Abstract
Mutations in human immunodeficiency virus type 1 (HIV-1) reverse transcriptase and protease that confer resistance to antiretroviral agents are usually accompanied by a reduction in the viral replicative capacity under drug-free conditions. Consequently, when antiretroviral treatment is interrupted in HIV-1-infected patients harboring drug-resistant virus, resistant quasi-species appear to be most often replaced within several weeks by wild-type virus. Using a real-time PCR-based technique for the selective quantification of resistant viral sequences in plasma, we have studied the kinetics of the switch from mutant to wild-type virus and evaluated the extent to which minority populations of resistant viruses not detected by genotyping persist in these individuals. Among 12 patients with viruses expressing the V82A or L90M resistance mutation who had undergone a 3-month interruption of therapy and for whom conventional genotyping had revealed an apparent total reconversion to wild-type virus, minority populations expressing these mutations, representing 0.1 to 21% of total virus, were still detectable in 9 cases. Kinetic studies demonstrated that viruses expressing resistance mutations could be detected for >5 months after the discontinuation of treatment in some patients. Most of the minority resistant genomes detected more than 3 months after the interruption of therapy carried only part of the mutations present in the resistant viruses prior to treatment interruption and appeared to result from the emergence of existing strains selected at earlier stages in the development of drug resistance. Thus, following the interruption of treatment, viral populations containing resistance mutations can persist for several months after the time when conventional genotyping techniques detect only wild-type virus. These populations include viral strains with only some of the resistance mutations initially present, strains that presumably express better fitness under drug-free conditions.
As is typical of infection with most RNA viruses, human immunodeficiency virus (HIV) infection is characterized by the high diversity and rapid evolution of viral genomes harbored by infected individuals. Thus, viral genomes recovered from plasma, peripheral blood mononuclear cells, or lymphoid tissues are composed of numerous more or less closely related viral quasi-species that are constantly competing and evolving (5, 12, 15, 26). The strains present in the plasma are believed to be representative of those recently released by productively infected cells (29) and therefore represent the quasi-species that are the most fit for active replication. In patients who fail antiretroviral therapy, the majority of plasma HIV genomes contain mutations in the protease and/or reverse transcriptase genes that promote HIV drug resistance (6, 23, 27). It is now well established that resistance mutations in HIV often decrease the overall activity of the mutated viral enzymes and consequently reduce the fitness of resistant variants compared to their wild-type counterparts under drug-free conditions (1, 9, 33). Therefore, when drug pressure is removed during structured treatment interruptions (STI), it would be expected that residual wild-type viruses or mutant viruses bearing a lighter mutation load would have a selective advantage and overtake highly resistant strains.
Using currently available HIV genotyping methods, only the initial phases of this mutant-to-wild-type transition is amenable to study, because these techniques cannot reliably detect viral populations representing less than 20% of total viruses (16). Thus, it is unclear how long and to what extent minor resistant quasi-species persist in the plasma of these patients. Furthermore, when such minority resistant populations are present, it remains to be established whether they result from the persistant proliferation of strains derived from the highly resistant viruses that predominate prior to beginning STI or from the emergence of existing viral variants with lower drug resistance but with presumably better replicative fitness under drug-free conditions. To address these questions, we have developed a technique based on real-time PCR that permits the detection of minority viral species containing key mutations involved in viral resistance to protease inhibitors and used this technique to evaluate the kinetics of the genotypic switch from mutant to wild-type virus during STI.
MATERIALS AND METHODS
Patients.
Peripheral blood was obtained from HIV-infected patients prior to and following interruption of all antiretroviral therapy, and plasma was stored at −80°C. Two cohorts were evaluated: patients at Hôpital Bichat-Claude Bernard, Paris, France (kinetic studies), and patients participating in a clinical trial of a 3-month STI performed at Centre Hospitalier Universitaire (CHU) Purpan, Toulouse, France. Clinical aspects and outcome of this trial have been presented elsewhere (20). All patients gave informed consent before undertaking STI.
Preparation of viral cDNA.
Plasma (0.5 to 1 ml) was centrifuged (23,500 × g, 1 h, 4°C), and RNA was extracted from the pellet (QIAamp viral RNA mini kit; Qiagen GmbH, Hilden, Germany), and eluted in 60 μl of water. Oligonucleotides were synthesized by Genset (Paris, France); nucleotide positions relative to the HXB2 strain are indicated in parentheses. Fifteen microliters of RNA used for reverse transcription (RT)-PCR (Titan One-Tube RT-PCR kit; Boehringer, Mannheim, Germany) following the manufacturer's instructions and using 0.4 μM (final concentration) each primers ProtF1-(2243–2265) (5′-CTTTAGCTTCCCTCAGATCACTC) and ProtR1-(2593–2570) (5′-CCTGGCTTTAATTTTACTGGTACA). Cycling parameters were 40 cycles of denaturation at 94°C for 30 s, annealing at 52°C for 30 s, and extension at 68°C for 45 s.
Quantification of viral populations containing resistance mutations by real-time PCR.
The V82A (GTC→GCC) and L90M (TTG→ATG) resistance mutations were introduced into plasmid pNL4-3 by site-directed mutagenesis as previously described (30) and verified by sequencing. Plasmid DNA from wild-type, V82A, and L90M plasmids was then amplified by PCR using primer pair ProtF1/ProtR1. These amplification products were used for standards, ensuring that samples and standards used for real-time PCR were present in the same form (e.g., amplification products generated with the same primers).
To quantify the proportion of sequences expressing V82A mutations, products obtained by RT-PCR were diluted in 10 mM Tris buffer (pH 7.4) containing 0.1 mM EDTA and 100 ng of herring sperm DNA (Sigma, St. Louis, Mo.) per ml (usually 1:104 to 1:106), and 10-μl aliquots were added to PCRs, permitting the nonselective amplification of all viral sequences or the preferential amplification of sequences containing the V82A mutation. For nonselective amplification, reaction conditions were (50 μl, final volume) 1× TaqMan buffer, 3 mM MgCl2, 100 nM each of the four deoxynucleoside triphosphates (dNTP), 100 nM each primers F3-(2469–2492) (5′-GGTACAGTATTAGTAGGACCTACA) and R1-(2576–2553) (5′-TGGTACAGTTTCAATAGGACTAAT), 50 nM TaqMan probe-(2524–2551) [5′-(6-FAM)CTCAGATTGGTTGCACTTTAAATTTTCC(TAMRA)(phosphate)], 0.5 U of uracil-n-glycosylase, and 1.25 U of AmpliTaq Gold Taq polymerase. For preferential amplification of V82A, conditions were identical except that the F3 primer was replaced by the M2-V82A-(2476–2496) primer (5′-TATTAGTAGGACCTACACCAGC). Reagents and materials were obtained from Perkin-Elmer (Foster City, Calif.). Samples were evaluated by real-time PCR (14, 17) using a Perkin-Elmer 7700 sequence detection system (PE Applied Biosystems), using the following cycling parameters: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 50°C for 1 min. The number of cycles required to reach threshold fluorescence (Ct) was determined, and the quantity of sequences initially present was calculated by extrapolation onto the standard curve. The same dilutions of standard (amplification products generated from the V82A plasmid) were used for both the nonselective and V82A-selective amplifications. The percentage of viral sequences containing V82A was then calculated as follows: % mutated sequences = [(quantity of mutated sequences in the sample)/(quantity of total sequences in the sample)] × 100. To quantify the proportion of sequences expressing L90M mutations, analogous techniques were used except that the primer M2-L90M-(2499–2520) (5′-AACATAATTGGAAGAAATCAGA) was used in place of the M2-V82A primer and dilutions of the amplification products generated from the L90M plasmid were used to generate the standard curve. Nonselective and selective amplifications were always performed at the same time. All reactions were performed in duplicate, and the mean of the two values was used for calculations.
Evaluation of cloned PCR products.
To validate the results obtained by sequence-selective PCR using an independent technique and to evaluate the genotype of minority viral populations expressing the V82A and L90M mutations, cloned viral sequences were analyzed. To obtain clones, serial 10-fold dilutions (10−2 to 10−6) of the original RT-PCR were prepared, and 10 μl of each dilution was amplified using primers F4-(2254–2272) (5′-CTCAGATCACTCTTTGGCA) and R1 in the following reaction conditions: 1× buffer, 3 mM MgCl2, 800 nM each dNTP, 200 nM each primer, and 2 U of AmpliTaq Gold Taq polymerase. Cycling parameters were 95°C for 10 min, 15 cycles at 95°C for 30 s, 50°C for 30 s, and 72°C for 3 min each; and a final step at 72°C for 10 min. After electrophoresis into agarose gels, ethidium bromide-stained products were examined, and the most dilute initial sample that contained a visible band was used for cloning. The PCR products were cloned into pCR4-TOPO (Invitrogen, Carlsbad, Calif.) and used to transfect Escherichia coli, and colonies were grown on Luria-Bertani (LB) plates.
To evaluate the proportion of clones containing mutated (V82A or L90M) and wild-type sequences, individual colonies were transferred to 200 μl of LB medium. Following overnight incubation at 37°C, cultures were resuspended and diluted 1:50 with water, and 10-μl aliquots were evaluated by real-time PCR as described above. Clones containing mutant and wild-type sequences were easily distinguished by comparing the Ct obtained for reactions performed using the nonselective and sequence-selective PCR conditions (mutant clones, ΔCt < 2 cycles; wild-type clones, ΔCt ≈ 10 cycles). Plasmid DNA from representative clones was purified, and the inserts were sequenced. When targets containing a mixture of related sequences are amplified by PCR, recombinants resulting from the annealing of incomplete amplification products can be formed (13, 21). As indicated above, reaction conditions were used to minimize this possibility (high nucleotide and enzyme concentrations, long extension times, avoidance of the plateau phase of the reaction). When mixtures containing 20% viral RNA with mutated sequences and 80% viral RNA with wild-type sequences were evaluated under these conditions, 10 of 10 clones identified as carrying the L90M mutation by real-time PCR also carried all the other mutations and polymorphisms initially present in the mutated sequence.
RESULTS
Selective amplification of viral DNA containing resistance mutations by real-time PCR.
The presence of primer-target mismatches at the 3′ end of the oligonucleotide impairs the efficiency of amplification by PCR (19, 22). Using real-time PCR, it is possible to accurately measure differences in the efficiency of amplification by determining the number of cycles (threshold cycle or Ct) required to generate a specific fluorescent signal resulting from cleavage of a probe recognizing the amplification product, cleavage permitting the physical separation of the fluorescent group (FAM) from the quencher (TAMRA) present in the uncleaved probe. This approach was used to quantify the proportion of viral sequences in clinical samples containing either of two well-recognized amino acid substitutions, V82A and L90M, which are frequently found in viruses that have developed resistance to protease inhibitors.
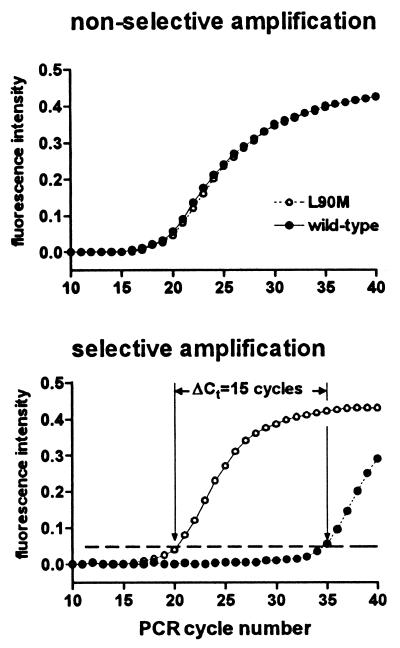
When an oligonucleotide primer perfectly matched for the sequence containing the resistance mutation (and therefore producing a mismatch at the 3′ end with the wild-type sequence) was used to amplify equal amounts of DNA standards with the wild-type and mutant sequences, the amplification of the wild-type sequence was only slightly retarded (data not shown). As previously reported (3, 24), the addition of an intentional mismatch at the −2 position relative to the 3′ terminus of the oligonucleotide considerably destabilized the amplification of wild-type sequences, whereas the presence of a single mutation at the −2 position had little impact on the amplification of the mutant sequence (Fig. 1). Using this approach, a ΔCt of 15 cycles was observed, comparing amplification of the L90M and wild-type standard, corresponding to a decrease in efficiency of amplification of the wild-type sequence of >30,000-fold (Fig. 1). Using the same approach, an oligonucleotide permitting the preferential amplification of sequences containing the V82A mutation was identified. Because the mismatch produced by the alignment of an oligonucleotide recognizing the V82A with the wild-type sequence (A-C) is intrinsically less destabilizing than that produced by the oligonucleotide recognizing L90M (C-C) (19, 22), the discrimination obtained for V82A (ΔCt = 10 cycles, corresponding to a >1,000-fold decrease in the efficiency of amplification of wild-type sequences) was somewhat less than that obtained for L90M.
FIG. 1.
Amplification of viral sequences containing the L90M mutation by sequence-selective real-time PCR. Equal amounts of DNA with the wild-type sequence (solid symbols) or containing the L90M mutation (T→A substitution in codon 90 of the protease) were amplified using a forward primer (F3) which hybridizes to a sequence that is identical in the two standards (nonselective amplification, top panel) or with the primer (L90M-M2) which is complementary to the L90M sequence but introduces a mismatch at the 3′ end with the wild-type sequence (selective amplification, bottom panel). Amplification products were quantified by real-time PCR, and the number of amplification cycles required to reach threshold fluorescence (dashed line in bottom panel) was determined. The reverse primer (R1) and probe were identical in the two amplifications. Viral DNA was obtained by amplifying plasmids containing the wild-type and L90M protease sequences using the primer pair ProtF1 and ProtR1; amplification products were quantified by real-time PCR in experiments independent of those shown in the figure.
Quantification of minority populations expressing mutated sequences in clinical samples.
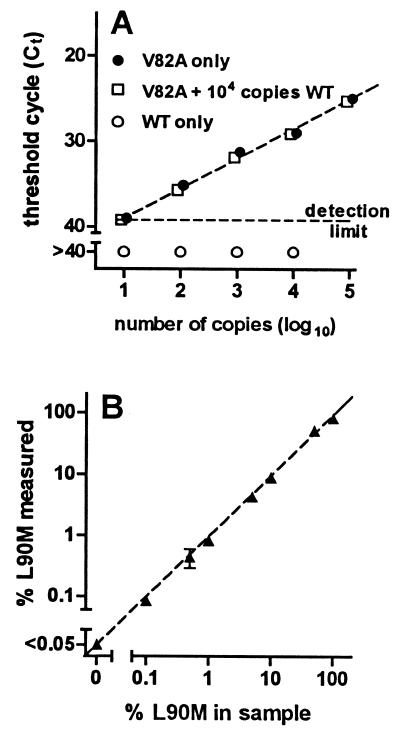
Using the sequence-selective oligonucleotides described above, amplification of mutant sequences was not influenced by the presence of a 1,000-fold excess of wild-type sequences (Fig. 2A). Thus, it was possible to quantify mutated sequences in samples in which they represented only a small proportion of total sequences. When DNA standards with the wild-type and L90M sequence were mixed in different proportions and the percentage of mutant sequences was determined by sequence-selective PCR as described in Materials and Methods, the results obtained by PCR were very close to the actual proportion of mutant sequences present in the sample (Fig. 2B). Mixtures containing 0.1% L90M sequences could be detected in these experiments.
FIG. 2.
Quantification of viral populations expressing resistance mutations by sequence-selective PCR. (A) Samples containing the indicated number of copies of DNA with the wild-type sequence (○), DNA with the V82A mutation (T→C substitution in codon 82 of the protease) alone (●), or DNA with the V82A mutation in the presence of 104 copies of DNA containing the wild-type sequence (□) were prepared and amplified for 40 cycles using the V82-M2/R1 primer pair, which preferentially amplifies viral sequences with the V82A mutation. Amplification products were quantified by real-time PCR, and the number of amplification cycles required to reach threshold fluorescence (Ct) was determined. If threshold fluorescence was not reached after 40 cycles of amplification, samples were considered to be below the detection limit. (B) DNA standards containing the L90M mutation and the wild-type protease sequence were mixed to prepare samples containing the indicated percentage of mutated sequences (% L90M in sample), and the proportion of mutated sequences was measured by sequence-selective PCR as described in Materials and Methods (% L90M measured). Mixtures containing <0.05% mutated sequences could not be distinguished from samples containing only wild-type sequences. Results are the mean ± SD for two (A) or four (B) independent determinations at each point. SDs are too small to be visible on the graphs for most points. The dashed line in panel B is the line of identity.
Although reactions performed using the M2-V82A and M2-L90M primers amplified sequences containing the corresponding mutation much more efficiently than wild-type sequences, amplification products resulting from the amplification of wild-type sequences were generated. When samples containing only wild-type sequences were evaluated as described above, results indicated the apparent presence of 0.6% ± 0.2% (mean ± standard deviation [SD], n = 10) mutated sequences using the V82A system and 0.01% ± 0.02% (mean ± SD, n = 10) mutated sequences using the L90M system. Based on these findings, the limit of sensitivity was defined as the mean + 2 SD of these values (1% for V82A and 0.05% for L90M).
To quantify minority populations in clinical samples, RNA was purified from plasma, and most of the protease gene was amplified by PCR before the proportion of mutated sequences was determined by sequence-selective PCR. This initial amplification step was required to ensure that sufficient mutated sequences were present to be accurately quantified by real-time PCR. Several findings indicated that the proportion of mutated sequences was not modified during this amplification. First, when viral RNAs with mutant and wild-type sequences were mixed in various proportions prior to the initial RT-PCR to produce samples containing <1 to 30% mutant sequences, the percentage of mutated sequences detected by sequence-selective PCR was quite reproducible and close to expected values (Table 1). Furthermore, when serial dilutions of a given mixture of viral RNAs were tested in parallel, the percentage of mutated sequences did not vary as a function of total RNA present as long as sufficient RNA was used to ensure that at least 100 copies of mutated viral RNA were present in the initial RT-PCR (data not shown). Consistent with prior studies (4), the evaluation of multiple samples from untreated patients and patients with virologic treatment failure, demonstrated by standard genotyping to contain viruses expressing multiple resistance mutations, indicated that under the reaction conditions used here, the presence of polymorphisms or additional resistance mutations encountered in most clinical samples had negligible impact on the efficiency of amplification of wild-type sequences or sequences with either the V82A or L90M mutation (data not shown). Additional validation of this procedure is provided by studies presented below comparing results obtained by sequence-selective PCR and those based on the evaluation of clonal populations.
TABLE 1.
Proportion of sequences with protease resistance mutations in mixtures of viral RNA containing different proportions of mutant and wild-type sequences: comparison of results based on evaluation of clones and sequence-selective PCRa
| Genotype | Relative amounts of RNA (mutant:wild type) | % Mutant sequences
|
No. of samples | |
|---|---|---|---|---|
| Expected | Measured | |||
| A (V82A) + B (WT) | 0.42:1 | 29.6 | 22.3 ± 0.6 | 2 |
| 0.085:1 | 7.8 | 6.0 | 1 | |
| 0.042:1 | 4.0 | 3.6 ± 0.7 | 3 | |
| 0.008:1 | 0.8 | <1 | 1 | |
| C (L90M) + D (WT) | 0.33:1 | 25.0 | 19.8 ± 1.6 | 2 |
| 0.044:1 | 4.2 | 2.6 ± 2.5 | 3 | |
Viral RNA was purified from plasma of patients, and RNAs from viruses containing resistance mutation V82A (patient A) or L90M (patient C) were mixed in various proportions with wild-type (WT) RNA from patients B and D, respectively. All genotypes were confirmed by sequencing using standard techniques. RT-PCR was performed using primers that do not distinguish mutant and wild-type sequences. The products obtained by RT-PCR for the mixtures containing the highest proportion of mutant viral RNA were reamplified using nonselective nested primers, the amplification products were cloned, and the proportion of clones containing mutant sequences were determined (mixture A+B, 25 of 84 clones with V82A; mixture C+D, 11 of 44 clones with L90M). The relative concentration of viral RNA in the two preparations used to prepare the mixtures, r = [RNAmutant]/[RNAwt], was determined using the following formula: 100 × {[(volume RNAmutant) × (r)]/[(volume RNAmutant) × (r) + (volume RNAwt × (1)]} = % mutant clones. Using this result, the relative amounts of mutant and wild-type RNA in the other mixtures was calculated using the same formula and expressed as percent mutant sequences expected. These results were compared to the percent mutant sequences measured by sequence-selective PCR. The number of mixtures containing the indicated proportions of mutant and wild-type RNA evaluated in parallel is also shown (number of samples).
Evaluation of kinetics of disappearance of resistant quasi-species during STI.
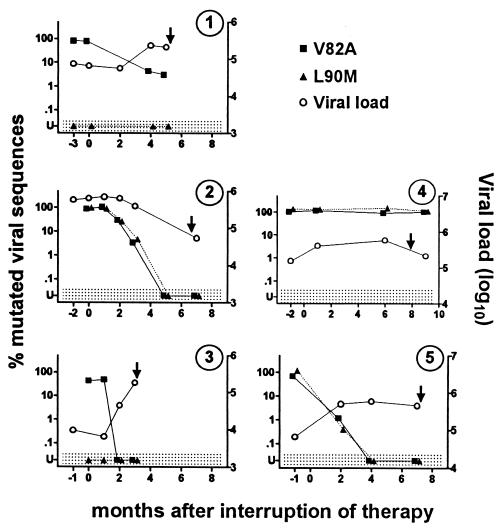
HIV-1-infected patients with highly drug-resistant viral strains who were undergoing STI were identified, and plasma samples obtained from these patients prior to and following interruption of all antiretroviral therapy were evaluated using our technique. The proportion of viral strains containing the L90M and/or V82A mutation as a function of time is shown in Fig. 3. Prior to stopping therapy, a high proportion of mutant strains were present. One month after stopping therapy, resistant strains remained predominant in all three individuals for whom samples were available. Subsequently, the proportion of resistant strains decreased in most patients. When samples obtained 3 months after stopping therapy were evaluated by standard genotyping, apparent conversion to wild-type virus was observed for four of five patients. When samples obtained at ≥3 months were evaluated by real-time PCR, however, residual resistant viral strains were still present in three of five patients, although only one of these patients had more than 5% resistant strains. Mutant viral strains did subsequently become undetectable by real-time PCR in three of the five patients, but the duration of the interruption of treatment required was quite variable (<2 months, 2 to 4 months, and 3 to 5 months, respectively, for patients 3, 5, and 2). In the remaining two patients, mutant viral sequences were still present in plasma when therapy was resumed after 5 and 6 months of interruption.
FIG. 3.
Kinetics of conversion from mutated to wild-type virus following STI. Plasma was obtained from five different patients at the indicated times before and after beginning STI, and the percentage of viral sequences expressing the V82A (■) and L90M (▴) resistance mutations was determined by sequence-selective PCR. Values in the shaded area (U) were below the limits of detection (V82A, <1%; L90M, <0.05%). Plasma viral load (○) was determined by Monitor (Roche). By standard genotyping, all patients had the V82A mutation and patients 1,4, and 5 had the L90M mutation prior to STI. V82A and L90M mutations were undetectable by standard genotyping at 4, 3, 2, and 2 months, respectively, for patients 1, 2, 3, and 5. The arrows indicate the time that antiretroviral therapy was reinitiated.
In four of the five patients studied, the viral load increased following discontinuation of therapy (Fig. 3). In this regard, it is noteworthy that the absolute number of mutant viruses present in the plasma decreased progressively in most patients. This finding indicates that the decrease in the proportion of mutant strains cannot be explained merely by the increase in the number of wild-type viruses that appear following discontinuation of treatment.
Genotyping of resistant quasi-species present during STI.
To confirm these findings and evaluate the genotype of minority viral populations expressing the V82A and L90M mutations, aliquots of the RT-PCRs performed on samples from two patients prior to and at various times after the discontinuation of therapy were reamplified using primers that do not distinguish wild-type and mutated sequences (F4 and R1), the amplification products were cloned, and the proportion of clones containing mutated sequences was determined. For all samples, the percentage of mutant sequences as determined by the evaluation of clones was concordant with the values obtained by sequence-selective real-time PCR (Table 2). In addition, representative mutant and wild-type clones were sequenced. All clones derived from samples obtained prior to discontinuing therapy contained all the resistance mutations identified by standard genotyping (Table 2). Conversely, all clones derived from samples obtained 5 months after the discontinuation of therapy that lacked both the V82A and L90M mutations had a strictly wild-type genotype in both patients.
TABLE 2.
Proportions and genotypes of clones expressing the V82A and L90M resistance mutations obtained at different times after interruption of antiretroviral therapya
| Patient | M after interruption | % Mutated (PCR) | No. of mutated clones/no. tested (95% CI) | No. sequenced | Relevant genotype (no. of clones) | Designation |
|---|---|---|---|---|---|---|
| 1 | −3 | 80 (V82A) | 12/12 V82A (76–100) | 4 (82A) | L10I, G48V, I54V, A71V, V82A (4) | A |
| 5 | 3 (V82A) | 3/176 V82A (1–7) | 3 (82A) | L10I, I54V, A71V, V82A (1) | B | |
| I54V, A71V, V82A (1) | C | |||||
| I54V, V82A (1) | D | |||||
| 4 (82V) | Wild type (4) | E | ||||
| 2 | 0 | 94 (L90M) | 21/21 L90M (86–100) | 5 (90M) | L10I, V32I, M46I, L63P, A71V, V82A, L90M (5) | F |
| 2 | 30 (V82A) | 14/34 V82A (26–62) | ||||
| 3 | 4 (L90M) | 5/131 L90M (2–11) | 5 (90M) | L10I, V32I, M46I, L63P, A71V, V82A, L90M (1) | G | |
| L10I, V32I, M46I, L63P, L90M (3) | H | |||||
| L90M (1) | I | |||||
| 5 | <0.05 (L90M) | 0/28 L90M (0–10) | 5 (90L) | Wild type (5) | J |
The percentage of viral sequences expressing the L90M and/or V82A mutation in samples from patients obtained at the indicated time after STI was determined by sequence-selective PCR (PCR) and by evaluation of clones derived from parallel PCRs (no. of mutated clones). Results for clones are presented as the number of clones containing the indicated mutation/total number of clones tested. The percent mutated clones expressed as a 95% confidence interval (CI) is indicated in parentheses.
Strikingly, the clones containing resistance mutations identified in samples obtained 5 months (patient 1) or 3 months (patient 2) after the discontinuation of therapy were not homogeneous in either patient, and most of these viruses had a genotype that was intermediate between that of the highly resistant population present prior to interruption of therapy and the majority wild-type population that emerged during STI. For patient 1, clones containing two (clone D), three (clone C), and four (clone B) of the original five mutations were identified. Similarly, 3 to 4% of the viral population present in patient 2 at 3 months after STI had the V82A and/or L90M mutation, but the results obtained by cloning indicate that these mutations were present in partially nonoverlapping populations. Among the five minority clones identified, one clone carried all seven mutations (clone G), three clones had retained five of seven mutations, including L90M but not V82A (H clones), and the remaining clone contained L90M in isolation (clone I).
Origin of viruses with intermediate resistance genotypes.
Both genetic analysis of these sequences and clinical-virological correlations strongly supported the conclusion that the viruses with intermediate resistance genotypes resulted from the emergence of viruses that had been selected at earlier stages of the development of drug resistance.
(i) Genetic analysis.
For patient 1, the sequences of the fully mutated pre-STI viruses and the wild-type post-STI viruses differed at 10 positions, five substitutions resulting in resistance mutations and five in neutral substitutions (one substitution producing a polymorphism and four silent third-position base changes). The intermediate viruses with two (clone D), three (clone C), and four (clone B) resistance mutations had, respectively, one, four, and five of the five neutral substitutions found in the fully mutated virus. The simultaneous modification of nucleotides producing resistance mutations and these neutral nucleotide substitutions would not be expected if the reversion of some mutations in the resistant pre-STI viruses had occurred after discontinuation of therapy. In contrast, the progressive accumulation of the neutral substitutions in parallel with the appearance of resistance mutations is consistent with the idea that viruses with intermediate resistance genotypes appeared sequentially during the development of drug resistance. Similarly, the fully mutated pre-STI viruses and the wild-type post-STI viruses from patient 2 differed at 16 positions, seven substitutions resulting in resistance mutations and nine in neutral nucleotide substitutions (one codon change, two substitutions producing polymorphisms, and four silent base changes). The intermediate viruses with one (clone I) and five (H clones) resistance mutations had, respectively, one and nine of the nine neutral substitutions found in the fully mutated virus.
(ii) Clinical-virological correlations.
Patient 1 had received ritonavir for 2 months and then indinavir for 10 months before being switched to a combination of drugs that included saquinavir. The combination of mutations V82A and I54V (clone D) is typical of early resistance to ritonavir and/or indinavir (31). The addition of A71V (clone C) both reinforces resistance to these two drugs and partially corrects drug-free virus fitness (18). On the other hand, mutations L10I (clone B) and G48V (found only in the most recent pre-STI population and clones) are more suggestive of resistance to saquinavir (31) and are likely to have been selected after switching to that drug. Patient 2 was treated with a regimen including indinavir (15 months), followed by switches to nelfinavir (5 months) and saquinavir plus low-dose ritonavir (15 months) prior to STI. No preferred order of appearance of substitutions leading to indinavir resistance has been described (6, 7), but L90M (clone I) can be observed in this context. The additional mutations observed in the H clones are typical of mutations that would improve resistance to indinavir and extend cross-resistance to nelfinavir and ritonavir.
Quantification of quasi-species expressing resistance mutations after STI of 3 months.
To further evaluate how frequently the commonly used STI duration of 3 months was sufficient to reduce the plasma levels of resistant virus to below the detection threshold, we quantified the proportion of viruses expressing the V82A and L90M mutations in patients who had undergone STI for 3 months and for whom conventional genotyping had revealed an apparent total conversion to wild-type virus. In the 12 patients with V82A or L90M mutations prior to the interruption of therapy, minority populations expressing these resistance mutations were still detectable after an STI of 3 months in nine cases (Table 3). Viruses with V82A or L90M mutations were never detected in individuals who did not harbor the corresponding mutation at baseline.
TABLE 3.
Detection of minority viral populations expressing V82A and/or L90M resistance mutation in plasma of patients after a 3-month STIa
| Technique | Time of evaluation | Genotype | Patient | % of sequences
|
Genotype | Patient | % of sequences
|
Genotype | Patient | % of sequences
|
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V82A | L90M | V82A | L90M | V82A | L90M | ||||||||
| Genotyping | Before STI | 82A 90M | 82A 90L | 82V 90M | |||||||||
| After 3 mo | 82V 90L | 82V 90L | 82V 90L | ||||||||||
| PCR | After 3 mo | I | 21 | 13 | D | 9 | bdl | L | bdl | 9 | |||
| B | 15 | 6 | F | 4 | bdl | A | bdl | 5 | |||||
| C | 9 | 0.1 | E | 3 | bdl | H | bdl | 2 | |||||
| K | bdl | bdl | J | bdl | bdl | G | bdl | bdl | |||||
HIV-infected patients followed at CHU Purpan (Toulouse, France) were evaluated after completing a 3-month STI, and patients were identified for whom standard genotyping revealed apparent reversion to wild-type virus. The proportion of viral sequences containing the V82A and/or L90M resistance mutation was determined by sequence-selective PCR for all patients whose genotype prior to STI contained the V82A and/or the L90M mutation. bdl, below detection limit (V82A, <1%; L90M, <0.05%).
DISCUSSION
Although the emergence of resistance mutations in the protease and RT of HIV-1 confers considerable selective advantage to the virus for its replication in the presence of antiretroviral agents (8, 18), it is now well established that these mutations can decrease its replicative capacity and reduce its fitness for competition with wild-type virus in the absence of drug (1, 9, 33). Correspondingly, it has been observed that in patients who undergo STI following the development of resistant virus, an apparent replacement of the resistant plasma quasi-species by wild-type genomes occurs in most cases (10, 11, 32). In this study, we have shown that in most patients undergoing STI, small proportions of residual resistant viruses are still detectable at times when conventional bulk-plasma virus genotyping detected only wild-type sequences. Our results demonstrate that the time required for mutant viruses to become undetectable is quite variable, but that the frequently used duration of 3 months for STI is clearly too short for washout of resistant viruses in most patients.
Interestingly, we found that resistant viruses recovered after more than 3 months of STI often carried fewer mutations in the protease gene than their pre-STI counterparts. Several arguments indicate that recombination occurring during PCR did not explain the detection of these sequences with “intermediate” genotype. First, the PCRs were performed under conditions that strongly disfavor such recombination (long extension times and avoidance of the plateau phase of the amplification reaction) (13, 21). Second, when mixtures of mutated and wild-type viral RNA were prepared and evaluated by these procedures, all clones expressing L90M and V82A resistance mutations also expressed all the other resistance mutations and polymorphisms expressed by the mutated viruses used to prepare these mixtures (e.g., no evidence of recombination was observed). Finally, in no case could the viruses with intermediate genotype observed in our patients be explained by the annealing of a portion of a wild-type sequence with the other extremity of a mutated sequence, as would be expected if the products were formed by recombination.
Rather, our results suggest that the appearance of viruses with an intermediate resistance genotype generally reflects the emergence of viral strains generated during earlier stages of the selection for drug resistance. Several independent lines of evidence support this conclusion. First, the therapeutic history of the patients is compatible with this idea. For example, patient 1 had initially failed on a therapeutic regimen including indinavir, followed by a switch to saquinavir. The mutations present in the fully resistant viruses that were missing in some of the intermediate viruses (e.g., G48V) are consistent with a selection that would have been exerted following these changes in therapy. Furthermore, phylogenetic analysis of the nucleotide sequences of intermediate viruses revealed that those with fewer resistance mutations were clearly more closely related to wild-type viral sequences, excluding the possibility of selective reversion of resistance mutations or recombination in these cases. The rapid emergence of fully wild-type virus detected by standard genotypic analysis in this and in prior studies (11, 32) and the rapid virus rebound that follows the interruption of an apparently fully suppressive antiretroviral therapy (28) further supports the conclusion that treated patients can continue to harbor strains lacking the full complement of resistance mutations.
Numerous groups have reported that following the appearance of drug resistance, additional mutations are subsequently added that can both extend drug resistance and improve viral replicative capacity as measured using in vitro systems (reviewed in reference 2). In this regard, our finding that viral strains with intermediate resistance genotypes persisted longer than the pre-STI viruses following the interruption of therapy is noteworthy. These results offer direct evidence that the in vivo fitness of the less mutated viruses appearing earlier in the development of resistance was higher than that of the highly resistant strains that ultimately emerged, indicating that drug resistance, not fitness, was the predominant force for selection in these patients. This finding is consistent with studies showing substantial impairment in the fitness of highly evolved resistant viruses (10), and recent findings by our group indicating that the sequential accumulation of mutations in the protease gene following treatment failure usually leads to a progressive decrease in viral fitness (A. Faye, E. Race, V. Obry, M. H. Prévot, V. Joly, S. Matheron, F. Damond, E. Dam, S. Paulous, and F. Clavel, Third International Workshop on HIV Drug Resistance and Treatment Strategies, abstr. 129, San Diego, Calif., June 1999). Thus, although increases in resistance (due to primary mutations) are likely to be accompanied by transitory improvements in fitness (due to compensatory mutations), over the long term, the deleterious effects of the progressive addition of new resistance mutations appears to be predominant in most patients and leads to a net loss in viral fitness. It is noteworthy that in patient 1, intermediate viruses were lacking the G48V mutation, which is known to significantly affect HIV fitness (25). Similarly, in patient 2, an intermediate virus expressing L90M but not V82A was found, and it has recently been shown that the association of these two mutations results in measurable loss of viral fitness (25). Taken together, our findings support the model in which the likelihood and the speed of the genotypic switch following STI are a function of two factors: (i) the difference in fitness between the resistant viruses present and their wild-type counterparts, and (ii) the relative abundance of these strains. Patients with a poorly fit resistant virus and an available pool of replication-competent wild-type virus will rapidly develop a genotypic switch. In patients who harbor resistant viruses with higher drug-free fitness or in whom prolonged replication of resistant viruses has led to a near extinction of wild-type or nearly wild-type species, recruitment of truly wild-type virus will be slow or may even not occur.
It remains unclear under what form(s) the reemerging viruses are present during treatment. One possibility is that these viruses are still actively replicating as minority species before STI. Indeed, strong differences in selective pressure exerted by drugs in different tissue compartments could result in the coexistence, during treatment failure, of viruses with radically different resistance profiles. Alternatively, inducible replication-competent proviruses harbored by long-lived cells could serve as a reservoir. In either case, the existence of these readily recruitable viral strains, which are likely to recapitulate many or all of the steps leading to full drug resistance, suggests that optimal post-STI treatment regimens will have to prevent their subsequent reemergence.
Finally, it is noteworthy that the changes in virus populations that followed STI were accompanied by a only a modest increase in plasma viral load. Since the sensitivity of our technique allowed us to accurately quantify the total number of mutant and wild-type viruses present, we could establish that the observed genotypic switches did not merely reflect the dilution of (persistent) resistant viruses by a surge of wild-type virus. Further studies will be required to identify the mechanisms through which the replication of resistant viruses is impaired following the appearance of wild-type virus. Our results are consistent with the possibility that the resistant and wild-type viruses directly compete for a common pool of infectable cells whose number is limited. Indirect mechanisms may also be responsible. The number of infected cells present at any time is the result of a dynamic steady state controlled by both the rate of infection and the death rate of infected cells (5, 26). It is possible that the rate of infection of susceptible cells increases following the emergence of a more fit virus but is largely balanced by an increase in the rate of death of infected cells (e.g., through stimulation of cytotoxic T-cell activity), resulting in only a small net increase in viral load. Because the rate of infection of cells by resistant virus would, at best, remain constant, the increased death of infected cells would result in a decrease in the total number of cells producing resistant viruses.
ACKNOWLEDGMENTS
V.L. was supported by a grant from the Fondation pour la Recherche Médicale. The study was supported in part by a grant from the Agence Nationale de Recherches sur le SIDA (ANRS).
REFERENCES
- 1.Back N K, Nijhuis M, Keulen W, Boucher C A, Oude Essink B O, van Kuilenburg A B, van Gennip A H, Berkhout B. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 1996;15:4040–4049. [PMC free article] [PubMed] [Google Scholar]
- 2.Bleiber G, Munoz M, Ciuffi A, Meylan P, Telenti A. Individual contributions of mutant protease and reverse transcriptase to viral infectivity, replication, and protein maturation of antiretroviral drug-resistant human immunodeficiency virus type 1. J Virol. 2001;75:3291–3300. doi: 10.1128/JVI.75.7.3291-3300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cha R S, Zarbl H, Keohavong P, Thilly W G. Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. PCR Methods Appl. 1992;2:14–20. doi: 10.1101/gr.2.1.14. [DOI] [PubMed] [Google Scholar]
- 4.Christopherson C, Sninsky J, Kwok S. The effects of internal primer-template mismatches on RT-PCR: HIV-1 model studies. Nucleic Acids Res. 1997;25:654–658. doi: 10.1093/nar/25.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 6.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, et al. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 7.Condra J H, Holder D J, Schleif W A, Blahy O M, Danovich R M, Gabryelski L J, Graham D J, Laird D, Quintero J C, Rhodes A, et al. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J Virol. 1996;70:8270–8276. doi: 10.1128/jvi.70.12.8270-8276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cozzi Lepri A, Sabin C A, Staszewski S, Hertogs K, Muller A, Rabenau H, Phillips A N, Miller V. Resistance profiles in patients with viral rebound on potent antiretroviral therapy. J Infect Dis. 2000;181:1143–1147. doi: 10.1086/315301. [DOI] [PubMed] [Google Scholar]
- 9.Croteau G, Doyon L, Thibeault D, McKercher G, Pilote L, Lamarre D. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J Virol. 1997;71:1089–1096. doi: 10.1128/jvi.71.2.1089-1096.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deeks S G, Wrin T, Liegler T, Hoh R, Hayden M, Barbour J D, Hellmann N S, Petropoulous C J, McCune J M, Hellerstein M K, Grant R M. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N Engl J Med. 2001;344:472–480. doi: 10.1056/NEJM200102153440702. [DOI] [PubMed] [Google Scholar]
- 11.Devereux H L, Loveday C, Youle M, Sabin C A, Burke A, Johnson M A. Reduction in human immunodeficiency virus type 1 mutations associated with drug resistance after initiating new therapeutic regimens in pretreated patients. J Infect Dis. 2000;181:1804–1807. doi: 10.1086/315444. [DOI] [PubMed] [Google Scholar]
- 12.Domingo E, Holland J J. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 13.Fang G, Zhu G, Burger H, Keithly J S, Weiser B. Minimizing DNA recombination during long RT-PCR. J Virol Methods. 1998;76:139–148. doi: 10.1016/s0166-0934(98)00133-5. [DOI] [PubMed] [Google Scholar]
- 14.Gibson U E, Heid C A, Williams P M. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 15.Goodenow M, Huet T, Saurin W, Kwok S, Sninsky J, Wain-Hobson S. HIV-1 isolates are rapidly evolving quasispecies: evidence for viral mixtures and preferred nucleotide substitutions. J Acquir Immune Defic Syndr. 1989;2:344–352. [PubMed] [Google Scholar]
- 16.Gunthard H F, Wong J K, Ignacio C C, Havlir D V, Richman D D. Comparative performance of high-density oligonucleotide sequencing and dideoxynucleotide sequencing of HIV type 1 pol from clinical samples. AIDS Res Hum Retroviruses. 1998;14:869–876. doi: 10.1089/aid.1998.14.869. [DOI] [PubMed] [Google Scholar]
- 17.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 18.Hirschel B, Opravil M. The year in review: antiretroviral treatment. AIDS. 1999;13:S177–S187. [PubMed] [Google Scholar]
- 19.Huang M M, Arnheim N, Goodman M F. Extension of base mispairs by Taq DNA polymerase: implications for single nucleotide discrimination in PCR. Nucleic Acids Res. 1992;20:4567–4573. doi: 10.1093/nar/20.17.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izopet J, Massip P, Souyris C, Sandres K, Puissant B, Obadia M, Pasquier C, Bonnet E, Marchou B, Puel J. Shift in HIV resistance genotype after treatment interruption and short-term antiviral effect following a new salvage regimen. AIDS. 2000;14:2247–2255. doi: 10.1097/00002030-200010200-00005. [DOI] [PubMed] [Google Scholar]
- 21.Judo M S B, Wedel A B, Wilson C. Stimulation and suppression of PCR-mediated recombination. Nucleic Acids Res. 1998;26:1819–1825. doi: 10.1093/nar/26.7.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwok S, Kellogg D E, McKinney N, Spasic D, Goda L, Levenson C, Sninsky J J. Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res. 1990;18:999–1005. doi: 10.1093/nar/18.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larder B A, Darby G, Richman D D. HIV with reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science. 1989;243:1731–1734. doi: 10.1126/science.2467383. [DOI] [PubMed] [Google Scholar]
- 24.Larder B A, Kellam P, Kemp S D. Zidovudine resistance predicted by direct detection of mutations in DNA from HIV-infected lymphocytes. AIDS. 1991;5:137–144. doi: 10.1097/00002030-199102000-00002. [DOI] [PubMed] [Google Scholar]
- 25.Mammano F, Trouplin V, Zennou V, Clavel F. Retracing the evolutionary pathways of human immunodeficiency virus type 1 resistance to protease inhibitors: virus fitness in the absence and in the presence of drug. J Virol. 2000;74:8524–8531. doi: 10.1128/jvi.74.18.8524-8531.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyerhans A, Cheynier R, Albert J, Seth M, Kwok S, Sninsky J, Morfeldt-Manson L, Asjo B, Wain-Hobson S. Temporal fluctuations in HIV quasispecies in vivo are not reflected by sequential HIV isolations. Cell. 1989;58:901–910. doi: 10.1016/0092-8674(89)90942-2. [DOI] [PubMed] [Google Scholar]
- 27.Molla A, Korneyeva M, Gao Q, Vasavanonda S, Schipper P J, Mo H M, Markowitz M, Chernyavskiy T, Niu P, Lyons N, Hsu A, Granneman G R, Ho D D, Boucher C A, Leonard J M, Norbeck D W, Kempf D J. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat Med. 1996;2:760–766. doi: 10.1038/nm0796-760. [DOI] [PubMed] [Google Scholar]
- 28.Neumann A U, Tubiana R, Calvez V, Robert C, Li T S, Agut H, Autran B, Katlama C. HIV-1 rebound during interruption of highly active antiretroviral therapy has no deleterious effect on reinitiated treatment. Comet Study Group. AIDS. 1999;13:677–683. doi: 10.1097/00002030-199904160-00008. [DOI] [PubMed] [Google Scholar]
- 29.Perelson A S, Neumann A U, Markowitz M, Leonard J M, Ho D D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 30.Petit C, Schwartz O, Mammano F. Oligomerization within virions and subcellular localization of human immunodeficiency virus type 1 integrase. J Virol. 1999;73:5079–5088. doi: 10.1128/jvi.73.6.5079-5088.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schinazi R F, Larder B A, Mellors J W. Mutations in retroviral genes associated with drug resistance: 2000–2001 update. Int Antiviral News. 2000;8:65–91. [Google Scholar]
- 32.Verhofstede C, Wanzeele F V, Van Der Gucht B, De Cabooter N, Plum J. Interruption of reverse transcriptase inhibitors or a switch from reverse transcriptase to protease inhibitors resulted in a fast reappearance of virus strains with a reverse transcriptase inhibitor-sensitive genotype. AIDS. 1999;13:2541–2546. doi: 10.1097/00002030-199912240-00007. [DOI] [PubMed] [Google Scholar]
- 33.Zennou V, Mammano F, Paulous S, Mathez D, Clavel F. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J Virol. 1998;72:3300–3306. doi: 10.1128/jvi.72.4.3300-3306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]