. 24:99-105. [DOI] [PubMed] [Google Scholar]
- 44.Sedarati, F., R. T. Javier, and J. G. Stevens. 1988. Pathogenesis of a lethal mixed infection in mice with two nonneuroinvasive herpes simplex virus strains. J. Virol. 62:3037-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sinzger, C., A. Grefte, B. Plachter, A. S. Gouw, T. H. The, and G. Jahn. 1995. Fibroblasts, epithelial cells, endothelial cells and smooth muscle cells are major targets of human cytomegalovirus infection in lung and gastrointestinal tissues. J. Gen. Virol. 76:741-750. [DOI] [PubMed] [Google Scholar]
- 46.Sinzger, C., and G. Jahn. 1996. Human cytomegalovirus cell tropism and pathogenesis. Intervirology 39:302-319. [DOI] [PubMed] [Google Scholar]
- 47.Smith, G. A., and L. W. Enquist. 2000. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc. Natl. Acad. Sci. USA 97:4873-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stoddart, C. A., R. D. Cardin, J. M. Boname, W. C. Manning, G. B. Abenes, and E. S. Mocarski. 1994. Peripheral blood mononuclear phagocytes mediate dissemination of murine cytomegalovirus. J. Virol. 68:6243-6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tarrago, D., C. Quereda, and A. Tenorio. 2003. Different cytomegalovirus glycoprotein B genotype distribution in serum and cerebrospinal fluid specimens determined by a novel multiplex nested PCR. J. Clin. Microbiol. 41:2872-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trincado, D. E., G. M. Scott, P. A. White, C. Hunt, L. Rasmussen, and W. D. Rawlinson. 2000. Human cytomegalovirus strains associated with congenital and perinatal infections. J. Med. Virol. 61:481-487. [DOI] [PubMed] [Google Scholar]
- 51.Wagner, M., A. Gutermann, J. Podlech, M. J. Reddehase, and U. H. Koszinowski. 2002. Major histocompatibility complex class I allele-specific cooperative and competitive interactions between immune evasion proteins of cytomegalovirus. J. Exp. Med. 196:805-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner, M., S. Jonjic, U. H. Koszinowski, and M. Messerle. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiland, F., G. M. Keil, M. J. Reddehase, and U. H. Koszinowski. 1986. Studies on the morphogenesis of murine cytomegalovirus. Intervirology 26:192-201. [DOI] [PubMed] [Google Scholar]
- 54.Ziegler, H., R. Thale, P. Lucin, W. Muranyi, T. Flohr, H. Hengel, H. Farrell, W. Rawlinson, and U. H. Koszinowski. 1997. A mouse cytomegalovirus glycoprotein retains MHC class I complexes in the ERGIC/cis-Golgi compartments. Immunity 6:57-66. [DOI] [PubMed] [Google Scholar]