Abstract
Mitogen-activated protein (MAP) kinases play distinct roles in a variety of cellular signaling pathways and are regulated through multiple mechanisms. In this study, a novel 61-kDa member of the MAP kinase family, termed extracellular signal-regulated kinase 7 (ERK7), has been cloned and characterized. Although it has the signature TEY activation motif of ERK1 and ERK2, ERK7 is not activated by extracellular stimuli that typically activate ERK1 and ERK2 or by common activators of c-Jun N-terminal kinase (JNK) and p38 kinase. Instead, ERK7 has appreciable constitutive activity in serum-starved cells that is dependent on the presence of its C-terminal domain. Interestingly, the C-terminal tail, not the kinase domain, of ERK7 regulates its nuclear localization and inhibition of growth. Taken together, these results elucidate a novel type of MAP kinase whereby interactions via its C-terminal tail, rather than extracellular signal-mediated activation cascades, regulate its activity, localization, and function.
Mitogen-activated protein (MAP) kinases constitute a superfamily of highly related proline-directed serine/threonine kinases which play critical roles in control of growth, differentiation, apoptosis, and stress responses (33, 46, 53). MAP kinases are regulated through phosphorylation and dephosphorylation involving upstream dual-specificity protein kinases, called MAP kinase kinases (MAPKKs) and specific protein phosphatases. MAPKKs phosphorylate tyrosine and threonine residues of a Thr-X-Tyr (TXY) motif within the activation loop of MAP kinases (12). Phosphorylation of the threonine and tyrosine residues is required and sufficient for full activation of MAP kinases (5).
The TXY activation motif can be used to divide the MAP kinase superfamily into three main families. The TEY (Thr-Glu-Tyr) family consists of extracellular signal-regulated kinases (ERKs) (41). ERK1 and ERK2 represent the most widely studied members of this family. They appear to mediate signals from growth factors leading to proliferation and/or differentiation in a variety of cell types (33, 46, 53). The TPY (Thr-Pro-Tyr) family encompasses the Jun N-terminal kinases (JNKs), which are analogous to the rat stress-activated protein kinases (SAPKs) (30). This family of MAP kinases transduces intracellular signals initiated by a wide variety of cytokines and stress stimuli such as tumor necrosis factor alpha, interleukins, and UV light (24, 55). The TGY (Thr-Gly-Tyr) family includes p38, a mammalian equivalent of the yeast high-osmolarity glycerol kinase, which is also activated by stress stimuli (21).
Many aspects of MAP kinase cascades are highly conserved in mammals, Drosophila, nematodes, and yeasts (23, 52, 53). In the budding yeast Saccharomyces cerevisiae, at least six distinct pathways involving MAP kinases have been identified (23). In haploid strains, MAP kinase signaling enzymes are involved in the regulation of the mating-pheromone response and invasive response. In diploid strains, they regulate pseudohypha and spore formation. MAP kinase cascades also regulate the response to high osmolarity and mediate the signals to maintain cell wall integrity. The existence of multiple MAP kinase pathways regulating diverse functions in yeast indicates that there may be additional members of the MAP kinase family in mammals that have not been identified.
Consistent with this notion is the recent identification of a new human MAP kinase termed Big MAP kinase 1 (BMK1) or ERK5 (31, 56). Initially BMK1/ERK5 was identified as a redox-sensitive kinase (2). Subsequently, it has been shown to activate the transcription factor MEF2C, a regulator of cardiac myogenesis (34), and to mediate early-gene expression following serum stimulation (25). BMK1/ERK5, like ERK1 and ERK2, has a TEY activation motif. However, unlike ERK1 and ERK2, BMK1/ERK5 has a unique 400-amino-acid C-terminal domain, which accounts for its higher molecular mass of 110 kDa (56). The purpose of the C-terminal domain or tail remains unknown.
Other members of MAP kinase signaling pathways contain regions outside of their kinase domains that have been determined to be critical to their function. Ste11p in yeast and c-Raf are members of the MAPKK family (40, 42) that are constitutively active following deletion of the N-terminal domain (7, 50). Analogously, deletion of a portion of the C-terminal domain of the recently described human germinal-center kinase-like kinase reduces its ability to activate JNK in vivo by 10- to 20-fold (16). Whether the potential SH3 binding sites within the proline-rich regions of the C-terminal domain of germinal-center kinase-like kinase have a role in this regulation remains to be studied. A MAPKK in S. cerevisiae, Pbs2p, has an N-terminal SH3-binding domain that functions as a scaffold for other members of the Sho1p-dependent high-osmolarity glycerol response pathway (44). This is one mechanism by which MAP kinase cascades can be assembled and activated with specificity. Thus, regions outside of the kinase domains of MAP kinases can regulate kinase activity and facilitate multiprotein complexes.
In this paper, we report the identification and characterization of a novel member of the MAP kinase family, termed ERK7. Although it contains a TEY activation motif, ERK7 has significant constitutive kinase activity. Surprisingly, we found that the C-terminal domain of ERK7, not kinase activity, is required for its cellular localization and function as a negative regulator of growth.
MATERIALS AND METHODS
Materials.
Phorbol 12-myristate 13-acetate (PMA), myelin basic protein (MBP), peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG), and peroxidase-conjugated goat anti-mouse IgG were purchased from Sigma Chemicals (St. Louis, Mo.). Okadaic acid was purchased from LC Laboratories (Woburn, Mass.). PHAS-1 was purchased from Stratagene (La Jolla, Calif.). GST-ATF2 was purchased from New England BioLabs (Beverly, Mass.). Anti-MAP kinase antiserum (Ab283) was developed as previously described (29). Anti-ERK7 rabbit antiserum (Cocalico Biologicals, Reamstown, Pa.) was raised against a peptide representing the last 14 amino acids of the C-terminal domain of ERK7 (Research Genetics, Huntsville, Ala.). Monoclonal antibody (12CA5) against the hemagglutinin (HA) epitope was purchased from BabCo (Emeryville, Calif.). Peroxidase-conjugated 12CA5 (12CA5-PC), high-affinity rat anti-HA monoclonal antibody (3F10), and peroxidase-conjugated, affinity purified sheep anti-rat Fab Ig were purchased from Boehringer Mannheim (Indianapolis, Ind.). Affinity-purified peroxidase-conjugated goat anti-rabbit IgG, and anti-mouse IgG were purchased from Transduction Laboratories (Lexington, Ky.). Anti-active MAP kinase polyclonal antibody was purchased from Promega (Madison, Wis.). Enhanced chemiluminenscence reagents, [35S]cysteine (>600 Ci/mmol), [α-32P]dCTP (3,000 Ci/mmol), and [γ-32P]ATP (6,000 Ci/mmol) were purchased from DuPont/NEN Research Products (Boston, Mass.). Degenerate PCR primers were obtained from Operon Technologies (Alameda, Calif.). Sequencing primers were synthesized by Integrated DNA Technologies (Coralville, Iowa) and Gibco/BRL (Grand Island, N.Y.). The pRSV–β-gal and pCMV–β-gal expression vectors were a gift from V. Sukhatme. The pSG5 MKP-1 plasmid was a gift from Lester Lau, and the control plasmid, pSG5, was purchased from Stratagene. Plasmid DNAs were prepared by CsCl-ethidium bromide gradient centrifugation as previously described (29) or by purification through columns as specified by the manufacturer (Qiagen, Chatsworth, Calif.). The unique site elimination mutagenesis kit was purchased from Pharmacia (Piscataway, N.J.).
Degenerate-primed PCR amplification.
The template for amplification was a 6-day-old rat brain cDNA library in λpCEV29 (19). Degenerate upstream and downstream oligonucleotide primers were synthesized based on regions of highly conserved amino acid sequences within MAP kinase subdomains II (VAIKKIS) [5′-CTG GAA TTC GTN GCN AT(T/A/C) AA(A/G) AA(A/G) AT-3′] and VII (TRWYRAP) [5′-GC TCT AGA TGG (A/G)GC NC(T/G) (A/G)TA CCA NC(T/G) NGT-3′], respectively. An EcoRI site (underlined) was attached to the 5′ end of the sense oligonucleotide and an XbaI site (underlined) was attached to 5′ end of the antisense oligonucleotide to allow subcloning of the PCR products into an Escherichia coli phagemid vector, pGEM-7Z(+) (Promega). The PCR buffer consisted of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 2 μM each primer, and 1 U of AmpliTaq DNA polymerase (Perkin-Elmer, Norwalk, Conn.) in a final volume of 50 μl. PCR was performed in a thermocycler (Perkin-Elmer) for 5 min at 97°C followed by 35 cycles of 30 s at 94°C, 60 s at 50°C, and 60 s at 72°C, and a final extension at 72°C for 5 min. The band of interest was approximately 450 bp. The PCR product was purified with the Wizard PCR prep DNA purification system as specified by the manufacturer (Promega) and subcloned into pGEM-7Z(+) with the EcoRI and XbaI restriction sites.
Screening of PCR products.
Southern blot screening was performed with an oligonucleotide probe [5′-GAG CA(C/T) CA(A/G) ACC TAC TGT CAG-3′] derived from the consensus amino acid sequence within subdomains II to VIII for rat ERK1 and ERK2, EHQTYCQ. DNA from colonies that did not hybridize to the ERK1 and ERK2 probe was sequenced (Sequenase version 2.0 kit; Amersham Life Science). The sequences were compared to the GenBank/EMBL Data Bank by using the Blastn program (4). Of the 20 colonies initially isolated, 3 were ERK1, 8 were ERK2, 1 was ERK3, 7 were BMK1/ERK5, and 1 was only moderately homologous to known ERKs.
Tissue Northern analysis.
cDNA or the PCR product was radiolabeled with 32P by using a nick translation labeling method and used to probe a poly(A)+ RNA rat multiple-tissue Northern blot (Clontech, Palo, Alto, Calif.). The blot was prehybridized for 6 h in 5× SSPE (0.75 M NaCl, 50 mM NaH2PO4, 5 mM EDTA [pH 7.4]) with 5× Denhardt’s solution (0.5% Ficoll, 0.5% polyvinylpyrrolidone, 0.5% bovine serum albumin), 50% formamide, 0.1% sodium dodecyl sulfate [SDS], and 100 μg of sheared salmon sperm DNA per ml, hybridized overnight in the prehybridization solution at 42°C with radiolabeled probe, washed extensively with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS at 65°C, and exposed to X-ray film at −80°C.
Screening of cDNA library and isolation of full-length ERK7.
The 32P-labeled ERK7 reverse transcription-PCR product was used to screen a rat testis library in λgt11 (106 recombinants: Clontech 5’ Stretch). Five positive plaques were isolated after secondary screening. cDNAs were isolated and subcloned into pBluescript SK(−) vector (Stratagene), at the EcoRI site. The isolated cDNAs were evaluated by Southern blotting to verify the identity of ERK7. Nucleotide sequences of these clones were determined, and a full-length (2.0-kb) cDNA clone was identified.
Plasmid construction and preparation.
The HA-tagged mouse ERK2 (29) was cloned into pcDNA3 (Invitrogen, Carlsbad, Calif.) at the BamHI and EcoRV sites. A hemagglutinin antigen tag, YPYDVPDY, was inserted immediately following the start methionine of ERK7 by using PCR and was verified by DNA sequencing. Unique site elimination mutagenesis (14) was performed to generate ERK7 mutants (K43R T175A Y177F and T175A Y177F). The ERK2/ERK7 chimera was generated by inserting the C-terminal region of ERK7 in frame with ERK2 at a XhoI site created by unique site elimination mutagenesis and at the EcoRV site in pcDNA3. In the ERK2/ERK7 chimera, the three amino acids, GYR, at the C terminus of ERK2 were replaced with REE, from ERK7. The truncated ERK7ΔT mutant was generated by a restriction site cleavage at a XhoI site, resulting in a 356-amino-acid protein where the last three residues were replaced with the sequence alanine-cysteine-isoleucine. The mutated sequences were verified by sequencing.
Cell culture.
COS, CV-1, NIH 3T3, and PC12 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and antibiotics (50 U of penicillin per ml and 50 μg of streptomycin per ml) at 37°C in a 95% air–5% CO2 atmosphere. Quiescent cells were obtained by incubating cells in serum-free medium for at least 24 h. The immortalized H19-7 cells were generated from embryonic rat hippocampal cells as previously described (18).
Transient transfections.
Unless otherwise noted, approximately 8 × 105 cells were seeded on 100-mm plates and incubated overnight. The medium was changed to serum-free OptiMEM (Gibco/BRL), and the cells were transfected with a total of 10 μg of plasmid DNA and 40 μl of TransIt LT-1 as specified by the manufacturer (PanVera Corp., Madison, Wis.). Of the total plasmid DNA, 10% consisted of either the pRSV–β-gal or pCMV–β-gal expression vector. β-Galactosidase activity was used to normalize for transfection efficiency between groups. Unless otherwise noted, cells were made quiescent in serum-free medium for 24 h prior to harvesting.
β-Galactosidase assay.
Cell lysate (10 μl) was diluted to a final volume of 300 μl in 0.1 M sodium phosphate buffer (pH 7.5)–0.05 M β-mercaptoethanol–1 mM MgCl2–0.9 mg of o-nitrophenyl-β-d-galactoside per ml. The samples were incubated at 37°C until a faint yellow color developed, at which time 500 μl of 1 M Na2CO3 was added and the optical density at 410 nm was measured. One unit of β-galactosidase enzyme was used as a positive control.
Preparation of cell and tissue extracts.
Culture cells were washed twice with ice-cold phosphate-buffered saline and lysed with 1% Triton-based lysis buffer (TLB) (1% Triton X-100, 50 mM Tris-HCl [pH 7.5], 40 mM β-glycerophosphate, 100 mM NaCl, 50 mM NaF, 2 mM EDTA, 1 mM sodium vanadate, 1 mM phenylmethylsulfonyl fluoride, 1 μg of aprotinin per ml, 1 μg of leupeptin per ml, 20 mM p-nitrophenyl phosphate). SDS was added to a final concentration of 0.1% where noted. Pulverized, frozen rat tissue (−80°C) was lysed in TLB containing 0.1% SDS. The cell debris was removed by centrifugation (16,000 × g for 10 min at 4°C). The protein concentration in the cleared supernatant was estimated as previously described (3).
Western analysis.
Cell extracts (10 to 60 μg of protein per lane, unless otherwise noted) were resolved on an 8% acrylamide separating gel by SDS-polyacrylamide gel electrophoresis (PAGE). Proteins were transferred to a nitrocellulose membrane. Membrane blocking, washing, antibody incubation, and detection by enhanced chemiluminescence were performed as previously described (3).
Immunoprecipitation and immune complex kinase assay.
Immune complex kinase assays were performed with ectopically expressed epitope-tagged protein kinases from COS and CV-1 cells as previously described (29), except that 2 μg of each substrate was used per reaction in kinase buffer (20 mM HEPES [pH 7.4], 10 mM MgCl2, 1 mM dithiothreitol, 0.2 mM sodium vanadate, 10 mM d-nitrophenylphosphate, 5 μCi of [γ-32P]ATP). Proteins were separated by SDS-PAGE on a 10% acrylamide separating gel, transferred to a nitrocellulose, and subjected to autoradiography. Quantitation of substrate phosphorylation was determined by digital optical scanning on an image analyzer (AMBIS, San Diego, Calif.). The presence of epitope-tagged proteins in the immunoprecipitates was verified by Western analysis with 12CA5, 12CA5-PC, or 3F10 monoclonal antibody. Immunoprecipitation assays were performed in a similar fashion; however, the immune complexes were eluted prior to the kinase reaction and subjected to Western analysis.
Immunocytochemistry.
CV-1 cells, about 60% confluent on coverslips, were transfected with either ERK7, K43R, or ERK7 ΔT and pCMV–β-gal with TransIt LT1 reagent. Following the transfection and overnight incubation in serum-containing medium, the medium was switched to serum-free DMEM for 24 h. The cells were then fixed in 10% formalin for 15 min, washed, blocked, and incubated simultaneously with rabbit polyclonal anti-β-galactosidase antibody (5 Prime-3 Prime, Boulder, Colo.) at a 1:300 dilution and anti-HA 12CA5 monoclonal antibody (1:500). After a 1-h incubation at room temperature, the cells were washed and then incubated with Texas Red-conjugated anti-mouse and fluorescein isothiocyanate-conjugated anti-rabbit antibodies (Molecular Probes, Inc., Eugene, Oreg.). The stained cells were analyzed under a Zeiss Axioplan fluorescence microscope, and images were captured with a Photometrics PLX cooled charge-coupled device digital camera controlled by OPENLAB software (Improvision Ltd.).
GST fusion proteins.
Plasmids encoding glutathione S-transferase (GST)–c-Fos (210 to 313), GST–c-Jun (1 to 93), GST–Elk-1 (307 to 428), and GST–c-Myc were generously provided by T. Deng, E. Wattenberg, R. Treisman, and N. Hay, respectively. GST fusion proteins were prepared by using a GST purification system (Pharmacia).
BrdU incorporation in CV-1 cells.
CV-1 cells plated in 12-well dishes at approximately 6 × 104 cells per well were transfected with either pcDNA3, ERK7, K43R, or ERK7 ΔT (1 μg/well) and pCMV–β-gal (0.5 μg/well) by using TransIt LT1 solution. Following the transfection and overnight incubation in serum-containing medium, the medium was switched to serum-free DMEM for 24 h. Subsequently, the cells were stimulated for 24 h with 10% fetal bovine serum (FBS) in DMEM containing BrdU (10 μM). Transfected cells were detected by an in situ β-galactosidase assay with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) as a substrate, as previously described (29). After the β-galactosidase assay, the cells were treated with 2 N HCl for 15 min, neutralized with 0.1 M borate buffer (pH 9.0), and washed with phosphate-buffered saline. BrdU incorporation was determined by immunocytochemistry with an anti-BrdU monoclonal antibody (Oncogene Research Products, Cambridge, Mass.). A colometric detection method, consisting of the Vectastain ABC kit with Vector VIP, a peroxidase substrate kit (Vector Laboratories, Burlingame, Calif.), was used. The stained cells were analyzed under a Leitz DMIRB microscope. The β-galactosidase-positive cells with or without BrdU incorporation were scored separately, and the percentage of BrdU-positive cells was calculated.
GenBank accession number.
The GenBank accession number for the ERK7 cDNA reported here is AF078798.
RESULTS
Molecular cloning and tissue distribution of ERK7.
Regions of highly conserved amino acid sequences within MAP kinase subdomains II (VAIKKIS) and VIII (TRWYRAP) were used to design degenerate upstream and downstream oligonucleotide primers. By using a 6-day old rat brain cDNA library in λpCEV29 as a template, a 455 bp clone representing a novel sequence was generated by PCR. The gene encoding this sequence was eventually termed ERK7. A rat testis library was screened with the PCR product to obtain the full-length clone.
Nucleotide sequence analysis of the ERK7 clone reveals a translation start site in good agreement with the Kozak consensus sequence (28) (Fig. 1A). The open reading frame extends 1,638 bp, encoding a protein of 546 amino acids with an estimated molecular mass of 61 kDa (Fig. 1A). The deduced amino acid sequence contains a putative ATP binding site, a highly conserved lysine in subdomain II, and a threonine-glutamine-tyrosine activation sequence within subdomain VIII, indicating that the cDNA encodes for a novel member of the ERK/MAP kinase family.
FIG. 1.
Primary structure of ERK7 and comparison with other members of the MAP kinase family. (A) The nucleotide and deduced protein sequence of ERK7 cDNA. The threonine and tyrosine residues within the activation motif are marked with asterisks. Putative SH3 binding motifs, PXXP, in the C-terminal region are underlined. The putative nuclear localization sequence, PARKRGP, is doubly underlined. (B) Computer-generated alignments (GeneWorks, Oxford Molecular Group, Inc., Campbell, Calif.) of ERK7, ERK1, ERK2, and ERK5/BMK1. The C-terminal regions of ERK7 and ERK5 were not aligned. Roman numerals indicate the 11 conserved protein kinase regions. The conserved threonine/tyrosine regulatory residues are marked with asterisks.
Alignment of the deduced amino acid sequence of the clone with other MAP kinases containing the TEY activation motif reveals a rather unique C-terminal extension of 195 amino acids, which is considerably shorter than that of ERK5/BMK1 (Fig. 1B). We therefore named this cDNA ERK7. This C-terminal region has two PXXP proline-rich segments thought to be ligands for binding to SH3 domains (43). In addition, the C-terminal region contains a putative nuclear localization signal (PARKRGP) (38). Apart from the C-terminal region, ERK7 is approximately 40% homologous to rat ERK1 and ERK2. Overall, ERK7 has low homology to ERK5/BMK1, suggesting that it is a distinct member of the ERK/MAP kinase family.
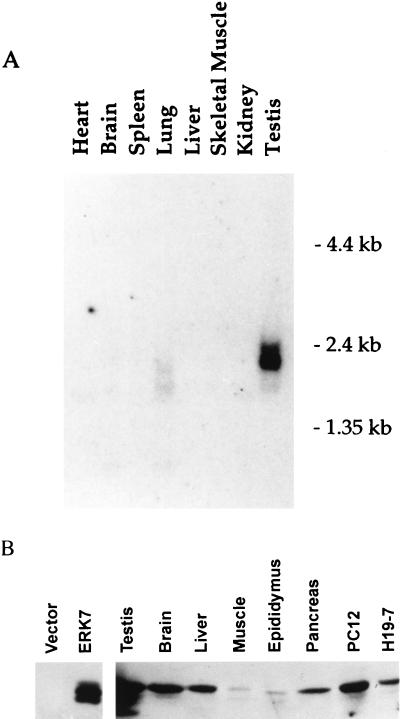
The tissue distribution of ERK7 transcripts was determined by Northern analysis of poly(A)+ RNA from multiple rat tissues (Fig. 2A). A 2.0-kb ERK7 mRNA was most highly expressed in testis followed by lung tissue (Fig. 2A). Both the testis and lung contain additional bands on Northern analysis which might represent splicing intermediates or mRNA encoding for homologous proteins. Although ERK7 was originally identified by using a 6-day-old rat brain library, the presence of ERK7 in adult rat brain could be observed only following extended exposure (data not shown). To further characterize the distribution of ERK7, antiserum was raised against a peptide corresponding to the last 14 amino acids of the C-terminal domain of ERK7. Western analysis of multiple rat tissues and cell types with this antiserum indicated that ERK7 is widely distributed, although at low levels (Fig. 2B). The relative abundance of ERK7 in testes compared to other tissues evaluated by Western analysis is consistent with the results of Northern analysis. These findings suggest that ERK7 is differentially expressed between tissue types.
FIG. 2.
Tissue distribution of ERK7. (A) A rat multiple tissue poly(A)+ RNA Northern blot (Clontech) was probed with radiolabeled ERK7. The positions of the RNA size markers in kilobases are illustrated on the right. (B) Lysates (100 μg) from multiple rat tissues, H19-7 and PC12 cells, were immunoblotted with the anti-ERK7 antiserum. Whole-cell lysates (50 μg) from COS cells transiently transfected with either pcDNA3 or HA-ERK7 are shown in the two left-hand lanes. The exposure time needed to detect endogenous ERK7 was considerably longer (4 min) than that needed to detect the ectopically expressed HA-ERK7 (<2 s).
Ectopic ERK7 is constitutively TEY phosphorylated in COS cells.
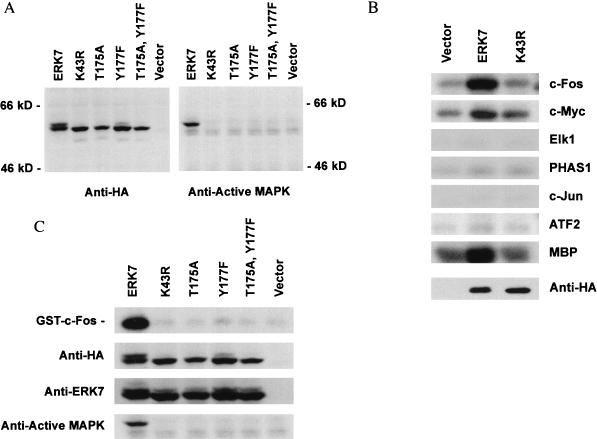
To study ERK7 in intact cells, ERK7 was epitope tagged with the 9-amino-acid immunodominant portion of the influenza virus HA by PCR and subcloned into the mammalian expression vector, pcDNA3. In addition, four mutant forms of epitope-tagged ERK7 were generated. Arginine was used instead of highly conserved lysine-43 in subdomain II to generate the kinase-inactive mutant, K43R. The TEY activation domain was mutated either by changing the threonine residue to alanine, T175A, changing the tyrosine residue to phenylalanine, Y177F, or combining both mutations, T175A Y177F. COS cells were transiently transfected with ERK7 and the mutant forms of ERK7, and a 61-kDa protein was recognized by Western analysis with the anti-HA antibody (Fig. 3A, left). The wild-type ERK7 is seen as a double band. To determine whether the band with reduced electrophoretic mobility represented the TEY phosphorylated form of ERK7, Western analysis was performed with the anti-active MAP kinase polyclonal antibody (Fig. 3A, right). This antibody specifically recognizes the dually but not the singly phosphorylated TEY motif of ERK1 and ERK2, which corresponds to the active form of ERK (26). It also cross-reacts with ERK5/BMK1, the only other known MAP kinase with a TEY activation motif (48). Only the wild-type ERK7 had a band at about 61 kDa, recognized by the anti-active MAP kinase antibody. This band corresponded to the band with reduced electrophoretic mobility seen in the anti-HA Western blot. Since this upper band represents about half of the total ectopically expressed ERK7, Western analysis suggests that as much as 50% of ERK7 may be TEY phosphorylated and active in serum-starved transfected cells. Since the K43R mutant is not recognized by the anti-active MAP kinase polyclonal antibody (Fig. 3A, right), it is possible that kinase activity, perhaps through autophosphorylation, is required for TEY phosphorylation and activation of ERK7.
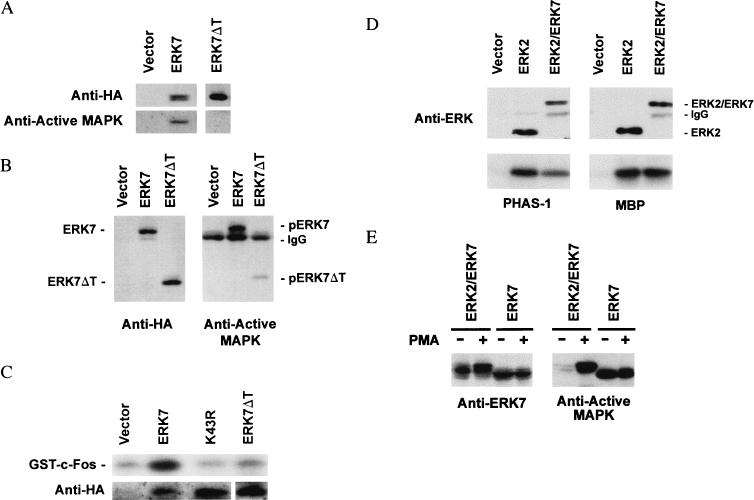
FIG. 3.
ERK7 is TEY phosphorylated and has constitutive activity in COS cells. (A) Ectopically expressed ERK7 in mammalian cells is constitutively TEY phosphorylated. COS cells were transfected with epitope-tagged ERK7, the empty mammalian expression vector pcDNA3, or various ERK7 mutants. A kinase-inactive ERK7 mutant, K43R, was made by replacing the highly conserved lysine in the ATP binding region with arginine. The TEY activation domain was mutated by either changing the threonine residue to alanine, T175A, changing the tyrosine residue to phenylalanine, Y177F, or using both mutations, T175A Y177F. Lysates from serum-starved transfected cells were prepared and analyzed by immunoblotting with either the anti-HA 12CA5 monoclonal antibody (left) or the anti-active MAP kinase antibody (right). The data shown are representative of three experiments. (B) COS cells were transiently transfected with either pcDNA3, HA-ERK7, or K43R, serum starved for 24 h, and lysed with 1% TLB. Following immunoprecipitation with the anti-HA 12CA5 monoclonal antibody, enzyme activity was measured by an immune complex kinase assay with 2 μg of either GST–c-Fos, GST–c-Myc, GST-Elk1, PHAS1, GST–c-Jun, GST-ATF2, or MBP as noted on the right. The immunoprecipitated proteins were analyzed by Western blotting with the anti-HA 3F10 monoclonal antibody. A representative blot of immunoprecipitated protein is displayed at the bottom. ERK7 is seen as a single band as a result of the shorter duration of electrophoresis and the higher percentage acrylamide gel used for the in vitro kinase assays. (C) COS cells were transiently transfected with pcDNA3, ERK7, K43R, or the TEY mutants, serum starved for 24 h, and lysed with 1% TLB. Following immunoprecipitation with anti-HA 12CA5 monoclonal antibody, enzyme activity was measured by an immune complex kinase assay with 2 μg of GST–c-Fos (top). Western analysis of the immunoprecipitated proteins was performed with the anti-HA 3F10 monoclonal antibody (second row), the anti-ERK7 antiserum (third row), and the anti-active MAP kinase antibody (bottom). The data shown are representative of three experiments.
ERK7 is constitutively active.
To further explore the possibility that ERK7 has constitutive kinase activity, potential substrates were evaluated by an in vitro kinase assay. Immunoprecipitated wild-type HA-ERK7, isolated from serum-starved transfected COS cells, phosphorylated GST-c-Fos, GST-c-Myc, and MBP in in vitro kinase assays (Fig. 3B). Immunoprecipitated K43R mutant failed to phosphorylate these substrates in vitro, indicating that this activity was specific for ERK7. Unlike other members of the MAP kinase family, ERK7 did not significantly phosphorylate GST-Elk1 (39), PHAS1 (35), GST–c-Jun (24), and GST-ATF2 (45). The latter results also suggest that the phosphorylation was on c-Fos, and c-Myc but not on GST. Similar in vitro kinase assays were performed with the TEY mutants (Fig. 3C). None of these mutants had significant kinase activity. These results confirm that ERK7 is a functional kinase with discrete activity even in serum-starved cells.
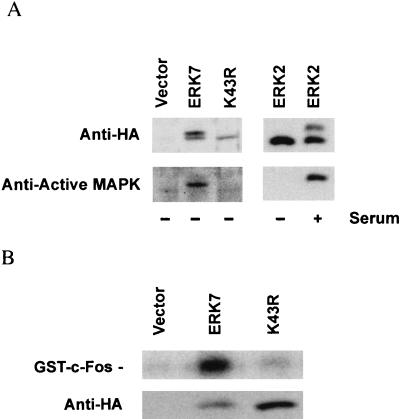
To test whether the simian virus 40 transformation of COS cells contributes to the constitutive activity of ERK7, similar experiments were performed with CV-1 cells. As seen in COS cells, the wild-type ERK7 and the kinase-inactive mutant were recognized by the anti-HA antibody as a double and a single band, respectively (Fig. 4A, top). Similarly, only the electrophoretically retarded band of the wild-type ERK7 was recognized by the anti-active MAP kinase antibody, indicative of TEY phosphorylation (Fig. 4A, bottom). An in vitro kinase assay with GST–c-Fos as a substrate confirmed that immunoprecipitated ERK7 is active in serum-starved CV-1 cells (Fig. 4B). For comparison, CV-1 cells were transiently transfected with HA-ERK2. In serum-starved cells, ERK2 appeared as a single band (Fig. 4A, top). After serum stimulation, a second band with reduced electrophoretic mobility appeared (Fig. 4A, top). Western analysis with the anti-active MAP kinase antibody demonstrated that the vast majority of the ectopically expressed ERK2 in serum-starved CV-1 cells was not TEY phosphorylated and therefore was not active as a kinase (Fig. 4A, bottom). The expression of constitutively activated ERK7 probably is not limited to monkey cells. Western analysis with the anti-HA antibody of NIH 3T3 cells transfected with HA-ERK7, like COS and CV-1 cells, also revealed a second band with reduced electrophoretic mobility corresponding to the active form (data not shown). Together, these data suggest that ERK7 is constitutively active even in serum-starved cells.
FIG. 4.
ERK7 is TEY phosphorylated and has constitutive activity in CV-1 cells. (A) CV-1 cells were transiently transfected either with pcDNA3, ERK7, K43R, or ERK2. Following serum starvation for 24 h, the cells were lysed with 1% TLB. Whole-cell lysates (left) were subjected to Western analysis with either anti-HA monoclonal antibody (top) or anti-active MAP kinase antibody (bottom). CV-1 cells transiently transfected with ERK2 (right) were treated with 10% FBS for 5 min or left untreated. Epitope-tagged ERK2 was immunoprecipitated with the anti-HA 12CA5 monoclonal antibody and analyzed by immunoblotting with either anti-HA monoclonal antibody (top) or anti-active MAP kinase antibody (bottom). (B) CV-1 cells were transiently transfected with pcDNA3, ERK7, or K43R, serum starved for 24 h, and lysed with 1% TLB. Following immunoprecipitation with the anti-HA 12CA5 monoclonal antibody, enzyme activity was measured by an immune complex kinase assay with 2 μg of GST–c-Fos. ERK7 is seen as a single band as a result of the shorter duration of electrophoresis and the higher-percentage acrylamide gel used for the in vitro kinase assays. The data shown are representative of three experiments.
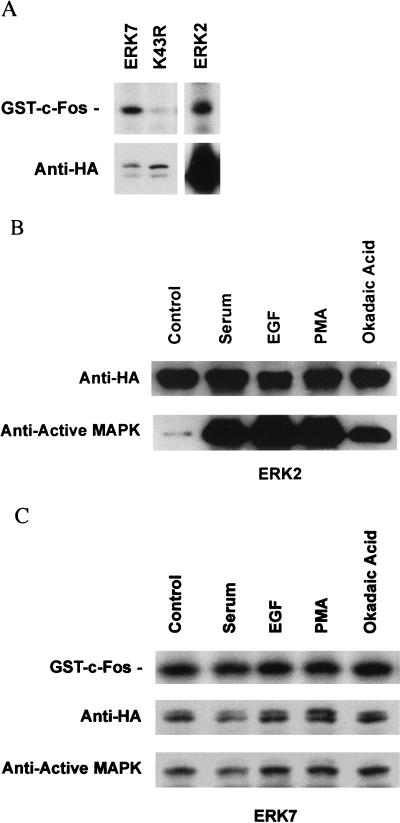
Consistent with the CV-1 results, the kinase activity of ERK7 is significantly greater than that of ERK2 in serum-starved COS cells. To assess the relative kinase activities of the two enzymes, COS cells were transfected with expression vectors for either ERK7 or ERK2. The ERKs were immunoprecipitated with anti-HA antibody and then either assayed directly for kinase activity under equivalent conditions with GST–c-Fos as a substrate (13) or quantitated by immunoblotting with anti-HA antibody. Surprisingly, the level of cytomegalovirus promoter-regulated ERK7 expression in COS cells was significantly lower than that of ERK2 (Fig. 5A, bottom). Differences in transfection efficiency as assessed by β-galactosidase activity (data not shown) could not explain this disparity. Over 140 times more ERK2 than ERK7 was isolated from serum-starved cells, but both enzymes had the same kinase activity as determined by scanning densitometry (Fig. 5A, top). These results demonstrate that ERK7 has significant constitutive kinase activity relative to ERK2.
FIG. 5.
ERK7 and ERK2 are differentially regulated. (A) The relative kinase activity of ERK7 in serum-starved COS cells is significantly greater than that of ERK2. COS cells were transiently transfected with equal amounts of either HA-ERK7 or HA-ERK2. Following serum starvation for 24 h, the cells were lysed with 1% TLB. The ERKs were immunoprecipitated with anti-HA 12CA5 monoclonal antibody and then either assayed directly for kinase activity under equivalent conditions with 2 μg of GST–c-Fos as a substrate (top) or immunoblotted with anti-HA 3F10 monoclonal antibody (bottom). ERK7 is seen as a single band as a result of the shorter duration of electrophoresis and the higher-percentage acrylamide gel used for the in vitro kinase assays. (B) Phosphorylation of the activation domain of ERK2 is stimulated by extracellular factors. COS cells were transiently transfected with ERK2. The cells were pooled and split equally. Following serum starvation for 24 h, the cells were treated with 20% FBS, 50 ng of EGF per ml, 200 nM PMA, or 200 nM okadaic acid or left untreated. The cells were lysed with 1% TLB, and the epitope-tagged proteins were immunoprecipitated with the anti-HA 12CA5 monoclonal antibody. Following separation by SDS-PAGE and transfer to nitrocellulose, ERK2 was subjected to immunoblotting with either anti-HA monoclonal antibody (top) or anti-active MAP kinase antibody (bottom). (C) Phosphorylation of the activation domain of ERK7 is unaffected by extracellular factors. COS cells were transiently transfected with ERK7. The cells were pooled and split equally. Following serum starvation for 24 h, the cells were treated with 20% FBS, 50 ng of EGF per ml, 200 nM PMA, or 200 nM okadaic acid or left untreated. The cells were lysed with 1% TLB, and the epitope-tagged proteins were immunoprecipitated with the anti-HA 12CA5 monoclonal antibody. Following immunoprecipitation, the enzyme activity of ERK7 was measured by an immune complex kinase assay with 2 μg of GST–c-Fos (top). Western analysis of the immunoprecipitated proteins was performed with the anti-HA 3F10 monoclonal antibody (middle). TEY phosphorylation of ERK7 was analyzed by immunoblotting with anti-active MAP kinase antibody (bottom).
Regulation of ERK7 differs from that of ERK2.
To determine if activators of ERK2 might also activate ERK7, COS cells were transiently transfected with HA-ERK7 and HA-ERK2. Cells within each transfection group were pooled and split to equalize transfection efficiency. After serum starvation for 24 h, the cells were stimulated with several known activators of ERK2. Cellular protein immunoprecipitated with the anti-HA antibody was subjected to Western analysis with either the anti-HA or the anti-active MAP kinase antibody.
As expected, serum, epidermal growth factor (EGF), PMA, and okadaic acid dramatically increase the amount of TEY-phosphorylated HA-ERK2 in comparison to quiescent cells (Fig. 5B, bottom). However, none of these agents significantly increases the amount of TEY-phosphorylated ERK7 (Fig. 5C, bottom). The lack of additional activation of ERK7 was confirmed in an immune-complex kinase assay with GST–c-Fos as a substrate (Fig. 5C, top). Consistent with the anti-active MAP kinase Western analysis, no increase in GST–c-Fos phosphorylation was seen following stimulation with serum, EGF, PMA, or okadaic acid. In addition, activation of ERK7 was not seen following stimulation with anisomycin or H2O2 (data not shown). In quiescent COS cells, only a very small fraction of HA-ERK2 is TEY phosphorylated (Fig. 5B, control). In contrast, a significant portion of HA-ERK7 appears to be TEY phosphorylated (Fig. 5C, control). Since TEY phosphorylation of the MAP kinase family is necessary and sufficient for activation (5), these results indicate that regulation of ERK7 differs from that of ERK2 and are consistent with constitutive activation of ERK7.
The C-terminal domain is required for ERK7 activity.
To determine whether the unique C-terminal region of ERK7 plays a role in regulating ERK7 activity, a form of HA-ERK7 truncated at Pro-353 of the C terminus, termed ERK7ΔT, was generated. When ERK2 and ERK5 are aligned by their homologous kinase domains, the carboxy terminus of ERK7ΔT is comparable in length to the carboxy terminus of ERK2. The anti-HA antibody recognizes both the full-length and truncated ERK7 in whole-cell lysates from transfected COS cells (Fig. 6A, top), whereas under the same conditions, the anti-active MAP kinase antibody recognizes only the full-length ERK7 (Fig. 6A, bottom). However, concentration by immunoprecipitation with the anti-HA antibody allows the truncated ERK7 to be detected by the anti-active MAP kinase antibody (Fig. 6B, right). Compared to the full-length ERK7, only a small fraction of the truncated form appears to be TEY phosphorylated. Detection by the anti-active MAP kinase antibody indicates that the truncated form of ERK7 is recognized and phosphorylated by its upstream activator, although perhaps with significantly reduced efficiency. An in vitro kinase assay with GST–c-Fos as a substrate confirmed that the kinase activity of the truncated ERK7 mutant in serum-starved COS cells is significantly reduced compared to the full-length ERK7 (Fig. 6C). In addition, ERK7ΔT, like the full-length ERK7, is not activated by serum stimulation (data not shown). To determine if the C-terminal domain is sufficient for activation of ERK2, a chimeric protein consisting of ERK2 with the C-terminal domain of ERK7 was constructed. Following serum stimulation, the chimeric protein was able to phosphorylate both PHAS1 and MBP in vitro like wild-type ERK2 (Fig. 6D), but the presence of the ERK7 C-terminal region was not sufficient to activate ERK2 in serum-starved COS cells (Fig. 6E). Together, these data indicate that the C-terminal tail is required for the full constitutive activity of ERK7 in serum-starved cells but is insufficient to activate ERK2. Reactivity of the truncated ERK7 with the anti-active MAP kinase antibody is substantially reduced in comparison to that of full-length ERK7, requiring concentration by immunoprecipitation to allow detection.
FIG. 6.
The C-terminal domain is required for ERK7 activity. COS cells were transiently transfected with either pcDNA3, ERK7, or an epitope-tagged ERK7 mutant lacking the C-terminal domain, ERK7ΔT. The cells were serum starved for 24 h and lysed with 1% TLB. (A) Whole-cell lysates were evaluated by Western analysis with either the anti-HA 12CA5 monoclonal antibody (top) or anti-active MAP kinase antibody (bottom). (B) Epitope-tagged proteins immunoprecipitated with the anti-HA 12CA5 monoclonal antibody were detected with either the peroxidase-conjugated anti-HA monoclonal antibody (left) or anti-active MAP kinase antibody (right). (C) The epitope-tagged wild-type ERK7, kinase-inactive K43R mutant, and truncated ERK7ΔT mutant were immunoprecipitated with the anti-HA 12CA5 monoclonal antibody. Immune complex in vitro kinase activity was measured with 2 μg of GST–c-Fos as a substrate. (D) COS cells were transiently transfected with ERK2 or ERK2/ERK7, a chimera consisting of ERK2 and the C-terminal region of ERK7. Following stimulation with serum, the epitope-tagged proteins were purified by immunoprecipitation with the anti-HA 12CA5 monoclonal antibody. Immune complex in vitro kinase activity was measured using 2 μg of either PHAS1 (left) or MBP (right) as a substrate. (E) COS cells were transiently transfected with ERK2/ERK7 or ERK7. Following serum starvation for 24 h, the cells were stimulated without or with PMA. Whole-cell lysates were subjected to Western analysis with either anti-ERK7 antiserum (left) or anti-active MAP kinase antibody (right). The data shown are representative of three experiments.
MKP-1 dephosphorylates ERK7.
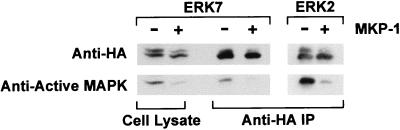
To explore the possibility that the C-terminal region of ERK7 contributes to the constitutive activity of ERK7 by protecting the TEY motif from dephosphorylation, cotransfections with the dual-specificity protein phosphatase, MKP-1, were performed. MKP-1 has been shown to inactivate ERK2 as well as other members of the MAP kinase family through specific dephosphorylation of the TEY activation motif (51). COS cells were cotransfected either with HA-ERK7 or HA-ERK2 and either MKP-1 or a control plasmid. When they were assayed by an electrophoretic mobility shift assay, reduced activation of both immunoprecipitated HA-ERK7 and HA-ERK2 from cells cotransfected with MKP-1 was observed compared to controls following serum stimulation (Fig. 7, top). Western analysis with the anti-active MAP kinase antibody confirmed that cotransfection with MKP-1 reduced the amount of the TEY-phosphorylated forms of both HA-ERK2 and HA-ERK7 (Fig. 7, bottom). A modest reduction in the HA-ERK7 gel shift was noted following immunoprecipitation, suggesting that ERK7 was further dephosphorylated during the immunoprecipitation process (Fig. 7, top). The effect of MKP-1 on ERK7 in serum-starved cells was identical to that in cells treated with serum (data not shown). These experiments indicate that the C-terminal region of ERK7 does not significantly protect ERK7 from TEY dephosphorylation. Therefore, it appears unlikely that the C-terminal region regulates the constitutive activity of ERK7 by protecting the activation motif against dephosphorylation.
FIG. 7.
MKP-1 dephosphorylates ERK7. COS cells were cotransfected with either the wild-type HA-ERK7 or HA-ERK2 and either MKP-1 or a control plasmid. Following serum starvation for 24 h, the cells were stimulated with 10% FBS for 5 min and cell lysates were prepared. Epitope-tagged ERK7 and ERK2 were immunoprecipitated with the 12CA5 monoclonal antibody and assessed by Western analysis with either anti-HA 3F10 monoclonal antibody (top) or anti-active MAP kinase antibody (bottom). Whole-cell lysate (left) from ERK7-transfected COS cells was also assessed by Western analysis with either anti-HA 3F10 monoclonal antibody (top) or anti-active MAP kinase antibody (bottom). The data shown are representative of three experiments.
Cellular localization of ERK7.
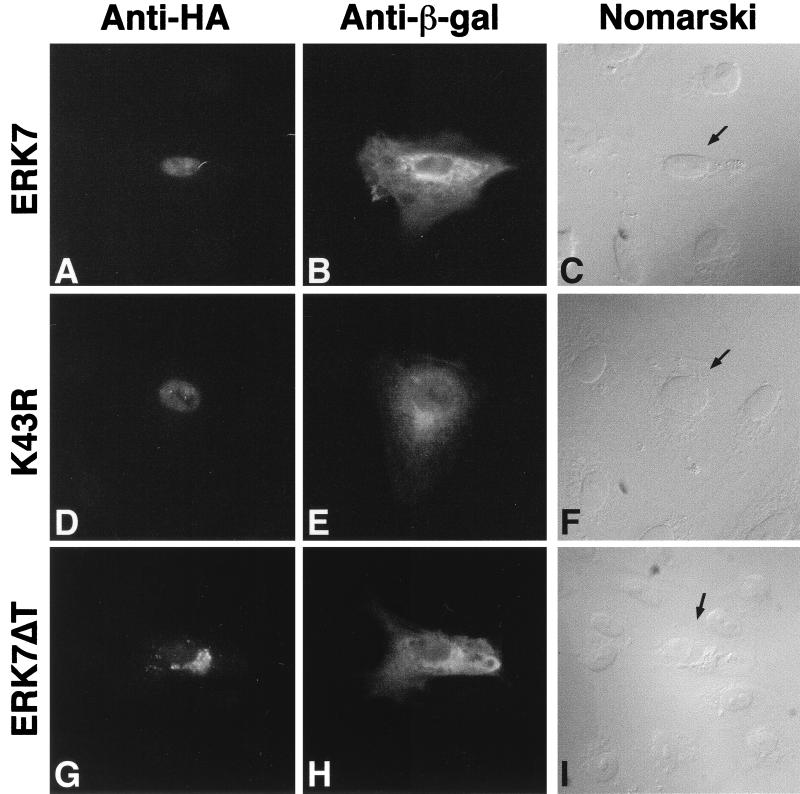
Another possible regulatory mechanism is that the C-terminal domain facilitates the activation of ERK7 by directing it to its activator within the cell. To explore this possibility, immunocytochemistry was performed to test whether the C-terminal domain influences the subcellular localization of ERK7. CV-1 cells were transiently transfected with either ERK7, K43R, or ERK7ΔT and pCMV–β-gal. With the anti-HA antibody, cells transfected either with ERK7 or K43R had predominantly nuclear staining (Fig. 8A and D). On the other hand, ERK7ΔT was localized outside the nucleus (Fig. 8G). Specific staining was not detected in adjacent, nontransfected cells. Staining for β-galactosidase in the transfected cells revealed a cytoplasmic pattern (Fig. 8B, E, and H). Unlike ERK1 and ERK2, which translocate to the nucleus upon serum stimulation (20, 32), ERK7 and K43R localize to the nucleus even in serum-starved cells. These findings suggest that localization of ERK7 to the nucleus is not dependent on its kinase activation state. The staining pattern of the truncated ERK7 mutant, on the other hand, suggests the C-terminal region plays a role in nuclear localization, consistent with the presence of a putative nuclear localization signal within this domain. Together, these results suggest that localization to the nucleus alone is insufficient for activation of ERK7 through TEY phosphorylation, since the majority of the kinase-inactive mutant is not recognized by the anti-active MAP kinase antibody (Fig. 3A). However, it remains possible that directed cellular localization by the C-terminal region of ERK7 is required for activation of ERK7.
FIG. 8.
Cellular localization of ectopically expressed ERK7, K43R, or ERK7ΔT in serum-starved CV-1 cells. CV-1 cells were cotransfected with either epitope-tagged ERK7 (A to C), K43R (D to F), or ERK7ΔT (G to I) and pCMV–β-gal. (A, D, and G) Immunofluorescent staining for HA-tagged proteins; (B, E, and H) the same cells after immunofluorescent staining for β-galactosidase; differential interference contrast (Nomarski) micrographs showing cells with double immunofluorescent staining (arrows) (C, F, and I). The data shown are representative of three experiments.
Inhibition of cell growth by ERK7 requires its C-terminal domain.
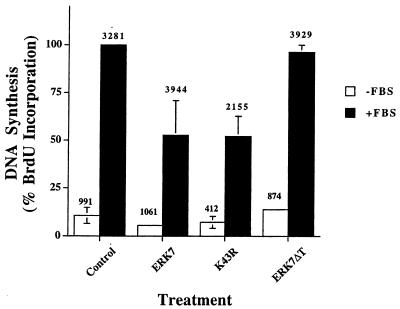
The classic MAP kinases ERK1 and ERK2 have been implicated in the regulation of cell growth by growth factors (33, 46, 53). To determine whether ERK7 can also influence cell growth, we measured the effect of ectopic expression of ERK7 on DNA synthesis as monitored by BrdU incorporation in CV-1 cells. CV-1 cells were cotransfected with either pcDNA3 alone or pcDNA3 containing ERK7, K34R, or ERK7ΔT cDNA along with a β-galactosidase expression vector to identify transfected cells. No effect of ERK7 on serum-starved cells was observed (Fig. 9). However, transient transfection of wild type ERK7 suppressed DNA synthesis in serum-treated CV-1 cells by approximately 50% compared to the control (P < 0.01) (Fig. 9). Surprisingly, this reduction appears to be independent of the kinase activity of ERK7, since the kinase-inactive mutant K43R has an identical effect (P < 0.002) (Fig. 9). Since β-galactosidase and ERK7 were coexpressed in only approximately 80% of the cells that were counted, it is likely that the actual inhibition of DNA synthesis was greater than 50%. In contrast to the kinase-inactive K43R mutant, the truncated ERK7Δ mutant that is also kinase impaired did not reduce BrdU incorporation, indicating that the C-terminal domain appears to be required. These data indicate that ERK7 can function as a negative regulator of growth. Surprisingly, it appears that the C-terminal domain, rather than kinase activity, is the primary component of ERK7 required for this function.
FIG. 9.
Effect of ERK7 on the proliferation of CV-1 cells. CV-1 cells were transfected with either pcDNA3, ERK7, K43R, or ERK7ΔT and pCMV–β-gal. In situ assay of β-galactosidase and immunostaining of BrdU incorporation were performed as described in Materials and Methods. The β-galactosidase-expressing cells were scored as proliferating (BrdU-positive) or nonproliferating (BrdU-negative) cells, without (□) and with (■) serum stimulation for 24 h. The total number of cells counted in each group is noted at the top of each column. The data represent the change in fractional BrdU labeling of serum-stimulated cells for each treatment group normalized to that of serum-stimulated control cells, which was defined as 100%. The data are derived from three independent experiments, and the means and standard deviations are indicated. Where not shown, the error bars were too small to be seen on the graph. Analysis by the one-tailed Student t test indicates that the inhibition by ERK7 (P < 0.01) and K43R (P < 0.002) is statistically significant.
DISCUSSION
In this report, we describe the cloning of the cDNA encoding a novel member of the MAP kinase family, which we have termed ERK7. Although it has the signature TEY activation motif of ERK1 and ERK2, ERK7 appears to be significantly different from other ERK family members. In contrast to previously reported ERKs, ERK7 is a constitutively activated enzyme that is targeted to the nucleus in both the active and inactive states and can function as a negative regulator of growth independent of its kinase activity. In all cases, these properties are dependent upon the presence of the C-terminal tail. Thus, ERK7 represents a new class of MAP kinase family members whose activity, localization, and function require interactions mediated by a C-terminal region rather than just response to extracellular signal activation cascades. Among this class of MAP kinases, regulation of protein expression may be as critical for function as regulation of the activation state of the enzyme.
Several lines of evidence suggest that ERK7 is constitutively active. Western blot analysis detected an electrophoretically shifted band consistent with phosphorylation of ERK7. The anti-active MAP kinase antibody recognizes only the shifted band of ERK7. Neither ERK7 dephosphorylated by MKP-1 nor the TEY mutants are recognized by the anti-active MAP kinase antibody, strongly suggesting that ERK7 contains a dually phosphorylated TEY activation motif. Phosphorylation of both threonine and tyrosine residues in this motif has been shown previously to be required and sufficient for ERK1 and ERK2 activation (5). Finally, constitutive kinase activity was confirmed through in vitro phosphorylation of substrates such as c-Fos, c-Myc, and MBP and by comparison to the basal activity of ERK2 under serum-starved conditions. The kinase activity is ERK7-specific as shown by comparative studies with the kinase-inactive mutant. These results suggest that approximately 50% of the ectopically expressed ERK7 is TEY phosphorylated and enzymatically active.
Although ERK7 is constitutively active, the level of in vitro substrate phosphorylation by ERK7 appears low. Direct comparison to ERK2 suggests that the low level of phosphorylation results primarily from limited ERK7 expression, since the amount of ERK7 is significantly smaller than that of ERK2. However, it is also possible that none of the proteins we tested represent the physiologic substrate of ERK7. While ERK5/BMK1 initially appeared to have poor in vitro kinase activity when MBP was used as a substrate (1), the extent of enzyme activation was significantly greater in recent studies with the transcription factor MEF2C as a target of both in vitro phosphorylation and in vivo activation (25). Therefore, the true measure of the kinase activity of ERK7 may await identification of a reporter system or a substrate reflective of the physiologic kinase function of ERK7.
The regulation of the constitutive activity of ERK7 appears to depend, at least in part, on two features of the enzyme, a functional kinase domain and the C-terminal region. The highly conserved lysine-43 facilitates ATP binding and is required for maximal enzyme activity (22). Since the kinase-inactive mutant K43R is not recognized by the anti-active MAPK antibody, inhibiting the ability of ERK7 to bind ATP apparently prevents dual phosphorylation of its TEY activation motif. Thus, the ability of ERK7 to bind ATP, rendering it a functional kinase, appears to be required for its own TEY phosphorylation and activation. One explanation is that ERK7 is activated by autophosphorylation. While ERK1 and ERK2 have been noted previously to autophosphorylate, the degree of autophosphorylation is limited to only a few percent (10). Although we have not been able to detect autophosphorylation of ERK7 in vitro, this failure may be related to low-level of ERK7 expression. Alternatively, it is possible that the upstream activator of ERK7 requires limited autophosphorylation of ERK7 or another binding protein for full activation of ERK7. In contrast, ERK2 can be TEY phosphorylated in the absence of its kinase activity (47). Whereas TEY phosphorylation of ERK2 leads to homodimerization and subsequent nuclear import (27), ERK7 can be translocated to the nucleus in the absence of TEY phosphorylation. Thus, the mechanism regulating ERK7 activation and cellular localization appears to differ from that of ERK2.
The second feature required for activation of ERK7 is the presence of the C-terminal region. Removal of the tail significantly reduces the TEY phosphorylation of ERK7, its ability to phosphorylate substrates in vitro, its nuclear localization, and its inhibitory effect on growth. Although the results indicate that the C-terminal region is required, more experiments must be done to determine whether the ERK7 tail is sufficient by itself to confer these properties on the enzyme. Interestingly, addition of the ERK7 tail to ERK2 did not alter ERK2 kinase activity, suggesting that the activation mechanism is specific for ERK7. However, the ERK2/ERK7 chimeric protein induced a small but reproducible reduction in BrdU incorporation compared to wild-type ERK2, consistent with a role for the C-terminal region of ERK7 as a negative regulator of growth (data not shown). It is likely that the effect of the ERK7 tail is masked or underestimated in these chimera experiments, since the properties of ERK2 with respect to growth regulation are contrary to those of ERK7. Thus, alternative approaches must be used to clearly elucidate the function of the ERK7 tail.
The possibility that truncation of the C-terminal region inactivates ERK7 through distortion of protein structure is a potential concern. However, several lines of evidence suggest that the truncated ERK7 is not significantly altered in conformation or stability. First, the truncated version of ERK7 is soluble and expressed at levels comparable to the wild-type ERK7. Since the truncated ERK7ΔT is comparable to ERK2 at the C terminus upon alignment of the homologous kinase domains, the catalytic domain of ERK7ΔT should be intact. Consistent with this observation, ERK7ΔT can be immunoprecipitated and detected by Western analysis with the anti-active MAP kinase antibody, although at low levels. Therefore, the structure of ERK7ΔT is intact enough to allow limited TEY phosphorylation despite deletion of the C-terminal region. In addition, similar C-terminal truncated mutants of other members of the MAP kinase pathways retain their kinase activity. Removal of the C-terminal domain of ERK5/BMK1 allowed a GST-ERK5/BMK1 fusion protein construct to autophosphorylate, whereas a full-length ERK5/BMK1 protein lacked detectable kinase activity (56). Similarly, truncation of the C-terminal domain of an ERK2/ERK5 chimera within a couple of residues of the comparable site in ERK7ΔT conferred kinase activity whereas the full-length ERK2/ERK5 chimera was inactive (17). This finding suggests that for ERK5/BMK1, truncation allowed the kinase domain to function. Removal of the 180 amino acids from the C terminal of ERK3, a related member of the MAP kinase family with a SEG activation motif, does not impair its ability to autophosphorylate (9). These results indicate that large C-terminal deletions do not inactivate other members of MAP kinase signaling pathways. Thus, the isolation of TEY-phosphorylated ERK7ΔT and the analogy to other ERKs with respect to activation state as well as structural features elucidated by the MAP kinase crystal structures (8) suggest that ERK7ΔT is a potentially active enzyme.
The exact mechanism by which the C-terminal region of ERK7 facilitates its activation is not entirely clear. The tail does not appear to provide significant protection against phosphatases such as MKP-1, an effective dual-specificity phosphatase that inactivates ERK1 and ERK2 (51). In cotransfection experiments, MKP-1 was able to dephosphorylate ERK7 almost as effectively as it dephosphorylated ERK2. One possibility is that the tail is required to direct ERK7 to its activator within the cell. Intact ERK7 is localized predominately in the nucleus, whereas loss of the tail confers an extranuclear localization. This is in contrast to ERK3, which, despite a large deletion of its C-terminal domain, remains constitutively nuclear (9). Thus, the C-terminal tail of ERK7 may facilitate activation through directed cellular localization. However, localization alone appears insufficient for activation, since the kinase-inactive mutant is not TEY phosphorylated yet localizes to the nucleus.
Alternatively, the C-terminal tail may act as a scaffold to bind activators of ERK7. An interesting feature of the tail of ERK7 is the existence of two potential SH3 binding motifs, PXXP, in the proline-rich C-terminal region of ERK7 (Fig. 1B). SH3 domains have been shown to facilitate the assembly of signaling complexes leading to enzyme activation (43). Recently, we have demonstrated that ERK7 binds specifically to the SH3 domains of Grb2 in vitro (data not shown). This finding demonstrates the potential for ERK7 to be regulated by or function through the association with SH3 domain-containing proteins such as Grb2. In general, SH3 interactions organize protein complexes, localize proteins within the cell, facilitate enzyme-substrate interactions, and regulate enzyme activity (43). In Saccharomyces cerevisiae, two proteins functioning as scaffolds for the components of both the mating/pheromone and osmoregulatory pathways have been recently identified. One of these proteins, Pbs2p, is a MAPKK in one of the high-osmolarity pathways (37). A proline-rich motif in its N-terminal domain localizes Pbs2p to the cytoplasmic SH3 domain of Sho1p, a transmembrane osmosensor. This interaction is essential for the activation of Pbs2p by its upstream activator, Ste11p (44). In addition, Pbs2p appears to function as a scaffolding protein binding other key members of the Sho1p-dependent high-osmolarity glycerol response pathway. Like that of Pbs2p, the SH3 binding motif on ERK7 provides a potential mechanism by which the C-terminal region can facilitate protein-protein interactions crucial for its activation and/or function.
The ability of ERK7 to function as a negative regulator of growth may also involve SH3 binding interactions, since this phenotype occurs independently of kinase activity. While we cannot rule out the possibility that this phenotype is a consequence of ERK7 overexpression, other examples of kinase-independent function exist. The most relevant example is that of the Rous sarcoma virus oncogene src, a prototypic signaling enzyme which has a tyrosine kinase catalytic domain as well as SH2 and SH3 binding domains (6). Kinase-independent function was recently demonstrated in Src−/− transgenic mice, whose one major phenotype is osteopetrosis as a result of an intrinsic defect in osteoclast function (49). Expression of kinase-deficient Src in Src−/− mice reduced osteopetrosis and partially restored a cytoskeletal organizational defect in the osteoclasts. Data from this transgenic-mouse model provides compelling evidence that certain signaling enzymes also have crucial kinase-independent functions. The SH2 and SH3 domains of Src are speculated to be responsible for the kinase-independent function, allowing Src to function as an adapter molecule and facilitate protein interaction or cellular localization. Recently, the ERK1 and ERK2 homolog, Kss1, involved in the mating/pheromone pathway in S. cerevisiae, was also shown to be a potent negative regulator of filamentation and/or invasive growth in its inactive state (11, 36). Activation of Kss1 through phosphorylation by Ste7 removed this repressive effect (11). Kss1 is the first example of a MAP kinase exhibiting regulatory function in the absence of kinase activation and is consistent with our finding that ERK7 can exhibit a growth-inhibitory function that is kinase-independent.
The regulation of ERK7 activity differs from that of other characterized MAPKs. The typical activators of ERK1, ERK2, JNK, and p38 do not activate ERK7 through TEY phosphorylation, and overexpression or inhibition of MEK1 has no effect on ERK7 activity (data not shown). In addition, our studies suggest that the level of expression of ERK7 appears to be very tightly regulated compared to ERK2. In similarly transfected cells, the level of ERK7 expression is markedly lower than that of ERK2. Since its ability to inhibit growth is not dependent on kinase activity, control of the level of expression of ERK7 is a possible regulatory mechanism. Consistent with this notion are preliminary studies indicating that extended stimulation with phorbol esters, treatment with hydrogen peroxide, or overexpression of constitutively active MEKK, an activator of the stress kinases (55), can lead to an increase in the relative amount of ERK7. These results raise the possibility that ERK7 contributes to the inhibition of growth during stressful conditions.
It seems unlikely that interference with growth is the only function of ERK7. Other cellular proteins appear to be multifunctional. Rac1, a member of the Rho family of Ras-related GDP-GTP-regulated proteins, has several functions including regulation of transformation, gene expression, and cytoskeletal organization (54). Recently, the cyclin-dependent kinase inhibitor p21Cip1/WAF1 was shown to inhibit keratinocyte differentiation independently of its ability to regulate cyclin–cyclin-dependent kinase complex formation (15). The ability of ERK7 to phosphorylate certain transcription factors in vitro and its localization primarily to the nucleus suggest that it may also play a role in the regulation of gene expression.
Cloning of this novel MAP kinase indicates that there are probably additional members of the MAP kinase family, perhaps with extended C-terminal regions, that remain unidentified. Certainly ERK7 appears to be significantly different from previously described MAP kinases. The initial characterization of ERK7 indicates that the extended C-terminal domain plays an important role in its regulation and function, perhaps through interaction with SH3 domain-containing proteins. Understanding the role of the C-terminal tail of ERK7 should provide further insight into the function of ERK7 and its integration into the increasingly complex cellular signaling pathways.
ACKNOWLEDGMENTS
We thank W. Li for his critical review of the manuscript, L. Hill for assistance with the manuscript, S. Gomes for technical assistance, G. Bowe for photographic assistance with the figures, and E. Wattenberg, R. Treisman, T. Deng, N. Hay, V. Sukhatme, and L. Lau for generously providing reagents.
This work was supported by National Institute of Health grants HL 03867 (M.K.A.), NS 33858 (M.R.R.), HL 54685 (M.B.H.), and HL 56399 (M.R.R. and M.B.H.) and a gift from the Cornelius Crane Trust Fund for Eczema Research (M.R.R.).
Mark K. Abe and Wen-Liang Kuo contributed equally to this work.
REFERENCES
- 1.Abe J, Kusuhara M, Ulevitch R J, Berk B C, Lee J-D. Big mitogen-activated protein kinase 1 (BMK1) is a redox-sensitive kinase. J Biol Chem. 1996;271:16586–16590. doi: 10.1074/jbc.271.28.16586. [DOI] [PubMed] [Google Scholar]
- 2.Abe J, Takahashi M, Ishida M, Lee J-D, Berk B C. c-Src is required for oxidative stress-mediated activation of big mitogen-activated protein kinase 1. J Biol Chem. 1997;272:20389–20394. doi: 10.1074/jbc.272.33.20389. [DOI] [PubMed] [Google Scholar]
- 3.Abe M K, Chao T S, Solway J, Rosner M R, Hershenson M B. Hydrogen peroxide stimulates mitogen-activated protein kinase in bovine tracheal myocytes: implications for human airway disease. Am J Respir Cell Mol Biol. 1994;11:577–585. doi: 10.1165/ajrcmb.11.5.7946386. [DOI] [PubMed] [Google Scholar]
- 4.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 5.Anderson N G, Maller J L, Tonks N K, Sturgill T W. Requirement for integration of signals from two distinct phosphorylation pathways for activation of MAP kinase. Nature. 1990;343:651–653. doi: 10.1038/343651a0. [DOI] [PubMed] [Google Scholar]
- 6.Brown M T, Cooper J A. Regulation, substrates and function of src. Biochim Biophys Acta. 1996;1287:121–149. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- 7.Cairns B R, Ramer S W, Kornberg R D. Order of action of components in the yeast pheromone response pathway revealed with a dominant allele of the STE11 kinase and the multiple phosphorylation of the STE7 kinase. Genes Dev. 1992;6:1305–1318. doi: 10.1101/gad.6.7.1305. [DOI] [PubMed] [Google Scholar]
- 8.Canagarajah B J, Khokhlatchev A, Cobb M H, Goldsmith E J. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell. 1997;90:859–869. doi: 10.1016/s0092-8674(00)80351-7. [DOI] [PubMed] [Google Scholar]
- 9.Cheng M, Boulton T G, Cobb M H. ERK3 is a constitutively nuclear protein kinase. J Biol Chem. 1996;271:8951–8958. doi: 10.1074/jbc.271.15.8951. [DOI] [PubMed] [Google Scholar]
- 10.Cobb M H, Xu S, Cheng M, Ebert D, Robbins D, Goldsmith E, Robinson M. Structural analysis of the MAP kinase ERK2 and studies of MAP kinase regulatory pathways. Adv Pharmacol. 1996;36:49–65. doi: 10.1016/s1054-3589(08)60576-1. [DOI] [PubMed] [Google Scholar]
- 11.Cook J G, Bardwell L, Thorner J. Inhibitory and activating functions of MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature. 1997;390:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- 12.Crews C M, Alessandrini A, Erikson R L. The primary structure of MEK, a protein kinase that phosphorylates the ERK gene product. Science. 1992;258:478–480. doi: 10.1126/science.1411546. [DOI] [PubMed] [Google Scholar]
- 13.Deng T, Karin M. c-Fos transcriptional activity stimulated by H-Ras-activated protein kinase distinct from Jnk and Erk. Nature. 1994;371:171–175. doi: 10.1038/371171a0. [DOI] [PubMed] [Google Scholar]
- 14.Deng W P, Nickoloff J A. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 15.Di Cunto F, Topley G, Calautti E, Hsiao J, Ong L, Seth P, Dotto G P. Inhibitory function of p21Cip/WAF1 in differentiation of primary mouse keratinocytes independent of cell cycle control. Science. 1998;280:1069–1072. doi: 10.1126/science.280.5366.1069. [DOI] [PubMed] [Google Scholar]
- 16.Diener K, Wang X S, Chen C, Meyer C F, Keesler G, Zukowski M, Tan T-H, Yao Z. Activation of the c-Jun N-terminal kinase pathway by a novel protein kinase related to human germinal center kinase. Proc Natl Acad Sci USA. 1997;94:9687–9692. doi: 10.1073/pnas.94.18.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.English J M, Pearson G, Baer R, Cobb M H. Identification of substrates and regulators of the mitogen-activated protein kinase ERK5 using chimeric protein kinases. J Biol Chem. 1998;273:3854–3860. doi: 10.1074/jbc.273.7.3854. [DOI] [PubMed] [Google Scholar]
- 18.Eves E M, Tucker M S, Roback J D, Downen M, Rosner M R, Wainer B H. Immortal rat hippocampal cell lines exhibit neuronal and glial lineages and neurotrophin gene expression. Proc Natl Acad Sci USA. 1992;89:4373–4377. doi: 10.1073/pnas.89.10.4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomes, I., W. Xiong, T. Miki, and M. R. Rosner. Unpublished data.
- 20.Gonzalez F A, Seth A, Raden D L, Bowman D S, Fay F S, Davis R J. Serum-induced translocation of mitogen-activated protein kinase to the cell surface ruffling membrane and the nucleus. J Cell Biol. 1993;122:1089–1101. doi: 10.1083/jcb.122.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han J, Lee J D, Bibbs L, Ulevitch R J. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science. 1994;265:808–811. doi: 10.1126/science.7914033. [DOI] [PubMed] [Google Scholar]
- 22.Hanks S K, Hunter T. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 23.Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 24.Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 25.Kato Y, Kravchenko V V, Tapping R I, Han J, Ulevitch R J, Lee J-D. BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. EMBO J. 1997;16:7054–7066. doi: 10.1093/emboj/16.23.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khokhlatchev A, Xu S, English J, Wu P, Schaefer E, Cobb M H. Reconstitution of mitogen-activated protein kinase phosphorylation cascades in bacteria. J Biol Chem. 1997;272:11057–11062. doi: 10.1074/jbc.272.17.11057. [DOI] [PubMed] [Google Scholar]
- 27.Khokhlatchev A V, Canagarajah B, Wilsbacher J, Robinson M, Atkinson M, Goldsmith E, Cobb M H. Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear translocation. Cell. 1998;93:605–615. doi: 10.1016/s0092-8674(00)81189-7. [DOI] [PubMed] [Google Scholar]
- 28.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuo W-L, Abe M, Rhee J, Eves E M, McCarthy S A, Yan M, Templeton D J, McMahon M, Rosner M R. Raf, but not MEK or ERK, is sufficient for differentiation of hippocampal neuronal cells. Mol Cell Biol. 1996;16:1458–1470. doi: 10.1128/mcb.16.4.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kyriakis J M, Banerjee P, Nikolakaki E, Dai T, Rubie E A, Ahmad M F, Avruch J, Woodgett J R. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 31.Lee J D, Ulevitch R J, Han J. Primary structure of BMK1: a new mammalian MAP kinase. Biochem Biophys Res Commun. 1995;213:715–724. doi: 10.1006/bbrc.1995.2189. [DOI] [PubMed] [Google Scholar]
- 32.Lenormand P, Sardet C, Pages G, L’Allemain G, Brunet A, Pouyssegur J. Growth factors induce nuclear translocation of MAP kinases (p42mapk and p44mapk) but not of their activator MAP kinase kinase (p45mapkk) in fibroblasts. J Cell Biol. 1993;122:1079–1088. doi: 10.1083/jcb.122.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis T S, Shapiro P S, Ahn N G. Signal transduction through MAP kinase cascades. Adv Cancer Res. 1998;74:49–139. doi: 10.1016/s0065-230x(08)60765-4. [DOI] [PubMed] [Google Scholar]
- 34.Lin Q, Schwarz J, Bucana C, Olson E N. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin T A, Kong X, Haystead T A, Pause A, Belsham G, Sonenberg N, Lawrence J C., Jr PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science. 1994;266:653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- 36.Madhani H D, Styles C A, Fink G R. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 37.Maeda T, Takekawa M, Saito H. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science. 1995;269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- 38.Makkerh J P S, Dingwall C, Laskey R A. Comparative mutagenesis of nuclear localization signals reveals the importance of neutral and acidic amino acids. Curr Biol. 1996;6:1025–1027. doi: 10.1016/s0960-9822(02)00648-6. [DOI] [PubMed] [Google Scholar]
- 39.Marais R, Light Y, Paterson H F, Marshall C J. Ras recruits Raf-1 to the plasma membrane for activation by tyrosine phosphorylation. EMBO J. 1995;14:3136–3145. doi: 10.1002/j.1460-2075.1995.tb07316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marshall C J. MAP kinase kinase kinase, MAP kinase kinase and MAP kinase. Curr Opin Genet Dev. 1994;4:82–89. doi: 10.1016/0959-437x(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 41.Marshall C J. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 42.Neiman A M, Herskowitz I. Reconstitution of a yeast protein kinase cascade in vitro: activation of the yeast MEK homologue STE7 by STE11. Proc Natl Acad Sci USA. 1994;91:3398–3402. doi: 10.1073/pnas.91.8.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pawson T, Scott J D. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 44.Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 45.Raingeaud J, Gupta S, Rogers J S, Dickens M, Han J, Ulevitch R J, Davis R J. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J Biol Chem. 1995;270:7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- 46.Robinson M J, Cobb M H. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 47.Rossomando A, Wu J, Weber M J, Sturgill T W. The phorbol ester-dependent activator of the mitogen-activated protein kinase p42mapk is a kinase with specificity for the threonine and tyrosine regulatory sites. Proc Natl Acad Sci USA. 1992;89:5221–5225. doi: 10.1073/pnas.89.12.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaefer, E. Personal communication.
- 49.Schwartzberg P L, Xing L, Hoffmann O, Lowell C A, Garrett L, Boyce B F, Varmus H E. Rescue of osteoclast function by transgenic expression of kinase-deficient Src in src−/− mutant mice. Genes Dev. 1997;11:2835–2844. doi: 10.1101/gad.11.21.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stanton V P J, Nichols D W, Laudano A P, Cooper G M. Definition of the human raf amino-terminal regulatory region by deletion mutagenesis. Mol Cell Biol. 1989;9:639–647. doi: 10.1128/mcb.9.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun H, Charles C H, Lau L F, Tonks N K. MKP-1 (3CH134), an immediate early gene product, is a dual specificity phosphatase that dephosphorylates MAP kinase in vivo. Cell. 1993;75:487–493. doi: 10.1016/0092-8674(93)90383-2. [DOI] [PubMed] [Google Scholar]
- 52.Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- 53.Waskiewicz A J, Cooper J A. Mitogen and stress response pathways: MAP kinase cascades and phosphatase regulation in mammals and yeast. Curr Opin Cell Biol. 1995;7:798–805. doi: 10.1016/0955-0674(95)80063-8. [DOI] [PubMed] [Google Scholar]
- 54.Westwick J K, Lambert Q T, Clark G J, Symons M, Van Aelst L, Pestell R G, Der C J. Rac regulation of transformation, gene expression, and actin organization by multiple, PAK-independent pathways. Mol Cell Biol. 1997;17:1324–1335. doi: 10.1128/mcb.17.3.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yan M, Dai T, Deak J C, Kyriakis J M, Zon L I, Woodgett J R, Templeton D J. Activation of stress-activated protein kinase by MEKK1 phosphorylation of its activator SEK1. Nature. 1994;372:798–800. doi: 10.1038/372798a0. [DOI] [PubMed] [Google Scholar]
- 56.Zhou G, Bao Z Q, Dixon J E. Components of a new human protein kinase signal transduction pathway. J Biol Chem. 1995;270:12665–12669. doi: 10.1074/jbc.270.21.12665. [DOI] [PubMed] [Google Scholar]