Abstract
Charcot-Marie-Tooth disease (CMT) with autosomal recessive (AR) inheritance is a heterogeneous group of inherited motor and sensory neuropathies. In some families from Japan and Brazil, a demyelinating CMT, mainly characterized by the presence of myelin outfoldings on nerve biopsies, cosegregated as an autosomal recessive trait with early-onset glaucoma. We identified two such large consanguineous families from Tunisia and Morocco with ages at onset ranging from 2 to 15 years. We mapped this syndrome to chromosome 11p15, in a 4.6-cM region overlapping the locus for an isolated demyelinating ARCMT (CMT4B2). In these two families, we identified two different nonsense mutations in the myotubularin-related 13 gene, MTMR13. The MTMR protein family includes proteins with a phosphoinositide phosphatase activity, as well as proteins in which key catalytic residues are missing and that are thus called “pseudophosphatases.” MTM1, the first identified member of this family, and MTMR2 are responsible for X-linked myotubular myopathy and Charcot-Marie-Tooth disease type 4B1, an isolated peripheral neuropathy with myelin outfoldings, respectively. Both encode active phosphatases. It is striking to note that mutations in MTMR13 also cause peripheral neuropathy with myelin outfoldings, although it belongs to a pseudophosphatase subgroup, since its closest homologue is MTMR5/Sbf1. This is the first human disease caused by mutation in a pseudophosphatase, emphasizing the important function of these putatively inactive enzymes. MTMR13 may be important for the development of both the peripheral nerves and the trabeculum meshwork, which permits the outflow of the aqueous humor. Both of these tissues have the same embryonic origin.
Introduction
Charcot-Marie-Tooth disease (CMT) is a heterogeneous group of inherited motor and sensory neuropathies characterized by chronic distal weakness with progressive muscular atrophy and sensory loss in the distal extremities (Dyck 1975). CMT is one of the most common hereditary disorders, affecting ∼1/2,500. Electrophysiological measures—in particular, median motor nerve conduction velocities (median MNCV)—distinguish axonal (MNCV >40 m/s) from demyelinating (MNCV <35 m/s) forms (Harding and Thomas 1980; Bouche et al. 1983). The mode of inheritance can be autosomal dominant, X-linked, or autosomal recessive (ARCMT). For the demyelinating forms of ARCMT, eight different loci have been reported, on chromosomes 5q (CMT4C [MIM 601596]) (LeGuern et al. 1996), 8q13-q21.1 (CMT4A [MIM 214400]) (Ben Othmane et al. 1993), 8q24 (HMSNL [MIM 601455]) (Kalaydjieva et al. 1996), 10q (ERG2 [MIM 129010]) (Warner et al. 1998), 11q23 (CMT4B1 [MIM 601382]) (Bolino et al. 1996), 11p15 (CMT4B2 [MIM 604563]) (Ben Othmane et al. 1999), and 19q (CMT4F [MIM 145900]) (Delague et al. 2000). Five genes have been identified, EGR2 (MIM 129010) (Warner et al. 1998), MTMR2 (MIM 603557; the CMT4B1 locus) (Bolino et al. 2000), NDRG1 (MIM 605262) (Kalaydjieva et al. 2000), PRX (MIM 605725) (Boerkoel et al. 2001; Guilbot et al. 2001), and GDAP1 (MIM 606598) (Baxter et al. 2002; Cuesta et al. 2002), corresponding to the 10q, CMT4B1, HMSNL, CMT4F, and CMT4A loci, respectively.
Glaucoma is characterized by progressive loss of visual fields and degeneration of the optic nerve, usually associated with increased intraocular pressure (IOP). Glaucoma in the first 40 years of life includes congenital/infantile glaucoma, which manifests before age 5 years; juvenile primary open-angle glaucoma (JOAG [MIM 137750]), in which the age at onset ranges from 5 to 40 years; and variants that are associated with abnormalities of the anterior chamber of the eye. These diseases are genetically heterogeneous. Seven loci have been reported for autosomal dominant forms, but only two genes have been identified: MYOC/TIGR (MIM 601652) (Stone et al. 1997), the GLC1A locus (MIM 601682) on chromosome 1q25; and OPTN (MIM 602432) on chromosome 10p14 (Rezaie et al. 2002). For autosomal recessive glaucoma, three genes have been identified: CYP1B1 (MIM 601771) (Stoilov et al. 1997), the GLC3A locus (MIM 231300) on chromosome 2p21; PITX2 (MIM 601542) (Semina et al. 1996), the IRID2 locus (MIM 137600) on 4q25; and FOXC1 (MIM 601090) (Nishimura et al. 1998), the IRID1 locus (MIM 601631) on 6p25.
Two groups (Arruda et al. 1999; Kiwaki et al. 2000) have reported the association between a demyelinating CMT, mainly characterized by myelin outfoldings on nerve biopsies, and early-onset glaucoma (EOG), segregating as an autosomal recessive trait. In two large consanguineous families with this phenotype from Tunisia and Morocco, we have mapped this syndrome to 11p15, in a region overlapping the locus for an isolated demyelinating ARCMT with myelin outfoldings (CMT4B2) (Ben Othmane et al. 1999), and we have identified two nonsense mutations in the causative gene, MTMR13, a member of the phosphatase-inactive subgroup within the myotubularin-related gene family.
Patients and Methods
Patients
The index patients in families TUN-RE and RBT-HAD and 23 at-risk relatives (fig. 1) were examined for the presence of motor and sensory loss, areflexia, foot deformities, scoliosis, and other associated signs, such as nerve hypertrophy, tremor, ataxia, pyramidal signs, cranial nerve involvement, and dementia. They also underwent an ophthalmologic examination. The electrophysiological examination was performed in all patients and at-risk relatives. Seven patients were clinically affected. All other family members examined, including both parents, were found to be normal on clinical and electrophysiological examination.
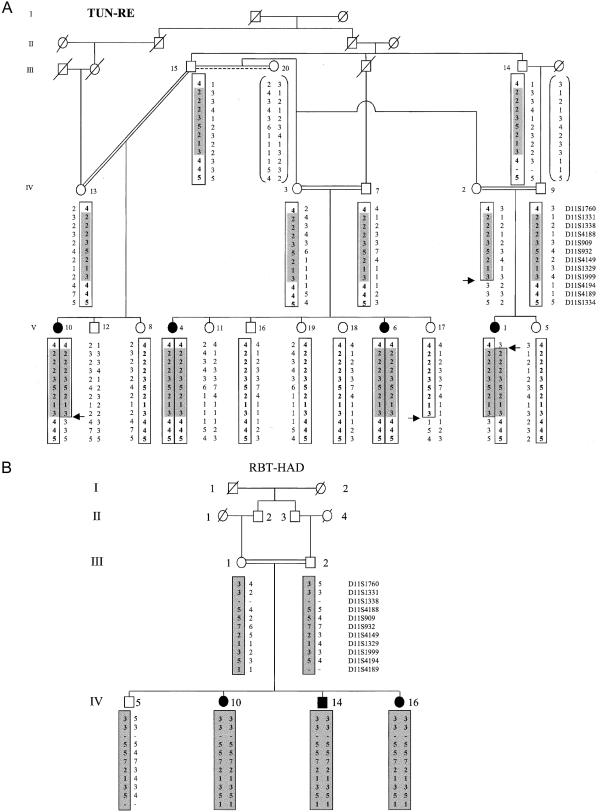
Figure 1.
Haplotype reconstruction of 14 markers in the Tunisian (A) and Moroccan (B) families with demyelinating ARCMT and glaucoma. Deduced haplotypes for individuals for whom DNA was not available are bracketed. Microsatellite markers are ordered, from telomere (top) to centromere (bottom), according to the Généthon genetic map. Boxes indicate the haplotype segregating with the disease, and shaded areas indicate the common region of homozygosity. Recombination events are indicated by arrows.
Genotyping
Blood samples from the 25 members from the Tunisian and Moroccan families were obtained after informed consent was given. Genomic DNA was extracted using standard procedures. Family members were genotyped with microsatellite markers, to test linkage to known demyelinating CMT and primary open-angle glaucoma (POAG) loci. The CMT loci are 17p11.2 (CMT1A [MIM 118220]) (Timmerman et al. 1990), 1q22-23 (CMT1B [MIM 118200], the MPZ gene [MIM 159440]) (Hayasaka et al. 1993), 10q23 (EGR2) (Warner et al. 1998), 5q23-q33 (CMT4C) (LeGuern et al. 1996), 8q13-q21 (the CMT4A locus, GDAP1) (Ben Othmane et al. 1993), 8q24 (the HMSNL locus, NDRG1 [MIM 605262]) (Kalaydjieva et al. 1996), 11q23 (the CMT4B1 locus, MTMR2) (Bolino et al. 1996); the POAG loci are 1q23-q25 (GLC1A) (Richards et al. 1994), 2cen-q13 (GLC1B [MIM 606669]) (Stoilova et al. 1996), 3q21-q24 (GLC1C [MIM 606669]) (Wirtz et al. 1997), 6p25 (IRID1) (Jordan et al. 1997), 8q23 (GLC1D [MIM 602429]) (Trifan et al. 1998), 10p15-p14 (GLC1E [MIM 137760]) (Sarfarazi et al. 1998), 7q35-q36 (GLC1F [MIM 603383]) (Wirtz et al. 1999), 2p21 (GLC3A [MIM 231300]) (Sarfarazi et al. 1995), and 1p36 (GLC3B [MIM 600975]) (Akarsu et al. 1996). PCRs were performed according to published procedures. The PCR products were pooled, combined with the GeneScan HD Rox 400 size standard, loaded onto a 4.8% PAGE-plus, and run on the ABI PRISM 377 DNA Sequencer (Perkin-Elmer/Applied Biosystems).
Linkage Analyses
Pairwise and multipoint LOD scores were calculated using the MLINK and LINKMAP programs of the FASTLINK package (Schaffer et al. 1994); a fully penetrant autosomal recessive trait with a disease-allele frequency of 0.0001 and an equal recombination fraction (θ) in males and females were assumed. We assigned equal frequencies to the alleles observed in the families studied. At-risk individuals who were clinically and electrophysiologically normal at age 15 years were considered to be unaffected. Haplotypes were constructed according to the principles of Thompson (1987). The order of the markers was that of the consensus CEPH/Généthon chromosome 11 linkage map in the Genome Database (fig. 1).
Sequencing
All coding exons of the MTMR13 gene, including exon-intron junctions, were amplified by PCR with primers designed from the genomic sequences available from GenBank (accession numbers XM_049218 and XM_113650, replaced with XM_208513). Both strands of the PCR products were sequenced with BigDye Terminators (Applied Biosystems) on an ABI 3100 sequencer. Sequence chromatograms were analyzed using SeqScape software version 1.1 (Applied Biosystems). In each family, the genotypes of at-risk members were determined by direct sequencing of the exon containing the mutation in the index patient.
RT-PCR
Total RNA was extracted from a human sciatic nerve, through use of standard procedures. Total RNA from the additional 10 human tissues used in the RT-PCR experiments (shown in fig. 3) were purchased from Becton Dickinson (Clontech). First-strand cDNA was prepared from 1 μg of RNA, through use of the Advantage RT-for-PCR kit (Becton Dickinson; Clontech). RT-PCR was performed using diluted cDNA, as recommended by the manufacturer’s instructions, with 2 mM MgCl2 and 30 cycles of amplification.
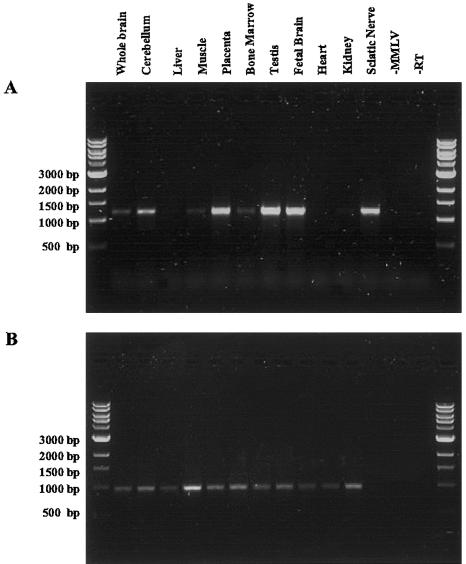
Figure 3.
RT-PCR analysis of MTMR13 expression in several human tissues. A, RT-PCR performed using primers amplifying a fragment corresponding to the first 1,250 bp of the MTMR13 ORF, starting from the ATG. A similar expression pattern was demonstrated for the remaining four overlapping fragments into which the ORF was divided (not shown). B, G3PDH was used as positive control for the quality of RNA and cDNA synthesis. −MMLV is a control reaction in which the MMLV reverse transcriptase was omitted. RT corresponds to the negative control of the PCR performed by omitting the cDNA thereafter.
Phylogenetic Analysis and Protein Domain Prediction
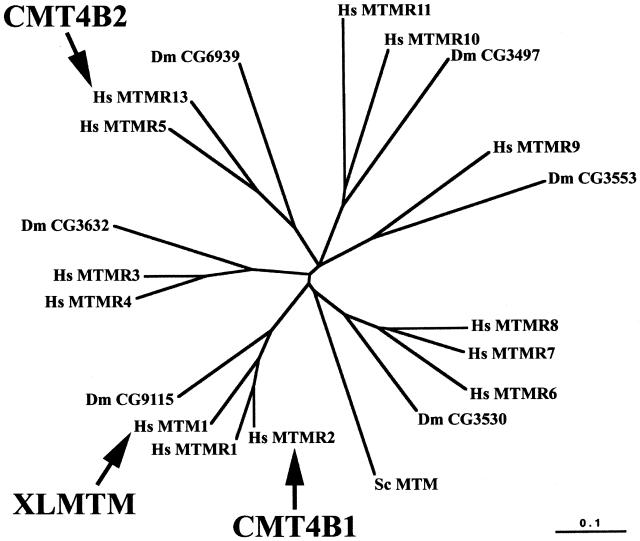
Protein sequences for members of the myotubularin family from human, Drosophila melanogaster, and Saccharomyces cerevisiae were aligned using Clustal X (Thompson et al. 1997), and the sequences encompassing the PTPc/DSPc homology domain and the SET interacting domain were realigned (corresponding to amino acids 1338–1520 in Hs MTMR13). The unrooted tree was generated with Clustal X, through use of the distance method based on amino acid identity. The protein sequences used in the tree and their NCBI or SwissProt (Entrez-Protein) protein accession numbers (in parentheses) are as follows: hMTM1 (Q13496), hMTMR1 (Q13613), hMTMR2 (Q13614), hMTMR3 (Q13615), hMTMR4 (AAH35609), hMTMR5 (O95248), hMTMR6 (Q9Y217), hMTMR7 (Q9Y216), hMTMR8 (AAH12399), hMTMR9 (CAC51114), hMTMR10 (AK000320), hMTMR11 (AY028703), ScMTM (P47147), DmCG3497 (AAF48390), DmCG3530 (AAF46997), DmCG3632 (AAF48581), DmCG6939 (AAK93570), DmCG3553 (AAF50343), DmCG9115 (AAF52327). The sequences used can also be found at the Institut de Génétique et de Biologie Moléculaire et Cellulaire Web site. MTMR5 (Cui et al. 1998; Laporte et al. 1998), MTMR10, and MTMR11 (Nandurkar and Huysmans 2002) are also called “Sbf1” (MIM 603560), “FLJ20313,” and “3-PAP,” respectively. Protein domain prediction was performed with full-length sequences by multiple sequence alignment and by using the SMART (Letunic et al. 2002; SMART Web site), Pfam (Bateman et al. 2002; Pfam Home Page), and PSORT (Nakai and Kanehisa 1992; PSORT WWW Server) databases.
Results
Mapping of the Gene Responsible for ARCMT Associated with EOG
Two families were studied: one Tunisian family with four affected members and one Moroccan family with three affected members. Sensory motor neuropathy was characterized by a mean age at onset of 8 years, a distal motor deficit affecting the four limbs, MNCV <20 m/s, and alteration of sensory nerve action potentials. Myelin outfoldings were observed at nerve biopsy in index patients. In the index patient of each family, visual impairment was precocious and severe, leading to a loss of vision. The ophthalmologic examination showed clear signs of congenital glaucoma with a buphthalmos, a megalocornea, and raised intraocular pressure. All the other affected members of both families had increasing intraocular pressure, which was in the upper normal limits (16–20 mmHg). Results of clinical electrophysiological and ophthalmologic examinations of all parents and examined at-risk subjects were normal (R.G. and H.A., unpublished data).
All loci for demyelinating ARCMT and AR glaucoma were excluded in both families, by means of haplotype reconstruction and linkage analysis, except the locus in 11p15.3 reported by Ben Othmane et al. (1999). Positive two-point LOD scores for both families reached a cumulative maximum LOD score (Zmax) of 6.25 at θ=0, for D11S4149 (table 1). In family TUN-RE, the centromeric boundary of the candidate interval corresponded to an inferred recombination between D11S1999 and D11S4194, which was observed in individuals V10, V1, and her mother IV2, and which conceivably occurred in individual III15 (fig. 1). The telomeric boundary of the region of homozygosity is delimited by the inferred recombination between D11S1760 and D11S1331, which was observed in patient V1. These results place the disease locus in a 4.6-cM interval between markers D11S1760 and D11S1329. Haplotype reconstruction in family RBT-HAD, which defined a homozygosity region from marker D11S1760 to D11S4189, did not help to refine the candidate region. The same alleles at markers D11S4149, D11S1329, and D11S1999 were found in patients from both families. Alleles were detected in patients from both families for the eight microsatellites, as well as for the 18 additional markers within the candidate region excluding a large genomic deletion (fig. 2A).
Table 1.
Combined Two-Point LOD Scores for Chromosome 11p15 Markers in the TUN-RE and RBT-HAD Families
|
LOD Score at θ = |
|||||||||
| Markera | .0 | .01 | .05 | .1 | .2 | .3 | .4 | Zmaxb | θmax |
| D11S4046 | −∞ | −4.39 | −2.75 | −1.64 | −.68 | −.29 | −.11 | −.11 | .40 |
| D11S1760 | .27 | 3.40 | 3.66 | 3.41 | 2.56 | 1.59 | .66 | 3.66 | .05 |
| D11S1331 | 5.67 | 5.54 | 5.05 | 4.43 | 3.18 | 1.96 | .83 | 5.67 | .00 |
| D11S1338 | 1.42 | 1.38 | 1.22 | 1.01 | .61 | .27 | .05 | 1.42 | .00 |
| D11S4188 | 5.61 | 5.48 | 4.98 | 4.34 | 3.05 | 1.81 | .73 | 5.61 | .00 |
| D11S909 | 4.88 | 4.78 | 4.35 | 3.81 | 2.71 | 1.63 | .66 | 4.88 | .00 |
| D11S932 | 6.19 | 6.06 | 5.54 | 4.88 | 3.55 | 2.23 | .98 | 6.19 | .00 |
| D11S4149 | 6.25 |
6.12 | 5.60 | 4.94 | 3.59 | 2.24 | .98 | 6.25 |
.00 |
| D11S1329 | 3.31 | 3.24 | 2.95 | 2.59 | 1.86 | 1.14 | .47 | 3.31 | .00 |
| D11S1999 | 5.06 | 4.95 | 4.51 | 3.96 | 2.83 | 1.73 | .73 | 5.06 | .00 |
| D11S4194 | −∞ | 2.44 | 3.35 | 3.32 | 2.66 | 1.76 | .84 | 3.35 | .00 |
| D11S4189 | −∞ | 1.43 | 2.40 | 2.47 | 2.00 | 1.31 | .62 | 2.47 | .00 |
| D11S1334 | 3.09 | 3.03 | 2.77 | 2.45 | 1.81 | 1.18 | .57 | 3.09 | .00 |
Markers are ordered according to the Marshfield linkage map (Center for Medical Genetics Web site). Markers in boldface italic type are from the ABI PRISM linkage mapping set version 2 (Applied Biosystems).
The highest Zmax value is underlined.
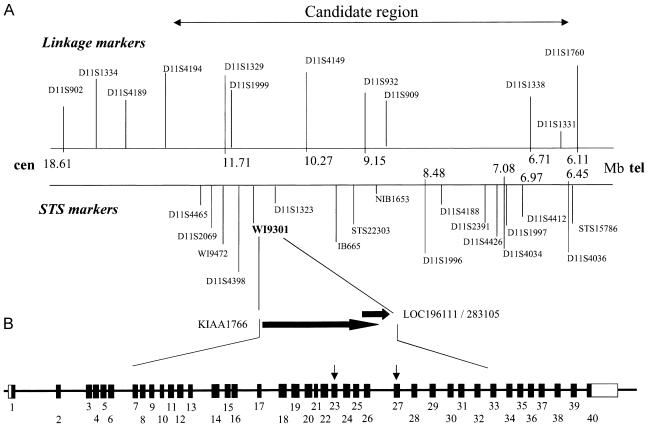
Figure 2.
Physical map of 11p15 and genomic structure of MTMR13. A, Physical map of the critical region, including the position of the linkage markers (top) and STS (bottom) tested. Physical distances (Ensembl Genome Browser; GenBank) are indicated in megabases. The marker closest to the gene is in bold. B, Location of the two ESTs, LOC19611/LOC283105 and KIAA1766, from which the coding sequence of MTMR13 (top) and intron-exon structure of MTMR13 (bottom) was constructed. This region was covered by four different genomic clones from contig NT_028309.7. Clone AC080023.6 contains only the first exon of MTMR13. AC100763.2 includes exons 2 and 3 and partially overlaps with clone AC011092.4, which extended from exons 3 to 17. The remaining portion of the gene is found in clone AC026250.16.
Identification of Mutations in MTMR13
At least 60 independent cDNA sequences were located in the 4.6-cM candidate region. Two of them showed homology to domains observed in members of the MTMR family of protein phosphatases. Transcript LOC196111 (GenBank accession number XM_113650), which was replaced by LOC283105 (GenBank accession number XM_208513), corresponded to an ORF of 1,482 bp, encoding a protein of 493 aa, sharing 53% identity and 70% similarity, at the protein level, with MTMR5 (also called “Sbf1,” for SET binding factor 1 [NCBI protein accession number O95248]) (fig. 2B). The transcript KIAA1766 (GenBank accession number XM_049218), containing an ORF of 3,459 bp coding for a protein of 1,152 aa, had 59% similarity and 73% identity, at the protein level, with a domain of MTMR5 closer to the N-terminus than LOC196111. This suggested that both mRNAs might belong to a transcript encoding the same protein with high homology with MTMR5. To confirm this hypothesis, we performed BLAST searches with the LOC196111 mRNA against EST databases (NCBI). An EST sequence (Agencourt_6446961 [GenBank accession number BM803570]) containing an ORF corresponding to the last 100 bp of the KIAA1766 coding sequence, an additional coding region of 198 bp, and the LOC196111 mRNA starting from its putative 5′ UTR were retrieved. This allowed the reconstruction of a 5,550-bp ORF encoding a 1,849-aa protein that we have named “MTMR13.”
At the genomic level, 40 exons were identified, spanning a region of ∼600 kb (fig. 2). The MTMR13 coding region has been confirmed by RT-PCR experiments performed using total RNA from 20 different tissues. The MTMR13 gene was found to be mainly expressed in cerebellum, placenta, testis, fetal brain, and sciatic nerve (fig. 3).
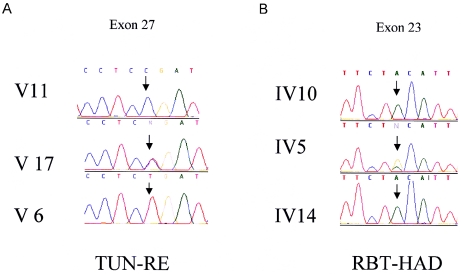
We initially tested one affected individual from each family and a normal control individual for mutations in MTMR13, by direct sequencing of all exons. We used intronic primers (sequences available upon request) to amplify the 40 coding exons and the exon-intron boundaries. In this way, two homozygous nonsense mutations, 2875C→T (Gln956Stop) in individual IV-10 from the RBT-HAD pedigree and 3586C→T (Arg1196Stop) in individual V-4 from the TUN-RE pedigree, were identified that were not present in the normal control (fig. 4). All available family members were analyzed, and segregation of the mutations with the disease was confirmed in both pedigrees. Both mutations, located in exons 23 and 27, respectively, were not detected by direct sequencing in 50 control individuals of northern African ancestry (100 chromosomes).
Figure 4.
Chromatograms of the mutation site sequence (arrow) are presented for both families. A, Family TUN-RE: a healthy homozygous sib (V11), a heterozygous sib (V17), and an affected sib (V6); N:C/T (forward strand sequence). B, Family RBT-HAD: a heterozygous sib (IV5) and two affected sibs (IV10 and IV14); N:A/G (reverse strand sequence).
Hs MTMR13 Is a Putative Inactive Phosphatase from the Myotubularin Family
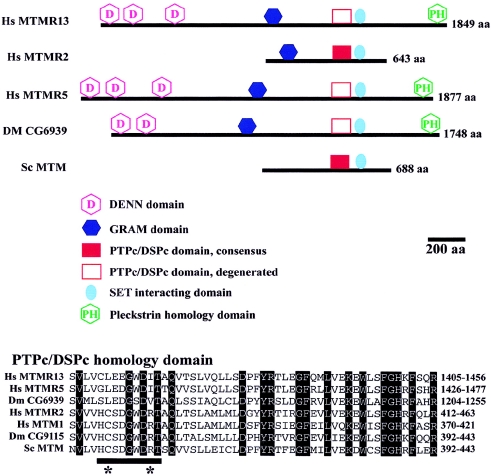
MTMR13 is a newly discovered protein that shares extensive homology with proteins from the myotubularin family (Laporte et al. 2001; Wishart et al. 2001), especially with MTMR5 (60% identity for the full-length sequence). We aligned the most conserved region of human, D. melanogaster, and S. cerevisiae myotubularin proteins, including MTMR13, and performed a phylogenetic analysis. The myotubularin family is divided into six subgroups. MTMR13 belongs to a subgroup that includes Hs MTMR5 and a D. melanogaster protein, Dm CG6939 (fig. 5). MTMR13, like MTMR5, has homology with the active site of tyrosine and dual-specificity phosphatases (PTPc/DSPc). Their consensus catalytic sequences, defined by CX5R in all active PTP/DSP phosphatases, are degenerate. In particular, the conserved cysteine and arginine residues are absent (fig. 6). MTMR13, like MTMR5, is thus a putative pseudophosphatase with no predicted enzymatic activity. Human MTM1 and MTMR2, which belong to a different subgroup of active phosphatases, are mutated in X-linked myotubular myopathy (Laporte et al. 1996) and in CMT type 4B1 (Bolino et al. 2000), respectively.
Figure 5.
Phylogenetic relationship of MTMR13 within the myotubularin family. The radial distance tree of the human, D. melanogaster, and S. cerevisiae protein is shown, illustrating the six subgroups, each defined by one protein in D. melanogaster. MTMR13 belongs to the same subgroup as MTMR5 (or Sbf1), its closest homologue (60% identity). MTMR2 belongs to the same subgroup as MTM1 and MTMR1 (70% identity with the latter). MTMR11 was also called “3-PAP” (Nandurkar and Huysmans 2002). While submitting this article, another MTMR was published that is not included in this analysis (CRAα/β) (Wishart and Dixon 2002a). The scale represents the percentage of divergence. Human genetic disorders associated with mutations in myotubularin protein are indicated: CMT4B1 (CMT type 4B1), CMT4B2 (CMT type 4B2 with early-onset glaucoma), and XLMTM (X-linked myotubular myopathy). The tree was generated using sequences encompassing the PTPc/DSPc homology domain and the SET interacting domain. Hs, Homo sapiens; Dm, D. melanogaster; Sc, S. cerevisiae.
Figure 6.
Domain structure of MTMR13 and homologous proteins. Top, Scaled representations of Hs MTMR2, Hs MTMR5 (Sbf1), and the three orthologous MTMR13 proteins in human (Hs MTMR13), D. melanogaster (Dm CG6939), and yeast (Sc MTM). The main protein domains are depicted. The DENN (differentially expressed in neoplastic versus normal cells) domain is formed by three subdomains that are always present together: uDENN, DENN, and dDENN (Levivier et al. 2001), all depicted as “DENN” here. In Dm CG6939, the first DENN subdomain is missing, suggesting incomplete prediction of the N-terminus of the protein. The GRAM domain is found in glucosyltransferase, rab-like GTPase activators and myotubularins and has no known function. The PTPc/DSPc homology domain (catalytic domain of tyrosine and dual-specificity phosphatases) is found in all members. However, the consensus sequence of the catalytic pocket, present in Hs MTMR2 and Sc MTM, is degenerated in Hs MTMR13. SET interacting domain (SID) was defined on the basis of interaction between Hs MTMR5 and proteins with SET (suvar 3–9, enhancer-of-zeste, trithorax) domains and is present in all members. The PH domain (pleckstrin homology) is a phosphoinositide-binding domain. Additional protein domains include a coiled-coil domain (amino acids 593–627) and a PDZ-binding domain (amino acids 640–643) at the C-terminus of Hs MTMR2, a protein kinase C conserved region in Dm CG6939 (C1 domain/diacyglycerol and phrobol ester binding; amino acids 1546–1593), and the RID (Rac1-induced recruitment domain), the boundaries of which have not been defined (around amino acid 283 in Hs MTMR2), and which is responsible for the plasma membrane recruitment of MTMR2 and other active and inactive homologues upon Rac 1 activation (Laporte et al. 2002). Bottom, Sequence alignment of the PTPc/DSPc homology domain in Hs MTMR13 and homologous proteins. Names of the proteins are on the left, and the sequence intervals are on the right. Dm CG9115 is the D. melanogaster orthologue of Hs MTMR2 and Hs MTM1. Shaded amino acids are conserved in all the depicted proteins. The underlined sequence HCSDGWDRT is conserved 100% in all active myotubularins from yeast to human and contains the compulsory consensus sequence CysX5Arg for the phosphatase activity. The catalytic cysteine and arginine are indicated by stars.
To gain insight into the putative function of MTMR13, protein domains were predicted by the SMART, PFAM, and PSORT databases and by comparison with known domains present in other myotubularin proteins. Not surprisingly, MTMR13 had the same organization as MTMR5 (fig. 6); in particular, the DENN (Levivier et al. 2001) and GRAM (Doerks et al. 2000) domains, which are also found in regulators of GTPase activity, although no specific functions have as yet been assigned to them, and the SID (SET-interacting) domain, which, in MTMR5, interacts with the SET domain of ALL-1, an epigenetic transcriptional regulator (Cui et al. 1998).
Discussion
We report the identification of the gene implicated for a demyelinating ARCMT associated with early-onset glaucoma. We first mapped the disease gene to chromosome 11p15. Haplotype reconstruction in both families placed the candidate interval in a 4.6-cM region flanked by markers D11S1760 and D11S4194. This locus overlaps the locus for CMT4B2 (Ben Othmane et al. 1999), in which glaucoma was not reported (fig. 1). Whether the same gene is responsible for the neuropathy with or without glaucoma can not be affirmed. However, the common North African origin (Tunisia and Morocco) of the families reported by both groups, their common neuropathological features (myelin outfoldings), and the well known phenotypic inter- and intrafamilial variability associated with neuropathies are compatible with the involvement of a same gene.
We first tested the hypothesis that a large deletion disrupts two different genes affecting, independently, the peripheral nervous system (PNS) and eye phenotypes. Indeed, many disorders, such as Di George syndrome (MIM 188400), Smith-Magenis syndrome (MIM 182290), Williams-Beuren syndrome (MIM 194050), and WAGR syndrome (MIM 194072), result from large genomic deletions causing a contiguous gene syndrome. However, a homozygous deletion was not detected in the families in our study when the eight microsatellites and 18 additional STS covering the candidate interval were used (fig. 2A), excluding the hypothesis of a contiguous gene syndrome.
We then analyzed the expressed sequences mapped in the candidate region. A single cDNA sequence was reconstructed from two apparently independent transcripts. It corresponds to a new gene we have named “MTMR13.” This gene, which is highly homologous to MTMR5, belongs to the MTMR gene family. Nonsense mutations in exons 23 and 27 were identified in the index patients of families RBT-HAD and TUN-RE. These stop mutations cosegregate with the disease in the families. At the protein level, these mutations are predicted to result in a truncated protein with deletion of the PTPc/DSPc and SID homology domains. Truncation of approximately half of the protein might also result in instability. These results strongly support the hypothesis that mutations in MTMR13 are responsible for the phenotype.
To our knowledge, MTMR13 is the first pseudophosphatase mutated in a human disorder. Loss of function of the pseudophosphatases STYX (Wishart and Dixon 2002b) and MTMR5 has recently been analyzed in the mouse. Loss of MTMR5, the closest homologue of MTMR13, causes a defect in late stages of spermatogenesis (Firestein et al. 2002). Mutations of MTM1, a close homologue of MTMR2, lead to myotubular myopathy (Laporte et al. 1996), a phenotype unrelated to demyelinating CMT. Thus, while loss-of-function mutations in MTMR2 (Bolino et al. 2000) and MTMR13 lead to related phenotypes, this is not the case for other myotubularins. Furthermore, there is apparently no complementation of MTMR13 loss of function by pseudophosphatase homologues in the target tissues, suggesting that MTMR13 has a specific function in the PNS and eye or that the level or pattern of expression of myotubularins differ. The fact that two forms of demyelinating CMT4B are caused by loss-of-function mutations in either an active phosphatase (MTMR2) and a pseudophosphatase (MTMR13) contradicts the hypothesis that pseudophosphatases act by competing with active phosphatases for their substrate (Cui et al. 1998). A common function for MTMR2 and MTMR13 might lie in other domains (DENN, GRAM, or SID, for example), and loss of these domains would be responsible for the phenotype. Another hypothesis is that the degenerate consensus phosphatase domain acquires a new function in the same transduction pathway as MTMR2. Myotubularin pseudophosphatases have been proposed to act as adaptators or modulators of phosphatases (Nandurkar et al. 2001).
Insight into the function of MTMR13 might be gained from the known functions of its closest homologue, MTMR5. The SET domain of ALL-1 was recently shown to have histone methyltransferase activity (Nakamura et al. 2002) that might be regulated through interaction with the SID domains of MTMR5 and MTMR13, affecting heterochromatin function. The PH domain of MTMR5 binds PI(3,4)P2 and PI(3,4,5)P3 (Isakoff et al. 1998), and PI3P and PI3,5P2 are preferential in vitro substrates for active myotubularins (Berger et al. 2002). MTMR13 might thus be involved in phosphoinositide metabolism or regulation of SET proteins and transcription.
The identification of a single gene involved in both a demyelinating peripheral neuropathy and a early-onset glaucoma is particularly interesting from a developmental point of view. Indeed, mutations in MTMR13 might cause dysfunction of both Schwann cells, the myelinating cells in peripheral nerves, and cells of the trabeculum meshwork that permits outflow of the aqueous humor. Both types of cells are derived from the neural crest, as are different types of neurons and glial cells, as well as melanocytes. Given the early onset of the disease, we can hypothesize that MTMR13 is involved in both the differentiation of Schwann cells during myelination and in the formation and development of the trabeculum.
Acknowledgments
We would like to convey our gratitude to the members of the TUN-RE and RBT-HAD families, for their participation in this study; to Merle Ruberg and Jean-Louis Mandel, for their critical reading of the manuscript; to Giovanni Stevanin, Alexandre Chojnowsky, and Pascal Fragner, for their kind help; and to the DNA and cell bank of the Institut Fédératif de Recherches en Neurosciences (IFR70). This work was supported by Association Française contre les Myopathies (AFM), the Assistance Publique des Hôpitaux de Paris, the Institut National de la Santé et de la Recherche Médicale (INSERM), the Centre National de la Recherche Scientifique, and the Hôpital Universitaire de Strasbourg. H.A. is supported by AFM; A.Bol. is supported by Telethon-Italy and is an Assistant Telethon Scientist from the Dulbecco Telethon Institute. H.A., A.Bol., R.G., N.B., A.Bou., A.Br., and E.L. are members of the French AFM/INSERM research network on autosomal recessive forms of CMT.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Center for Medical Genetics, Marshfield Clinic, http://research.marshfieldclinic.org/genetics/
- Ensembl Genome Browser, http://www.ensembl.org/ (for Ensembl database)
- Entrez-Protein, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=protein (for hMTM1 [accession number Q13496], hMTMR1 [accession number Q13613], hMTMR2 [accession number Q13614], hMTMR3 [accession number Q13615], hMTMR4 [accession number AAH35609], hMTMR5 [accession number O95248], hMTMR6 [accession number Q9Y217], hMTMR7 [accession number Q9Y216], hMTMR8 [accession number AAH12399], hMTMR9 [accession number CAC51114], hMTMR10 [accession number AK000320], hMTMR11 [accession number AY028703], ScMTM [accession number P47147], DmCG3497 [accession number AAF48390], DmCG3530 [accession number AAF46997], DmCG3632 [accession number AAF48581], DmCG6939 [accession number AAK93570], DmCG3553 [accession number AAF50343], and DmCG9115 [accession number AAF52327])
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for LOC196111 [accession number XM_113650], LOC283105 [accession number XM_208513], KIAA1766 [accession number XM_049218], Agencourt_6446961 [accession number BM803570, contig [accession number NT_028309.7], and genomic clones [accession numbers AC080023.6, AC100763.2, AC011092.4, and AC026250.16])
- Genome Database, http://www.gdb.org/
- Institut de Génétique et de Biologie Moléculaire et Cellulaire, http://www1-igbmc.u-strasbg.fr/THEMES/Theme_IX/Publi/Table_paper1.html (for IGBMC) [PMC free article] [PubMed]
- Online Mendelian Inheritance in Man (OMIM): http://www.ncbi.nlm.nih.gov/Omim/ (for CMT4C, CMT4A, HMSNL, ERG2, CMT4B1, CMT4B2, CMT4F, MTMR2, NDRG1, PRX, GDAP1, JOAG, MYOC/TIGR, GLC1A, OPTN, CYP1B1, GLC3A, PITX2, IRID2, FOXC1, IRID1, CMT1A, CMT1B, MPZ, NDRG1, GLC1B, GLC1C, GLC1D, GLC1E, GLC1F, GLC3A, GLC3B, Sbf1, Di George syndrome, Smith-Magenis syndrome, Williams-Beuren syndrome, and WAGR)
- Pfam Home Page, http://www.sanger.ac.uk/Software/Pfam (for Pfam)
- PSORT WWW Server, http://psort.nibb.ac.jp (for PSORT)
- Simple Modular Architecture Research Tool (SMART), http://smart.embl-heidelberg.de/ [DOI] [PMC free article] [PubMed]
References
- Akarsu AN, Turacli ME, Aktan SG, Barsoum-Homsy M, Chevrette L, Sayli BS, Sarfarazi M (1996) A second locus (GLC3B) for primary congenital glaucoma (buphthalmos) maps to the 1p36 region. Hum Mol Genet 5:1199–1203 [DOI] [PubMed] [Google Scholar]
- Arruda WO, Comerlato EA, Scola RH, Silvado CE, Werneck LC (1999) Hereditary motor and sensory neuropathy with congenital glaucoma. Report on a family. Arq Neuropsiquiatr 57:190–194 [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL (2002) The Pfam protein families database. Nucleic Acids Res 30:276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter RV, Ben Othmane K, Rochelle JM, Stajich JE, Hulette C, Dew-Knight S, Hentati F, Ben Hamida M, Bel S, Stenger JE, Gilbert JR, Pericak-Vance MA, Vance JM (2002) Ganglioside-induced differentiation-associated protein-1 is mutant in Charcot-Marie-Tooth disease type 4A/8q21. Nat Genet 30:21–22 [DOI] [PubMed] [Google Scholar]
- Ben Othmane K, Hentati F, Lennon F, Ben Hamida C, Blel S, Roses AD, Pericak-Vance MA, Ben Hamida M, Vance JM (1993) Linkage of a locus (CMT4A) for autosomal recessive Charcot-Marie-Tooth disease to chromosome 8q. Hum Mol Genet 2:1625–1628 [DOI] [PubMed] [Google Scholar]
- Ben Othmane K, Johnson E, Menold M, Graham FL, Hamida MB, Hasegawa O, Rogala AD, Ohnishi A, Pericak-Vance M, Hentati F, Vance JM (1999) Identification of a new locus for autosomal recessive Charcot-Marie-Tooth disease with focally folded myelin on chromosome 11p15. Genomics 62:344–349 [DOI] [PubMed] [Google Scholar]
- Berger P, Bonneick S, Willi S, Wymann M, Suter U (2002) Loss of phosphatase activity in myotubularin-related protein 2 is associated with Charcot-Marie-Tooth disease type 4B1. Hum Mol Genet 11:1569–1579 [DOI] [PubMed] [Google Scholar]
- Boerkoel CF, Takashima H, Stankiewicz P, Garcia CA, Leber SM, Rhee-Morris L, Lupski JR (2001) Periaxin mutations cause recessive Dejerine-Sottas neuropathy. Am J Hum Genet 68:325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolino A, Brancolini V, Bono F, Bruni A, Gambardella A, Romeo G, Quattrone A, Devoto M (1996) Localization of a gene responsible for autosomal recessive demyelinating neuropathy with focally folded myelin sheaths to chromosome 11q23 by homozygosity mapping and haplotype sharing. Hum Mol Genet 5:1051–1054 [DOI] [PubMed] [Google Scholar]
- Bolino A, Muglia M, Conforti FL, LeGuern E, Salih MA, Georgiou DM, Christodoulou K, Hausmanowa-Petrusewicz I, Mandich P, Schenone A, Gambardella A, Bono F, Quattrone A, Devoto M, Monaco AP (2000) Charcot-Marie-Tooth type 4B is caused by mutations in the gene encoding myotubularin-related protein-2. Nat Genet 25:17–19 [DOI] [PubMed] [Google Scholar]
- Bouche P, Gherardi R, Cathala HP, Lhermitte F, Castaigne P (1983) Peroneal muscular atrophy. Part 1. Clinical and electrophysiological study. J Neurol Sci 61:389–399 [DOI] [PubMed] [Google Scholar]
- Cuesta A, Pedrola L, Sevilla T, Garcia-Planells J, Chumillas MJ, Mayordomo F, LeGuern E, Marin I, Vilchez JJ, Palau F (2002) The gene encoding ganglioside-induced differentiation-associated protein 1 is mutated in axonal Charcot-Marie-Tooth type 4A disease. Nat Genet 30:22–25 [DOI] [PubMed] [Google Scholar]
- Cui X, De V, I, Slany R, Miyamoto A, Firestein R, Cleary ML (1998) Association of SET domain and myotubularin-related proteins modulates growth control. Nat Genet 18:331–337 [DOI] [PubMed] [Google Scholar]
- Delague V, Bareil C, Tuffery S, Bouvagnet P, Chouery E, Koussa S, Maisonobe T, Loiselet J, Megarbane A, Claustres M (2000) Mapping of a new locus for autosomal recessive demyelinating Charcot-Marie-Tooth disease to 19q13.1-13.3 in a large consanguineous Lebanese family: exclusion of MAG as a candidate gene. Am J Hum Genet 67:236–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerks T, Strauss M, Brendel M, Bork P (2000) GRAM, a novel domain in glucosyltransferases, myotubularins and other putative membrane-associated proteins. Trends Biochem Sci 25:483–485 [DOI] [PubMed] [Google Scholar]
- Dyck PJ (1975) Inherited neuronal degeneration and atrophy affecting peripheral motor, sensory, and autonomic neurons. In: Dyck PJ, Thomas PK, Lambert EH (eds) Peripheral neuropathy. Saunders, Philadelphia, pp 825–867 [Google Scholar]
- Firestein R, Nagy PL, Daly M, Huie P, Conti M, Cleary ML (2002) Male infertility, impaired spermatogenesis, and azoospermia in mice deficient for the pseudophosphatase Sbf1. J Clin Invest 109:1165–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilbot A, Williams A, Ravise N, Verny C, Brice A, Sherman DL, Brophy PJ, LeGuern E, Delague V, Bareil C, Megarbane A, Claustres M (2001) A mutation in periaxin is responsible for CMT4F, an autosomal recessive form of Charcot-Marie-Tooth disease. Hum Mol Genet 10:415–421 [DOI] [PubMed] [Google Scholar]
- Harding AE, Thomas PK (1980) Genetic aspects of hereditary motor and sensory neuropathy (types I and II). J Med Genet 17:329–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayasaka K, Himoro M, Wang Y, Takata M, Minoshima S, Shimizu N, Miura M, Uyemura K, Takada G (1993) Structure and chromosomal localization of the gene encoding the human myelin protein zero (MPZ). Genomics 17:755–758 [DOI] [PubMed] [Google Scholar]
- Isakoff SJ, Cardozo T, Andreev J, Li Z, Ferguson KM, Abagyan R, Lemmon MA, Aronheim A, Skolnik EY (1998) Identification and analysis of PH domain-containing targets of phosphatidylinositol 3-kinase using a novel in vivo assay in yeast. EMBO J 17:5374–5387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan T, Ebenezer N, Manners R, McGill J, Bhattacharya S (1997) Familial glaucoma iridogoniodysplasia maps to a 6p25 region implicated in primary congenital glaucoma and iridogoniodysgenesis anomaly. Am J Hum Genet 61:882–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaydjieva L, Gresham D, Gooding R, Heather L, Baas F, de Jonge R, Blechschmidt K, Angelicheva D, Chandler D, Worsley P, Rosenthal A, King RH, Thomas PK (2000) N-myc downstream-regulated gene 1 is mutated in hereditary motor and sensory neuropathy-Lom. Am J Hum Genet 67:47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaydjieva L, Hallmayer J, Chandler D, Savov A, Nikolova A, Angelicheva D, King RH, Ishpekova B, Honeyman K, Calafell F, Shmarov A, Petrova J, Turnev I, Hristova A, Moskov M, Stancheva S, Petkova I, Bittles AH, Georgieva V, Middleton L, Thomas PK (1996) Gene mapping in Gypsies identifies a novel demyelinating neuropathy on chromosome 8q24. Nat Genet 14:214–217 [DOI] [PubMed] [Google Scholar]
- Kiwaki T, Umehara F, Takashima H, Nakagawa M, Kamimura K, Kashio N, Sakamoto Y, Unoki K, Nobuhara Y, Michizono K, Watanabe O, Arimura H, Osame M (2000) Hereditary motor and sensory neuropathy with myelin folding and juvenile onset glaucoma. Neurology 55:392–397 [DOI] [PubMed] [Google Scholar]
- Laporte J, Blondeau F, Buj-Bello A, Mandel JL (2001) The myotubularin family: from genetic disease to phosphoinositide metabolism. Trends Genet 17:221–228 [DOI] [PubMed] [Google Scholar]
- Laporte J, Blondeau F, Buj-Bello A, Tentler D, Kretz C, Dahl N, Mandel JL (1998) Characterization of the myotubularin dual specificity phosphatase gene family from yeast to human. Hum Mol Genet 7:1703–1712 [DOI] [PubMed] [Google Scholar]
- Laporte J, Blondeau F, Gansmuller A, Lutz Y, Vonesch JL, Mandel JL (2002) The PtdIns3P phosphatase myotubularin is a cytoplasmic protein that also localizes to Rac1-inducible plasma membrane ruffles. J Cell Sci 115:3105–3117 [DOI] [PubMed] [Google Scholar]
- Laporte J, Hu LJ, Kretz C, Mandel JL, Kioschis P, Coy JF, Klauck SM, Poustka A, Dahl N (1996) A gene mutated in X-linked myotubular myopathy defines a new putative tyrosine phosphatase family conserved in yeast. Nat Genet 13:175–182 [DOI] [PubMed] [Google Scholar]
- LeGuern E, Guilbot A, Kessali M, Ravise N, Tassin J, Maisonobe T, Grid D, Brice A (1996) Homozygosity mapping of an autosomal recessive form of demyelinating Charcot-Marie-Tooth disease to chromosome 5q23–q33. Hum Mol Genet 5:1685–1688 [DOI] [PubMed] [Google Scholar]
- Letunic I, Goodstadt L, Dickens NJ, Doerks T, Schultz J, Mott R, Ciccarelli F, Copley RR, Ponting CP, Bork P (2002) Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res 30:242–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levivier E, Goud B, Souchet M, Calmels TP, Mornon JP, Callebaut I (2001) uDENN, DENN, and dDENN: indissociable domains in Rab and MAP kinases signaling pathways. Biochem Biophys Res Commun 287:688–695 [DOI] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M (1992) A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14:897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E (2002) ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell 10:1119–1128 [DOI] [PubMed] [Google Scholar]
- Nandurkar HH, Caldwell KK, Whisstock JC, Layton MJ, Gaudet EA, Norris FA, Majerus PW, Mitchell CA (2001) Characterization of an adapter subunit to a phosphatidylinositol (3)P 3-phosphatase: identification of a myotubularin-related protein lacking catalytic activity. Proc Natl Acad Sci USA 98:9499–9504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandurkar HH, Huysmans R (2002) The myotubularin family: novel phosphoinositide regulators. IUBMB Life 53:37–43 [DOI] [PubMed] [Google Scholar]
- Nishimura DY, Swiderski RE, Alward WL, Searby CC, Patil SR, Bennet SR, Kanis AB, Gastier JM, Stone EM, Sheffield VC (1998) The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat Genet 19:140–147 [DOI] [PubMed] [Google Scholar]
- Rezaie T, Child A, Hitchings R, Brice G, Miller L, Coca-Prados M, Heon E, Krupin T, Ritch R, Kreutzer D, Crick RP, Sarfarazi M (2002) Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science 295:1077–1079 [DOI] [PubMed] [Google Scholar]
- Richards JE, Lichter PR, Boehnke M, Uro JL, Torrez D, Wong D, Johnson AT (1994) Mapping of a gene for autosomal dominant juvenile-onset open-angle glaucoma to chromosome Iq. Am J Hum Genet 54:62–70 [PMC free article] [PubMed] [Google Scholar]
- Sarfarazi M, Akarsu AN, Hossain A, Turacli ME, Aktan SG, Barsoum-Homsy M, Chevrette L, Sayli BS (1995) Assignment of a locus (GLC3A) for primary congenital glaucoma (Buphthalmos) to 2p21 and evidence for genetic heterogeneity. Genomics 30:171–177 [DOI] [PubMed] [Google Scholar]
- Sarfarazi M, Child A, Stoilova D, Brice G, Desai T, Trifan OC, Poinoosawmy D, Crick RP (1998) Localization of the fourth locus (GLC1E) for adult-onset primary open-angle glaucoma to the 10p15-p14 region. Am J Hum Genet 62:641–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer AA, Gupta SK, Shriram K, Cottingham RW Jr (1994) Avoiding recomputation in linkage analysis. Hum Hered 44:225–237 [DOI] [PubMed] [Google Scholar]
- Semina EV, Reiter R, Leysens NJ, Alward WL, Small KW, Datson NA, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel BU, Carey JC, Murray JC (1996) Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet 14:392–399 [DOI] [PubMed] [Google Scholar]
- Stoilov I, Akarsu AN, Sarfarazi M (1997) Identification of three different truncating mutations in cytochrome P4501B1 (CYP1B1) as the principal cause of primary congenital glaucoma (Buphthalmos) in families linked to the GLC3A locus on chromosome 2p21. Hum Mol Genet 6:641–647 [DOI] [PubMed] [Google Scholar]
- Stoilova D, Child A, Trifan OC, Crick RP, Coakes RL, Sarfarazi M (1996) Localization of a locus (GLC1B) for adult-onset primary open angle glaucoma to the 2cen-q13 region. Genomics 36:142–150 [DOI] [PubMed] [Google Scholar]
- Stone EM, Fingert JH, Alward WL, Nguyen TD, Polansky JR, Sunden SL, Nishimura D, Clark AF, Nystuen A, Nichols BE, Mackey DA, Ritch R, Kalenak JW, Craven ER, Sheffield VC (1997) Identification of a gene that causes primary open angle glaucoma. Science 275:668–670 [DOI] [PubMed] [Google Scholar]
- Thompson EA (1987) Crossover counts and likelihood in multipoint linkage analysis. IMA J Math Appl Med Biol 4:93–108 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman V, Raeymaekers P, De Jonghe P, De Winter G, Swerts L, Jacobs K, Gheuens J, Martin JJ, Vandenberghe A, Van Broeckhoven C (1990) Assignment of the Charcot-Marie-Tooth neuropathy type 1 (CMT 1a) gene to 17p11.2-p12. Am J Hum Genet 47:680–685 [PMC free article] [PubMed] [Google Scholar]
- Trifan OC, Traboulsi EI, Stoilova D, Alozie I, Nguyen R, Raja S, Sarfarazi M (1998) A third locus (GLC1D) for adult-onset primary open-angle glaucoma maps to the 8q23 region. Am J Ophthalmol 126:17–28 [DOI] [PubMed] [Google Scholar]
- Warner LE, Mancias P, Butler IJ, McDonald CM, Keppen L, Koob KG, Lupski JR (1998) Mutations in the early growth response 2 (EGR2) gene are associated with hereditary myelinopathies. Nat Genet 18:382–384 [DOI] [PubMed] [Google Scholar]
- Wirtz MK, Samples JR, Kramer PL, Rust K, Topinka JR, Yount J, Koler RD, Acott TS (1997) Mapping a gene for adult-onset primary open-angle glaucoma to chromosome 3q. Am J Hum Genet 60:296–304 [PMC free article] [PubMed] [Google Scholar]
- Wirtz MK, Samples JR, Rust K, Lie J, Nordling L, Schilling K, Acott TS, Kramer PL (1999) GLC1F, a new primary open-angle glaucoma locus, maps to 7q35–q36. Arch Ophthalmol 117:237–241 [DOI] [PubMed] [Google Scholar]
- Wishart MJ, Dixon JE (2002a) PTEN and myotubularin phosphatases: from 3-phosphoinositide dephosphorylation to disease. Phosphatase and tensin homolog deleted on chromosome ten. Trends Cell Biol 12:579–585 [DOI] [PubMed] [Google Scholar]
- ——— (2002b) The archetype STYX/dead-phosphatase complexes with a spermatid mRNA-binding protein and is essential for normal sperm production. Proc Natl Acad Sci USA 99:2112–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart MJ, Taylor GS, Slama JT, Dixon JE (2001) PTEN and myotubularin phosphoinositide phosphatases: bringing bioinformatics to the lab bench. Curr Opin Cell Biol 13:172–181 [DOI] [PubMed] [Google Scholar]