Abstract
Incorporation of the 21st amino acid, selenocysteine, into proteins is specified in all three domains of life by dynamic translational redefinition of UGA codons. In eukarya and archaea, selenocysteine insertion requires a cis-acting selenocysteine insertion sequence (SECIS) usually located in the 3′UTR of selenoprotein mRNAs. Here we present comparative sequence analysis and experimental data supporting the presence of a second stop codon redefinition element located adjacent to a selenocysteine-encoding UGA codon in the eukaryal gene, SEPN1. This element is sufficient to stimulate high-level (6%) translational redefinition of the SEPN1 UGA codon in human cells. Readthrough levels further increased to 12% when tested in the presence of the SEPN1 3′UTR SECIS. Directed mutagenesis and phylogeny of the sequence context strongly supports the importance of a stem loop starting six nucleotides 3′ of the UGA codon. Sequences capable of forming strong RNA structures were also identified 3′ adjacent to, or near, selenocysteine-encoding UGA codons in the Sps2, SelH, SelO, and SelT selenoprotein genes.
Keywords: readthrough, redefinition, selenocysteine, selenoprotein, termination
Introduction
Dynamic reprogramming of the genetic code redefines a subset of stop codons to encode an amino acid. Translation beyond the stop codon results in a fusion protein derived from information in two adjacent open reading frames. In some cases, continued translation beyond the stop codon is the relevant feature and the identity of the specified amino acid unimportant (Namy et al, 2004). In other cases, the identity of the specified amino acid is critical. Redefinition of UGA codons to specify the 21st amino acid, selenocysteine, directs insertion of this highly reactive amino acid, which is often a required residue for protein activity (for reviews, see Hatfield and Gladyshev, 2002; Driscoll and Copeland, 2003). The recoding of stop codons in select mRNAs discussed in this manuscript is to be clearly distinguished from the genome-wide reassignment of stop codons found in mycoplasma, ciliates, and some mitochondria, where the reassigned codon is directly decoded in all mRNAs as a sense codon.
The extension of the genetic code to specify UGA decoding as selenocysteine is found in a subset of genes with diverse functions in all three domains of life (reviewed in Hatfield and Gladyshev, 2002). In eukarya and archaea, decoding UGA as selenocysteine requires a selenocysteine insertion sequence (SECIS) located in the 3′UTR of most selenoprotein-encoding mRNAs (Berry et al, 1991; Rother et al, 2001). In contrast, bacterial bSECIS elements are located immediately adjacent to the UGA codon (Zinoni et al, 1990; Hüttenhofer et al, 1996). The eukaryal SECIS structure consists of an elongated stem loop (Martin and Berry, 2001; Korotkov et al, 2002) with an internal loop and non-Watson–Crick quartet tandem (G.A/A.G), which may form a kink-turn motif (Walczak et al, 1996, 1998; Allmang et al, 2002). In addition to this specialized cis-acting element, at least two trans-acting factors, the SECIS binding protein 2 (SBP2) (Copeland et al, 2000, 2001; Low et al, 2000), and a specialized elongation factor (mSelB) (Fagegaltier et al, 2000; Tujebajeva et al, 2000a) are required to achieve redefinition of the UGA codon to selenocysteine by tRNASec decoding. The bacterial elongation factor SelB and associated tRNASec, in contrast, bind directly to the bSECIS located directly adjacent to the stop codon. This implies an important mechanistic distinction, as bSECIS elements act locally at the site of UGA decoding, whereas the positioning of the eukaryal and archaeal SECIS elements to the 3′UTR allows for selenocysteine insertion at one or more UGA codons within the mRNA.
Despite the identification of several key components of selenocysteine incorporation, the mechanism by which a ribosome is ‘informed' by the 3′UTR SECIS element to direct the appropriate level of selenocysteine incorporation remains unknown. Measurements of selenocysteine incorporation efficiency in bacteria (Suppmann et al, 1999) and in eukaryaea using transfected cells or partially purified translation systems show relatively low-level selenocysteine insertion (Berry et al, 1994; Kollmus et al, 1996; Mehta et al, 2004). Paradoxically, the PHGPx selenoprotein is made at particularly high levels in testis (Ursini et al, 1999), and expression of the selenoprotein P gene, containing 10–17 UGA codons depending upon the species (Hill et al, 1991; Tujebajeva et al, 2000b), would appear to demand efficient incorporation. Incorporation efficiency for specific selenocysteine-encoding mRNAs may be influenced by tissue-specific or other general accessory factors, additional cis-acting elements either within or outside the coding region, or even subcellular localization—complicating measurements of redefinition efficiency in experimental systems. It seems certain that additional cis- and trans-acting factors remain to be identified that modulate the interaction between translational termination and selenocysteine insertion at specific UGA codons to affect redefinition efficiency and regulation in vivo.
In the absence of SECIS elements, stop codon redefinition by directed insertion of standard amino acids is commonly achieved by readthrough stimulators located adjacent to the stop codon (Gesteland and Atkins, 1996; Namy et al, 2004). Although it has been suggested that only the single base 3′ of a stop codon can be sufficient to direct programmed readthrough (Li and Rice, 1993), more commonly up to six nucleotides downstream of the redefined codon are involved. Two surveys of the stop codon sequence contexts from nearly 100 known viral examples of readthrough illustrated that a limited number of variants of a 3′ adjacent readthrough motif, CARYYA (Skuzeski et al, 1991), are utilized (Beier and Grimm, 2001; Harrell et al, 2002). In addition, H-type RNA pseudoknot structures (ten Dam et al, 1990) have been shown to direct readthrough in several mammalian retroviruses (Wills et al, 1991; Feng et al, 1992). A well-studied example is gag-pol expression in the murine leukemia virus (MuLV) where the gag UAG stop codon is redefined with approximately 5–10% efficiency (Philipson et al, 1978; Yoshinaka et al, 1985). Both the sequence identity of the eight nucleotides 3′ of the UAG codon (Wills et al, 1994) and key features of the pseudoknot are required activators of readthrough (Alam et al, 1999). Recently, MuLV readthrough levels were shown to be dynamically regulated by binding of the Pol product of MuLV to the eukaryal translational release factor 1 (Orlova et al, 2003). Enhanced readthrough levels attained by this interaction were shown to be important for viral replication. Another example of regulatory stop codon redefinition comes from studies of kelch expression during Drosophila development (Robinson and Cooley, 1997). In this study, the ratio of the termination to readthrough product was suggested to be regulated in a tissue-specific manner. These findings illustrate that not only can redefinition levels be specified by local sequence context for proper gene expression, but also, in some cases, readthrough efficiency is dynamically regulated to achieve optimal gene expression.
The occurrence of stop codon readthrough stimulators located adjacent to stop codons, which are redefined as ‘standard' amino acids (most often glutamine for UAG and tryptophan for UGA), and selenocysteine-encoding codons in bacteria (with 3′ adjacent bSECIS elements) prompted a re-examination of local eukaryal selenocysteine codon sequence contexts for potential effectors of redefinition. The local sequence contexts of eukaryal selenocysteine-encoding UGA codons were examined for the occurrence of conserved downstream RNA secondary structures. Here we present comparative sequence analysis and experimental data, which supports the existence of a phylogenetically conserved stop codon redefinition element located adjacent to the SEPN1 selenocysteine-encoding UGA codon. Although the biological function of SEPN1 is unknown, mutations in SEPN1 are associated with several early-onset myopathies including rigid spinal muscular dystrophy, classical multiminicore disease, and Mallory body-like form of desmin-related myopathies (Moghadaszadeh et al, 2001; Ferreiro et al, 2002, 2004).
Results
RNA secondary structure predictions and comparative sequence analysis
Sequences located downstream of 36 selenocysteine-encoding UGA codons from 25 human selenoprotein genes, including the selenoprotein P gene, which contains 10 selenocysteine-encoding UGA codons, were examined for potential RNA secondary structures using mfold version 3. 1 (for specific folding parameters and accession numbers, see Materials and methods). Mfold predicts the minimum free energy, ΔG, for multiple RNA foldings of an input RNA sequence (Mathews et al, 1999; Zuker, 2003). The 60 nucleotides located just 3′ of the UGA codon were used as input and then ranked according to the most favorable predicted ΔG value for each RNA sequence. For the 36 selenocysteine insertion sites analyzed, ΔG values for the best RNA folding varied from −2.2 to −36.4 kcal/mol. RNA structures with ΔG values lower than −25 kcal/mol were predicted downstream of five UGA codons; SelO ΔG=−25.8, SEPN1 UGA1 ΔG=−30.5, SEPN1 UGA2 ΔG=−29.5, SelH ΔG=−32.4, and Sps2 ΔG=−36.4 (see Discussion). Analysis of sequences downstream of the 10 selenocysteine-encoding UGA codons in the human selenoprotein P gene reveals a range of ΔG values from −2.2 to −16.6. The identification of several selenocysteine-encoding UGA codons with little potential to form downstream RNA secondary structure suggests that, unlike the case in bacteria, a stem loop near the UGA codon is not required for selenocysteine insertion in eukarya. However, the possibility of structures formed by long-range RNA interactions cannot be ruled out.
Based on promising phylogenetic conservation and the identification of sequence variations that maintain base-pairing potential (see below), the sequence surrounding the selenocysteine-encoding codon located in exon 10 of the SEPN1 gene, SEPN1 UGA2, was selected for further examination in this study. SEPN1 genes were identified by BLAST analysis of publicly available protein and nucleotide databases using the human SEPN1 protein sequence as query. Genes with significant similarity to the human SEPN1 gene were identified in 14 vertebrates and two urochordates. Additional BLAST analysis using the distantly related sequence from Ciona intestinalis as query did not identify additional genes with significant similarity to SEPN1. Sequence alignments were performed using ClustalW (Thompson et al, 1994) (Figure 1), revealing a high degree of conservation surrounding the selenocysteine-encoding UGA2 codon. The first UGA codon, UGA1, in the human SEPN1 gene is not phylogenetically conserved and occurs only in an alternately processed transcript with uncertain biological significance. The codon preceding the UGA2 codon and five nucleotides downstream are universally conserved in all SEPN1 genes identified. The potential for a stem loop secondary structure starting seven nucleotides downstream of the UGA codon is predicted and supported by sequence variations that maintain base-pairing potential (Figure 2). The first five base pairs and the size of the predicted stems (17 nucleotides) are preserved. The length of each stem assumes that A:U and G:U pairs are formed at the apical end. These pairings may be nonexistent or only transiently formed in vivo. In addition, three nucleotides in the loop are identical for all predicted structures and a conserved C–A bulge is found after the ninth base pair in the stem of seven out of 13 predicted structures.
Figure 1.
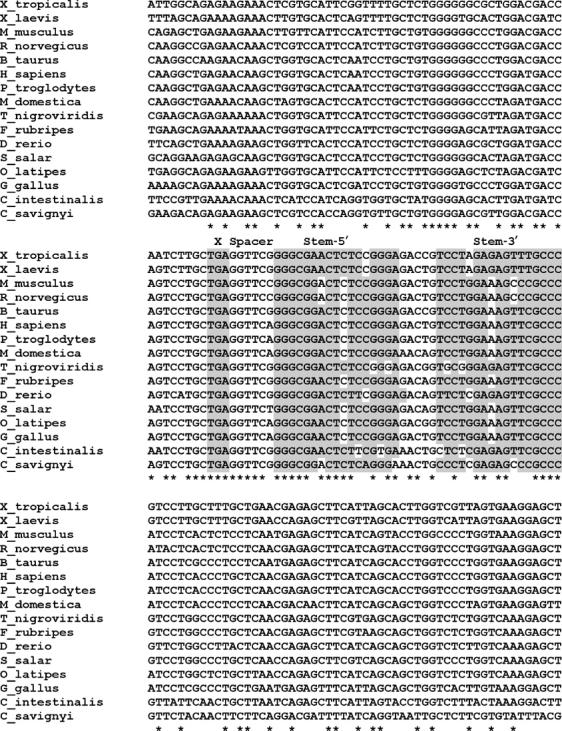
Sequence alignment of SEPN1 UGA sequence context. The sequence of the selenocysteine insertion site and 64 nucleotides upstream and 101 nucleotides downstream of 16 SEPN1 genes are shown. The location of the UGA codon (X), spacer, and the 5′ (Stem-5′) and 3′ (Stem-3′) strands of the predicted stem loop are indicated above the middle panel of sequences. The UGA codon and nucleotides predicted to be base paired are highlighted in gray. Asterisks indicate positions that are 100% conserved.
Figure 2.
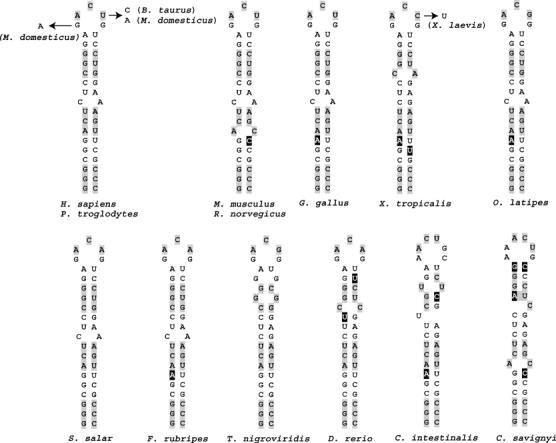
Secondary structures predicted downstream of SEPN1 UGA codons. The predicted stem loop structures from the 16 SEPN1 genes described in Figure 1 are shown in a two-dimensional representation. Light gray shading is used to indicate codons and black shading with white letters denotes sequence variations that maintain base-pairing potential.
An important stem loop
The effect of the surrounding sequence context on decoding of the UGA2 codon located in the human SEPN1 gene was examined using a dual luciferase reporter assay in cultured human embryonic kidney cell line (HEK293). A total of 35 nucleotides located 5′ and 46 nucleotides 3′ of the UGA codon were cloned between the Renilla and firefly luciferase reporter genes in plasmid p2luc (Grentzmann et al, 1998) to create the reporter construct UGA2. The firefly luciferase gene lacks an initiation codon and can only be expressed as a fusion protein with Renilla luciferase if translational stop codon readthrough occurs. Readthrough efficiency is calculated as a ratio of the firefly to Renilla luciferase activities standardized to an in-frame control in which the intervening stop codon has been altered to a sense codon (see Materials and methods; Grentzmann et al, 1998).
Readthrough efficiency was calculated to be 6% for UGA2 (Figure 3B). An equivalent sequence context was tested for all 10 of the human selenoprotein P UGA codons. Translational readthrough levels were measured at 1% or less for all stop codons with the exception of the first UGA codon from the selenoprotein P gene, which revealed translational readthrough levels of approximately 1.7% (data not shown). The importance of the predicted stem loop in SEPN1 UGA2 for stimulation of translational readthrough was addressed by systematic mutagenesis of the stem loop region. Three nucleotides at a time were changed beginning at the bottom of the stem to interrupt base pairing (Figure 3A). Each disruption mutation reduced stop codon readthrough levels to less than 1% (Figure 3B). Pairing potential was restored at each position by making compensatory mutations such that for each consecutive block of three base pairs in the stem, G:C, A:U, and G:U pairs were altered to C:G, U:A, and U:G, respectively (Figure 3A, SC1-3, SC2-3, SC3-3, SC4-3, SC5-3). Altering the first three base pairs from G:C to C:G (SC1-3) failed to restore readthrough levels. However, restoring base–pairing potential at the second (SC2-3), third (SC3-3), fourth (SC4-3), and fifth (SC5-3) set of three base pairs resulted in partial restoration of stop codon readthrough levels to 5, 1, 2.5, and 4.5%, respectively (Figure 3B).
Figure 3.
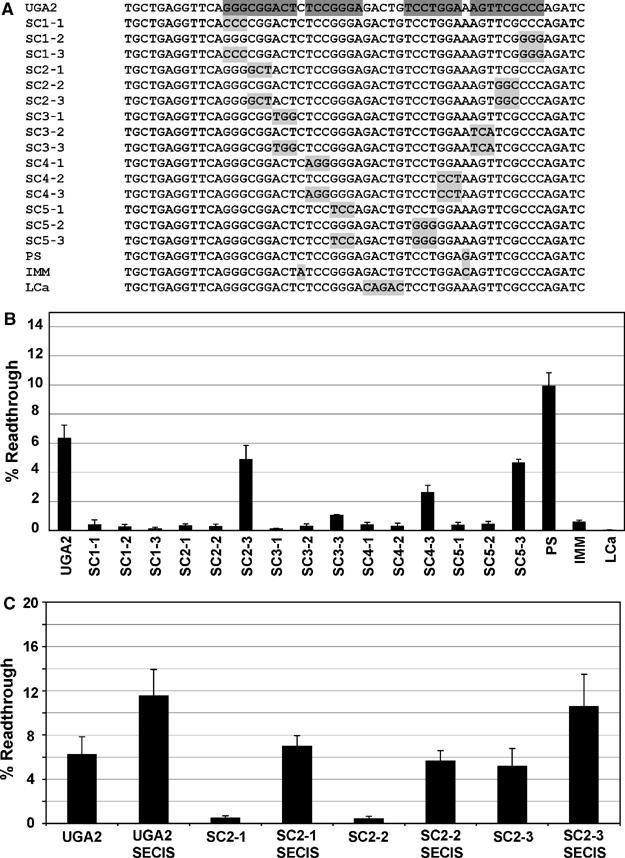
Mutagenic analysis of the human SEPN1 stem loop. The human SEPN1 UGA codon located in exon 10 and surrounding sequence context was cloned into a dual luciferase reporter vector, p2luc, to produce the wild-type construct UGA2. Selective mutations were made in the stem loop region and each construct was expressed in human HEK293 cells and percent readthrough determined. (A) The sequence of the wild-type (UGA2) stem loop region is shown, with base-paired regions highlighted in dark gray (see Supplementary data for complete SEPN1 sequence analyzed). Each subsequent sequence shown corresponds to a unique construct that is identical with the exception of the region highlighted in light gray. (B) Histogram of the calculated percent readthrough for each dual luciferase construct shown in panel A. Error bars indicate standard deviation from the mean. (C) Histogram of the percent readthrough for the dual luciferase constructs containing the UGA2, SC2-1, SC2-2, and SC2-3 sequences in the presence (SECIS) or absence of the SEPN1 3′UTR SECIS element.
The stem loop structure predicted for the human SEPN1 gene contains a single C:A mismatch following the ninth base pair of the stem. Converting the C:A mismatch to a C:G base pair increased readthrough levels to 10% (Figure 3B, PS), whereas inverting the C:A mismatch to an A:C mismatch reduced readthrough levels to less than 1% (Figure 3B, IMM). Finally, changing the sequence of the loop reduced readthrough levels to background (Figure 3B, LCa), implying that, in addition to the stem, sequences contained in the loop are important for readthrough stimulation.
To determine the ability of this stem loop readthrough element to induce stop codon readthrough in the presence of the natural SEPN1 3′UTR SECIS element, the human SEPN1 SECIS element was cloned into the 3′UTR of the dual luciferase vectors containing the UGA2 stem loop element, or the stem sequence variants SC2-1, SC2-2, and SC2-3 (see Materials and methods). UGA2 readthrough levels increased to approximately 12% with the addition of the SECIS element to the 3′UTR (Figure 3C, UGA2 SECIS). Mutations that disrupt the stem, thereby inactivating the readthrough activity of the stem loop structure (SC2-1 and SC2-2), reduced frameshifting levels to approximately 6% when tested in the presence of the SECIS (Figure 3C, SC2-1 SECIS, SC2-2 SECIS). Restoring the base-pairing potential (SC-3) revealed levels of readthrough (11%) approximating those observed for the wild-type UGA2 in the presence of the SECIS structure (Figure 3C, SC-3 SECIS). These results demonstrate that, in this reporter system, the SEPN1 SECIS element and the stem loop structure each contribute approximately equally to readthrough efficiency at the UGA2 selenocysteine-encoding codon.
Stop codon and upstream sequences
The ability of the SEPN1 sequences to induce stop codon readthrough of UAA and UAG stop codons was examined in HEK293 cells as described above. When the UGA2 stop codon was replaced with UAG, only a slight reduction in readthrough to 4.5% was observed (Figure 4B, UAG). However, low-level readthrough (<1%) was obtained when UGA was replaced with the UAA stop codon (Figure 4B, UAA).
Figure 4.
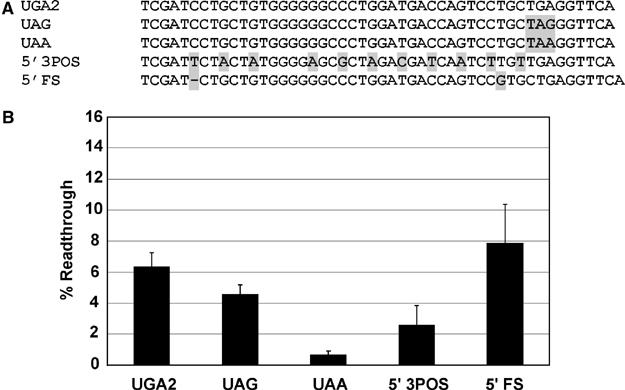
Mutagenic analysis of the human SEPN1 UGA codon and upstream flanking sequence. The human SEPN1 UGA codon located in exon 10 and surrounding sequence context was cloned into a dual luciferase reporter vector, p2luc, to produce the wild-type construct UGA2. The UGA codon was changed to UAG and UAA to test readthrough efficiency for each stop codon and selective mutations were made in the upstream region. Each construct was expressed in human HEK293 cells and percent readthrough determined. (A) The relevant sequence of the wild-type (UGA2) stem loop region is shown. Each subsequent sequence shown corresponds to a unique construct that is identical with the exception of the region highlighted in light gray. (B) Histogram of the calculated percent readthrough for each dual luciferase construct. Error bars indicate standard deviation from the mean.
The contribution of SEPN1 upstream RNA sequence to stop codon readthrough efficiency was examined by altering the third position of each codon (with the exception of the single Trp codon, which is encoded by a single codon, UGG) such that the amino-acid sequence encoded was maintained (Figure 4A, 5′ 3POS). Stop codon efficiency was reduced approximately two-fold by these changes (Figure 4B, 5′ 3POS). In a second construct, a single base was deleted from the 5′ end of the readthrough cassette and a single base was inserted three nucleotides prior to the stop codon (Figure 4A, 5′ FS). In this case, the RNA sequence is maintained essentially intact but the sequence of the nascent peptide is changed due to the shift in reading frame. Changing the composition of the nascent peptide did not significantly alter readthrough efficiency.
A significant spacer sequence
The RNA sequences located between the stop codon and the stem loop were changed to examine the effect of this spacer region on stop codon readthrough efficiency (Figure 5A). Mutagenesis of the six-nucleotide spacer, GGUUCA, was tested using the dual luciferase reporter system in HEK293 cells as described above. Comparison of the six-nucleotide SEPN1 spacer region GGUUCA to the previously identified CARYYA readthrough motif revealed a match for the last three nucleotides. Altering the last three nucleotides to AGU or ACA resulted in a modest, approximately two-fold, decrease in readthrough efficiency (Figure 5B, Spa, Spd). However, alterations that retain the YYA composition at this site, CUA or UUA, maintained readthrough levels near wild-type efficiency (Figure 5B, Spb, Spc). When the first three nucleotides of the spacer were changed to CAG and CAA to match the CARYYA motif, readthrough efficiency increased to 12 and 17%, respectively (Figure 5B, Spe, Spf). The contribution of the first three nucleotides of the spacer to readthrough efficiency was further examined by changing each nucleotide independently to a C (Figure 5A, Spg, Sph, Spi). Changing either of the first two nucleotides to a C reduced readthrough levels to 0.2 and 3%, respectively (Figure 5B, Spg, Sph), whereas changing the third nucleotide had no appreciable effect on readthrough (Figure 5B, Spi). The role of spacer length was examined by changing the length to eight nucleotides (Figure 5A, Spj, Spk, Spl) or four nucleotides (Figure 5A, Spm). Each resulted in a reduction in readthrough efficiency to approximately 2% (Figure 5B).
Figure 5.
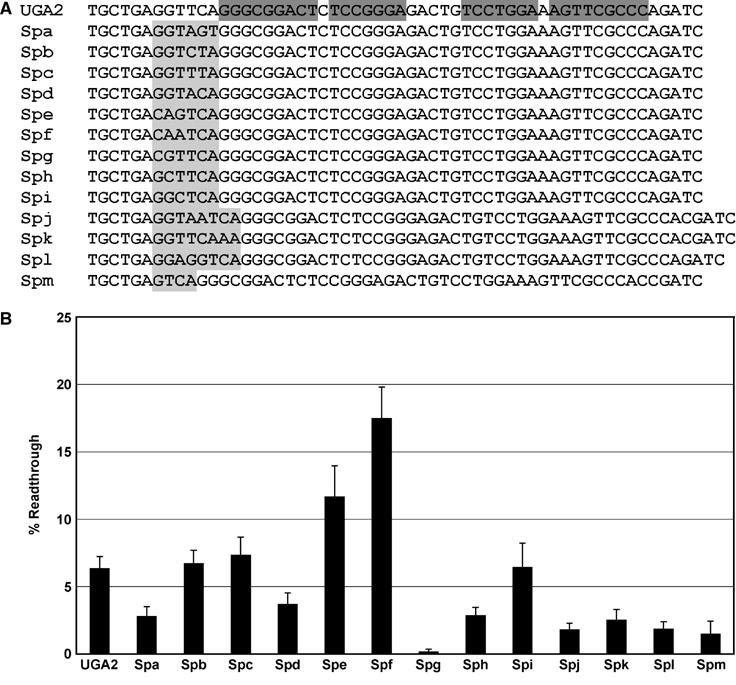
Mutagenic analysis of the human SEPN1 spacer sequence. Selective mutations were made to the spacer sequence in the wild-type construct UGA2 and each construct was expressed in human HEK293 cells and percent readthrough determined. (A) The relevant sequence of the wild-type (UGA2) stem loop region is shown, with base-paired regions highlighted in dark gray. Each subsequent sequence shown corresponds to a unique construct that is identical with the exception of the region highlighted in light gray. (B) Histogram of the calculated percent readthrough for each dual luciferase construct shown in panel A. Error bars indicate standard deviation from the mean.
The RNA element described here contains a key sequence separating the UGA codon from the essential downstream stem loop structure. Mutations designed to conform the spacer to the previously identified CARYYA motif (Skuzeski et al, 1991; Beier and Grimm, 2001; Harrell et al, 2002) resulted in higher readthrough levels, with the exception of changing the first G of the SEPN1 spacer to C, which eliminated readthrough. Those changes that were designed to reduce similarity to the known readthrough motif lowered readthrough efficiency. The spacer length is also critical and likely serves to position the structure at the predicted distance to be near the mRNA entrance site (Yusupova et al, 2001) and the ribosome mRNA unwinding center (‘helicase') (Takyar et al, 2005).
MuLV gag stop codon readthrough stimulation by the SEPN1 stem loop
Comparison of the spacer sequence of SEPN1 to that of the spacer region known to be important for MuLV readthrough reveals that the first two and last three nucleotides are identical; MuLV spacer=GGAGGUCA (Figure 5). A dual luciferase reporter was constructed that contains the MuLV stop codon (UAG) and downstream pseudoknot (Figure 6A, MuLVWT). The ability of the SEPN1 stem loop to stimulate readthrough of the MuLV stop codon was tested by replacing the MuLV pseudoknot with the SEPN1 stem loop (Figure 6A, MuLV1). Readthrough efficiency promoted by the wild-type MuLV pseudoknot was 7% and replacing the pseudoknot with the SEPN1 stem loop reduced readthrough to 1% (Figure 6B, MuLVwt, MuLV1). Reducing the spacer length to six nucleotides by replacing it with the SEPN1 spacer or deleting the first and fourth Gs of the MuLV spacer partially restored readthrough efficiency to approximately 3% (Figure 6B, MuLV2, MuLV3). Adding two nucleotides to the end or between the GGU and UCA of the SEPN1 spacer reduced readthrough of the UAG to 1 and 0.5% respectively (Figure 6B, MuLV4, MuLV7), and 2% readthrough was observed with an eight-nucleotide spacer, GGGUGUCA (Figure 6B, MuLV8). Finally, the MuLV and SEPN1 spacer regions do not induce high-level readthrough in the absence of downstream structures, as readthrough efficiency dropped to approximately 0.3% when the MuLV or SEPN1 structures were deleted entirely (Figure 6B, MuLV5, MuLV6).
Figure 6.
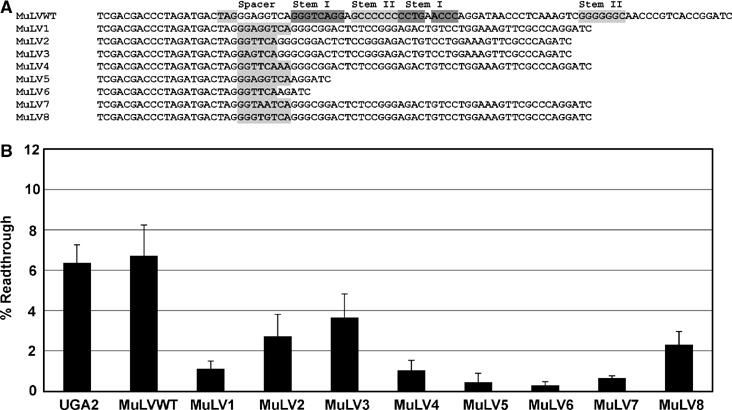
Comparison of MuLV and SEPN1 readthrough stimulators. The MuLV readthrough sequences were cloned into a dual luciferase reporter vector, p2luc, to produce the MuLVWT construct. The MuLV pseudoknot was replaced by the SEPN1 stem loop and selective mutations were made to the spacer sequences and each construct was expressed in human HEK293 cells by transient transfection and percent readthrough determined. (A) The sequence of the MuLV readthrough region (MuLVWT) is shown with the stop codon in light gray, and regions of each stem of the readthrough pseudoknot that are predicted to be base paired are shown highlighted in dark gray (Stem I) and light gray (Stem II). The MuLV pseudoknot was deleted for MuLV5/6, and replaced by the SEPN1 stem loop for MuLV1, 2, 3, 4, 7, and 8. Each subsequent sequence shown corresponds to a unique construct, with changes to the spacer region highlighted in light gray. (B) Histogram of the calculated percent readthrough for each dual luciferase construct shown in panel A. Error bars indicate standard deviation from the mean.
The strong sequence bias for the six nucleotides located downstream of redefined stop codons, experimental evidence that the spacer region in MuLV contains key sequence stimulators of gag-pol readthrough (Wills et al, 1994), and mutagenesis of the SEPN1 spacer region further illustrate that sequences located adjacent to stop codons can be utilized as a means to facilitate stop codon readthrough for gene expression purposes. In addition, structure ‘swapping' experiments here demonstrate that the SEPN1 stem loop can induce stop codon readthrough of the MuLV gag stop codon, albeit at a lower level than the wild-type MuLV pseudoknot structure.
Discussion
Phylogenetically conserved potential RNA structures have been found 3′ of a subset of eukaryal selenocysteine-specifying UGA codons. Detailed experimental characterization of one, located near the selenocysteine-encoding UGA in the SEPN1 mRNA, demonstrates it to be a potent stimulator of readthrough in a cell-based reporter system. The possibility of a stop codon redefinition element located adjacent to a eukaryal UGA/selenocysteine codon is unexpected when viewed from the context of existing models for selenocysteine incorporation, which have focused almost exclusively on the 3′UTR SECIS as the critical cis-acting element directing selenocysteine incorporation. The extent of conservation implies an important function for this element, and enhanced readthrough of selenocysteine-encoding UGA codons induced by local redefinition elements is certain to have direct consequences for protein expression from selenocysteine-encoding genes.
The SEPN1 stem loop element, when tested in the absence of the 3′UTR SECIS, induces readthrough of both UGA and UAG codons. The simplest explanation implies that inhibition of translational termination by the stem loop element, either by impeding access of termination factors or directly interacting with the ribosome, allows for near cognate tRNAs to compete more effectively for decoding of the UGA codon. Several possible tRNA species may participate in decoding UGA codons. Intermediates in the synthesis of Sec-tRNASec (Carlson et al, 2004) can decode UGA (Hatfield et al, 1982; Jung et al, 1994; Chittum et al, 1998). In addition, at low levels, some tRNATrp isoacceptors (Cordell et al, 1980; Keith and Heyman, 1990) can decode UGA codons, and cysteine and arginine can also be specified by UGA in certain contexts (Feng et al, 1990). Our observation that, in the absence of the SECIS element, a UAG codon can be redefined with nearly the same efficiency as the UGA codon argues against the idea that there is a unique standard or nonstandard aminoacyl tRNA that is recruited by the SEPN1 stem loop element.
Selenocysteine incorporation is in competition with translational termination at the UGA codon. One mechanism by which the SEPN1 stem loop structure may alter selenocysteine incorporation efficiency is indirectly through interference with translational termination as discussed above. In this scenario, selenocysteine incorporation efficiency would benefit from a reduction in translational termination at the UGA codon. Alternatively, the stem loop structure may interact directly with components of the selenocysteine incorporation machinery to affect selenocysteine incorporation rates. A recent crystal structure of selenocysteine-specific archaeal translation elongation factor SelB reveals many key features of tRNASec recognition and binding (Leibundgut et al, 2005). Of relevance to this study is the observation that domain IV of SelB, when modeled to the pretranslocation state of the ribosome, is pointed to the mRNA entrance cleft and consequently may be positioned to interact directly with the mRNA sequences or structures located downstream of the UGA codon. This observation suggests a mechanism by which the SEPN1 stem loop or other structures occurring near selenocysteine-encoding UGA codons could potentially interact directly with selenocysteine insertion factors. The bacterial bSECIS stem loop is located directly 3′ adjacent to selenocysteine-encoding UGA codons (Zinoni et al, 1990; Hüttenhofer et al, 1996), and is recognized directly by SelB (Kromayer et al, 1996; Fourmy et al, 2002). Binding of SelB to the bSECIS structure causes ribosomal pausing and can interfere with the EF-Tu decoding of the UGA codon by a suppressor tRNA (Suppmann et al, 1999). By analogy, it is possible that the SEPN1 stem loop structure facilitates selenocysteine insertion by interacting directly with mSelB, thus favoring decoding by tRNASec over other near cognate tRNAs or eukaryotic release factors.
Analysis of readthrough levels measured in the presence or absence of the 3′UTR SECIS demonstrates that each element alone can induce approximately 6% readthrough, and when both are present readthrough levels rise to 12% (Figure 3C). From these results, we can conclude that each element is contributing substantially to overall readthrough efficiency. However, as readthrough levels determined in these reporter assays do not discriminate between selenocysteine and standard amino-acid incorporation, it is premature to conclude whether the newly identified stem loop structure is modulating selenocysteine incorporation efficiency, acting independently to increase levels of a second protein product, or both. In addition, all results obtained by the use of reporter genes or transfected cDNAs must be interpreted with care, as forced expression patterns may limit trans-acting factors, or otherwise disturb critical features required for the natural role of recoding elements during selenocysteine incorporation.
The biosynthesis of selenocysteine (Leinfelder et al, 1990) does not occur free in solution but proceeds from its precursor, serine, in a tRNA-bound state (Ser-tRNASec). One of the steps is catalyzed by the SelD gene product, selenophosphate synthetase (Leinfelder et al, 1990). Eukarya have a pair of homologous genes. While the product of one of them, Sps1, does not itself contain selenocysteine (Low et al, 1995), it is interesting that the other, Sps2, does (Guimarães et al, 1996; Tamura et al, 2004). An early report looking at Sps2 cDNA expression revealed that in the absence of the 3′UTR SECIS, significant levels of full-length protein were observed (approximately 5% of those obtained with the wild-type cDNA containing the SECIS with a possible hint of 75Se labeling) (Guimarães et al, 1996). The role of SECIS-independent readthrough during Sps2 decoding was not followed up in those initial experiments. The sequence surrounding the Sps2 selenocysteine-encoding UGA was identified in this study as having the potential for a strong RNA secondary structure, in addition to the SelH, SelO, and SelT genes (see Results). Predicted structures for these additional candidates are shown in Figure 7.
Figure 7.

Predicted stem loop structures are shown for sequences locate 3′ of the selenocysteine-encoding UGA codon for each human gene. The selenocysteine-encoding UGA is boxed. Given the limitations of RNA folding algorithms, more elaborate structures, such as pseudoknots, not shown here are possible. Weak pairing at the base of SelH and SelO structures makes it difficult to predict the exact spacing distance from the UGA codon to the beginning of the stem loop.
The potential structures shown for Sps2, SelH, SelO, and SelT have varying degrees of phylogenetic conservation, with the gene and the associated structure for Sps2 being even more widely phylogenetically distributed than SEPN1 (manuscript in preparation). In contrast with SEPN1, the Sps2 structure has an extended stem and contains a rather large 12-nucleotide loop. Potential interactions between these RNA elements and downstream coding sequences, trans-acting factors, and/or the UTRs are possible mediators of function that are being investigated. The variation between these structures in distance from the UGA codon, stem length, bulges, and loop size begs the question of whether these structures play similar or diverse roles during selenoprotein expression.
Materials and methods
RNA folding and comparative sequence analysis of readthrough regions
Accession numbers for the 25 Homo sapien selenoprotein genes are as follows: Selenoprotein P, NM_005410; Sps2, NM_012248; SelW, NM_003009; SelV, NM_182704; 15Kda, NM_004261, SelM, NM_080430; TR1, NM_003330; TR2, XM_051264; TR3, NM_006440; Gpx1, NM_000581; Gpx2, NM_002083, Gpx3, NM_002084; Gpx4, NM_002085; Gpx6, NM_182701; DI1, NM_000792; DI2, NM_013989; DI3, NM_001362; SelR, NM_016332; SelT NM_016275; SelN, NM_020451; SelH, NM_170746; SelK, NM_021237; SelS, NM_203472; SelI, NM_033505; SelO, NM_031454. RNA folding parameters for mfold v3.1 are as follows: 37°C, 1 M NaCl, percent suboptimality=5, window=2, maximum interior bulge loop size=30, maximum asymmetry of an interior/bulge loop=30.
Sequences with significant similarity to SEPN1 genes were identified by BLAST analysis. Accession numbers used for multiple alignments are as follows: H. sapiens, NM_206926; Danio rerio, NM_001004294; Mus musculus, BI555369; Gallus gallus, XM_417734; Xenopus tropicalis, BC063905; Ciona intestinalis, AK112355; Rattus norvegicus, CK600139; Xenopus laevis, CF288737; Salmo salar, CK887829; Oryzias latipes, BJ734513; Bos taurus, BF073924; Strongylocentrotus purpuratus, CD330282; Pan troglodytes, AACZ01296920; Fugu rubripes, CAAB01000005; Monodelphis domesticus, AAFR01073613; Tetraodon nigroviridis, CAAE01015019; Ciona savignyi, AACT01005174. Multiple alignments of sequences were generated using ClustalW (Thompson et al, 1994) and refined manually.
Luciferase reporter constructs
Complimentary oligonucleotides, to construct the sequences utilized in this manuscript (Supplementary data), were synthesized at the University of Utah DNA/Peptide Core Facility so that when annealed they would have appropriate ends to ligate into the SalI/BamHI restriction sites of the dual luciferase vector, p2luc (Grentzmann et al, 1998). Control constructs were designed such that the UGA codon was changed to CGA. Constructs containing the SECIS element were produced by insertion of the SEPN1 SECIS element into the NotI site of the dual luciferase vector. SEPN1 SECIS insert=GGCCGCAGTGGCTTCCCCGGCAGCAGCCCCATGAT GGCTGAATCCGAAATCCTCGATGGGTCCAGCTTGA TGTCTTTGCAGCTGCACCTATGGGGCGGCC. All dual luciferase constructs were sequence verified.
Cell culture and transfections
HEK293 cell line was obtained from ATCC and maintained in DMEM supplemented with 5% FBS in the absence of antibiotics. Cells used in these studies were subcultured at 70% confluence and used between passages 7 and 15. Cells were transfected using Lipofectamine 2000 reagent (Invitrogen), using the 1-day protocol in which suspension cells are added directly to the DNA complexes in 96-well plates. In all, 25 ng DNA and 0.2 μl Lipofectamine 2000 per well in 25 μl Opti-Mem (Gibco) were incubated and plated in opaque 96-well half-area plates (Costar). Cells were trypsinized, washed, and added at a concentration of 4 × 104 cells/well in 50 μl DMEM and 10% FBS. Transfected cells were incubated overnight at 37°C in 5% CO2, then 75 μl DMEM+10% FBS were added to each well, and the plates were incubated for an additional 48 h.
Dual luciferase assay of ribosomal readthrough efficiency
Luciferase activities were determined using the Dual Luciferase Reporter Assay System (Promega). Relative light units were measured on an MLX microplate luminometer (Dynex). Transfected cells were lysed in 12.5 μl lysis buffer and light emission was measured following injection of 25 μl of luminescence reagent. Percent readthrough was calculated by comparing firefly:Renilla luciferase ratios of experimental constructs with those of control constructs: (firefly experimental RLUs/Renilla experimental RLUs)/(firefly control RLUs/Renilla control RLUs) × 100. The total number of independent experiments for each construct varies between three and 11, and for each experiment, 3–12 independent data points were obtained for each construct. For each construct, all data points were then averaged and the standard deviation calculated. Data points that fell greater than 2 standard deviations from the mean were discarded as outliers.
Supplementary Material
Supplementary data
Acknowledgments
We thank Raymond Gesteland and Norma Wills for helpful discussions. This work was supported by GM48152 to JFA who was also supported by Science Foundation Ireland and NIH R01 NS43264 (KMF and JFA). MTH was supported by an MDA Development grant.
References
- Alam SL, Wills NM, Ingram JA, Atkins JF, Gesteland RF (1999) Structural studies of the RNA pseudoknot required for readthrough of the gag-termination codon of murine leukemia virus. J Mol Biol 288: 837–852 [DOI] [PubMed] [Google Scholar]
- Allmang C, Carbon P, Krol A (2002) The SBP2 and 15.5 kD/Snu13p proteins share the same RNA binding domain: identification of SBP2 amino acids important to SECIS RNA binding. RNA 8: 1308–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier H, Grimm M (2001) Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res 29: 4767–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry MJ, Banu L, Chen YY, Mandel SJ, Kieffer JD, Harney JW, Larsen PR (1991) Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature 353: 273–276 [DOI] [PubMed] [Google Scholar]
- Berry MJ, Harney JW, Ohama T, Hatfield DL (1994) Selenocysteine insertion or termination: factors affecting UGA codon fate and complementary anticodon:codon mutations. Nucleic Acids Res 22: 3753–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson BA, Xu XM, Kryukov GV, Rao M, Berry MJ, Gladyshev VN, Hatfield DL (2004) Identification and characterization of phosphoseryl-tRNA[Ser]Sec kinase. Proc Natl Acad Sci USA 101: 12848–12853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittum HS, Lane WS, Carlson BA, Roller PP, Lung FD, Lee BJ, Hatfield DL (1998) Rabbit beta-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry 37: 10866–10870 [DOI] [PubMed] [Google Scholar]
- Copeland PR, Fletcher JE, Carlson BA, Hatfield DL, Driscoll DM (2000) A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J 19: 306–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland PR, Stepanik VA, Driscoll DM (2001) Insight into mammalian selenocysteine insertion: domain structure and ribosome binding properties of Sec insertion sequence binding protein 2. Mol Cell Biol 21: 1491–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell B, DeNoto FM, Atkins JF, Gesteland RF, Bishop JM, Goodman HM (1980) The forms of tRNATrp found in avian sarcoma virus and uninfected chicken cells have structural identity but functional distinctions. J Biol Chem 255: 9358–9368 [PubMed] [Google Scholar]
- Driscoll DM, Copeland PR (2003) Mechanism and regulation of selenoprotein synthesis. Annu Rev Nutr 23: 17–40 [DOI] [PubMed] [Google Scholar]
- Fagegaltier D, Hubert N, Yamada K, Mizutani T, Carbon P, Krol A (2000) Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J 19: 4796–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng YX, Copeland TD, Oroszlan S, Rein A, Levin JG (1990) Identification of amino acids inserted during suppression of UAA and UGA termination codons at the gag–pol junction of Moloney murine leukemia virus. Proc Natl Acad Sci USA 87: 8860–8863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng YX, Yuan H, Rein A, Levin JG (1992) Bipartite signal for read-through suppression in murine leukemia virus mRNA: an eight-nucleotide purine-rich sequence immediately downstream of the gag termination codon followed by an RNA pseudoknot. J Virol 66: 5127–5132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreiro A, Ceuterick-de Groote C, Marks JJ, Goemans N, Schreiber G, Hanefeld F, Fardeau M, Martin JJ, Goebel HH, Richard P, Guicheney P, Bonnemann CG (2004) Desmin-related myopathy with Mallory body-like inclusions is caused by mutations of the selenoprotein N gene. Ann Neurol 55: 676–686 [DOI] [PubMed] [Google Scholar]
- Ferreiro A, Quijano-Roy S, Pichereau C, Moghadaszadeh B, Goemans N, Bonnemann C, Jungbluth H, Straub V, Villanova M, Leroy JP, Romero NB, Martin JJ, Muntoni F, Voit T, Estournet B, Richard P, Fardeau M, Guicheney P (2002) Mutations of the selenoprotein N gene, which is implicated in rigid spine muscular dystrophy, cause the classical phenotype of multiminicore disease: reassessing the nosology of early-onset myopathies. Am J Hum Genet 71: 739–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourmy D, Guittet E, Yoshizawa S (2002) Structure of prokaryotic SECIS mRNA hairpin and its interaction with elongation factor SelB. J Mol Biol 324: 137–150 [DOI] [PubMed] [Google Scholar]
- Gesteland RF, Atkins JF (1996) Recoding: dynamic reprogramming of translation. Annu Rev Biochem 65: 741–768 [DOI] [PubMed] [Google Scholar]
- Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF (1998) A dual-luciferase reporter system for studying recoding signals. RNA 4: 479–486 [PMC free article] [PubMed] [Google Scholar]
- Guimarães MJ, Peterson D, Vicari A, Cocks BG, Copeland NG, Gilbert DJ, Jenkins NA, Ferrick DA, Kastelein RA, Bazan JF, Zlotnik A (1996) Identification of a novel selD homolog from eukaryotes, bacteria, and archaea: is there an autoregulatory mechanism in selenocysteine metabolism? Proc Natl Acad Sci USA 93: 15086–15091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell L, Melcher U, Atkins JF (2002) Predominance of six different hexanucleotide recoding signals 3′ of read-through stop codons. Nucleic Acids Res 30: 2011–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield D, Diamond A, Dudock B (1982) Opal suppressor serine tRNAs from bovine liver form phosphoseryl-tRNA. Proc Natl Acad Sci USA 79: 6215–6219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield DL, Gladyshev VN (2002) How selenium has altered our understanding of the genetic code. Mol Cell Biol 22: 3565–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill KE, Lloyd RS, Yang JG, Read R, Burk RF (1991) The cDNA for rat selenoprotein P contains 10 TGA codons in the open reading frame. J Biol Chem 266: 10050–10053 [PubMed] [Google Scholar]
- Hüttenhofer A, Westhof E, Bock A (1996) Solution structure of mRNA hairpins promoting selenocysteine incorporation in Escherichia coli and their base-specific interaction with special elongation factor SELB. RNA 2: 354–366 [PMC free article] [PubMed] [Google Scholar]
- Jung JE, Karoor V, Sandbaken MG, Lee BJ, Ohama T, Gesteland RF, Atkins JF, Mullenbach GT, Hill KE, Wahba AJ, Hatfield DL (1994) Utilization of selenocysteyl-tRNA[Ser]Sec and seryl-tRNA[Ser]Sec in protein synthesis. J Biol Chem 269: 29739–29745 [PubMed] [Google Scholar]
- Keith G, Heyman T (1990) Heterogeneities in vertebrate tRNAs(Trp) avian retroviruses package only as a primer the tRNA(Trp) lacking modified m2G in position 7. Nucleic Acids Res 18: 703–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmus H, Flohe L, McCarthy JE (1996) Analysis of eukaryotic mRNA structures directing cotranslational incorporation of selenocysteine. Nucleic Acids Res 24: 1195–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkov KV, Novoselov SV, Hatfield DL, Gladyshev VN (2002) Mammalian selenoprotein in which selenocysteine (Sec) incorporation is supported by a new form of Sec insertion sequence element. Mol Cell Biol 22: 1402–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kromayer M, Wilting R, Tormay P, Bock A (1996) Domain structure of the prokaryotic selenocysteine-specific elongation factor SelB. J Mol Biol 262: 413–420 [DOI] [PubMed] [Google Scholar]
- Leibundgut M, Frick C, Thanbichler M, Bock A, Ban N (2005) Selenocysteine tRNA-specific elongation factor SelB is a structural chimaera of elongation and initiation factors. EMBO J 24: 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinfelder W, Forchhammer K, Veprek B, Zehelein E, Bock A (1990) In vitro synthesis of selenocysteinyl-tRNA(UCA) from seryl-tRNA(UCA): involvement and characterization of the selD gene product. Proc Natl Acad Sci USA 87: 543–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Rice CM (1993) The signal for translational readthrough of a UGA codon in Sindbis virus RNA involves a single cytidine residue immediately downstream of the termination codon. J Virol 67: 5062–5067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low SC, Grundner-Culemann E, Harney JW, Berry MJ (2000) SECIS–SBP2 interactions dictate selenocysteine incorporation efficiency and selenoprotein hierarchy. EMBO J 19: 6882–6890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low SC, Harncy JW, Berry MJ (1995) Cloning and functional characterization of human selenophosphate synthetase, an essential component of selenoprotein synthesis. J Biol Chem 270: 21659–21664 [DOI] [PubMed] [Google Scholar]
- Martin GW III, Berry MJ (2001) SECIS elements. In Selenium. Its Molecular Biology and Role in Human Health, Hatfield D (ed) pp 45–54. Boston: Kluwer [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol 288: 911–940 [DOI] [PubMed] [Google Scholar]
- Mehta A, Rebsch CM, Kinzy SA, Fletcher JE, Copeland PR (2004) Efficiency of mammalian selenocysteine incorporation. J Biol Chem 279: 37852–37859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghadaszadeh B, Petit N, Jaillard C, Brockington M, Roy SQ, Merlini L, Romero N, Estournet B, Desguerre I, Chaigne D, Muntoni F, Topaloglu H, Guicheney P (2001) Mutations in SEPN1 cause congenital muscular dystrophy with spinal rigidity and restrictive respiratory syndrome. Nat Genet 29: 17–18 [DOI] [PubMed] [Google Scholar]
- Namy O, Rousset JP, Napthine S, Brierley I (2004) Reprogrammed genetic decoding in cellular gene expression. Mol Cell 13: 157–168 [DOI] [PubMed] [Google Scholar]
- Orlova M, Yueh A, Leung J, Goff SP (2003) Reverse transcriptase of Moloney murine leukemia virus binds to eukaryotic release factor 1 to modulate suppression of translational termination. Cell 115: 319–331 [DOI] [PubMed] [Google Scholar]
- Philipson L, Andersson P, Olshevsky U, Weinberg R, Baltimore D, Gesteland R (1978) Translation of MuLV and MSV RNAs in nuclease-treated reticulocyte extracts: enhancement of the gag-pol polypeptide with yeast suppressor tRNA. Cell 13: 189–199 [DOI] [PubMed] [Google Scholar]
- Robinson DN, Cooley L (1997) Examination of the function of two kelch proteins generated by stop codon suppression. Development 124: 1405–1417 [DOI] [PubMed] [Google Scholar]
- Rother M, Resch A, Gardner WL, Whitman WB, Bock A (2001) Heterologous expression of archaeal selenoprotein genes directed by the SECIS element located in the 3′ non-translated region. Mol Microbiol 40: 900–908 [DOI] [PubMed] [Google Scholar]
- Skuzeski JM, Nichols LM, Gesteland RF, Atkins JF (1991) The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J Mol Biol 218: 365–373 [DOI] [PubMed] [Google Scholar]
- Suppmann S, Persson BC, Bock A (1999) Dynamics and efficiency in vivo of UGA-directed selenocysteine insertion at the ribosome. EMBO J 18: 2284–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takyar S, Hickerson RP, Noller HF (2005) mRNA helicase activity of the ribosome. Cell 120: 49–58 [DOI] [PubMed] [Google Scholar]
- Tamura T, Yamamoto S, Takahata M, Sakaguchi H, Tanaka H, Stadtman TC, Inagaki K (2004) Selenophosphate synthetase genes from lung adenocarcinoma cells: Sps1 for recycling L-selenocysteine and Sps2 for selenite assimilation. Proc Natl Acad Sci USA 101: 16162–16167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dam EB, Pleij CW, Bosch L (1990) RNA pseudoknots: translational frameshifting and readthrough on viral RNAs. Virus Genes 4: 121–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tujebajeva RM, Copeland PR, Xu XM, Carlson BA, Harney JW, Driscoll DM, Hatfield DL, Berry MJ (2000a) Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep 1: 158–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tujebajeva RM, Ransom DG, Harney JW, Berry MJ (2000b) Expression and characterization of nonmammalian selenoprotein P in the zebrafish, Danio rerio. Genes Cells 5: 897–903 [DOI] [PubMed] [Google Scholar]
- Ursini F, Heim S, Kiess M, Maiorino M, Roveri A, Wissing J, Flohe L (1999) Dual function of the selenoprotein PHGPx during sperm maturation. Science 285: 1393–1396 [DOI] [PubMed] [Google Scholar]
- Walczak R, Carbon P, Krol A (1998) An essential non-Watson–Crick base pair motif in 3′UTR to mediate selenoprotein translation. RNA 4: 74–84 [PMC free article] [PubMed] [Google Scholar]
- Walczak R, Westhof E, Carbon P, Krol A (1996) A novel RNA structural motif in the selenocysteine insertion element of eukaryotic selenoprotein mRNAs. RNA 2: 367–379 [PMC free article] [PubMed] [Google Scholar]
- Wills NM, Gesteland RF, Atkins JF (1991) Evidence that a downstream pseudoknot is required for translational read-through of the Moloney murine leukemia virus gag stop codon. Proc Natl Acad Sci USA 88: 6991–6995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills NM, Gesteland RF, Atkins JF (1994) Pseudoknot-dependent read-through of retroviral gag termination codons: importance of sequences in the spacer and loop 2. EMBO J 13: 4137–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaka Y, Katoh I, Copeland TD, Oroszlan S (1985) Murine leukemia virus protease is encoded by the gag-pol gene and is synthesized through suppression of an amber termination codon. Proc Natl Acad Sci USA 82: 1618–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupova GZ, Yusupov MM, Cate JH, Noller HF (2001) The path of messenger RNA through the ribosome. Cell 106: 233–241 [DOI] [PubMed] [Google Scholar]
- Zinoni F, Heider J, Bock A (1990) Features of the formate dehydrogenase mRNA necessary for decoding of the UGA codon as selenocysteine. Proc Natl Acad Sci USA 87: 4660–4664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data


