Abstract
Thiol proteins are important in cellular antioxidant defenses and redox signalling. It is postulated that reactive oxidants cause selective thiol oxidation, but relative sensitivities of different cell proteins and critical targets are not well characterized. We exposed Jurkat cells to H2O2 for 10 min and measured changes in reversibly oxidized proteins by labelling with iodoacetamidofluorescein and two-dimensional electrophoresis. At 200 μM H2O2, which caused activation of the MAP (mitogen-activated protein) kinase ERK (extracellular-signal-regulated kinase), growth arrest and apoptosis, relatively few changes were seen. A total of 28 spots were reversibly oxidized (increased labelling intensity) and 24 decreased. The latter included isoforms of peroxiredoxins 1 and 2, which were irreversibly oxidized. Oxidation of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was striking, and other affected proteins included glutathione S-transferase P1-1, enolase, a regulatory subunit of protein kinase A, annexin VI, the mitotic checkpoint serine/threonine-protein kinase BUB1β, HSP90β (heat-shock protein 90β) and proteosome components. At 20 μM H2O2, changes were fewer, but GAPDH and peroxiredoxin 2 were still modified. Dinitrochlorobenzene treatment, which inhibited cellular thioredoxin reductase and partially depleted GSH, caused reversible oxidation of several proteins, including thioredoxin 1 and peroxiredoxins 1 and 2. Most changes were distinct from those with H2O2, and changes with H2O2 were scarcely enhanced by dinitrochlorobenzene. Relatively few proteins, including deoxycytidine kinase, nucleoside diphosphate kinase and a proteosome activator subunit, responded only to the combined treatment. Thus most of the effects of H2O2 were not linked to thioredoxin oxidation. Our study has identified peroxiredoxin 2 and GAPDH as two of the most oxidant-sensitive cell proteins and has highlighted how readily peroxiredoxins undergo irreversible oxidation.
Keywords: glutathione, hydrogen peroxide (H2O2)-sensitive thiol proteins, oxidative stress, redox signalling, thioredoxin, two-dimensional electrophoresis
Abbreviations: 2D, two-dimensional; ACTH, adrenocorticotropic hormone; BrdU, 5-bromo-2′-deoxyuridine; BUB, budding uninhibited by benzimidazole; DEVD-AMC, Asp-Glu-Val-Asp-7-amino-4-methylcoumarin; DNCB, 1-chloro-2,4-dinitrobenzene; DTT, dithiothreitol; ERK, extracellular-signal-regulated kinase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HSP90β, heat-shock protein 90β; IAF, 5-iodoacetamidofluorescein; JNK, c-Jun N-terminal kinase; MALDI-TOF MS, matrix-assisted laser desorption–ionization time-of-flight MS; MAP kinase, mitogen-activated protein kinase; NEM, N-ethylmaleimide; PKA, cAMP-dependent kinase; PTP, protein-tyrosine phosphatase
INTRODUCTION
Cells respond to oxidants by activating a number of stress-induced pathways. These may lead to increased gene expression, proliferation, growth arrest or apoptosis [1–3]. Various antioxidant pathways protect or regulate cellular responses to oxidative stress. Thiol redox systems involving GSH and thioredoxin are important for antioxidant protection, and the weight of evidence points to thiol proteins also acting as sensors and transmitters of oxidant signals [4,5]. Many regulatory proteins contain critical cysteine residues that are sensitive to oxidation to sulphenic acids, intra- and inter-molecular disulphides or mixed disulphides with GSH. These transformations are readily reversible through interaction with GSH, glutaredoxin or thioredoxin systems, making them well suited for signal transduction [6,7]. Although redox signalling is well accepted as a cellular mechanism, there are still uncertainties about how signals are transmitted, and the critical targets of oxidant action are not well characterized.
Several thiol proteins, such as the peroxiredoxins [8–11], GAPDH (glyceraldehyde-3-phosphate dehydrogenase) [12,13] and tyrosine phosphatases such as PTP1B (protein-tyrosine phosphatase 1B [14–16]) have been shown to undergo oxidation in cells treated with peroxides. The phosphatases have attracted considerable attention as potential targets for activating signalling pathways. Isolated thiol proteins vary considerably in their susceptibility to oxidation [17]. However, most cellular studies have focussed on a particular protein without establishing whether its oxidation is selective or part of more global oxidative changes.
As a step towards characterizing relative susceptibilities of cell proteins to oxidation and identifying what changes are important in signalling or metabolic regulation, we have taken a proteomic approach to identifying thiol proteins that become oxidized in cells treated with H2O2. Most intracellular protein thiol groups are kept reduced, so it is easier to detect formation of oxidized proteins than changes in amounts of the reduced forms [18]. Therefore, we monitored oxidized thiols by a technique that involves labelling with the fluorescent probe IAF (5-iodoacetamidofluorescein) [14] and separating the proteins by 2D (two-dimensional) electrophoresis [19]. This detects reversibly oxidized thiols and, as we shall demonstrate, also picks up some irreversible oxidation. Our objectives were to establish whether oxidation of cellular thiol proteins is widespread or selective at low H2O2 concentrations that have functional effects on the cells, and to determine how the pattern of protein oxidation is affected by disrupting thiol reduction pathways with DNCB (1-chloro-2,4-dinitrobenzene). We show a considerable degree of selectivity and have identified a number of the sensitive proteins by peptide mass fingerprinting using MALDI-TOF MS (matrix-assisted laser desorption/ionization time-of-flight MS).
EXPERIMENTAL
Materials
Cell-culture materials were from Gibco BRL supplied by Invitrogen New Zealand Ltd. (Auckland, New Zealand). Complete™ protease cocktail tablets, sequencing-grade trypsin, Tris and CHAPS were from Roche Diagnostics (Mannheim, Germany). The caspase substrate DEVD-AMC (Asp-Glu-Val-Asp-7-amino-4-methylcoumarin) was purchased from Peptide Institute Inc. (Osaka, Japan). A phospho-MAP kinase (mitogen-activated protein kinase) antibody kit was obtained from Cell Signaling Technology (Beverly, MA, U.S.A.), and peroxiredoxin antibodies were from LabFrontier (Seoul, Korea). IAF was from Molecular Probes Inc. (Eugene, OR, U.S.A.). Micro Bio-Spin® 6 chromatography columns, acrylamide/bisacrylamide solution, Bio-Rad Dc protein assay, ReadyStrip™ (17 cm, pH 3–10) IPG (immobilized pH gradient) strips, Bio-Lyte® 3/10 ampholytes and PDQuest™ 2D gel analysis software were from Bio-Rad Laboratories (Hercules, CA, U.S.A.). Other reagents were from Sigma Chemical Co. (St Louis, MO, U.S.A.). ZipTip® C18 pipette tips were from Millipore (Billerica, MA, U.S.A.).
Cell culture
The Jurkat T-lymphocyte cell line was obtained from the A.T.C.C. [American Type Culture Collection, Rockville, MD, U.S.A. (now at Manassas, VA, U.S.A)]. Cells were maintained in RPMI-1640 supplemented with 10% (v/v) heat-inactivated fetal-bovine serum, 2 mM glutamine, 100 units/ml penicillin and 100 μg/ml streptomycin at 37 °C in a humidified atmosphere with 5% CO2. Before use, cells were suspended at 1×106 cells/ml in fresh media.
Proliferation assay
A colorimetric BrdU (5-bromo-2′-deoxyuridine) ELISA kit available from Roche Diagnostics (Mannheim, Germany) was used to measure cell proliferation. Jurkat cells (1×106 cells/ml) were treated for 1 h with a range of H2O2 concentrations, then diluted 1:1 with fresh medium and incubated at 37 °C for another 1 h to optimize the rate of cell proliferation. BrdU solution was added to each sample and the DNA labelling reaction carried out for 2 h at 37 °C. Cells were then harvested, washed in PBS and transferred to 96-well plates (5×104 cells/well). The cells were sedimented by 10 min centrifugation at 500 g and BrdU-labelled DNA was detected as described in the manufacturer's instructions.
Caspase activity assay
Jurkat cells were harvested by centrifugation after 6 h treatment with H2O2. The activity of caspase 3-like enzymes was measured by cleavage of the caspase substrate DEVD-AMC [3].
MAP kinase activity and peroxiredoxin Western blotting
After H2O2 treatment for various times, Jurkat cells were washed and extracted with 1 ml of 40 mM Hepes buffer, pH 7.4, containing 1% (w/v) CHAPS, 50 mM NaCl, 1 mM EDTA, 1 mM EGTA, and Complete™ protease inhibitors per 5×106 cells. Cell extracts (50 μg of protein) were separated by SDS/PAGE and Western-blotted with an antibody against either the dually phosphorylated form of ERK (extracellular-signal-regulated kinase) 1/2, peroxiredoxin 2 or overoxidized peroxiredoxin (anti-Prx-SO3). Peroxidase-conjugated secondary antibodies and chemiluminescence were used for detection.
Assesment of cell viability
After treatment, cells (approx. 2×105) were harvested and incubated for 5 min with 2 μg of propidium iodide. Cell fluorescence was analysed with a bivariate flow cytometer (Becton Dickinson, Mountain View, CA, U.S.A.). Cells that did not stain with propidium iodide were classified as viable, and the viability was expressed as a percentage of the total cells analysed.
Thioredoxin reductase assay
Thioredoxin reductase activity was estimated indirectly as NADPH-dependent reduction of DTNB [5,5′-dithiobis-(2-nitrobenzoic acid)] on cell extracts containing approx. 200 μg of protein [20]. Enzyme activity was estimated as the difference in rate of absorbance increase at 412 nm before and after the addition of NADPH.
Glutathione assay
The intracellular concentration of GSH was measured by HPLC with fluorescence detection following derivatization with monobromobimane [21].
Fluorescent labelling of oxidized thiol proteins and 2D electrophoresis
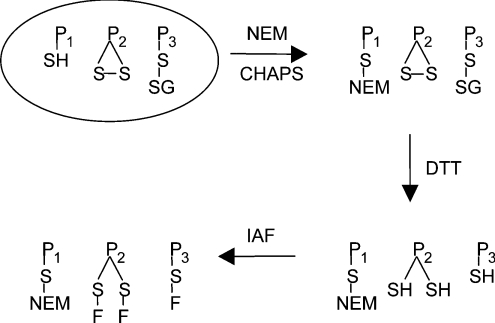
Reversibly oxidized thiol proteins were monitored as described previously [19], and summarized in Scheme 1. Briefly, reduced protein thiols were blocked by resuspending Jurkat cells in buffer containing NEM (N-ethylmaleimide) and incubating them at room temperature for 15 min [22]. After lysing the cells, excess NEM was removed and the oxidized thiols were reduced with DTT (dithiothreitol). The unblocked protein thiols were then labelled with IAF. Samples were desalted and excess IAF was removed before 2D electrophoresis, which was performed as described in [19], except that both SDS/10%- and 15%-(w/v)-PAGE gels were used for the second dimension.
Scheme 1. Principles for the fluorescence labelling of reversibly oxidized thiol proteins.
P1SH represents reduced thiol proteins, P2S-S represents intramolecular disulphides and P3S-SG represents mixed disulphides with glutathione. NEM blocks reduced thiol proteins (P-S-NEM), which are not seen on the gel. DTT reduces reversibly oxidized thiol proteins, which, after treatment with IAF, become fluorescently labelled (P-S-F). Irreversibly oxidized proteins are not labelled. Reversible thiol oxidation gives greater fluorescence labelling of the particular protein spot in oxidant-treated compared with control cells [19].
MALDI-TOF MS
Fluorescence-labelled thiol proteins were isolated from 2D SDS/PAGE gels for identification prior to staining the gels for total protein. The spots that decreased in fluorescence intensity were excised from untreated gels and those that increased in intensity were excised from H2O2-treated gels. Gels were immediately scanned for fluorescence at the conclusion of electrophoresis. An actual size image of the gel was printed and the gel was placed on a glass plate and carefully oriented over the gel image to visualize the fluorescent protein spots. The protein spots of choice were excised and the gels re-scanned to confirm that the desired protein spots had been accurately obtained.
The gel pieces were washed in 100 mM ammonium bicarbonate before being dehydrated in 100% acetonitrile and dried in a vacuum concentrator at 45 °C. The gel pieces were rehydrated in 50 mM ammonium bicarbonate solution containing 0.25 μg of trypsin for 45 min on ice. Unabsorbed trypsin solution was replaced with 50 mM ammonium bicarbonate solution and in-gel enzymatic proteolysis was performed overnight at 37 °C. Peptides were eluted from the gel pieces by repeated incubations in 1:1 (v/v) solution of acetonitrile and 0.5% (v/v) trifluoroacetic acid. The eluted peptides were dried in a vacuum concentrator at 60 °C and then resuspended in 0.5% trifluoroacetic acid [23]. C18 reversed-phase ZipTip® pipette tips were used to concentrate and desalt the samples according the manufacturer's instructions. The peptides were eluted from the ZipTips® with 0.5% (w/v) α-cyano-4-hydroxycinnamic acid in 50% (v/v) acetonitrile and 0.25% (v/v) trifluoroacetic acid. Spectra were acquired in positive-ion reflector mode with accelerating voltage 20000 V, grid voltage 75%, guide-wire voltage 0.002% and extraction delay time 180 ns. Masses were calibrated with external standards [angiotensin, 1 m/z 1296.6853; ACTH (adrenocorticotropic hormone) fragment 1–17, m/z 2093.0876; and ACTH fragment 18–39, m/z 2465.1989]. The peptide mass fingerprints were used to search online protein databases found at: http://129.85.19.192/profound_bin/WebProFound.exe [24].
RESULTS
Cellular responses to H2O2
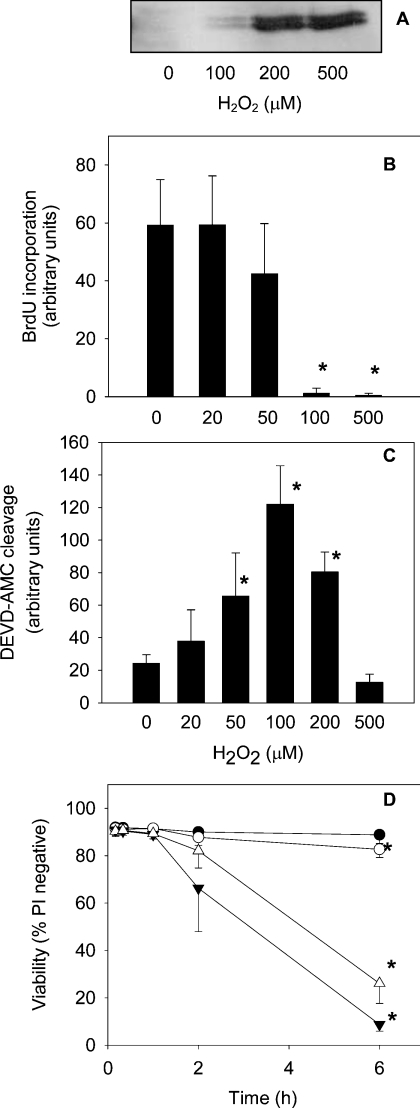
H2O2 triggers a range of cellular responses, including proliferation, growth arrest, induction of apoptosis and MAP kinase activation [3,15,25]. The effect of H2O2 treatment on these processes in Jurkat cells was investigated in order to establish the concentration range over which signalling pathways were affected. To detect activation of ERK 1/2, the dually phosphorylated form was visualized by Western blotting (Figure 1A). Slight activation was observed after 20 min with 100 μM H2O2, and maximal activation at 200–500 μM H2O2. Cell proliferation, measured at 2 h, was almost completely inhibited by 100 μM H2O2 (Figure 1B). These cells subsequently underwent apoptosis, as determined by an increase in activity of caspase 3-like enzymes 6 h after H2O2 treatment (Figure 1C). Some apoptosis was also detected with 50 μM H2O2.
Figure 1. Jurkat cell responses to H2O2.
(A) MAP kinase activity: Jurkat cells (1×106/ml) were treated with H2O2 for 20 min and the dually phosphorylated form of ERK 1/2 was detected in total cell lysates. The Western blot is representative of two independent experiments. (B) Growth arrest: Jurkat cells were treated with H2O2 for 2 h and cell proliferation was measured using the BrdU assay. Results are means±S.D. for at least three experiments. (C) Caspase activation: cells were treated with H2O2 for 6 h and the activity of caspase-3-like enzymes was monitored by cleavage of DEVD-AMC. Data are means and standard deviations from four experiments. (D) Jurkat cell viability as measured by propidium iodide (PI) uptake after treatment with 20 μM H2O2 (●), 200 μM H2O2 (○), 30 μM DNCB (▼) or with a combination of both 30 μM DNCB and 200 μM H2O2 (△). Results are means±S.D. for three experiments. An asterisk (*) indicates a significant difference (P<0.05) when compared with control cells (one-way repeated-measures ANOVA using Bonferroni's method for multiple comparisons).
We selected two concentrations of H2O2 for investigating thiol-protein oxidation. A low concentration of 20 μM was used to detect proteins that undergo oxidation without long-term changes in cell physiology. A higher dose of 200 μM H2O2 was used to detect thiol protein changes that occur with more severe oxidative stress that eventually leads to growth arrest and cell death. The cells were treated for 10 min. As previously [3], approx. 50% of the peroxide was consumed in this time. There was no loss of viability, measured as propidium iodide uptake (Figure 1D). Only after 6 h was there a slight decrease with 200 μM H2O2. DNCB, which was used to disrupt thiol homoeostasis (see below), had no effect at 10 min, but subsequently caused a progressive loss in viability that was not enhanced by H2O2 (Figure 1D).
Detection of H2O2-sensitive thiol proteins
Using a protocol that labels reversibly oxidized thiol proteins with IAF (Scheme 1), we have previously shown that only a minor fraction of the cysteine residues in proteins of resting Jurkat cells are oxidized [19]. Nevertheless, approx. 400 proteins with oxidized cysteine residues can be detected by 2D electrophoresis. These represent proteins normally present as disulphides, those that undergo redox cycling and exist in a partially oxidized form, and any that were incompletely blocked with NEM. In the present study, cells were treated with H2O2 for 10 min so as to focus on redox changes rather than changes in protein expression. Each treatment was compared with control cells, and spots that doubled in intensity or decreased by at least half in at least three out of the four experiments were deemed to reflect genuine changes.
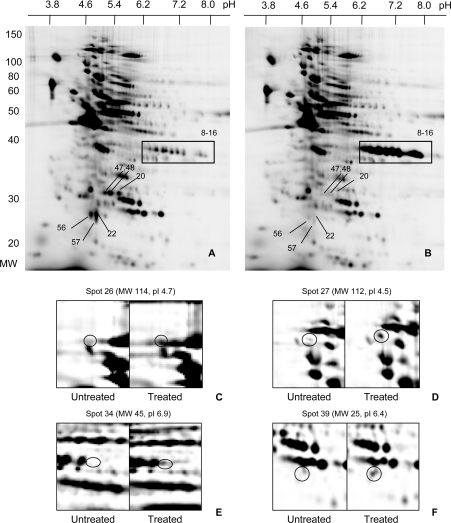
When Jurkat cells were treated with 200 μM H2O2, the IAF labelling intensity of the vast majority of the spots was unchanged (Figures 2A and 2B). However, there were selective changes to specific proteins. Some of these, including those highlighted in the 37, 29 and 26 kDa regions, stood out when comparing the whole gels (Figures 2A and 2B). Others, such as those in Figures 2C–2F, became evident in zoomed images. Treatment with H2O2 increased labelling of 28 spots. Over half of the increases well exceeded the 2-fold threshold. Unexpectedly, H2O2 also caused decreased labelling of 24 spots, including the 26 and 29 kDa clusters (as shown in Figures 2A and 2B). In most of these cases, the spots became scarcely detectable.
Figure 2. 2D electrophoresis of oxidized thiol proteins in Jurkat cells.
Oxidized thiol proteins from untreated cells (A) and cells treated with 200 μM H2O2 (B) were labelled with IAF and resolved by 2D electrophoresis (10% gel). Boxed regions highlight where major changes were evident. (C)–(F) are enlargements of selected regions showing spots that had undergone major changes. Left-hand panels are from controls and right-hand panels are from treated cells. The numbering of the spots is arbitrary. The identities of all spots that changed, on the basis of molecular mass (MW), pI and, where available, MS analysis, are given in Figure 4 and Table 1.
The molecular masses and pI values of the peroxide-sensitive spots are given in Figure 3. The identities of those that could be excised from the gels and characterized by MALDI-TOF mass fingerprinting are in Table 1. With a few exceptions, there is good agreement between the pI values and molecular masses of the identified proteins predicted from their sequences and estimates from their positions on the 2D gels. The identified proteins have a range of structural and metabolic functions. The nine 37 kDa spots that increased in intensity are isoforms of GAPDH. One cluster of 26 kDa proteins (pI≈5) that showed a marked decrease includes several spots identified as peroxiredoxin 2. Proteins in the peroxiredoxin 1 region (pI≈7) also decreased, but not as strikingly. The 29 kDa spots that almost disappeared could not be identified. Other proteins that showed marked increases included ubiquitin thiolesterase, proteosome activator complex subunit 1, HSP90β (heat-shock protein 90β) and enolase. A substantial decrease in labelling was seen for PKA (cAMP-dependent kinase) type II-α regulatory chain. One spot, identified as the actin-binding protein moesin, decreased, whereas another moesin spot at a higher molecular mass increased, suggesting possible modification in the H2O2-treated cells.
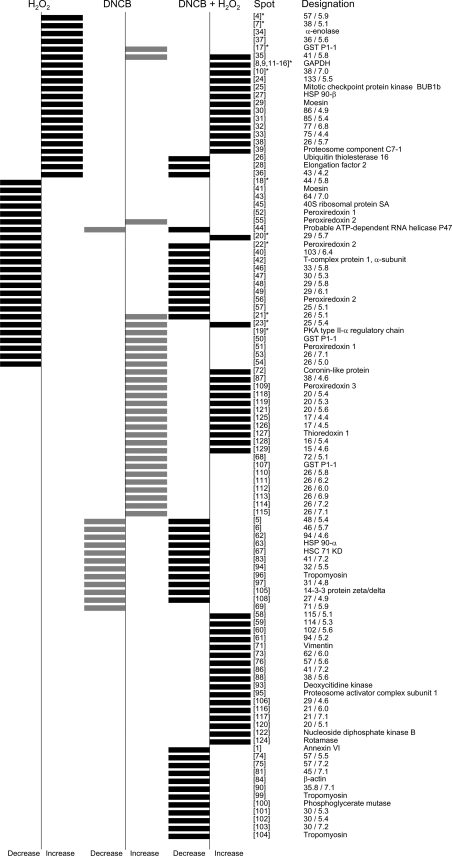
Figure 3. Cluster diagram summarizing all the thiol-protein changes observed when cells were treated with H2O2, DNCB or with a combination of both DNCB and H2O2.
Bars to the left indicate a consistent decrease in IAF labelling intensity on treatment and those to the right indicate an increase. The data shown for H2O2 are with 200 μM H2O2; spots designated * also changed with 20 μM H2O2. There were five spots [numbers 1, 5, 6, and spots at molecular mass 76 kDa/pI 6.1 and at molecular mass 64 kDa/pI 6.2) that increased with 20 μM, but not 200 μM H2O2. The numbering of the spots is arbitrary. Abbreviation: GST, glutathione S-transferase; HSC, heat-shock cognate.
Table 1. Details of the H2O2-sensitive proteins identified by MALDI-TOF MS.
H2O2-sensitive protein spots were excised from the 2D gels and their tryptic digests were analysed by MALDI-TOF MS. The 44 identified spots, which include 31 distinct proteins, all contain cysteine residues. A number of other spots were excised, but they could not be identified. They were usually faintly IAF-stained, suggesting low abundance, or gave complex spectra, indicating protein mixtures. Identities of proteins with low sequence coverage, a low Z-score (measure of the statistical significance of the result relative to an alignment of random structures) and deviation from the predicted molecular mass and pI should be treated cautiously. However, even though the pI values of the spots identified as peroxiredoxin 1 are lower than theoretical, they ran in a 2D region similar to that reported for peroxiredoxin 1 [35,37,45,52]. The designation ‘Up’ or ‘Down’ refers to changes in IAF spot intensity. Molecular mass (kDa) and pI data estimated from positions of spots in 2D gels are shown in the columns marked ‘2D’. ‘Pred’ indicates molecular mass (MM) and pI predicted from the protein sequences. Abbreviation: GST, glutathione S-transferase.
| Spot code | 20 μM H202 | 200 μM H202 | DNCB | DNCB+H202 | MM (2D) | pI (2D) | MM (pred) | pI (pred) | Coverage (%) | Peptide match | Z-score | Identity (Swiss-Prot/TrEMBL accession number) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Energy metabolism | ||||||||||||
| 8 | Up | Up | – | Up | 39 | 6.8 | 36 | 8.7 | 24 | 6 | 0.79 | GAPDH (P04406) |
| 9 | Up | Up | – | Up | 38 | 6.9 | 36 | 8.7 | 14 | 5 | 1.23 | GADPH |
| 11 | Up | Up | – | Up | 38 | 7.0 | 36 | 8.7 | 30 | 11 | 2.18 | GADPH |
| 12 | Up | Up | – | Up | 38 | 7.1 | 36 | 8.7 | 31 | 11 | 2.33 | GADPH |
| 13 | Up | Up | – | Up | 37 | 7.2 | 36 | 8.7 | 37 | 13 | 2.38 | GADPH |
| 14 | Up | Up | – | Up | 37 | 7.3 | 36 | 8.7 | 31 | 13 | 2.32 | GADPH |
| 15 | Up | Up | – | Up | 37 | 7.5 | 36 | 8.7 | 29 | 11 | 2.37 | GADPH |
| 16 | Up | Up | – | Up | 37 | 7.5 | 36 | 8.7 | 52 | 19 | 2.35 | GADPH |
| 34 | – | Up | – | – | 45 | 6.9 | 47 | 7.0 | 37 | 13 | 2.20 | α-Enolase (P06733) |
| 100 | – | – | – | Up | 30 | 6.0 | 29 | 6.7 | 29 | 7 | 2.11 | Phosphoglycerate mutase, brain form (P18669) |
| Cell structure | ||||||||||||
| 29 | – | Up | – | Up | 96 | 6.8 | 68 | 6.1 | 23 | 15 | 2.18 | Moesin (P26038) |
| 41 | – | Up | – | – | 79 | 6.2 | 68 | 6.1 | 23 | 14 | 2.37 | Moesin |
| 84 | – | – | – | Up | 43 | 5.0 | 40 | 5.6 | 23 | 6 | 2.26 | β-Cytoskeletal actin (P02570) |
| 96 | – | – | Up | Up | 31 | 4.7 | 29 | 4.7 | 26 | 8 | 0.87 | Tropomyosin, cytoskeletal type (P12324) |
| 99 | – | – | – | Up | 30 | 5.0 | 33 | 4.7 | 23 | 7 | 0.43 | Tropomyosin α-chain, skeletal muscle type (P06753) |
| 104 | – | – | – | Up | 29 | 4.6 | 29 | 4.7 | 23 | 8 | 2.25 | Tropomyosin, fibroblast non-muscle type (P07226) |
| Protein folding | ||||||||||||
| 27 | – | Up | – | Up | 112 | 4.5 | 83 | 5.0 | 22 | 12 | 2.31 | HSP90β (P08238) |
| 42 | – | Up | – | Up | 71 | 5.5 | 60 | 5.8 | 22 | 11 | 2.25 | T-complex protin 1, α-subunit (P17987) |
| 63 | – | – | Up | Up | 91 | 5.2 | 85 | 4.9 | 18 | 13 | 1.71 | Heat-shock 90 kDa protein 1α (P07900) |
| 67 | – | – | Up | Up | 78 | 5.5 | 71 | 5.4 | 21 | 11 | 2.39 | Heat-shock cognate 71 kDa protein (P11142) |
| 124 | – | – | – | Up | 18 | 6.8 | 18 | 8.0 | 29 | 5 | 1.42 | Rotamase (P05092) |
| Stress–related proteins | ||||||||||||
| 17 | Up | Up | Up | – | 28 | 5.2 | 24 | 5.7 | 32 | 4 | 0.87 | Chain A, GST P1-1 (P09211) |
| 50 | – | Up | Up | – | 28 | 5.5 | 24 | 5.7 | 22 | 3 | 0.56 | Chain A, GST P1-1 |
| 107 | – | – | Up | – | 27 | 5.2 | 23 | 5.7 | 44 | 5 | 1.33 | Chain A GST P1-1 |
| 22 | Up | Up | – | Up | 25 | 5.2 | 22 | 5.7 | 29 | 6 | 2.29 | Peroxiredoxin 2 (P32119) |
| 55 | – | Up | Up | – | 26 | 5.3 | 22 | 5.7 | 24 | 5 | 1.14 | Peroxiredoxin 2 |
| 56 | – | Up | – | Up | 25 | 5.1 | 22 | 5.7 | 18 | 3 | 0.44 | Peroxiredoxin 2 |
| 51 | – | Up | Up | – | 27 | 6.4 | 22 | 8.7 | 21 | 4 | 0.76 | Peroxiredoxin 1 (Q06830) |
| 52 | – | Up | – | – | 26 | 6.9 | 22 | 8.7 | 16 | 3 | 1.01 | Peroxiredoxin 1 |
| 109 | – | – | Up | Up | 27 | 5.6 | 28 | 7.1 | 28 | 6 | 1.27 | Peroxiredoxin 3 (P30048) |
| 127 | – | – | Up | Up | 17 | 4.7 | 12 | 4.8 | 49 | 6 | 2.05 | Thioredoxin 1 (P10599) |
| Protein degradation | ||||||||||||
| 26 | – | Up | – | Up | 114 | 4.7 | 93 | 6.5 | 16 | 13 | 1.68 | Ubiquitin thiolesterase 16 (Q9Y5T5) |
| 39 | – | Up | – | Up | 25 | 6.4 | 23 | 6.5 | 22 | 5 | 0.94 | Proteosome component C7-1 (P49721) |
| 95 | – | – | – | Up | 31 | 5.3 | 29 | 5.8 | 33 | 8 | 2.21 | Proteosome activator complex subunit 1 (Q06323) |
| Cell signalling | ||||||||||||
| 1 | Up | – | – | Up | 78 | 5.5 | 76 | 5.4 | 20 | 17 | 2.03 | Annexin VI (P08133) |
| 19 | Up | Up | Up | – | 40 | 5.9 | 46 | 5.0 | 12 | 4 | 0.86 | PKA type II, α regulatory chain (P13861) |
| 25 | – | Up | – | Up | 129 | 5.4 | 120 | 5.2 | 15 | 12 | 1.46 | Mitotic checkpoint protein kinase BUB1β (O60566) |
| 72 | – | – | Up | Up | 64 | 5.9 | 51 | 6.3 | 12 | 6 | 1.63 | Coronin-like protein P57 (P31146) |
| 93 | – | – | – | Up | 33 | 5.1 | 31 | 5.1 | 14 | 4 | 1.17 | Deoxycytidine kinase (P27707) |
| 105 | – | – | Up | Up | 29 | 4.5 | 28 | 4.7 | 33 | 8 | 1.38 | 14-3-3 protein zeta/delta (P29312) |
| 122 | – | – | – | Up | 19 | 7.1 | 17 | 8.8 | 44 | 6 | 2.28 | Nucleoside diphosphate kinase B (P22392) |
| Translation and RNA processing | ||||||||||||
| 28 | – | Up | – | Up | 97 | 6.4 | 93 | 6.4 | 17 | 13 | 2.15 | Elongation factor 2 (P13639) |
| 44 | – | Up | Up | Up | 58 | 5.4 | 49 | 5.5 | 21 | 9 | 1.85 | Probable ATP-dependent RNA helicase P47 (Q9BV96) |
| 45 | – | Up | – | – | 44 | 4.6 | 33 | 4.8 | 18 | 5 | 2.09 | 40S ribosomal protein SA (P08865) |
At 20 μM, H2O2 still caused a dramatic increase in labelling of the GAPDH cluster (results not shown), but there were many fewer changes than at the higher concentration. These were evident only on zoomed images, where eight consistent but modest increases were detected along with six decreases, including three (two identified) in the peroxiredoxin 2 region and PKA type IIα regulatory subunit (Figure 3).
Thiol-protein oxidation in cells treated with DNCB
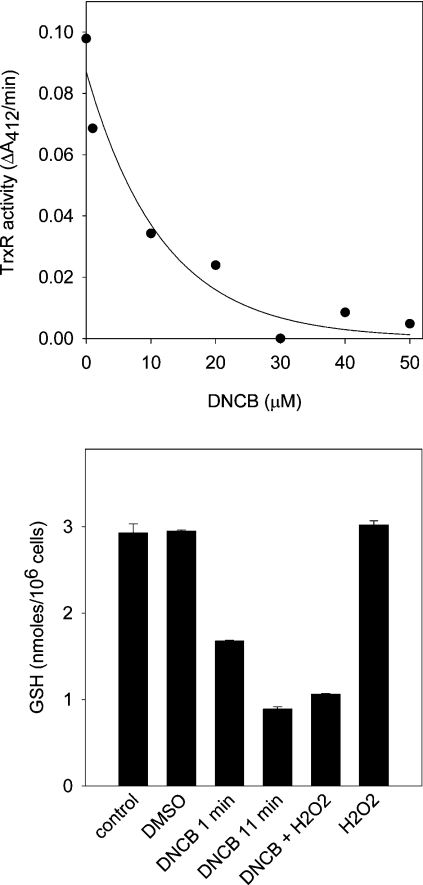
GSH and thioredoxin can regulate the redox state of thiol proteins either by reducing oxidized cysteine residues or by consuming oxidants such as H2O2 via the glutathione peroxidase/glutathione reductase and peroxiredoxin/thioredoxin reductase pathways [5,6]. Their influence on the pattern of protein oxidation induced by H2O2 was probed by treating Jurkat cells with DNCB [26] to inhibit thioredoxin reductase and deplete GSH. Treatment for 10 min inhibited thioredoxin reductase with an IC50 of 8 μM (Figure 4, upper panel). Almost complete inhibition was observed with 30 μM, which was used for all following experiments. Intracellular GSH content decreased after 1 min incubation with DNCB to 57%, and after 11 min to 31%, of that measured in untreated cells. When cells were incubated with 200 μM H2O2 for 10 min, there was no decrease in GSH, and the decrease produced by a combination of H2O2 and DNCB was comparable with that produced by DNCB alone (Figure 4, lower panel).
Figure 4. Thioredoxin reductase inhibition (A) and cellular GSH depletion (B) by DNCB.
Upper panel: thioredoxin reductase (TrxR) activity after treating cells with DNCB for 10 min. Lower panel: intracellular GSH concentration. Cells were treated with DMSO for 11 min, with 30 μM DNCB for 1 min or 11 min or with 200 μM H2O2 for 10 min. Where cells were exposed to both reagents, the H2O2 was added 1 min after DNCB and incubation was continued for another 10 min. Error bars represent the range of duplicates from one experiment.
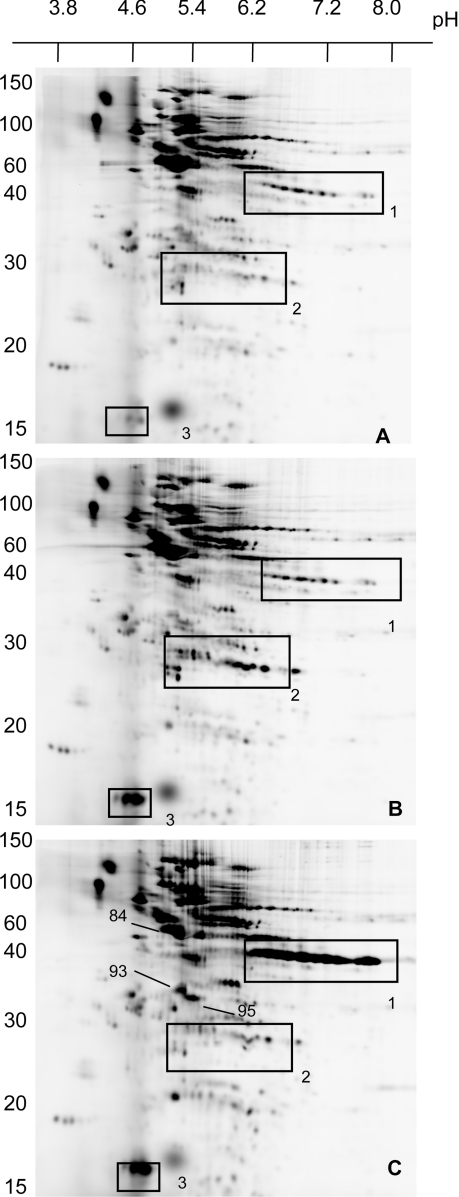
Even in the absence of H2O2, incubation of Jurkat cells with DNCB caused changes in IAF-labelling pattern of oxidized proteins (Figures 5A and 5B). A total of 29 spots increased in intensity and 13 decreased (Figure 3). The most obvious changes were the increase in labelling of three proteins in the 14 kDa region (box 3 in Figure 5), one of which was identified as thioredoxin 1, and increases in clusters of spots around 25 kDa were shown to include members of the peroxiredoxin 1 and 2 families (box 2). The accumulation of oxidized thioredoxin and peroxiredoxins is consistent with inhibition of thioredoxin reductase. Three spots with mass fingerprints corresponding to glutathione S-transferase also increased in intensity.
Figure 5. 2D electrophoresis of oxidized thiol proteins in control Jurkat cells (A), and cells treated with 30 μM DNCB (B) or with a combination of 200 μM H2O2 and 30 μM DNCB (C).
H2O2 was added 1 min after the addition of DNCB, and cells were incubated for another 10 min. Electrophoresis was performed on 15% gels. Boxes correspond to regions where major changes were evident for GAPDH (box 1), peroxiredoxins (box 2) and thioredoxin (box 3). The values on the extreme left are molecular masses in kDa.
As illustrated in the cluster diagram (Figure 3), the set of spots that changed with DNCB was largely independent of the set that changed with H2O2. In particular, labelling of the GAPDH series (box 1, Figure 5) increased only with H2O2, and oxidized thioredoxin increased only with DNCB. There were also several spots, including the peroxiredoxins, GST P1-1 and the PKA regulatory subunit, which were reversibly oxidized with DNCB, but decreased in intensity with H2O2.
When the cells were treated with a combination of DNCB and 200 μM H2O2 (Figure 5C), many of the changes seen with either treatment alone (Figures 2B and 5B) were evident. However, the peroxiredoxin and GST spots that increased in intensity with DNCB and decreased with H2O2 were unchanged compared with controls. The overall pattern (summarized in the cluster diagram in Figure 3) was largely a summation of the changes seen with either treatment alone. There was little evidence of DNCB sensitizing the cells to H2O2, as relatively few spots changed only with the combination. Most obvious of these were the increases in intensity seen for deoxycytidine kinase and proteosome activator complex subunit 1 and the decrease in actin (marked with arrows in Figure 5C). These proteins may have cysteine residues that are sensitive to oxidation and rapidly reduced by the glutathione and thioredoxin systems or become oxidant-sensitive only when these pathways are compromised. Although not assessed quantitatively, the effect of H2O2 on spots such as GAPDH did not appear to be greater in DNCB-treated cells.
Detection of irreversible oxidation of peroxiredoxins
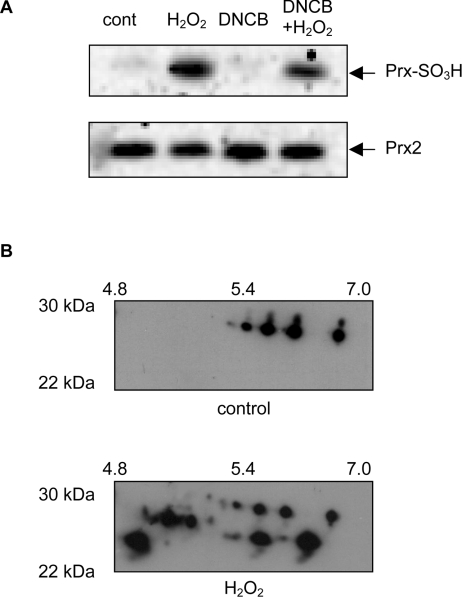
An unexpected finding was that IAF labelling of a number of proteins including peroxiredoxins was less after treatment with H2O2. This implies a decrease in reversibly oxidized thiols. To test whether this represented irreversible oxidation, we investigated the fate of the peroxiredoxins, which have active-site cysteine residues that can be oxidized to sulphinic acids by excess H2O2 [9–11]. Western blotting was performed using an antibody that recognizes the overoxidized proteins [9]. Control Jurkat cells gave little staining, but cells treated with H2O2 in the presence or absence of DNCB gave a strongly positive band at about 25 kDa (Figure 6A), which co-localized with peroxiredoxin 2. These treatments gave either a decrease (Figure 2B) or no change (Figure 5C) in IAF labelling of the peroxiredoxins. DNCB alone, which increased IAF labelling (Figure 5B), did not cause irreversible oxidation (Figure 6A). Blotting for overoxidized peroxiredoxins on 2D gels of proteins from H2O2-treated cells showed labelling of several spots in the peroxiredoxin region (Figure 6B). These findings are consistent with irreversible oxidation being one cause of decreased IAF labelling.
Figure 6. Overoxidation of peroxiredoxins by H2O2.
(A) Western blot using a polyclonal antibody that recognizes the sulphinic and sulphonic acid forms of peroxiredoxins 1–3 (Prx-SO3H). Equal loading was confirmed by probing the blots with anti-(peroxiredoxin 2). Jurkat cells were treated with 200 μM H2O2, 30 μM DNCB or a combination of both, and extracted proteins separated by SDS/PAGE. Abbreviation: Prx2, peroxiredoxin 2. (B) Western blot of equivalent regions of 2D gels from control (cont) and 200 μM H2O2-treated cells, showing an increase in the number of bands recognized by the antibody to overoxidized peroxiredoxins.
DISCUSSION
Using a fluorescence labelling procedure, we have detected changes in the pattern of reversibly oxidized thiol proteins in Jurkat cells after treatment with H2O2. A striking observation was the selective nature of the protein oxidation. Even with concentrations that caused functional changes and led eventually to apoptosis, only a small minority of spots changed in intensity. The sensitive proteins had diverse functions, including energy metabolism, protein degradation, structure maintenance and signalling (summarized in Table 1). Many have not been previously identified as redox-sensitive.
Although it was anticipated that most of the oxidation would be reversible and result in increased IAF labelling, almost half of the observed changes were decreases. Increases can be explained by conversion of cysteine residues either into internal disulphides and mixed disulphides with glutathione, or into sulphenic acids and sulphenyl–amide linkages [7,27]. One mechanism that could account for decreased labelling is irreversible thiol oxidation. Direct evidence for this was obtained for the peroxiredoxins. The IAF method would not detect irreversible oxidation of proteins that were fully reduced in control cells, however, as neither the free thiol nor overoxidized products are labelled. Other explanations for decreased spot intensity could include oxidation resulting in modification at a different site (e.g. phosphorylation) or cleavage to give a change in electrophoretic mobility. There were a few losses (e.g. moesin and glutathione S-transferase 1-1) that might have been due to this, but no obvious changes on protein-stained gels. With 10 min experiments, changes due to altered expression or turnover would not be expected.
GAPDH stood out as one of the most H2O2-sensitive proteins. Reversible oxidation was maximal even at 20 μM H2O2, where eight of the 22 spots that changed were GAPDH isoforms. GAPDH has an active-site cysteine residue and is well recognized as being sensitive to inactivation and glutathionylation in oxidant-treated cells [12,13,28,29]. Nevertheless, its selective sensitivity in cells is surprising when it is considered that its rate constant with H2O2 is only about 100 M−1·s−1 [30], which is a 1000-fold less than for the peroxiredoxins [17]. This raises the possibility that reaction with the cellular enzyme is indirect. It would seem likely that the extreme sensitivity of GAPDH has a purpose. The possibility has been raised that inhibition of its glycolytic activity may be advantageous for a cell facing oxidative stress, by diverting glucose through the pentose phosphate pathway and increasing the availability of NADPH for antioxidant enzyme pathways. However, GAPDH has a number of non-glycolytic actions [31], some of which may be influenced by its oxidation state [32]. For example, it forms associations with other proteins and may play a part in apoptosis [33]. However, we saw GAPDH oxidation at lower H2O2 concentrations than those causing growth arrest or apoptosis, and the consequences for the cell are as yet unknown.
Other predominant cell targets of H2O2 were the peroxiredoxins, which underwent irreversible oxidation. Peroxiredoxin 2 appeared most sensitive, with decreased IAF labelling apparent at 20 μM H2O2. At 200 μM H2O2, losses of peroxiredoxin 2 were more marked, with decreases in two additional isoforms being identified by mass fingerprinting. There was also decreased labelling of a cluster of spots in the peroxiredoxin 1 region, two of which were identified. Peroxiredoxins reduce H2O2 as well as alkyl hydroperoxides and peroxynitrite, and could be direct targets for some of the peroxide that enters cells [17,34]. They all have a cysteine residue at the active site. For peroxiredoxins 1–4, oxidation gives a probable sulphenic acid intermediate that forms an intermolecular disulphide with a second cysteine residue on the other identical subunit. The disulphide is reduced by thioredoxin. Peroxiredoxins can become overoxidized to sulphinic or sulphonic acids [8–10,35]. The proposed mechanism involves a fraction of the sulphenic acid being further oxidized during each catalytic cycle rather than forming the disulphide, with turnover by thioredoxin required for efficient conversion into overoxidized products [8]. Recent studies suggest that the sulphinic forms of peroxiredoxins can be reduced enzymatically [36], and it has been postulated that overoxidation and recycling may control peroxide levels and redox signalling [34,35].
Others have shown loss of peroxiredoxin thiols and formation of overoxidized products in oxidant-treated cells [8–10,35,37]. Our results agree with these studies and, in addition, show that there is little if any accumulation of the disulphides. We saw peroxiredoxin overoxidation by H2O2 both in the absence of any accumulation of oxidized thioredoxin and in association with thioredoxin oxidation in DNCB-treated cells. This goes against suggestions that the mechanism requires depletion of reduced thioredoxin [37].
Several other identified proteins that changed with H2O2 may have roles in oxidant metabolism and regulation. These include glutathione S-transferase P1-1, a protein with several functions in addition to its glutathione transferase activity. It can be inactivated by H2O2, possibly through glutathionylation [38], and it has been proposed to catalyse glutathionylation of 1-cys peroxiredoxin [39]. It has also been shown to bind to the MAP kinase JNK (c-Jun N-terminal kinase), with H2O2 treatment causing dissociation and allowing JNK phosphorylation and activation [40]. A spot identified as type II-α regulatory chain of the serine/threonine kinase PKA was consistently lost even with the lower H2O2 concentration, possibly indicating irreversible oxidation. Other proteins that were sensitive to peroxide were annexin VI, a protein that may be involved in Ca2+ regulation, the mitotic checkpoint serine/threonine-protein kinase BUB1β (budding uninhibited by benzimidazole 1β) and the HSP90β, whose activity is known to be impaired by oxidation [41]. A component of the proteasome system and ubiquitin thiolesterase were also reversibly oxidized. APC11 (anaphase-promoting complex 11), ubiquitin-protein ligase that targets cell cycle regulators, has been proposed as a target for H2O2 [42]. Our results suggest that de-ubiquitination may also be oxidant-sensitive.
We did not detect oxidation of creatine kinase or tyrosine phosphatases such as PTP1B and PTEN (phosphatase and tensin homologue deleted on chromosome 10) [14–16], even though they have been reported to be H2O2-sensitive and implicated in signalling. One possibility is that these were among the low-copynumber proteins that could not be characterized by mass fingerprinting. Indeed, we detected only very low PTP1B expression in Jurkat cells by Western blotting (results not shown). Most other studies of specific protein oxidation were carried out with higher H2O2 concentrations than we used, and an alternative explanation is that these proteins do not rank among the most oxidant-sensitive. More information is needed, including characterization of low-copy-number proteins and spots in crowded regions before any changes can be linked to redox-regulated events. It should also be noted that our extraction buffer may have selected against membrane proteins, so the method is likely to favour detection of cytoplasmic changes.
Other groups are also using proteomics to monitor cellular redox changes, with different approaches to the technique described here. Most have focussed on glutathionylation [43–46], with others detecting interchain disulphides [47,48] or sulphenic acids [49]. Our technique detects all these changes, and the approaches are complementary for assessing the extent and nature of thiol-protein oxidation. With different cell types and stresses used, observed patterns of oxidation vary, but in all cases proteins with a wide variety of functions were oxidized. Some common themes emerge, including oxidation of GAPDH and enolase [43,44,46–48], peroxiredoxins [43,46–48], thioredoxin [50], nucleoside diphosphate kinase [44,48], the regulatory subunit of PKA [47], HSP90 [47,48], ubiquitin thiolesterase [48] and various structural proteins [43,44,48,49,51]. Enolase α, peroxiredoxin 1, T complex protein 1 and HSP90β were among the proteins that were glutathionylated in T-lymphocytes [43] and ECV304 endothelial cells [45] treated with diamide, although not with H2O2.
The glutathione and thioredoxin systems regulate responses to H2O2 and other oxidative stresses. Thioredoxin is the preferred reductant for protein disulphides, whereas glutaredoxin catalyses the reduction of glutathionylated proteins [5]. We examined the effects of DNCB, which in principle could inhibit these processes, as well as H2O2 removal, through the glutathione/glutathione peroxidase or peroxiredoxin/thioredoxin pathways. Although more specific approaches to distinguish the two pathways are needed for confirmation, the almost complete inhibition of thioredoxin reductase compared with a 30% decrease in cellular GSH makes it more likely that the influence of DNCB was on thioredoxin-dependent pathways. Several changes in thiol-protein labelling were evident with DNCB alone. One of the most prominent was the emergence of two strong, reversibly oxidized low-molecular-mass spots, one of which was identified as thioredoxin 1. The other, although not characterized, could be mitochondrial thioredoxin 2, which from its sequence should have a lower pI than the cytoplasmic form. Several spots in the regions where peroxiredoxins 1 and 2 were located decreased in intensity, with one in each region characterized by mass fingerprinting. Oxidation of the mitochondrial form, peroxiredoxin 3, was also seen. In contrast with H2O2, DNCB caused reversible oxidation of the peroxiredoxins.
The rapid accumulation of oxidized thioredoxin after inhibition of thioredoxin reductase indicates that this pathway must be continually cycling in unstressed cells. The resultant inability of thioredoxin to reduce reversibly oxidized peroxiredoxins 1–3 would explain why these forms accumulated. These findings imply that the peroxiredoxins also undergo rapid redox cycling. The same may be the case for other proteins that became oxidized with DNCB. Our findings are consistent with the proposal that peroxiredoxins having a redox regulatory role [34,35], even though, on the basis of rate constants and abundance, they would not be expected to out-compete glutathione peroxidase for H2O2 [17].
The pattern of oxidation with DNCB was largely distinct from that caused by H2O2, implying that most of the H2O2-sensitive proteins are not those that readily oxidize when the thioredoxin system is inhibited. It also suggests that they are not oxidized indirectly via oxidized GSH or thioredoxin. When H2O2 was added to DNCB-treated cells, the spots that changed in intensity were largely a combination of those that changed with either reagent alone. Also, H2O2 caused similar changes in intensity of individual spots, e.g. the GAPDH isoforms, regardless of whether the cells were treated with DNCB. This may be because the proteins were already completely oxidized, or that GSH depletion and/or inhibition of the thioredoxin system did not enhance their peroxide-sensitivity. Some spots did change only with a combination of H2O2 and DNCB. These were remarkably few, but did include key enzymes in nucleoside phosphorylation and several components of the cytoskeletal network. These could be proteins that were normally oxidized by H2O2 and recycled by GSH or thioredoxin. Alternatively, intracellular H2O2 concentrations could have risen due to inhibition of removal pathways, and caused additional proteins to be oxidized.
In conclusion, we have described a technique for assessing redox changes to the whole thiol proteome. Most other studies have focused on a particular cell protein, often with considerably higher concentrations of peroxide than we used and without a perception of whether oxidation is selective. Comparing changes seen with both methods is now needed for relative sensitivities to be established. Our study has highlighted isoforms of GAPDH and peroxiredoxin 2 as among the most sensitive proteins. It has also demonstrated the ease at which peroxiredoxins are over-oxidized by H2O2 without inhibition of the thioredoxin system and has shown that inhibition of thioredoxin reductase and partial GSH depletion does not greatly enhance the effects of H2O2.
Acknowledgments
This work was supported by the Marsden Fund and the Health Research Council of New Zealand, including a Sir Charles Hercus Health Research Fellowship to M.B.H. We are grateful to Dr T. William Jordan, School of Biological Sciences, Victoria University of Wellington, Wellington, New Zealand, for advice and for the use of the Victoria University of Wellington MS facility.
References
- 1.Allen R. G., Tresini M. Oxidative stress and gene regulation. Free Radicals Biol. Med. 2000;28:463–499. doi: 10.1016/s0891-5849(99)00242-7. [DOI] [PubMed] [Google Scholar]
- 2.Burdon R. H. Superoxide and hydrogen peroxide in relation to mammalian cell proliferation. Free Radicals Biol. Med. 1995;18:775–794. doi: 10.1016/0891-5849(94)00198-s. [DOI] [PubMed] [Google Scholar]
- 3.Hampton M. B., Orrenius S. Dual regulation of caspase activity by hydrogen peroxide: implications for apoptosis. FEBS Lett. 1997;414:552–556. doi: 10.1016/s0014-5793(97)01068-5. [DOI] [PubMed] [Google Scholar]
- 4.Finkel T. Redox-dependent signal transduction. FEBS Lett. 2000;476:52–54. doi: 10.1016/s0014-5793(00)01669-0. [DOI] [PubMed] [Google Scholar]
- 5.Holmgren A. Antioxidant function of thioredoxin and glutaredoxin systems. Antioxid. Redox Signal. 2000;2:811–820. doi: 10.1089/ars.2000.2.4-811. [DOI] [PubMed] [Google Scholar]
- 6.Dickinson D. A., Forman H. J. Cellular glutathione and thiols metabolism. Biochem. Pharmacol. 2002;64:1019–1026. doi: 10.1016/s0006-2952(02)01172-3. [DOI] [PubMed] [Google Scholar]
- 7.Poole L. B., Karplus P. A., Claiborne A. Protein sulfenic acids in redox signaling. Annu. Rev. Pharmacol. Toxicol. 2004;44:325–347. doi: 10.1146/annurev.pharmtox.44.101802.121735. [DOI] [PubMed] [Google Scholar]
- 8.Yang K. S., Kang S. W., Woo H. A., Hwang S. C., Chae H. Z., Kim K., Rhee S. G. Inactivation of human peroxiredoxin I during catalysis as the result of the oxidation of the catalytic site cysteine to cysteine-sulfinic acid. J. Biol. Chem. 2002;277:38029–38036. doi: 10.1074/jbc.M206626200. [DOI] [PubMed] [Google Scholar]
- 9.Woo H. A., Kang S. W., Kim H. K., Yang K. S., Chae H. Z., Rhee S. G. Reversible oxidation of the active site cysteine of peroxiredoxins to cysteine sulfinic acid: immunoblot detection with antibodies specific for the hyperoxidized cysteine-containing sequence. J. Biol. Chem. 2003;278:47361–47364. doi: 10.1074/jbc.C300428200. [DOI] [PubMed] [Google Scholar]
- 10.Rabilloud T., Heller M., Gasnier F., Luche S., Rey C., Aebersold R., Benahmed M., Louisot P., Lunardi J. Proteomics analysis of cellular response to oxidative stress. Evidence for in vivo overoxidation of peroxiredoxins at their active site. J. Biol. Chem. 2002;277:19396–19401. doi: 10.1074/jbc.M106585200. [DOI] [PubMed] [Google Scholar]
- 11.Chevallet M., Wagner E., Luche S., Van Dorsselaer A., Leize-Wagner E., Rabilloud T. Regeneration of peroxiredoxins during recovery after oxidative stress: only some overoxidized peroxiredoxins can be reduced during recovery after oxidative stress. J. Biol. Chem. 2003;278:37146–37153. doi: 10.1074/jbc.M305161200. [DOI] [PubMed] [Google Scholar]
- 12.Brodie A. E., Reed D. J. Cellular recovery of glyceraldehyde-3-phosphate dehydrogenase activity and thiol status after exposure to hydroperoxides. Arch. Biochem. Biophys. 1990;276:212–218. doi: 10.1016/0003-9861(90)90028-w. [DOI] [PubMed] [Google Scholar]
- 13.Schuppe-Koistinen I., Moldeus P., Bergman T., Cotgreave I. S-thiolation of human endothelial cell glyceraldehyde-3-phosphate dehydrogenase after hydrogen peroxide treatment. Eur. J. Biochem. 1994;221:1033–1037. doi: 10.1111/j.1432-1033.1994.tb18821.x. [DOI] [PubMed] [Google Scholar]
- 14.Wu J. F., Kwon K. S., Rhee S. G. Probing cellular protein targets of H2O2 with fluorescein-conjugated iodoacetamide and antibodies to fluorescein. FEBS Lett. 1998;440:111–115. doi: 10.1016/s0014-5793(98)01415-x. [DOI] [PubMed] [Google Scholar]
- 15.Lee K., Esselman W. J. Inhibition of PTPs by H2O2 regulates the activation of distinct MAPK pathways. Free Radicals Biol. Med. 2002;33:1121–1132. doi: 10.1016/s0891-5849(02)01000-6. [DOI] [PubMed] [Google Scholar]
- 16.Cho S. H., Lee C. H., Ahn Y., Kim H., Ahn C. Y., Yang K. S., Lee S. R. Redox regulation of PTEN and protein tyrosine phosphatases in H2O2 mediated cell signaling. FEBS Lett. 2004;560:7–13. doi: 10.1016/s0014-5793(04)00112-7. [DOI] [PubMed] [Google Scholar]
- 17.Hofmann B., Hecht H. J., Flohe L. Peroxiredoxins. Biol. Chem. 2002;383:347–364. doi: 10.1515/BC.2002.040. [DOI] [PubMed] [Google Scholar]
- 18.Gitler C., Mogyoros M., Kalef E. Labeling of protein vicinal dithiols: role of protein-S2 to protein-(SH)2 conversion in metabolic regulation and oxidative stress. Methods Enzymol. 1994;233:403–415. doi: 10.1016/s0076-6879(94)33047-6. [DOI] [PubMed] [Google Scholar]
- 19.Baty J. W., Hampton M. B., Winterbourn C. C. Detection of oxidant-sensitive thiol proteins by fluorescent labeling and two dimensional electrophoresis. Proteomics. 2002;2:1261–1266. doi: 10.1002/1615-9861(200209)2:9<1261::AID-PROT1261>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 20.Arner E. S., Zhong L., Holmgren A. Preparation and assay of mammalian thioredoxin and thioredoxin reductase. Methods Enzymol. 1999;300:226–239. doi: 10.1016/s0076-6879(99)00129-9. [DOI] [PubMed] [Google Scholar]
- 21.Cotgreave I. A., Moldeus P. Methodologies for the application of monobromobimane to the simultaneous analysis of soluble and protein thiol components of biological systems. J. Biochem. Biophys. Methods. 1986;13:231–249. doi: 10.1016/0165-022x(86)90102-8. [DOI] [PubMed] [Google Scholar]
- 22.Gitler C., Zarmi B., Kalef E. General method to identify and enrich vicinal thiol proteins in intact cells in the oxidized, disulfide state. Anal. Biochem. 1997;252:48–55. doi: 10.1006/abio.1997.2294. [DOI] [PubMed] [Google Scholar]
- 23.Rosenfeld J., Capdevielle J., Guillemot J. C., Ferrara P. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- 24.Zhang W., Chait B. T. ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal. Chem. 2000;72:2482–2489. doi: 10.1021/ac991363o. [DOI] [PubMed] [Google Scholar]
- 25.Whisler R. L., Goyette M. A., Grants I. S., Newhouse Y. G. Sublethal levels of oxidant stress stimulate multiple serine/threonine kinases and suppress protein phosphatases in Jurkat T cells. Arch. Biochem. Biophys. 1998;319:23–35. doi: 10.1006/abbi.1995.1263. [DOI] [PubMed] [Google Scholar]
- 26.Arner E. S., Bjornstedt M., Holmgren A. 1-Chloro-2,4-dinitrobenzene is an irreversible inhibitor of human thioredoxin reductase. Loss of thioredoxin disulfide reductase activity is accompanied by a large increase in NADPH oxidase activity. J. Biol. Chem. 1995;270:3479–3482. doi: 10.1074/jbc.270.8.3479. [DOI] [PubMed] [Google Scholar]
- 27.Salmeen A., Andersen J. N., Myers M. P., Meng T. C., Hinks J. A., Tonks N. K., Barford D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature (London) 2003;423:769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 28.Hyslop P. A., Hinshaw D. B., Halsey W. A. J., Schraufstatter I. U., Sauerheber R. D., Spragg R. G., Jackson J. H., Cochrane C. G. Mechanisms of oxidant-mediated cell injury. The glycolytic and mitochondrial pathways of ADP phosphorylation are major intracellular targets inactivated by hydrogen peroxide. J. Biol. Chem. 1988;263:1665–1675. [PubMed] [Google Scholar]
- 29.Baker M. S., Feigan J., Lowther D. A. The mechanism of chondrocyte hydrogen peroxide damage. Depletion of intracellular ATP due to suppression of glycolysis caused by oxidation of glyceraldehyde-3-phosphate dehydrogenase. J. Rheumatol. 1989;16:7–14. [PubMed] [Google Scholar]
- 30.Little C., O'Brien P. J. Mechanism of peroxide-inactivation of the sulphydryl enzyme glyceraldehyde-3-phophate dehydrogenase. Eur. J. Biochem. 1969;10:533–538. doi: 10.1111/j.1432-1033.1969.tb00721.x. [DOI] [PubMed] [Google Scholar]
- 31.Sirover M. A. New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim. Biophys. Acta. 1999;1432:159–184. doi: 10.1016/s0167-4838(99)00119-3. [DOI] [PubMed] [Google Scholar]
- 32.Arutyunova E. I., Danshina P. V., Domnina L. V., Pleten A. P., Muronetz V. I. Oxidation of glyceraldehyde-3-phosphate dehydrogenase enhances its binding to nucleic acids. Biochem. Biophys. Res. Commun. 2003;307:547–552. doi: 10.1016/s0006-291x(03)01222-1. [DOI] [PubMed] [Google Scholar]
- 33.Berry M. D., Boulton A. A. Glyceraldehyde-3-phosphate dehydrogenase and apoptosis. J. Neurosci. Res. 2000;60:150–154. doi: 10.1002/(SICI)1097-4547(20000415)60:2<150::AID-JNR3>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 34.Wood Z. A., Schroder E., Robin H. J., Poole L. B. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 35.Woo H. A., Chae H. Z., Hwang S. C., Yang K. S., Kang S. W., Kim K., Rhee S. G. Reversing the inactivation of peroxiredoxins caused by cysteine sulfinic acid formation. Science. 2003;300:653–656. doi: 10.1126/science.1080273. [DOI] [PubMed] [Google Scholar]
- 36.Budanov A. V., Sablina A. A., Feinstein E., Koonin E. V., Chumakov P. M. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 37.Wagner E., Luche S., Penna L., Chevallet M., Van Dorsselaer A., Leize-Wagner E., Rabilloud T. A method for detection of overoxidation of cysteines: peroxiredoxins are oxidized in vivo at the active-site cysteine during oxidative stress. Biochem. J. 2002;366:777–785. doi: 10.1042/BJ20020525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen H. X., Tamai K., Satoh K., Hatayama I., Tsuchida S., Sato K. Modulation of class Pi glutathione transferase activity by sulfhydryl group modification. Arch. Biochem. Biophys. 1991;286:178–182. doi: 10.1016/0003-9861(91)90025-e. [DOI] [PubMed] [Google Scholar]
- 39.Manevich Y., Feinstein S. I., Fisher A. B. Activation of the antioxidant enzyme 1-CYS peroxiredoxin requires glutathionylation mediated by heterodimerization with pi GST. Proc. Natl. Acad. Sci. U.S.A. 2004;101:3780–3785. doi: 10.1073/pnas.0400181101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adler V., Yin Z., Fuchs S. Y., Benezra M., Rosario L., Tew K. D., Pincus M. R., Sardana M., Henderson C. J., Wolf C. R., et al. Regulation of JNK signaling by GSTp. EMBO J. 1999;18:1321–1334. doi: 10.1093/emboj/18.5.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nardai G., Sass B., Eber J., Orosz G., Csermely P. Reactive cysteines of the 90-kDa heat shock protein, Hsp90. Arch. Biochem. Biophys. 2000;384:59–67. doi: 10.1006/abbi.2000.2075. [DOI] [PubMed] [Google Scholar]
- 42.Chang T. S., Jeong W., Lee D. Y., Cho C. S., Rhee S. G. The RING-H2-finger protein APC11 as a target of hydrogen peroxide. Free Radicals Biol. Med. 2004;37:521–530. doi: 10.1016/j.freeradbiomed.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 43.Fratelli M., Demol H., Puype M., Casagrande S., Eberini I., Salmona M., Bonetto V., Mengozzi M., Duffieux F., Miclet E., et al. Identification by redox proteomics of glutathionylated proteins in oxidatively stressed human T lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3505–3510. doi: 10.1073/pnas.052592699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eaton P., Byers H. L., Leeds N., Ward M. A., Shattock M. J. Detection, quantitation, purification, and identification of cardiac proteins S-thiolated during ischemia and reperfusion. J. Biol. Chem. 2002;277:9806–9811. doi: 10.1074/jbc.M111454200. [DOI] [PubMed] [Google Scholar]
- 45.Lind C., Gerdes R., Hamnell Y., Schuppe-Koistinen I., von Lowenhielm H. B., Holmgren A., Cotgreave I. A. Identification of S-glutathionylated cellular proteins during oxidative stress and constitutive metabolism by affinity purification and proteomic analysis. Arch. Biochem. Biophys. 2002;406:229–240. doi: 10.1016/s0003-9861(02)00468-x. [DOI] [PubMed] [Google Scholar]
- 46.Sullivan D. M., Wehr N. B., Fergusson M. M., Levine R. L., Finkel T. Identification of oxidant-sensitive proteins: TNF-α induces protein glutathiolation. Biochemistry. 2000;39:11121–11128. doi: 10.1021/bi0007674. [DOI] [PubMed] [Google Scholar]
- 47.Brennan J. P., Wait R., Begum S., Bell J. R., Dunn M. J., Eaton P. Detection and mapping of widespread intermolecular protein disulfide formation during cardiac oxidative stress using proteomics with diagonal electrophoresis. J. Biol. Chem. 2004;279:41352–41360. doi: 10.1074/jbc.M403827200. [DOI] [PubMed] [Google Scholar]
- 48.Cumming R. C., Andon N. L., Haynes P. A., Park M., Fischer W. H., Schubert D. Protein disulfide bond formation in the cytoplasm during oxidative stress. J. Biol. Chem. 2004;279:21749–21758. doi: 10.1074/jbc.M312267200. [DOI] [PubMed] [Google Scholar]
- 49.Saurin A. T., Neubert H., Brennan J. P., Eaton P. Widespread sulfenic acid formation in tissues in response to hydrogen peroxide. Proc. Natl. Acad. Sci. U.S.A. 2004;101:17982–17987. doi: 10.1073/pnas.0404762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gitler C., Zarmi B., Kalef E., Meller R., Zor U., Goldman R. Calcium-dependent oxidation of thioredoxin during cellular growth initiation. Biochem. Biophys. Res. Commun. 2002;290:624–628. doi: 10.1006/bbrc.2001.6214. [DOI] [PubMed] [Google Scholar]
- 51.Fratelli M., Demol H., Puype M., Casagrande S., Villa P., Eberini I., Vandekerckhove J., Gianazza E., Ghezzi P. Identification of proteins undergoing glutathionylation in oxidatively stressed hepatocytes and hepatoma cells. Proteomics. 2003;3:1154–1161. doi: 10.1002/pmic.200300436. [DOI] [PubMed] [Google Scholar]