Abstract
In higher eukaryotes the biogenesis of spliceosomal UsnRNPs involves a nucleocytoplasmic shuttling cycle. After the m7G-cap-dependent export of the snRNAs U1, U2, U4 and U5 to the cytoplasm, each of these snRNAs associates with seven Sm proteins. Subsequently, the m7G-cap is hypermethylated to the 2,2,7-trimethylguanosine (m3G)-cap. The import adaptor snurportin1 recognises the m3G-cap and facilitates the nuclear import of the UsnRNPs by binding to importin-β. Here we report the crystal structure of the m3G-cap-binding domain of snurportin1 with bound m3GpppG at 2.4 Å resolution, revealing a structural similarity to the mRNA-guanyly-transferase. Snurportin1 binds both the hypermethylated cap and the first nucleotide of the RNA in a stacked conformation. This binding mode differs significantly from that of the m7G-cap-binding proteins Cap-binding protein 20 (CBP20), eukaryotic initiation factor 4E (eIF4E) and viral protein 39 (VP39). The specificity of the m3G-cap recognition by snurportin1 was evaluated by fluorescence spectroscopy, demonstrating the importance of a highly solvent exposed tryptophan for the discrimination of m7G-capped RNAs. The critical role of this tryptophan and as well of a tryptophan continuing the RNA base stack was confirmed by nuclear import assays and cap-binding activity tests using several snurportin1 mutants.
Keywords: 5′-cap, crystallography, nuclear transport, snurportin1, UsnRNP
Introduction
The transport of macromolecules between the nuclear and cytoplasmic compartments in eukaryotic cells is achieved by transport factors, most of them belonging to the importin-β superfamily (Görlich and Kutay, 1999; Chook and Blobel, 2001; Weis, 2003). Nuclear import of karyophilic proteins bearing a canonical nuclear localisation signal (NLS) requires the recognition by the adaptor importin-α, which in turn binds to importin-β via an N-terminal importin-β-binding (IBB) domain (Görlich et al, 1996). The interaction with and translocation through the nuclear pore complex is mediated by importin-β. Besides the importin-α/β-dependent nuclear import, several alternative pathways have been identified (Görlich and Kutay, 1999; Weis, 2003). Among these pathways, there is an exceptional strategy that has evolved in higher eukaryotes for the transport of spliceosomal UsnRNPs, namely U1, U2, U4 and U5, as their biogenesis involves a nucleocytoplasmic shuttling cycle (Will and Lührmann, 2001).
Transcription and modification of the 3′ and 5′ ends leads to m7G-capped UsnRNAs that are recognised by the cap-binding proteins Cap-binding protein 20 (CBP20)/80. Three additional proteins, phosphorylated adaptor for RNA export (PHAX), the actual export receptor chromosome region maintenance 1 (CRM1) or Exportin1 (XpoI; Ohno et al, 2000) and Ras-related nuclear antigen (RanGTP; Ohno et al, 2000; Segref et al, 2001) are required for the export of the UsnRNAs into the cytoplasm.
In the cytoplasm, seven Sm proteins bind to the Sm site common to these UsnRNAs, forming the snRNP core complex. This assembly process is mediated by the SMN complex (Meister et al, 2002; Gubitz et al, 2004; Yong et al, 2004). Proper assembly of the core UsnRNPs and the SMN complex bound to it is a prerequisite for the hypermethylation of the m7G-cap to the 2,2,7-trimethyl-guanosine (m3G)-cap (Mattaj, 1986; Massenet et al, 2002). Initial studies on the cap hypermethylation of human U1snRNP showed that the snRNA-(guanosine-N2)-methyltransferase is an S-adenosylmethionine-dependent enzyme that binds to the SmB/B′ proteins (Plessel et al, 1994). Further in vitro reconstitution experiments revealed that the presence of the SmB/B′protein in the snRNP core complex is essential for the cap hypermethylation (Raker et al, 1996). Finally, the cap hypermethylase was identified in Saccharomyces cerevisiae and denoted TGS1 (for Trimethyl-Guanosine Synthase) (Mouaikel et al, 2002). A sequence database search revealed that the putative mammalian orthologs are significantly larger, with their C-terminal domain harbouring the conserved methyltransferase domain (Mouaikel et al, 2002).
The m3G-cap is part of a bipartite NLS specific for UsnRNP nuclear import (Fischer and Lührmann, 1990; Fischer et al, 1991). The second signal is located on the Sm-core RNP complex (Fischer et al, 1993). The functionality of both import signals has been shown to depend on importin-β (Palacios et al, 1997; Huber et al, 1998; Narayanan et al, 2004). The m3G-cap is specifically recognised by snurportin1, which acts as an adaptor between the UsnRNP cargo and importin-β (Huber et al, 1998). Snurportin1 consists of two functional domains, the N-terminal IBB domain and the m3G-cap-binding domain. The IBB domain comprises amino acids 1–65 and exhibits high homology to the IBB domains of other transport adaptors, such as importin-α and RIP-α, that use importin-β as a transport receptor (Görlich et al, 1996; Jullien et al, 1999; Huber et al, 2002). The m3G-cap-binding domain, ranging from amino acids 95 to 300, shares no significant sequence similarities with other cap-binding proteins or with any other protein (Huber et al, 1998).
The three-dimensional structures of several m7G-cap-binding proteins, namely CBP20 of the cap-binding complex (CBC) involved in nuclear export of UsnRNA (Calero et al, 2002; Mazza et al, 2002), the viral nucleoside 2′-O-methyltransferase VP39 (Hodel et al, 1997, 1998; Hu et al, 2002) and the eukaryotic translation initiation factor 4E (eIF4E) (Marcotrigiano et al, 1997; Matsuo et al, 1997; Niedzwiecka et al, 2002; Tomoo et al, 2003) have been determined. Despite the lack of structural similarity, they all reveal a common strategy in the specific binding of m7G-cap-bearing RNAs. These proteins interact with the m7G-cap by sandwiching the monomethylated guanine base between two aromatic side chains. However, to date there has been no experimental clue as to whether snurportin1 shares that canonical binding mode of the m7G-cap-binding proteins, or whether a different strategy is used for specific binding of the m3G-cap. Hence, the crystal structure determination of snurportin1 in complex with m3G-cap has been of particular interest in order to reveal and understand the structural basis for the specific recognition of the m3G-cap by snurportin1. After all attempts to crystallise full-length snurportin1 either by itself or in complex with m3G-cap oligo and/or importin-β had failed, we focused on the structure determination of the m3G-cap-binding domain of human snurportin1 identified by limited proteolysis experiments (Strasser et al, 2004). Although a 37 kDa fragment of snurportin1 was used for crystallisation, the obtained single crystals contain just a 26 kDa m3G-cap-binding fragment, indicating a continuation of proteolysis within the crystallisation droplet (Strasser et al, 2004).
We have determined the crystal structure of the 26 kDa m3G-cap-binding domain of human snurportin1 in complex with an m3G-cap dinucleotide. Snurportin1 binds the m3G-base and the first base of the UsnRNA in a stacked conformation. This is in contrast to the structurally characterised m7G-cap-binding proteins, where only the cap base is sandwiched between two aromatic side chains. In order to investigate this novel binding mode in more detail, binding constants of several cap oligonucleotides to snurportin1 were determined by fluorescence spectroscopy, and the role of the tryptophan residues in the binding pocket was analysed by nuclear import assays and spectroscopic cap-binding affinity tests for various tryptophan mutants.
Results and discussion
Structure of the snurportin1–m3G-cap complex
The crystal structure of the m3G-cap-binding domain of human snurportin1 was determined by means of MIRAS using several heavy-atom derivatives and refined at a resolution of 2.4 Å (Table I). The overall structure is composed of five α-helices and 10 β-strands that form two almost coplanar β-sheets linked by two crossing β-strands (Figure 1). The m3G-cap-binding pocket is located between the two β-sheets; several residues of strands β1, β3, β10 and adjacent loop regions interact with the entire m3G-cap dinucleotide (Figure 2A, see Supplementary Figure 1). Both bases of the bound m3GpppG are in a nearly coplanar orientation 3.5–3.8 Å apart, but slightly displaced with respect to a perfect base stacking (Figure 2B). The six-membered ring of the nonmethylated guanine is almost perfectly centered on the dimethylated N2 atom (Figure 2B). The base-stacking of the dinucleotide is continued by the side chain of Trp276 on the side of the trimethylated guanine, and is flanked by Leu104 on the side of the nonmethylated guanine (Figure 2A). Additionally, the m3G-base is in hydrophobic contact with the side chains of Glu106 and Trp107, the latter in an almost perpendicular orientation to the stack with a distance of 4.3 Å. Remarkably, the protein forms only two hydrogen bonds with the trimethylated guanine base. The hydroxyl group of Ser105 donates a hydrogen to O6 and accepts one from N1 (Figure 2A). The nonmethylated guanine also forms hydrogen bonds with Ser105 as its NH2 group at position 2, N1 is hydrogen-bonded to the main-chain carbonyl oxygen, and O6 accepts the hydrogen of the main-chain amide. The interaction is further stabilised by hydrogen bonds between several basic residues, the triphosphate and the riboses. The side chain of Lys128 and the main-chain amide of Arg129 interact with phosphate oxygens, and Lys144 is in contact with the cyclic oxygen of the ribose of the first snRNA nucleotide, as well as with phosphate oxygens (Figure 2A, see Supplementary Figure 1). A database search revealed that all residues involved in the interaction with the m3GpppG dinucleotide are highly conserved among all known snurportin1 sequences. The only exception was found in Arabidopsis thaliana, where Ser105 is replaced by a proline residue. This replacement is structurally possible, but appears very unlikely due to the loss of hydrogen bonds involved in ligand binding. Since only a single base exchange is necessary to change serine into proline, an error of the database entry could also be plausible.
Table 1.
Crystallographic data and refinement statistics
| Data collection | ||||
|---|---|---|---|---|
| Data set | Native | MIRAS |
||
| Hg | U | Pt | ||
| Wavelength (Å) | 0.98 | 1.54 | 1.54 | 1.54 |
| Resolution range (Å) | 20–2.4 (2.5–2.4) | 100–3.5 (3.6–3.5) | 100–3.5 (3.6–3.5) | 100–3.5 (3.6–3.5) |
| Space group | P41212 | P41212 | P41212 | P41212 |
| Cell dimensions (Å) | a=b=57.47 | a=b=57.43 | a=b=57.09 | a=b=57.12 |
| c=130.09 | c=130.83 | c=130.5 | c=131.47 | |
| No. of reflections | 249994 (8674) | 57589 (3113) | 54724 (2993) | 109333 (3087) |
| Completeness (%) | 95.4 (66.3) | 100.0 (100.0) | 97.5 (99.3) | 99.7 (100.0) |
| Average I/δ | 18.7 (3.0) | 18.1 (9.3) | 11.7 (4.6) | 16.2 (3.6) |
| Rmerge | 0.09 (0.25) | 0.112 (0.3) | 0.124 (0.27) | 0.132 (0.55) |
| MIRAS phasing | ||||
| Heavy-atom sites | 4 | 3 | 4 | |
| Phasing power | ||||
| Isomorphous | 1.631 | 1.005 | 0.782 | |
| Anomalous | 0.627 | 0.845 | 0.312 | |
| R-cullis | ||||
| Isomorphous | 0.644 | 0.799 | 0.834 | |
| Anomalous | 0.922 | 0.914 | 0.976 | |
| Overall figure of merit | ||||
| Before solvent flattening | 0.23 | |||
| After solvent flattening | 0.75 | |||
| Refinement statistics | ||||
| R-factor (%) | 22.7 (32.1) | |||
| Rfree (%) | 27.6 (40.3) | |||
| No. of protein atoms | 1604 | |||
| No. of ligand atoms | 53 | |||
| No. of water molecules | 77 | |||
| Ramachandran plot statistics | ||||
| Most favourable regions (%) | 87.6 | |||
| Additionally allowed regions (%) | 11.2 | |||
| Generously allowed regions (%) | 1.2 | |||
| Disallowed regions (%) | 0.0 | |||
| r.m.s. deviations from ideal values | ||||
| Bond distances (Å) | 0.0074 | |||
| Angles (deg) | 1.75 | |||
| Average B-value protein (Å2) | 46.7 | |||
| Average B-value m3GpppG (Å2) |
75.5 |
|
|
|
| Values in parenthesis refer to the highest resolution shells. | ||||
| Rmerge=∑hkl∑i∣Ii(hkl)−〈Ii(hkl)〉∣/∑hkl∑i〈Ii(hkl)〉, where the sum i is over all separate measurements of the unique reflection hkl. | ||||
| R-factor=∑hkl∣∣Fobs∣−∣Fcalc∣∣/∑hkl∣Fobs∣. | ||||
| Rfree as R-factor, but summed over a 5% test set of reflections. | ||||
Figure 1.
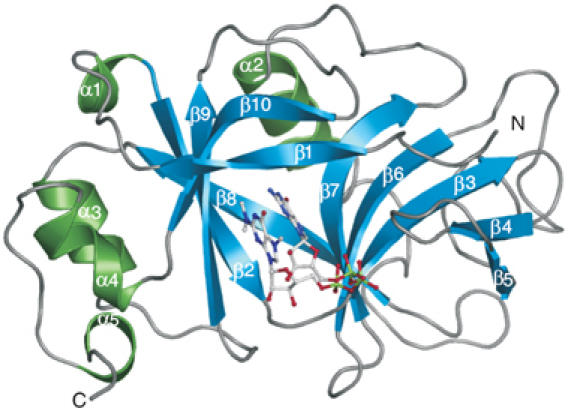
Structure of human snurportin1 (residues 97–300) with bound m3GpppG-cap dinucleotide (PDB accession code 1XK5). Ribbon plot of the m3G-cap-binding domain with β-sheets coloured in blue, α-helices in green and loop regions in grey. Secondary structure motifs are numbered consecutively from the N-terminus to the C-terminus. The m3GpppG dinucleotide is depicted in ball-and-stick mode with nitrogen atoms in blue, carbons in grey, oxygens in red and phosphorus atoms in green.
Figure 2.
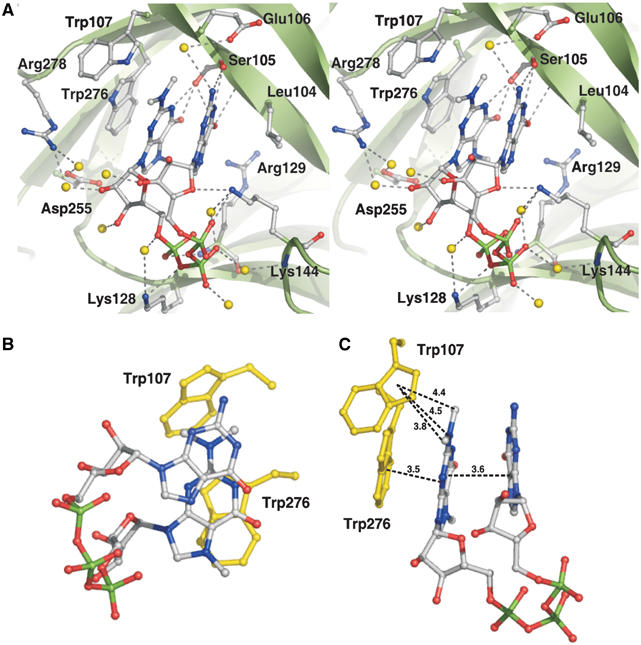
The m3G-cap-binding pocket. Snurportin1 is shown in ribbon presentation and coloured in green. Side chains of residues interacting with the m3G-cap dinucleotide are shown in ball-and-stick mode. The m3G-cap dinucleotide is coloured as described in Figure 1. (A) Stereo view of the cap-binding pocket. Side chains forming hydrogen bonds or hydrophobic contacts with the cap are coloured as the m3G-cap, and water molecules involved in the interaction are shown as yellow balls. Hydrogen bonds are depicted as dashed grey lines. (B) Base stack of the m3GpppG dinucleotide, whereas Trp276 and Trp107 are coloured yellow. (C) Distances between both bases of the cap dinucleotide, between the m3G-base and Trp276 and between the atoms of the dimethylamine of the cap base and Trp107 are depicted as dashed black lines. Numbers indicate the distances in Å.
The binding mode of the m3G-cap by snurportin1 differs significantly from that observed for the m7G-cap-binding proteins CBP20, eIF4E and VP39, as these proteins always intercalate the m7G-base between two aromatic side chains and keep the bound di- or oligonucleotides in an extended conformation (Figure 3) (Hodel et al, 1997, 1998; Calero et al, 2002; Mazza et al, 2002; Niedzwiecka et al, 2002; Tomoo et al, 2003).
Figure 3.
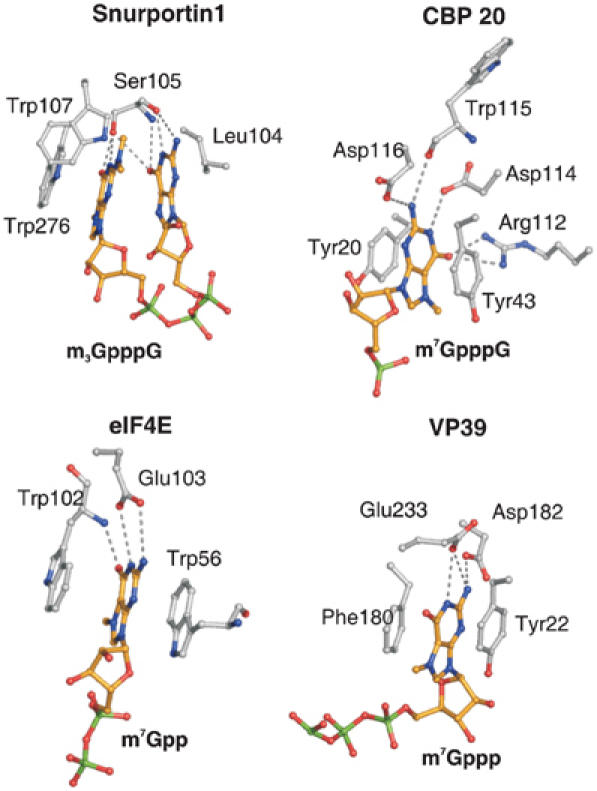
Comparison of cap-binding pockets. m7G-cap-binding pockets of CBP20, eIF4E and the viral nucleoside 2′-O-methyltransferase (VP39) are presented in comparison to the m3G-cap-binding pocket of snurportin1. Side chains of residues interacting with the caps are depicted in ball-and-stick mode. Atoms of the caps and the interacting side chains are coloured as described in Figure 1, with the exception of carbon atoms of the dinucleotide, which are shown in orange. In all presented cases, the residues stacking the bases and those forming hydrogen bonds with the cap bases are depicted. Hydrogen bonds are shown as dashed grey lines.
So far only crystals of the m3G-cap-binding domain with bound m3GpppG have been obtained. All attempts to crystallise full-length snurportin1 have failed, probably due to a high degree of conformational flexibility between the N-terminal IBB domain and the m3G-cap-binding domain. Also, no crystals of the m3G-cap-binding domain could be obtained in the absence of m3GpppG, indicating an effect of the dinucleotide on the structural integrity of the protein. Furthermore, it was not possible to crystallise a complex using a larger oligonucleotide, like synthetic m3GpppAmpUmpA, which resembles the UsnRNAs more closely. The inspection of the crystal packing revealed that a larger cap oligonucleotide could not be accommodated in this crystal form of the snurportin1–m3GpppG complex.
Mode of interaction between snurportin1 and m3G-cap
During UsnRNP biogenesis, the hypermethylation of the m7G-cap occurs after assembly of the Sm-core UsnRNP complex and initiates the import of this complex into the nucleus. Therefore, the specificity of m3G-cap binding by snurportin1 is of particular interest with respect to the discrimination of m7G-cap-bearing UsnRNAs and mRNAs, preventing their accidental reimport into the nucleus. The physiological cargoes of human snurportin1 share the common sequence m3GpppAm at the 5′ end of the UsnRNA. Previous studies have already demonstrated that an m3GpppG dinucleotide is sufficient to be bound by snurportin1, and that the binding of m7GpppG to snurportin1 is two to three orders of magnitude less efficient than that of m3GpppG (Huber et al, 1998). In order to quantify the interaction of snurportin1 with different 5′-caps, we determined the binding constants of various 5′-oligonucleotides by means of fluorescence spectroscopy (Table II). For these experiments the full-length snurportin1 was used, as the crystallised 26 kDa domain formed in situ during the co-crystallisation of the 37 kDa fragment and m3GpppG (Strasser et al, 2004). Attempts to obtain the 26 kDa fragment either by preparative proteolysis or as recombinant protein failed, since this fragment precipitates due to unfolding or incorrect folding, respectively, in the absence of m3GpppG and is therefore not suitable for biochemical experiments. The Kd-value for the m3GpppG dinucleotide (which is present in the crystal structure) was determined to be 1.0 μM, while it is 12.1 μM for the m3GpppA dinucleotide. The crystal structure of the complex containing the m3GpppG dinucleotide reveals that the adenosine of m3GpppA could be harboured in the binding pocket as well, since there is no strong discrimination between the purines commonly caused by polar interactions. The higher Kd-value of the m3GpppA appears to be related to the lack of two hydrogen bonds with Ser105. It turned out to be impossible to determine the exact Kd-value of the m7GpppA dinucleotide by fluorescence spectroscopy, owing to an extremely large inner filter effect caused by the high dinucleotide concentrations required, but it can be estimated that the Kd-value for m7GpppA is greater than 170 μM. Obviously, the difference in Kd-values between m3GpppA and m7GpppA must be related to the two methyl groups on N2 of the m3G-base. These two methyl groups are in van der Waals (VDW) contact with Trp107, but the difference in affinities between m3GpppA/G and m7GpppA cannot be caused exclusively by the contribution of these two VDW contacts to the free binding energy. Hence, the difference in affinity should be related to the binding process itself. Upon binding to snurportin1 the cap is dehydrated, and hydrogen bonds to water molecules are replaced by hydrogen bonds to the protein. Obviously, the m7G- and m3G-caps differ in solution with respect to their hydration shells, as the m7G-cap is capable to bind two water molecules via its NH2 group. These two water molecules have to be released for the binding of the m7G-cap requiring a substantial amount of energy, as these hydrogen bonds are not replaced by hydrogen bonds to the protein. The dimethylated N2 of the m3G-cap has no bound water molecules; hence its binding to snurportin1 and the hydrophobic interaction with the Trp107 side chain is much more favourable. These differences could already cause the observed differences in Kd, but other molecular properties might additionally contribute to the discrimination.
Table 2.
Binding constants of various m3G-cap oligoribonucleotides for human snurportin1
| Cap oligo | Kd values (μM) | Standard deviation of curve fit (μM) |
|---|---|---|
| m3GpppG | 1.00±0.03 | 0.12 |
| m3GpppA | 12.1±0.55 | 2.72 |
| m7GpppA | ⩾170±12.9 | 17 |
| m3GpppAmpUmpA | 0.23±0.02 | 0.08 |
The discrimination between m7G- and m3G-caps by snurportin1 might also be related to the strength of the cation–π interaction. With respect to the pKa-value of 7.46 of m3GpppG (Wieczorek et al, 1995), the m3G-base is expected to be positively charged under crystallisation conditions (pH 5.5). As suggested for m7G-cap-binding proteins (Quiocho et al, 2000), a cation–π interaction between the positively charged trimethylated guanine and the coplanar aromatic side chain of Trp276 is expected. The presence of the two methyl groups bound to N2 probably changes the distribution of the positive charge within m3G in comparison to m7G and therefore significantly affects the cation–π interaction. Interestingly, Trp107 is only 4.5 Å distant to the N2 atom of the m3G-base (Figure 2C); hence, a cation–π interaction appears reasonable assuming a significant positive partial charge on N2 stabilised by the two methyl groups.
Consistent with a previous report (Huber et al, 1998), the binding of the synthetic tetranucleotide m3GpppAmpUmpA, which resembles more closely UsnRNAs in vivo, to full-length snurportin1 is even stronger (Kd=0.23 μM) than that of m3GpppG, which should be related to the presence of additional nucleotides and/or due to the 2′O-methylated riboses. Inspection of the electrostatic surface potential of the m3G-cap-binding domain gives no hint on binding sites for the additional nucleotides (data not shown). However, the crystallised domain lacks the C-terminal 61 residues, which could extend the RNA-binding surface. To clarify the role of these C-terminal residues, the determination of the Kd-value for the crystallised snurportin1 fragment would be necessary, but the 26 kDa fragment aggregates rapidly in the absence of m3GpppG (see above) and could therefore not be used for biochemical experiments.
The crystallised domain is also lacking the N-terminal 78 residues, raising the question whether the IBB domain could affect the m3G-cap-binding analogous to importin-α. This appears possible since the IBB domain of importin-α is known to compete with the NLS of a cargo for binding to importin-α (Kobe, 1999). To examine this potential autoinhibitory effect of the snurportin1 IBB domain on m3G-cap binding, the affinity of the m3GpppG was evaluated in the presence of increasing amounts of IBB domain (residues 1–65). Remarkably, even at 100-fold excess of IBB domain over snurportin1 (and a five-fold excess of m3GpppG over snurportin1) the affinity remains unaltered (see Supplementary Table Ia). In a second approach, an N-terminally truncated snurportin1 comprising residues 66–360 was used. The Kd-value for m3GpppG turned out to be 1.24 μM (±0.3 μM), which is close to the value of 1.0 μM (±0.03 μM) observed for the full-length protein. Both experiments demonstrate that the IBB domain of snurportin1 does not exhibit an autoinhibitory effect on cargo binding as reported for importin-α.
Role of tryptophan residues Trp276 and Trp107 in m3G-cap binding
An obvious feature of the m3G-cap-binding pocket is the presence of the two tryptophan residues Trp107 and Trp276 (Figure 2). In order to understand their functional significance, these tryptophan residues were substituted by alanine, either singly or together. The resulting mutated proteins were purified and characterised with regard to m3G-cap-binding affinity using fluorescence spectroscopy and an in vitro nuclear import assay. However, an exact determination of Kd-values for the mutated proteins was not possible, as the strongly reduced affinities (see below) would require high concentrations of m3GpppG, leading to a very large internal filter effect. Therefore, the change in affinity was estimated comparing the changes in fluorescence upon addition of m3GpppG.
Binding of m3GpppG to wild-type snurportin1 leads to a decrease in fluorescence of 62.7% (±3.9%), indicating a strong interaction. Taking the changed fluorescence properties of each mutant into account, the binding of m3GpppG decreases the fluorescence for the W276A and W107A mutants only by 20.0% (±6.1%) and 25.0% (±6.3%), respectively, and even less (15.8% (±4.2%) for the double mutant, corresponding to a reduced cap-binding activity. The effect of the W276A substitution is expected, as this tryptophan stacks on the m3G-base and forms a strong cation–π interaction. More interestingly, the W107A mutation shows almost the same reduction in fluorescence, suggesting an important role in m3G-cap binding. Since the two methyl groups of N2 are pointing towards Trp107, this tryptophan should have a substantial impact on the discrimination of m7G-cap-bearing RNAs. Indeed, the binding affinity of m7GpppA to the W107A mutant is identical to that of m3GpppA, as determined by the decrease in tryptophan flourescence (see Supplementary Table Ib). Hence, the fully solvent-exposed Trp107 plays an important role in the process of cap-binding and discrimination of m7G-caps.
Furthermore, the β-strand (β1) harbouring Trp107 exhibits a highly twisted conformation and might adopt a different, more relaxed conformation in the absence of the cap, with the side chain of Trp107 located inside the cap-binding pocket. However, there is currently no experimental evidence for such a conformational flexibility of the β-strand β1 or for an induced fold upon cap binding as observed for CBP20 (Calero et al, 2002; Mazza et al, 2002).
The essential role of these two tryptophan residues, Trp276 and Trp107, was also observed in the in vitro nuclear import assay using intact U1snRNPs as transport cargo. The decrease in import observed for the three mutants correlates quantitatively with the reduced cap-binding affinities evaluated by fluorescence spectroscopy (Figure 4). Therefore, it appears unlikely that snurportin1 forms other strong contacts, like protein–protein interactions, with the U1snRNP. However, the nuclear import of UsnRNPs in vivo was reported to occur by means of a larger complex containing snurportin1, importin-β, the SMN protein and presumably other components of the SMN complex (Narayanan et al, 2002, 2004).
Figure 4.
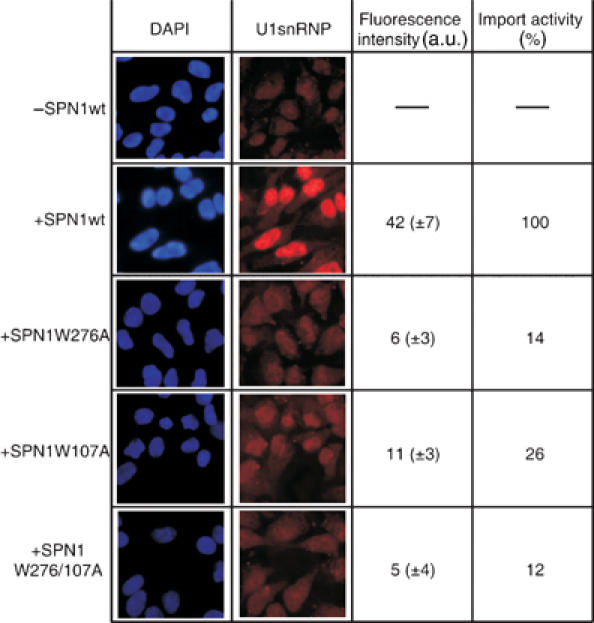
In vitro nuclear import assay of snurportin1. Nuclear import of fluorescently labelled U1snRNPs in the presence of importin-β, RanGDP, NTF2, an energy regenerating system, and either snurportin1 or one of the mutated proteins (W107A, W276A, and W107A/W276/A). In the left panel, cell nuclei stained with DAPI are shown. The middle panel shows the import of Cy3-labelled U1snRNPs. The quantification of the import rates is given in the right panel (a.u.: arbitrary units for intensity per pixel).
Structural homology
Searching the PDB for structurally related proteins revealed that the overall fold of the m3G-cap-binding domain exhibits high structural similarity to the GTP-binding domain of the mRNA-guanylyltransferase indicated by an r.m.s.d. of 2.6 Å for 204 common Cα atoms. Interestingly, this enzyme is involved in the formation of the 5′-cap as it transfers GMP to the 5′end of mRNAs and UsnRNAs via a 5′–5′-triphosphate bridge (Hakansson et al, 1997). This domain is also found in DNA ligases, where it is involved in ATP binding (Subramanya et al, 1996). Superposing the structures of snurportin1 and mRNA-guanylyltransferase, which shares the highest structural similarity to snurportin1, reveals that the binding sites for GTP and the m3G-cap are in a similar area of the protein surface, but the positions of the nucleotides and the residues forming the binding pocket, and therefore all interactions, are completely different (Figure 5A/B). Furthermore, the GTP-binding pocket of the mRNA-guanylyltransferase penetrates much more deeply between the β-sheets than the m3G-cap-binding pocket of snurportin1.
Figure 5.
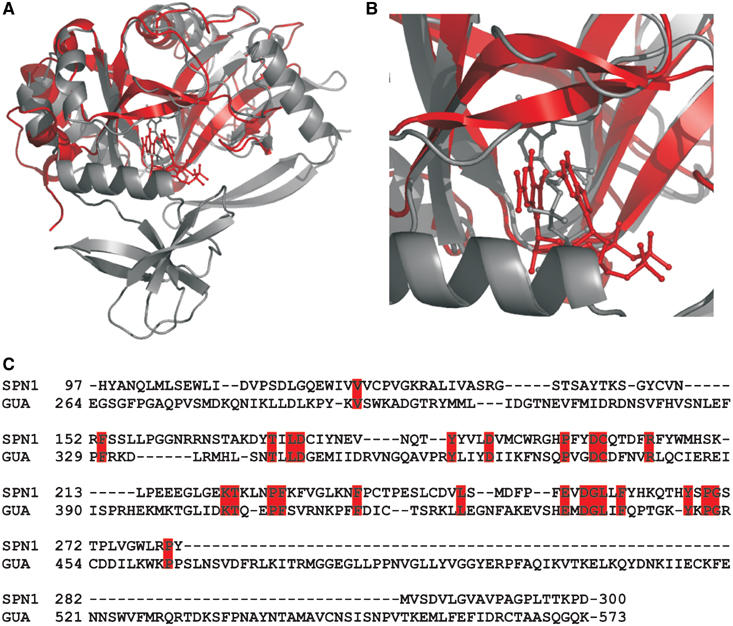
Structural similarity of snurportin1 with bound m3GpppG to the mRNA-guanylyltransferase with bound GTP. (A) Superimposed protein structures presented as ribbon diagrams and the bound nucleotides in ball-and-stick representation. Human snurportin1–m3GpppG complex (coloured red) and the mRNA-guanylyltransferase–GTP complex (coloured grey) of the Paramecium bursaria chlorella virus 1 (PDB accession code 1CKM) share an amino-acid sequence identity of 8.3% for the structurally homologous regions. (B) Close-up view of the nucleotide-binding pockets. In the mRNA-guanylyltransferase, the bound GTP protrudes much deeper into the cleft between the β-sheets. (C) Structure-based sequence alignment of human snurportin1 and human mRNA-guanylyltransferase reveals a sequence identity of 12.7%.
The structural homology between the m3G-cap-binding domain and the mRNA-guanylyltransferase was unexpected, since all searches for sequences homologous to the m3G-cap-binding domain gave no significant hint to any other protein (Huber et al, 1998). Indeed, the amino-acid sequence identity between the human m3G-cap-binding domain and the human mRNA-guanylyltransferase is only 12.7% (Figure 5C). A more elaborate database search recently identified the m3G-cap-binding domain of snurportin1 as an inactive paralogue of the mRNA-guanylyltransferase (Mans et al, 2004). The structural homology suggests that the m3G-cap-binding domain shares a common ancestor with the GTP-binding domain of the mRNA-guanylyltransferase that was linked to an IBB domain by shuffling and fusion of the corresponding exons.
Materials and methods
Structure determination
Purification, m3G-cap-binding affinity test and crystallisation strategy are described elsewhere (Strasser et al, 2004). Shortly, crystals of a 26 kDa m3G-cap-binding domain of snurportin1 were only obtained in the presence of the synthetic m3GpppG-cap dinucleotide. Single crystals for X-ray data collection were grown in sitting drops using 8–10% (w/v) PEG 10 k and 200 mM sodium citrate (pH 5.5) as precipitant. Diffraction data of a native crystal were collected at the PSF beamline BL1 (BESSY, Berlin) to a resolution of 2.4 Å. Three different heavy-atom derivative data sets were collected on a Micromax 007 rotating anode generator (Rigaku/MSC, USA) operating at 40 kV and 20 mA, equipped with Osmic focusing mirrors and a Mar345dtb detector system (Xray-Research, Germany) in order to solve the phase problem. Data were processed using Denzo/Scalepack (HKL-research) and CCP4 programs (Bailey, 1994). Heavy-atom sites were found and initial phases were improved by AutoSHARP (Globalphasing Ltd, UK). The graphics program O (Jones et al, 1991) was used for model-building and the structure was refined with CNS (Brünger et al, 1998). A 37 kDa fragment of snurportin1 was used for crystallisation, but the crystals contain only a 26 kDa fragment. Inspection of possible GluC-protease cleavage sites and preliminary mass spectroscopic data indicates that this fragment consists of residues 79–300. The refined model comprises residues 97–300. The missing residues are not defined in the electron densisty map due to conformational flexibility. The m3GpppG-cap dinucleotide was well defined in the electron density map.
Topology and parameter files for the m3GpppG-cap dinucleotide were created with the help of the Dundee PRODRG2 Server (van Aalten et al, 1996). Secondary structure elements were assigned by STRIDE (Frishman and Argos, 1995) and figures were generated with Pymol (www.pymol.org).
Expression constructs
For all expression constructs described, full-length snurportin1 in pGEX6P1 (Amersham Biosciences) was used as a PCR template (Strasser et al, 2004). The coding region of the deletion mutant of snurportin1 (residues 66–360) was amplified using primers with restriction sites for BamHI and XhoI and inserted into pGEX6P1. The IBB domain (residues 1–65) was cloned in pET28b (Novagen) via NcoI and XhoI restriction sites. Tryptophan mutants W276A, W107A and W276A/W107A in pGEX6P1 were generated using the QuickChange™ Site Directed Mutagenesis Kit (Stratagene).
Protein expression and purification
Expression and purification of snurportin1 wild-type, truncated snurportin1 (residues 66–360) and the tryptophan mutants were performed as described previously (Strasser et al, 2004). The IBB domain (residues 1–65) was expressed as a His-tagged protein at 30°C for 5 h using 1 mM IPTG and purified at first by Ni-NTA agarose. Therefore, the supernatant was applied to the resin in 300 mM NaCl, 20 mM Tris, pH 7.5, 2 mM 2-mercaptoethanol, and eluted with the loading buffer containing 300 mM imidazole. The fractions containing the IBB domain were pooled, concentrated and further purified by gel filtration on Superdex 75 media (100 mM NaCl, 50 mM HEPES, pH 7.5, 2 mM DTT).
Fluorescence spectroscopy
Fluorescence measurements were performed on a Fluoromax3™ spectrofluorimeter (Jobin Yvon) at 20°C in 100 mM NaCl, 50 mM HEPES/NaOH (pH 7.5), using cuvettes with 0.5 × 1.0 cm2 section (Hellma). Emission at 315 or 340 nm, used for the calculation of dissociation constants, was recorded during excitation at 295 nm (1 nm bandwidth) for 60 s with time constants of 0.5 s. Equilibrium dissociation constants were obtained by fitting the solutions of a quadratic function, assuming a 1:1 stoichiometry and taking into account the fluorescence of the cap oligonucleotides (Graphit 3.1, Erithacus Software). Standard deviations were calculated from three independent experiments. To determine the cap-binding affinity of wild-type snurportin1 and of the mutants W107A, W276A and W107A/W276A, emission spectra (310–500 nm) were collected after excitation at 295 nm for the proteins alone and in the presence of m3GpppG in a 20-fold, or in case of m7GpppA and m3GpppA in a 50-fold, molar excess. Spectra were corrected for the fluorescence of the cap dinucleotides and buffer contributions. The decrease in fluorescence upon ligand binding to snurportin1 was determined in three independent measurements. For competition experiments with the IBB domain, the decrease in tryptophan fluorescence after addition of m3GpppG in a five-fold molar excess to snurportin1 full length was compared to the decrease after addition of increasing amounts of the IBB domain (residues 1–65). The IBB domain was titrated up to a 100-fold molar excess to snurportin1 wild type.
Nuclear import assay
Nuclear import assays were performed as described by Huber et al (2002). Each transport reaction mixture contained 100 nM Cy3-labelled human U1snRNPs, and, except for the negative control, 500 nM importin-β, 300 nM of snurportin1 or the mutated snurportin1 proteins (W107A, W276A and W107A/W276/A). The transport was performed on permeabilised HeLa cells for 10 min at room temperature in a humidified chamber. After extensive washing with 1 × PBS, the cells were fixed with paraformaldehyde (5% in 1 × PBS) and mounted on coverslips. Images were taken at 400-fold magnification. Rates of U1snRNP import were calculated with the help of the program ImageJ (http://rsb.info.nih.gov/ij/), by averaging nuclear fluorescence intensities of 50 cells. In order to compare the results, the intensities of each experiment were normalised against wild-type snurportin1 intensities.
Supplementary Material
Supplementary Information
Acknowledgments
We thank the staff of PSF beamline BL1 at BESSY (Berlin) for excellent support during data collection. We are grateful to our colleague Markus Rudolph for help with fluorescence spectroscopy experiments and to Jörg Kahle and Detlef Doenecke for the opportunity to perform nuclear import assays. We thank M Sekine for providing a synthetic m3G-cap-tetranucleotide. This work was supported by the Deutsche Forschungsgemeinschaft (SFB523). The coordinates and structure factors have been deposited in the Protein Data Base (PDB accession code 1XK5).
References
- Bailey S (1994) The CCP4 Suite—programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Calero G, Wilson KF, Ly T, Rios-Steiner JL, Clardy JC, Cerione RA (2002) Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat Struct Biol 9: 912–917 [DOI] [PubMed] [Google Scholar]
- Chook YM, Blobel G (2001) Karyopherins and nuclear import. Curr Opin Struct Biol 11: 703–715 [DOI] [PubMed] [Google Scholar]
- Fischer U, Darzynkiewicz E, Tahara SM, Dathan NA, Lührmann R, Mattaj IW (1991) Diversity in the signals required for nuclear accumulation of U snRNPs and variety in the pathways of nuclear transport. J Cell Biol 113: 705–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Lührmann R (1990) An essential signaling role for the m3G cap in the transport of U1 snRNP to the nucleus. Science 249: 786–790 [DOI] [PubMed] [Google Scholar]
- Fischer U, Sumpter V, Sekine M, Satoh T, Lührmann R (1993) Nucleo-cytoplasmic transport of U snRNPs: definition of a nuclear location signal in the Sm core domain that binds a transport receptor independently of the m3G cap. EMBO J 12: 573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frishman D, Argos P (1995) Knowledge-based protein secondary structure assignment. Proteins 23: 566–579 [DOI] [PubMed] [Google Scholar]
- Görlich D, Henklein P, Laskey RA, Hartmann E (1996) A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J 15: 1810–1817 [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Kutay U (1999) Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15: 607–660 [DOI] [PubMed] [Google Scholar]
- Gubitz AK, Feng W, Dreyfuss G (2004) The SMN complex. Exp Cell Res 296: 51–56 [DOI] [PubMed] [Google Scholar]
- Hakansson K, Doherty A, Shuman S, Wigley D (1997) X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell 89: 545–553 [DOI] [PubMed] [Google Scholar]
- Hodel AE, Gershon PD, Quiocho FA (1998) Structural basis for sequence-nonspecific recognition of 5′-capped mRNA by a cap-modifying enzyme. Mol Cells 1: 443–447 [DOI] [PubMed] [Google Scholar]
- Hodel AE, Gershon PD, Shi X, Wang SM, Quiocho FA (1997) Specific protein recognition of an mRNA cap through its alkylated base. Nat Struct Biol 4: 350–354 [DOI] [PubMed] [Google Scholar]
- Hu G, Oguro A, Li C, Gershon PD, Quiocho FA (2002) The ‘cap-binding slot' of an mRNA cap-binding protein: quantitative effects of aromatic side chain choice in the double-stacking sandwich with cap. Biochemistry 41: 7677–7687 [DOI] [PubMed] [Google Scholar]
- Huber J, Cronshagen U, Kadokura M, Marshallsay C, Wada T, Sekine M, Lührmann R (1998) Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J 17: 4114–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber J, Dickmanns A, Lührmann R (2002) The importin-beta binding domain of snurportin1 is responsible for the Ran- and energy-independent nuclear import of spliceosomal U snRNPs in vitro. J Cell Biol 156: 467–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Jullien D, Görlich D, Laemmli UK, Adachi Y (1999) Nuclear import of RPA in Xenopus egg extracts requires a novel protein XRIPalpha but not importin alpha. EMBO J 18: 4348–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B (1999) Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat Struct Biol 6: 388–397 [DOI] [PubMed] [Google Scholar]
- Mans BJ, Anantharaman V, Aravind L, Koonin EV (2004) Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle 3: e99–e123 [DOI] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK (1997) Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89: 951–961 [DOI] [PubMed] [Google Scholar]
- Massenet S, Pellizzoni L, Paushkin S, Mattaj IW, Dreyfuss G (2002) The SMN complex is associated with snRNPs throughout their cytoplasmic assembly pathway. Mol Cell Biol 22: 6533–6541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo H, Li H, McGuire AM, Fletcher CM, Gingras AC, Sonenberg N, Wagner G (1997) Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat Struct Biol 4: 717–724 [DOI] [PubMed] [Google Scholar]
- Mattaj IW (1986) Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell 46: 905–911 [DOI] [PubMed] [Google Scholar]
- Mazza C, Segref A, Mattaj IW, Cusack S (2002) Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J 21: 5548–5557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Eggert C, Fischer U (2002) SMN-mediated assembly of RNPs: a complex story. Trends Cell Biol 12: 472–478 [DOI] [PubMed] [Google Scholar]
- Mouaikel J, Verheggen C, Bertrand E, Tazi J, Bordonne R (2002) Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol Cells 9: 891–901 [DOI] [PubMed] [Google Scholar]
- Narayanan U, Achsel T, Lührmann R, Matera AG (2004) Coupled in vitro import of U snRNPs and SMN, the spinal muscular atrophy protein. Mol Cells 16: 223–234 [DOI] [PubMed] [Google Scholar]
- Narayanan U, Ospina JK, Frey MR, Hebert MD, Matera A (2002) SMN, the spinal muscular atrophy protein, forms a pre-import snRNP complex with snurportin1 and importin beta. Hum Mol Genet 11: 1785–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiecka A, Marcotrigiano J, Stepinski J, Jankowska-Anyszka M, Wyslouch-Cieszynska A, Dadlez M, Gingras AC, Mak P, Darzynkiewicz E, Sonenberg N, Burley SK, Stolarski R (2002) Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J Mol Biol 319: 615–635 [DOI] [PubMed] [Google Scholar]
- Ohno M, Segref A, Bachi A, Wilm M, Mattaj IW (2000) PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 101: 187–198 [DOI] [PubMed] [Google Scholar]
- Palacios I, Hetzer M, Adam SA, Mattaj IW (1997) Nuclear import of U snRNPs requires importin beta. EMBO J 16: 6783–6792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessel G, Fischer U, Lührmann R (1994) m3G cap hypermethylation of U1 small nuclear ribonucleoprotein (snRNP) in vitro: evidence that the U1 small nuclear RNA-(guanosine-N2)-methyltransferase is a non-snRNP cytoplasmic protein that requires a binding site on the Sm core domain. Mol Cell Biol 14: 4160–4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiocho FA, Hu G, Gershon PD (2000) Structural basis of mRNA cap recognition by proteins. Curr Opin Struct Biol 10: 78–86 [DOI] [PubMed] [Google Scholar]
- Raker VA, Plessel G, Lührmann R (1996) The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J 15: 2256–2269 [PMC free article] [PubMed] [Google Scholar]
- Segref A, Mattaj IW, Ohno M (2001) The evolutionarily conserved region of the U snRNA export mediator PHAX is a novel RNA-binding domain that is essential for U snRNA export. RNA 7: 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser A, Dickmanns A, Schmidt U, Penka E, Urlaub H, Sekine M, Lührmann R, Ficner R (2004) Purification, crystallization and preliminary crystallographic data of the m(3)G cap-binding domain of human snRNP import factor snurportin 1. Acta Crystallogr D 60: 1628–1631 [DOI] [PubMed] [Google Scholar]
- Subramanya HS, Doherty AJ, Ashford SR, Wigley DB (1996) Crystal structure of an ATP-dependent DNA ligase from bacteriophage T7. Cell 85: 607–615 [DOI] [PubMed] [Google Scholar]
- Tomoo K, Shen X, Okabe K, Nozoe Y, Fukuhara S, Morino S, Sasaki M, Taniguchi T, Miyagawa H, Kitamura K, Miura K, Ishida T (2003) Structural features of human initiation factor 4E, studied by X-ray crystal analyses and molecular dynamics simulations. J Mol Biol 328: 365–383 [DOI] [PubMed] [Google Scholar]
- van Aalten DM, Bywater R, Findlay JB, Hendlich M, Hooft RW, Vriend G (1996) PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J Comput Aided Mol Des 10: 255–262 [DOI] [PubMed] [Google Scholar]
- Weis K (2003) Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 112: 441–451 [DOI] [PubMed] [Google Scholar]
- Wieczorek Z, Stepinski J, Jankowska M, Lönnberg H (1995) Fluorescence and absorption spectroscopic properties of RNA 5′-cap analogues derived from 7-methyl-, N2, 7-dimethyl- and N2, N2, 7-trimethyl-guanosines. J Photochem Photobiol 28: 57–63 [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R (2001) Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol 13: 290–301 [DOI] [PubMed] [Google Scholar]
- Yong J, Wan L, Dreyfuss G (2004) Why do cells need an assembly machine for RNA–protein complexes? Trends Cell Biol 14: 226–232 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


