Abstract
Similar to that of other herpesviruses, Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) lytic replication destroys the host cell, while the virus can persist in a latent state in synchrony with the host. During latency only a few genes are transcribed, and the question becomes one of what determines latent versus lytic gene expression. Here we undertake a detailed analysis of the latency-associated nuclear antigen (LANA [orf73]) promoter (LANAp). We characterized a minimal region that is necessary and sufficient to maintain high-level transcription in all tissues tested, including primary endothelial cells and B cells, which are the suspected natural host for KSHV. We show that in transient-transfection assays LANAp mimics the expression pattern observed for the authentic promoter in the context of the KSHV episome. Unlike other KSHV promoters tested thus far, LANAp is not affected by tetradecanoyl phorbol acetate or viral lytic cycle functions. It is, however, subject to control by LANA itself and cellular regulatory factors, such as p53. This is in contrast to the K14/vGCR (orf74) promoter, which overlaps LANAp and directs transcription on the opposite strand. We isolated a minimal cis-regulatory region sufficient for K14/vGCR promoter activity and show that it, too, mimics the regulation observed for the authentic viral promoter. In particular, we demonstrate that its activity is absolutely dependent on the immediate-early transactivator orf50, the KSHV homolog of the Epstein-Barr virus Rta transactivator.
Using representational difference analysis Chang et al. (6) demonstrated the presence of a novel human virus in Kaposi's sarcoma (KS) biopsy samples: Kaposi's sarcoma-associated herpesvirus (KSHV), also called human herpesvirus 8. KSHV has since been detected in all manifestations of KS as well as in two lymphoproliferative disorders: primary effusion lymphoma (4) and multicentric Castleman's disease (53). On the basis of the complete sequence of the 137-kbp double-stranded DNA genome, KSHV is classified as a gamma-2 herpesvirus, a member of the lymphotropic subgroup of the Herpesviridae (17, 36, 45).
The epidemiological evidence implicating KSHV as a causative agent for KS is compelling (reviewed in reference 51). (i) KSHV DNA is found in >90% of KS biopsy samples. (ii) KSHV latent mRNAs and proteins are detectable in every KS spindle cell by in situ methods. (iii) Antibodies to KSHV exist in ≥80% of KS patients, and multiple viral antigens have been identified as targets of this response. (iv) Increases in peripheral-blood viral load as well as anti-KSHV antibody titer precede the onset of disease and correlate with increased risk for KS. These observations establish KSHV as a necessary cofactor for KS.
KSHV, like all herpesviruses, displays two modes of replication: lytic replication, during which the host cell is destroyed and viral progeny are released, and latent replication, during which the viral genome persists indefinitely and no viral progeny are released. In KS, KSHV persists latently in ≥90% of tumor cells (40, 54). Only a subset of viral genes is transcribed during KSHV latency (47, 63), while lytic gene expression and replication are induced in response to outside stimuli (5, 31, 42). In the related Epstein-Barr virus (EBV), latency-associated genes are essential for episome maintenance and host cell transformation (reviewed in reference 43). The KSHV latent proteins LANA (latency-associated nuclear antigen [also known as orf73]) and k-cyclin (orf72) similarly have been implicated in KSHV episome persistence and oncogenesis (2, 12, 14, 22, 32).
We and others previously identified two 3′-coterminal, coregulated latency-associated transcripts that encode open reading frames (ORFs) with sequence homology to cellular growth regulatory proteins (9, 40, 47, 56). These are v-FLIP/orf71 (a putative apoptosis inhibitor), k-cyclin/orf72 (the viral cyclin D homolog), and the KSHV LANA orf73. The larger, 5,400-nucleotide (nt) mRNA contains all three ORFs, while the smaller, spliced, 1,700-nt mRNA contains k-cyclin and v-FLIP. Thus far no mRNA encoding just v-FLIP has been described, suggesting that v-FLIP might be translated by internal ribosome entry. Both mRNAs were detected in every KS spindle cell by in situ hybridization (8, 9, 40). These very same transcripts were also identified after experimental infection of SCID-hu Thy/Liv mice (10), indicating that their pattern of transcription typifies KSHV latent infection in vivo, rather than that of a particular tumor cell line in culture. Unlike other latent mRNAs analyzed to date, these latency-associated transcripts are not induced during KSHV lytic replication (9, 47, 56). Therefore, they are likely to play a unique role in viral pathogenesis, which motivates our analysis of their promoter.
Here, we present a detailed analysis of the LANA promoter (LANAp) and its regulation by viral (LANA) and cellular (p53) trans-acting factors. We also identify the cis-regulatory region for the K14/vGCR. The start site of the bicistronic K14/vGCR mRNA partly overlaps with LANA mRNA and originates on the opposite strand in opposite orientation (19). We demonstrate that the K14/vGCR promoter (K14p) is responsive to the KSHV immediate-early transactivator orf50 and that orf50 alone is sufficient for high-level K14p activity in KSHV-negative cells. Together LANAp and K14p exemplify all the regulatory features of KSHV gene regulation within a 1,000-bp stretch of the viral genome. Hence, their analysis represents a unique opportunity to delineate the fundamental mechanism of KSHV lytic versus latent gene regulation.
MATERIALS AND METHODS
Cell lines.
HeLa, CV-1, SLK, NIH 3T3, and 293 cells (all from the American Type Culture Collection) were maintained in Dulbecco's modified Eagle medium (DMEM) (Cellgrow, Inc.) supplemented with 10% calf serum (Gibco-BRL), 2 mM l-glutamine, penicillin (0.05 μg/ml), and streptomycin (5 U/ml; Gibco-BRL) at 37°C under 5% CO2. SLK cells are KS tumor-derived cells that grow indefinitely in culture and exhibit cobblestone, i.e., endothelial cell, morphology. They do not contain KSHV. SAOS-2, (10)1 (courtesy of G. Zambetti, St. Jude Children's Hospital), and BHK (American Type Culture Collection) cells were maintained in DMEM supplemented with 15% fetal bovine serum (Gibco-BRL), penicillin (0.05 μg/ml), and streptomycin (5 U/ml; Gibco-BRL) at 37°C under 5% CO2. Cells were passaged at subconfluency (approximately every 3 days) in order to maintain a 3T3-like phenotype (16, 57). A fresh aliquot was thawed every 30 passages. LnCAP, BJAB, and BCBL-1 cells were cultured in a solution containing RPMI-1860, 25 mM HEPES (pH 7.55), 10% fetal bovine serum, 0.05 mM 2-mercaptoethanol, 1 mM sodium pyruvate, 2 mM l-glutamine, penicillin (0.05 μg/ml), and streptomycin (50 U/ml) (all Gibco-BRL) at 37°C under 5% CO2. Cells were split every 5 days to 2 · 105 cells/ml.
Real-time RT-PCR.
Quantitative DNA analysis and reverse transcription (RT)-PCR were carried out in duplicate using Taqman RT and Taqman PCR with Amplitaq Gold reagents (PE Biosystems Inc.). RT was carried out using a 2.5 μM concentration of random hexamers at 25°C for 10 min, 48°C for 30 min, and 95°C for 5 min. Real-time PCR was carried out using universal cycle conditions (2 min at 50°C, 10 min at 95°C, and then 40 cycles of 15 s at 95°C and 1 min at 60°C) on an ABI PRIZM 7000 sequence detector (18). Primers are described in Table 1. To prevent contamination, all PCRs were assembled in a segregated space in which neither KSHV virions nor cloned KSHV DNA was handled. Carryover of the amplification product was avoided using positive displacement pipettes and UNGglycosylase in the amplification reaction mixture (24).
TABLE 1.
Primers used for the detection of latent and lytic transcripts by quantitative RT-PCR
| Target and primer | Sequence |
|---|---|
| orf29a | |
| tac29-5F | 5′-CCCGGAGGACGGTCCA |
| tac29-68R | 5′-CCCCGAATGCTCTGTTCTTATT |
| tac29-22T | 5′-FAM-CTCGCTGATGTGCGCAACATGCT-TAMRA |
| k-cyclina | |
| lat-335R | 5′-CAGGTTCTCCCATCGACGA |
| TAQ-F1 | 5′-AGGCAGCTGCGCCACGAAGCA |
| lat-294T | 5′-FAM-TAGCGTACTCTCGCGGCCCAGC-TAMRA |
| GAPDHa | |
| gapdhF | 5′-GAAGGTGAAGGTCGGAGTC |
| gapdhR | 5′-GAAGATGGTGATGGGATTTC |
| gapdhP | 5′-VIC-CAAGCTTCCCGTTCTCAGCC-TAMRA |
| orf50b | |
| orf50s1 | 5′-CACAAAAATGGCGCAAGATGA |
| orf50a4 | 5′-TGGTAGAGTTGGGCCTTCAGTT |
| orf50p | 5′-FAM-AGAAGCTTCGGCGGTCCTG-TAMRA |
| vGCR | |
| gerF | 5′-TGGCCCAAACGGAGGATCCTAG |
| gerR | 5′-AGTTTCATTCCAGGATTCATCATC |
| gerP | 5′-FAM-AGAAGATGGTTAGGAAATCCTCGGC-TAMRA |
Plasmids.
The source of genomic DNA for all clones was a KSHV genomic lambda library derived from a KS lesion (63). All nucleotide sequence positions are according to the numbering of Russo et al. (45). pDD130B contains a 1,879-bp PCR fragment (nt 127300 to 129179) amplified with primer 7326 (5′-TCGGGAAAGCTTGTCTGACA; nt 127300 to 127319 with an engineered HindIII site [underlined]) and with primer 7327 (5′-ctcgagCGGCCGCTAGCTTGTCACTCCCCTGA; nt 129161 to 129179 with XhoI [lowercase letters], NotI [underlined], and NheI [boldface type] sites) and inserted into pCR 2.1 (Invitrogen, Inc.). PCR was performed using Ready-To-Go PCR beads (Amersham) under the following conditions: 30 cycles of 30 s at 94°C, 1 min at 56°C, and 2 min at 74°C followed by 10 min at 74°C. pDD121B (nt 127539 to 128164) is derived from pDD130B by internal deletion of a 1,013-bp NheI fragment. pDD124 was constructed by subcloning the 1,879-bp HindIII/NotI fragment of pDD130B into pβgeo (courtesy of Limin Li, University of California—San Francisco). pDD125 was derived from pDD124 by an internal NheI deletion. pDD41 was constructed by subcloning the 1,763-bp NcoI fragment (nt 127607 to 129370) into the NcoI site of pGLbasic (Promega); pDD43 contains the same fragment in the opposite orientation. pDD53 and pDD83 were derived from pDD41 by an internal SmaI and an internal NheI deletion, respectively. pDD213, pDD268, and pDD270 were constructed by PCR amplification of a 373-bp fragment (nt 127546 to 127919), a 389-bp fragment (nt 127546 to 127935), and a 422-bp fragment (nt 127546 to 127968) from pDD39 using primer 7304 (5′-AGTCCTGGTGGCTCACCTGCC) and primer 7306 (5′-GCGGCGCCCGGGAC AATC), primer 7304 and primer 7331 (5′-CTCCGCCCTCCACTAC), and primer 7304 and primer 7332 (5′-AGCTGCCTCCAAATGATACACA), respectively. PCR products were gel purified and cloned into pCR2.1 (Invitrogen, Inc.). pDD271 contains a 349-bp NcoI/XhoI fragment of pDD213 in pGLbasic; pDD272 contains a 365-bp NcoI/XhoI fragment of pDD268 in pGLbasic. pDD274 contains a 398-bp NcoI/XhoI fragment of pDD270 in pGLbasic. pDD154 contains a 586-bp PCR fragment (nt 127297 to 127883) in pCRII-topo (Invitrogen, Inc.). pDD159 contains a 583-bp PCR fragment (nt 127300 to 127883) in pCRII-topo in the opposite direction. pDD163 and pDD168 were derived from pDD154 and pDD159 by subcloning the respective HindIII/XhoI fragments into pGL3basic (Promega, Inc.). pDD383 is a SacI deletion mutant of pDD163, with nucleotides 127300 to 127394 removed. pDD395 is an NheI/AvrII deletion mutant of pDD163, with nt 127300 to 27616 removed. All plasmids were sequenced in both orientations at the Oklahoma University Health Sciences Center sequencing core facility.
Transfection.
At day 1, cells were seeded to 2 · 105 cells/10 ml/100-mm-diameter dish, 105 cells/3 ml/35-mm-diameter dish (6-well plate), or 2 · 104 cells/0.5 ml/well (12-well plate) to reach 50% confluency after 24 h. At day 2, 2,000 ng of total DNA together with 400 ng of pDD173 (pCDNA3.1-hislacZ; Invitrogen) was suspended in 200 μl of DMEM (no serum, no antibiotics) and 7.5 μl of Superfect (Qiagen, Inc.) was added. To minimize variability all plasmids were kept as 100-ng/μl stock solutions in Tris-EDTA, and normalizing plasmid was added to bulk DMEM before aliquoting for single transfection. The total DNA concentration was held constant to 2,400 ng by adding appropriate amounts of filler plasmid (pBluescript KS). We verified (data not shown) that our transfection data fell within the linear range of the assay using pDD83 reporter plasmid. Increasing amounts of reporter resulted in a commensurate increase in luciferase activity (normalized for transfection efficiency using 400 ng of cotransfected lacZ reporter plasmid). As little as 500 ng of pDD83 was sufficient to detect significant luciferase activity, while overall transfection efficiency dropped when more than 2,500 ng of DNA was added. The transfection mixture was incubated for 30 min at room temperature. For transfection in 35-mm-diameter dishes, the volume was adjusted to 1 ml with complete medium (DMEM, 10% fetal calf serum [FCS]). Cells were washed once, and 1 ml of transfection mixture per 35-mm-diameter dish was added. For transfection in 12-well plates, the volume was adjusted to 1.5 ml with complete medium (DMEM, 10% FCS). Cells were washed once with DMEM and 0.5 ml of transfection mixture was added per triplicate well. For transfection in 10-cm dishes, the volume was adjusted to 3 ml with complete medium (DMEM, 10% FCS). Cells were washed once with DMEM, and transfection mixture was added. Cells were incubated overnight at 37°C under 5% CO2, and complete medium was exchanged. Luciferase activity was measured at 72 h after transfection using the Promega luciferase kit in a Turner TD20/20 luminometer according to the manufacturers' instructions.
RESULTS
Real-time quantitative RT-PCR distinguishes between KSHV latent and lytic mRNAs.
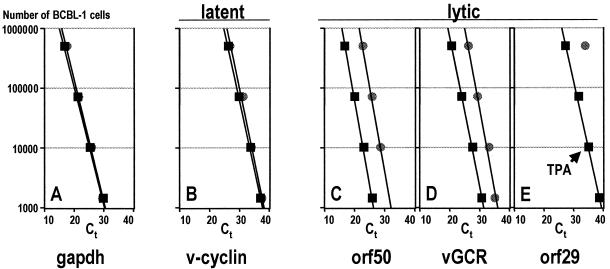
The distinction between lytic and latent mRNAs is fundamental to herpesvirus gene regulation. For KSHV Sarid et al. (47) described three classes of differentially transcribed messages in KSHV-infected BC-1 cells. Class I mRNAs can be detected in untreated (latent) cells and are not increased after 12-O-tetradecanoylphorbol-13-acetate (TPA) treatment, which reactivates KSHV. Class II mRNAs can be detected in untreated cells and are greatly increased after TPA treatment. Finally, class III mRNAs can only be detected in TPA-induced cells. Translated into the customary herpesvirus classification (44), class III mRNAs include lytic (alpha, beta, and gamma) and class I latent transcripts. Class II mRNAs present a conundrum, since it is not clear whether the mRNA detected in uninduced cells stems from the approximately 3% of cells that at any given time undergo spontaneous lytic replication or whether they represent latent mRNAs that are also induced during lytic replication. Here we employ quantitative real-time RT-PCR to assign KSHV mRNAs to their respective class: lytic or latent. Real-time quantitative PCR measures the amount of PCR product that is generated at each PCR cycle using a fluorescently labeled oligonucleotide which only fluoresces upon annealing to the amplified product. So-called Ct values indicate the cycle at which the fluorescence crosses a particular threshold. Hence, Ct values indicate the abundance of any given mRNA on a log scale. A low Ct value represents a highly abundant target mRNA. BCBL-1 cells are latently infected with KSHV and can be induced to release KSHV by TPA (42). Cells were either treated with TPA or mock treated, and total RNA was isolated at 72 h after induction. We then performed real-time quantitative RT-PCR on a dilution series of cells using primers listed in Table 1. It is important to note that all KSHV primers used in this experiment cross intron-exon borders and therefore do not amplify viral DNA. Figure 1 shows the result of our analysis. For all primers the real-time quantitative RT-PCR signal was linear over 4 orders of magnitude. The amount of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA remained unchanged in the presence or absence of TPA (Fig. 1A). This is indicated by the corresponding Ct values. The GAPDH Ct values for TPA-treated cells overlap the Ct values for mock-treated cells at every cell concentration tested. Hence, the input RNA concentration is the same. An identical pattern was observed using primers specific for the KSHV k-cyclin mRNA (Fig. 1B). Again, the Ct values and regression lines for TPA- and mock-treated samples superimpose. This establishes, quantitatively, that k-cyclin mRNA is transcribed independently of TPA or KSHV lytic genes, corroborating its classification as a latent transcript. Next, we tested the transcription pattern of orf50 and K14/vGCR, which are the subject of our subsequent analysis (Fig. 1C and D). Their real-time PCR analysis revealed a marked difference to the k-cyclin data. Ct values obtained from TPA-treated cells were much lower than those for mock-treated cells, indicating that orf50 and vGCR were induced upon TPA treatment. At the extreme end, quantitative real-time RT-PCR for orf29 (Fig. 1E) failed to detect any mRNA unless at least 500,000 latent BCBL-1 cells were used as input, while orf29 mRNA was readily detected in 1,000 TPA-treated cells. This corroborates earlier data (5, 10), which showed orf29 to be a true gamma-2 transcript. We calculated the induction to be 500-fold (calculated as exp[Ct(TPA) − Ct(mock)], where Ct(TPA) and Ct(mock) are Ct values for TPA- and mock-treated cells, respectively). Previous reports showed that about 2% of BCBL-1 cells undergo spontaneous lytic reactivation (63). Consistent with this observation, 106 mock-treated BCBL-1 cells are needed to give a Ct value equivalent to 104 TPA-induced cells in the orf29, orf50, and vGCR assay (Fig. 1C). Note that a quantitative comparison is only valid when the same primer pair is used but not between different probes. This is because the amplification efficiency may be different for each primer pair. In sum, these experiments represent the first quantitative analysis of LANA mRNA expression and introduce a new assay to classify KSHV mRNAs. Important to our studies, it quantitatively establishes K14/vGCR as a lytic (TPA-inducible) and LANA/v-cyclin as a latent (resistant to TPA induction) message.
FIG. 1.
Real-time quantitative RT-PCR analysis of endogenous (gapdh), latent (v-cyclin), and lytic (orf50, vGCR, orf29) mRNAs in TPA-treated (squares) or mock-treated (circles) BCBL-1 cells. The number of cells per reaction is indicated on the vertical axis, and the Ct values at which PCR products accumulated 5 standard deviations above background are indicated on the horizontal axis.
Fine mapping of LANAp.
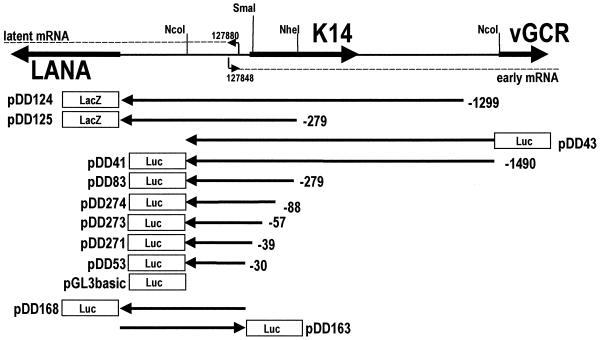
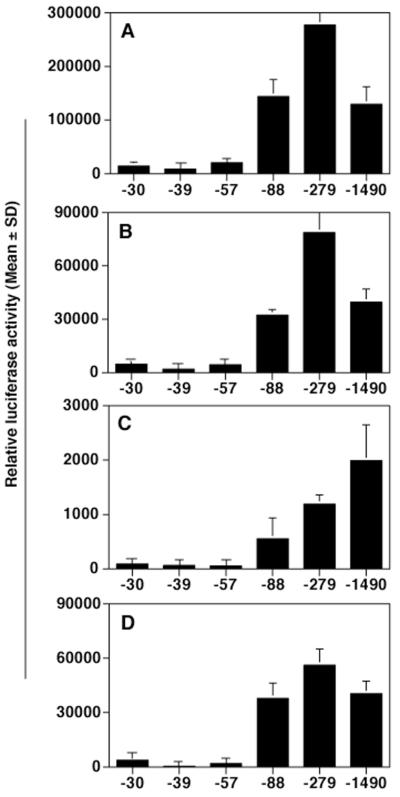
We and others previously identified the transcription start site for the LANA (orf73) and k-cyclin (orf72) mRNAs (9, 47, 56). The major transcription start site as determined by nuclease protection analysis and 5′ rapid amplification of cDNA ends is at nt 127880, with less-prominent transcripts initiating at nt 127900 and 127948 (47). Since no consensus TATA box was evident 30 bp upstream of nt 127880 but at nt 127934, the hypotheses were proposed either that the LANAp is TATA independent (like EBV EBNA Qp [37, 49]) or that the start site represents premature termination of the reverse transcriptase reaction. To identify the minimal cis sequence necessary for basal promoter activity we undertook an extended deletion analysis. Figure 2 schematizes the structure of our deletion clones. Plasmids pDD124 and pDD125 contain the entire 5′ untranslated region (5′-UTR) from the LANA translation initiation site at nt 127296 fused to lacZ and extend out to position −1299 (nt 129179) and position −279, respectively (nucleotide positions refer to GenBank accession number U75698 [45]). Both plasmids exhibited promoter activity (data not shown). We therefore used our previously published plasmids (9) together with additional deletion mutants to define the minimal LANAp cis sequence. Plasmids pDD41 to pDD53 contain progressive deletions and use the Ncol site at position +271 (nt 127609) to drive a luciferase reporter gene. Luciferase assays were performed in triplicate, and transfection efficiency was controlled by cotransfection with a constitutive lacZ expression plasmid. Figure 3 depicts the result of our promoter analysis in 293, SLK, (10)1, and BJAB cells. In all cell lines tested, deletion clones up to plasmid pDD274 extending just to position −88 exhibited significant promoter activity. In contrast, pDD273 extending to position −55 had lost all ability to drive reporter gene transcription. Reporter pDD83, extending to position −273, showed maximal expression in the three fibroblast cell lines, while in BJAB cells we observed a gradual decrease in promoter activity proportional to the length of the upstream region. These experiments establish that cis elements between positions −55 (nt 127935) and −88 (nt 127968) are essential for minimal LANAp activity, whereas sequences up to position −273 are needed for robust expression in some instances.
FIG. 2.
Overview of the LANA/vGCR promoter region and the constructs that were employed in this study.
FIG. 3.
Minimal region of the LANAp necessary for high level transcription. 293 (A), (10)1 (B), BJAB (C), and SLK (D) cells were transfected with the indicated plasmids. Data represent the mean relative luciferase activity after 72 h of triplicate experiments normalized for transfection efficiency using a cotransfected β-galactosidase reporter (error bars, standard deviations).
To determine whether additional, perhaps KSHV-specific, sequence elements were located further upstream, we compared the promoter activity of pDD83 (position −273) to that of pDD44 (position −1490) in several additional cell lines: HeLa, LnCAP, or KSHV-positive BCBL-1 and BC-3 cells (Table 2). Independent of the cell line, pDD41 and pDD83 showed 5- to 20-fold higher activity than vector alone or pDD53. Plasmid pDD43, which contains the reporter in the opposite direction of pDD41, showed no activity. Overall, LANAp activity was significantly higher than that of a simian virus 40 minimal promoter (pGL3 promoter; Invitrogen). Except in BC-3 cells no significant differences could be discerned between LANAp extending to positions −279 (pDD83) and −1490 (pDD41), suggesting that pDD83 contains all the cis elements that are necessary for LANAp function. Therefore, we decided to use pDD83 in all subsequent experiments.
TABLE 2.
LANAp activity in various cell lines
| Plasmid | Mean fold induction ± SD in cell linea:
|
|||
|---|---|---|---|---|
| BCBL-1 | BC-3 | LnCAP | HeLa | |
| pDD41 (−1490) | 28 ± 6 | 27 ± 4 | 100 ± 30 | 40 ± 10 |
| pDD43 (rev) | 8 ± 1 | 1b | 10 ± 10 | 1 |
| pDD83 (−279) | 22 ± 3 | 7 ± 2 | 160 ± 20 | 50 ± 10 |
| pDD53 (−30) | 2 ± 1 | 1 | 1 | 1 |
| Simian virus 40 promoter | 4 ± 1 | 2 ± 1 | 1 | 20 ± 20 |
| Vector | 1 | 1 | 1 | 1 |
Shown are fold inductions ± SDs compared to vector control for triplicate experiments normalized to a cotransfected secreted alkaline phosphatase vector for various cell lines.
The numeral 1 indicates luciferase activity at or below that of the vector control.
LANAp activity is independent of other KSHV functions.
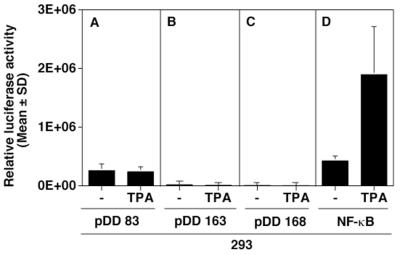
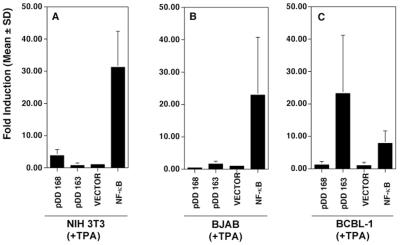
Unlike other KSHV mRNAs, LANA message is not upregulated after induction of the KSHV lytic cycle by phorbol ester, butyrate, or gamma interferon (5, 9, 47, 48, 56). To determine whether the isolated LANAp maintained this property, we transfected 293 cells in the presence or absence of TPA. Figure 4A shows that the activity of the pDD83 promoter fragment remained the same regardless of the presence or absence of TPA. This is in contrast to the activity of an NF-κB reporter (Fig. 4D) which was dramatically induced by TPA. Figure 4B and C show negative controls and indicate the lack of discernible luciferase activity when the LANA 5′-UTR fragment (pDD168) or K14/vGCR promoter (pDD163) fused to luciferase was transfected. This establishes that LANAp is not affected by cellular signaling pathways, which induce KSHV immediate early proteins.
FIG. 4.
Unresponsiveness of the LAT promoter to TPA. 293 cells were transfected with the indicated plasmids in either the absence or the presence of TPA (20 ng/ml). Data represent the mean fold luciferase activity relative to vector alone after 72 h of triplicate experiments normalized for transfection efficiency using a cotransfected β-galactoside reporter (error bars, standard deviations). Shown are the activity of the LANAp promoter (pDD83) (A) and those of the vGCR promoter (pDD163) and antisense control (pDD168), respectively (B and C). (D) TPA-induced induction of the NF-κB reporter, which is used as a positive control.
To verify that these results also apply to B cells, we performed a similar experiment in BJAB cells, which are negative for EBV and KSHV. Figure 5C shows fold induction of LANAp with increasing concentrations of TPA. LANAp activity did not change. In this experiment we also transfected the KSHV gB promoter (26) into BJAB cells as a control (Fig. 5D). Since the KSHV gB promoter is a true late promoter it was similarly not induced by TPA in this KSHV-negative cell line. Note that the activity of LANAp in the absence of TPA is 10-to 100-fold higher than that of gB in either cell line (data not shown). Hence, we calculated fold induction in either case.
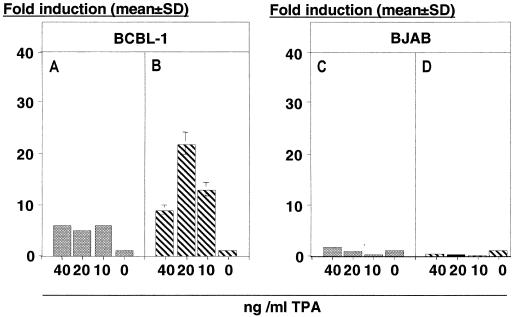
FIG. 5.
Activity of the LANA promoter under conditions of latent or lytic KSHV replication. Bars represent the fold induction, 72 h after TPA treatment, of transfected BCBL-1 cells (A and B) or BJAB cells (C and D). Neither LANAp nor a lytic promoter (gB) is induced in the absence of KSHV (C and D). As expected, the lytic promoter is induced by TPA in a dose-dependent manner in BCBL-1 (B), but the latent promoter LANAp is not (A).
What would happen in the presence of the virus and its complete set of immediate-early, early, and late proteins? To address this question, we transfected BCBL-1 cells. BCBL-1 cells harbor latent, episomal KSHV that can be induced to undergo lytic replication in response to TPA (42). This viral isolate is fully functional, since virions isolated from BCBL-1 cells are infectious in culture (11, 20, 29, 33, 41) as well as in vivo (10). TPA at 20 ng/ml represents the optimal dose for the induction of lytic KSHV replication (5, 42). The KSHV gB promoter exhibited a TPA dose-dependent increase in activity (Fig. 5B), demonstrating (i) that our TPA was biologically active and (ii) that KSHV transactivating functions are induced in BCBL-1 cells. In contrast, LANAp showed no significant increase in activity (Fig. 5A). Although suboptimal amounts of TPA upregulated the LANAp somewhat, no dose-response curve was obtained. The twofold induction by TPA might be the result of the well-documented increase in transfection efficiency in the presence of phorbol esters or butyrate (28). This establishes that in transient-transfection assays LANAp is indifferent to cellular or viral immediate-early, early, or late functions, mimicking the behavior of LANA mRNA.
Identification of K14/vGCR promoter.
The LANA mRNA +1 site is located within the K14 ORF, which is oriented in the opposite direction (Fig. 2). Recently, Kirshner et al. (19) showed that K14 and vGCR are contained on a bicistronic, spliced message that initiates within the LANA 5′-UTR at nt 127848. In other words, nt 127848 to 127880 are part of the LANA mRNA as well as—in antisense orientation—part of the K14/vGCR mRNA. The K14/vGCR mRNA, however, is an early transcript in KSHV and was never detected in latently infected BCBL-1 cells or KS tumors (19). To determine the minimal cis-acting region necessary to direct K14/vGCR transcription and regulation, we cloned the complete intervening region (nt 127296 to 127883) in both orientations upstream of a luciferase reporter. We then transfected 293 cells with the putative K14/vGCR 5′-proximal region (pDD163, vGCRp) containing the K14/vGCR start site at nt 127848 in the presence or absence of TPA. We did not detect any luciferase activity (Fig. 4B). In the same experiment transfection of a reporter under control of NF-κB response elements exhibited TPA-inducible activity, establishing the functionality of our assay (Fig. 4D). As expected, we failed to observe promoter activity when we transfected the construct containing the same region in the antisense orientation (pDD168, antisense vGCR, Fig. 4C). This evidences (i) that there is no cryptic latent promoter located in the LANA 5′-UTR downstream of +1 and (ii) that the 5′-proximal fragment of the K14/v-GCR promoter does not function in 293 cells, despite the presence of a predicted TATA box 30 bp upstream of +1.
To test the hypothesis that the K14/vGCR promoter might be tissue specific, we transfected NIH 3T3 cells (Fig. 6A) and BJAB cells (Fig. 6B) in the presence of TPA. The K14/vGCR promoter (pDD163) failed to exhibit any significant activity in either cell line compared to the antisense orientation control (pDD168) or vector (pGL3basic). This is in contrast to the human immunodeficiency virus (HIV) long terminal repeat (LTR), which served as a positive control. The result changed drastically when we assayed these constructs in KSHV-positive BCBL-1 cells (Fig. 6C). The putative K14/vGCR promoter fragment showed approximately 20-fold higher reporter activity compared to the antisense control or vector. In TPA-treated BCBL-1 cells this promoter was even stronger than the HIV LTR. This identifies a cis-regulatory region that is sufficient to function as a K14/vGCR promoter. Furthermore, it demonstrates that vGCR promoter activity is absolutely dependent on KSHV transactivating functions, which is consistent with the regulation of authentic K14/vGCR mRNA.
FIG. 6.
The activity of the lytic vGCR promoter in the presence of TPA in different cell lines. NIH 3T3 (A), BJAB (B), and BCBL-1 (C) cells were transfected with the indicated plasmids containing the vGCR promoter in either the sense (pDD163) or antisense (pDD168) orientation with respect to the reporter gene and stimulated with TPA (20 ng/ml). Bars represent the mean fold induction after 72 h normalized for transfection efficiency using a cotransfected β-galactoside reporter in triplicate (error bars, standard deviations). In panel C the difference in mean between pDD163 and pDD168 is significant (P ≤ 0.05).
orf50 transactivates the K14/vGCR promoter.
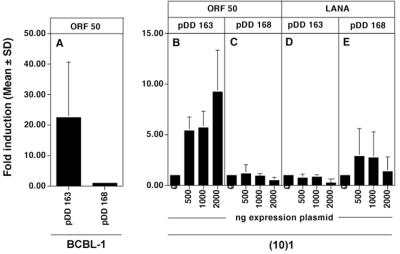
Having established that, on the one hand, the LANAp fragment recapitulated the constitutive expression pattern previously observed for LANA mRNA and that, on the other hand, the K14/vGCR promoter was critically dependent on KSHV transactivation functions, we tried to identify transactivating factors specific for either promoter. First, we asked the question, is the K14/vGCR inducible by KSHV orf50? The immediate-early protein encoded by orf50 is the EBV R homolog of KSHV (25, 51, 55, 64). KSHV orf50 is necessary and sufficient to initiate complete lytic viral replication (15, 26, 58). We transfected BCBL-1 cells (Fig. 7A) with an orf50 expression plasmid and the K14/vGCR promoter directed toward the reporter gene (pDD163) or directed away from it (pDD168). orf50 induced expression from the K14/vGCR reporter more then 20-fold, whereas the same fragment located in the opposite direction had no activity (Fig. 7A). This implies that KSHV orf50 induces the vGCR promoter either directly or through other KSHV transactivating functions.
FIG. 7.
Regulation of the K14/vGCR promoter by orf50. (10)1 and BCBL-1 cells were transfected with plasmid pDD267 expressing orf50 together with reporter plasmids containing the vGCR promoter in sense (pDD163) or antisense (pDD168) orientation relative to the luciferase gene. Bars represent the mean fold induction after 72 h of three transfections (error bars, standard deviations). (A) Positive regulation of K14/vGCR promoter by orf50 in BCBL-1 cells. (B) Dose-dependent activity of K14/vGCR promoter by orf50. (C) Antisense orientation control. (D and E) Results of cotransfection with a LANA expression plasmid. In panel A the difference in mean between pDD163 and pDD168 is significant (P ≤ 0.05).
To determine whether orf50 alone was sufficient to activate the vGCR promoter we repeated the previous experiment in KSHV-negative cells. Shown are the results of one such experiment performed in (10)1 fibroblasts (Fig. 7); similar data were obtained in SLK cells (data not shown). Cotransfection of increasing amounts of orf50 expression plasmids increased K14/vGCR promoter activity in a dose-dependent manner (Fig. 7B). At maximum, 10-fold induction could be observed in the presence, compared to the absence, of orf50, while no effect was observed on the antisense orientation control plasmid (Fig. 7C). In the same experiment cotransfection of LANA had no effect (Fig. 7D and E). Since these experiments were performed in fibroblasts, which do not harbor KSHV, this establishes that orf50 alone is sufficient to activate the K14/vGCR promoter.
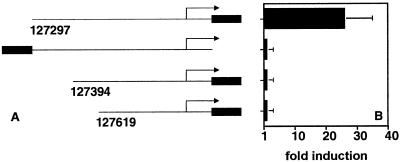
To determine whether orf50 activates the K14/vGCR promoter through interaction with the basal transcription machinery or whether the orf50 response is mediated through a separate cis site, we undertook a deletion analysis. The results are shown in Fig. 8. Deletion of the distal-most 97 bp (nt 127297 to 127394) completely abolished orf50 responsiveness, as did deletion of a larger (nt 127297 to 127619) part of the promoter. This maps the orf50 element to a 97-bp region at position −454 of the transcription start site for the K14/vGCR message.
FIG. 8.
Identification of the orf50 response element in the K14/vGCR promoter. 293 cells were transfected with plasmid pDD267 expressing orf50 together with reporter plasmids containing the vGCR promoter in the sense (pDD163) or antisense (pDD168) orientation relative to the luciferase gene as well as the indicated deletion constructs. Bars represent the mean fold induction after 72 h of three transfections (error bars, standard deviations).
Regulation of LANAp by LANA and p53.
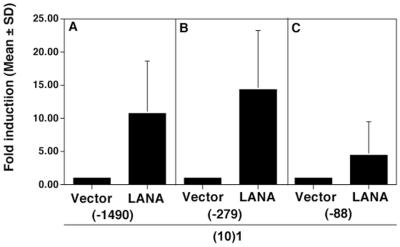
Renne et al. present preliminary evidence that LANA positively autoregulates its own promoter, while at the same time inhibiting the HIV LTR (40a). We extend these experiments here, by narrowing the region for the presumed LANA responsive element. Figure 9 shows the results of transfecting LANA with the LANAp deletion constructs. Compared to vector alone, LANA increases transcription of plasmids pDD41 (position −1490) and pDD83 (position −279). In contrast, LANA did not significantly affect the minimal latent promoter pDD274 (position −88). As shown in Fig. 7D and E, LANA did not activate or repress the K14/vGCR promoter. This demonstrates promoter specificity for LANA transactivation and locates the LANA responsive element to a 200-nt region between position −88 and −279.
FIG. 9.
Regulation of LANAp by LANA. (10)1 cells were cotransfected with the indicated plasmids and an empty expression vector or a LANA expression vector. Bars represent the mean fold induction after 72 h of three experiments (error bars, standard deviations).
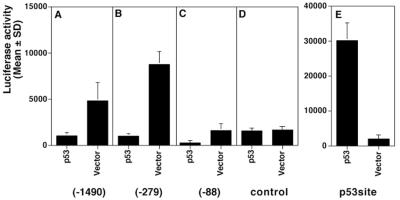
Recently, Friborg et al. showed that LANA could bind to the p53 tumor suppressor protein and could suppress the p53 transactivating function (12). Here we asked the converse question, namely, whether p53 could regulate the LANAp. Figure 10 shows that human p53 suppressed LANAp (position −1490 or −279) activity (Fig. 10A and B) compared to the vector control (Fig. 10D). This holds true to a lesser extent for the minimal promoter fragment pDD274 (position −89 [Fig. 10C]), suggesting that p53 exerts its effect on the basal transcriptional apparatus. Unfortunately, in (10)1 cells pDD274 basal activity is much lower than for the two larger promoter fragments. Consequently, fold repression in this case is marginal. These transfections were conducted in the established p53-negative (10)1 cell line (61). Following the protocol described in the initial reports on p53-mediated suppression (23, 27, 52), we chose to normalize for total protein concentration rather than rely on a cotransfected reporter. This was mainly for two reasons: (i) a wide range of promoters is suppressed or activated by p53, including the cytomegalovirus promoter-enhancer, which we used as transfection control in all other experiments; (ii) any additional promoter-enhancer could influence our results by binding cellular factors. We did not observe toxicity in transfected cells, and we verified the functionality of our p53 expression plasmid using a consensus p53 reporter construct (Fig. 10E). These experiments raise the possibility for p53's involvement in the regulation of KSHV latency.
FIG. 10.
Inhibition of the LANA promoter by p53. (10)1 cells were cotransfected with the indicated promoter plasmids and an empty expression vector or a p53 expression vector. Bars represent the mean fold repression after 72 h of three experiments (error bars, standard deviations).
DISCUSSION
This paper takes a close look at the cis-regulatory elements of the major KSHV latency promoter, namely, LANAp. According to Sarid et al. (47), LANA and v-cyclin mRNAs are the only class I message in the entire KSHV genome. They are the only mRNAs not induced by signals that induce the viral lytic cycle, such as TPA (42), butyrate (60), or gamma interferon (5). Other transcripts are present during latency such as Kaposin (34, 46), K15 (38), interleukin 6 (35) (in some instances), and vIRF-1 (3, 21). Yet their transcription is induced after induction of the KSHV lytic cycle, while we showed using quantitative real-time RT-PCR that LANA/v-cyclin mRNA is not (Fig. 1). Although only in situ analysis can assess whether even LANA/v-cyclin mRNA might be induced in a subset of PEL cells, this induction would be much less in magnitude than that observed for class II transcripts. This points to a unique mode of regulation of the LANA promoter LANAp, which warrants a detailed analysis.
Here we show that a minimal region starting at position −88 from the transcription start site at nt 127880 is necessary and sufficient for constitutive LANAp activity in all tissues tested (Fig. 3; Table 2), while elements up to position −273 are needed for robust expression. LANAp showed no responsiveness to TPA, neither in KSHV-negative cells nor in KSHV-positive cells (Fig. 4 and 5). As such, our data agree with other studies (9, 47, 56). Within our panel of cell lines we find no evidence for tissue-specific differences in basal LANAp activity. However, LANAp activity in BJAB cells is proportional to the amount of upstream sequence, which is not the case in the three other cell lines. This suggests the possibility of B-cell-specific enhancer elements located distal to position −279. The identification of such putative elements is currently ongoing. Since TPA induces proliferation in lymphoid cells (13) and in the BCBL-1 cell line induces cellular as well as KSHV replication, we conclude that in this cell type the LANAp promoter is not cell cycle regulated.
Whether herpesvirus cis sequences have the same regulatory propensity in isolation as in the context of the viral genome is much debated. While alpha- and betaherpesvirus gamma-2 promoters seem to be strictly dependent on the contextual information of the entire genome (44), that requirement is relaxed for gammaherpesviruses (30, 39). For KSHV, we demonstrated that a number of cis sequences in isolation faithfully mimicked the regulatory pattern of the corresponding viral gene in the context of the KSHV genome. In particular, the latent LANA promoter is unaffected by any lytic KSHV gene products, whereas the late (gamma) gB promoter-construct is critically dependent on them (Fig. 5). This is in agreement with Lukac et al. (25), who showed that gB is not induced by orf50 alone, the principal KSHV immediate-early transactivator. It suggests that additional viral factors are needed for gB promoter activity. In contrast, we isolated the promoter for the KSHV K14/vGCR ORFs. Not much is known about the product of K14 other than that it possesses predicted homology to the human OX-2 protein (36, 45). The vGCR/orf74 protein, however, has been the subject of intense scrutiny because of its transforming activity (1, 59). It has been particularly difficult to reconcile vGCR's oncogenic phenotype with its expression pattern. By in situ analysis (19) vGCR mRNA is present only in lytically infected cells. Like v-FLIP(orf71) the vGCR ORF is preceded by an upstream ORF (K14) on a bicistronic mRNA, calling into question whether vGCR protein is made at all and how its translation would be regulated. Like for v-FLIP(orf71), an internal ribosomal entry site might also operate between K14 and vGCR. In BCBL-1 cells the bicistronic K14/vGCR mRNA is induced with early kinetics (19), and data presented here show that the K14/vGCR promoter is absolutely dependent on viral transactivators (Fig. 6) and KSHV orf50 in particular (Fig. 7). We mapped the orf50 response element to a 97-bp region (nt 127297 to 127394) distal to the K14/vGCR transcription initiation site (Fig. 8). In sum, this report establishes three cis sequences, representing latent (LANAp), early (vGCRp), and late (gB) regulation. This will allow us to dissect the corresponding regulatory pathways and identify relevant trans-acting factors.
We investigated three transactivating proteins that are relevant to the regulation of LANAp and the overlapping vGCR promoter: LANA, p53, and orf50. So far the only known KSHV sequences that can bind LANA protein are located near the terminal repeats and are implicated in episomal maintenance (2, 7). We show that LANA autoregulates its own promoter by narrowing the region of LANA-mediated regulation to nt 127969 to 128159. Whether LANA binds directly to DNA in this region is the subject of current investigations. By contrast, no effect of LANA on the K14/vGCR promoter could be observed (Fig. 7). This argues for a role of LANA in regulating the transcription of specific genes, rather than modulating overall promoter activity in transfected cells. Recently, LANA has been shown to bind to and inactivate the human tumor suppressor protein p53 (12), suggesting that it might block p53-dependent signaling pathways. We find that p53 suppresses the LANA promoter and localized the suppression to or very near the transcription initiation site (Fig. 10). Although the suppressive effect is small, it is entirely consistent with the magnitude of p53-TATA binding protein-mediated suppression of cellular promoters (23, 27, 52). Presumably, the exact ratio of transcriptionally competent p53 to LANA will determine the outcome of this regulatory loop in latently infected cells.
Finally, in vGCRp we added another promoter to the list of KSHV early promoters (25) which are dependent on orf50 (Fig. 7 and 8). Ectopic expression of orf50 induces KSHV early gene transcription as well as complete lytic replication (15, 25, 51, 55, 58, 62, 64), whereas a dominant-negative orf50 counteracts induction of the lytic cycle (25). This establishes orf50 as necessary and sufficient to induce KSHV replication, a property that it shares with MHV68 orf50 (58). Although a consensus binding site for LANA or orf50 cannot yet be readily extracted from the cis regions discussed here, the question of how two differently expressed, overlapping mRNAs (LANA and K14/vGCR) initiate within 32 bp of each other on the opposite strand is now open to experimental analysis.
ACKNOWLEDGMENTS
This project was supported by a beginning-grant-in-aid from the American Heart Association.
K. Smith is acknowledged for expert technical help. We thank R. Renne, D. Ganem, D. Lukac, and G. Zambetti for plasmids and cell lines and E. Howard, R. Renne, and Don Ganem for critical reading of the manuscript.
REFERENCES
- 1.Bais C, Santomasso B, Coso O, Arvanitakis L, Raaka E G, Gutkind J S, Asch A S, Cesarman E, Gershengorn M C, Mesri E A, Gerhengorn M C. G-protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature. 1998;391:86–89. doi: 10.1038/34193. . (Erratum, 392:210.) [DOI] [PubMed] [Google Scholar]
- 2.Ballestas M E, Chatis P A, Kaye K M. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 3.Burysek L, Yeow W S, Lubyova B, Kellum M, Schafer S L, Huang Y Q, Pitha P M. Functional analysis of human herpesvirus 8-encoded viral interferon regulatory factor 1 and its association with cellular interferon regulatory factors and p300. J Virol. 1999;73:7334–7342. doi: 10.1128/jvi.73.9.7334-7342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 5.Chang J, Renne R, Dittmer D, Ganem D. Inflammatory cytokines and the reactivation of Kaposi's sarcoma-associated herpesvirus lytic replication. Virology. 2000;266:17–25. doi: 10.1006/viro.1999.0077. [DOI] [PubMed] [Google Scholar]
- 6.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 7.Cotter M A, II, Robertson E S. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology. 1999;264:254–264. doi: 10.1006/viro.1999.9999. [DOI] [PubMed] [Google Scholar]
- 8.Davis M A, Sturzl M A, Blasig C, Schreier A, Guo H G, Reitz M, Opalenik S R, Browning P J. Expression of human herpesvirus 8-encoded cyclin D in Kaposi's sarcoma spindle cells. J Natl Cancer Inst. 1997;89:1868–1874. doi: 10.1093/jnci/89.24.1868. [DOI] [PubMed] [Google Scholar]
- 9.Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J Virol. 1998;72:8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dittmer D, Stoddart C, Renne R, Linquist-Stepps V, Moreno M E, Bare C, McCune J M, Ganem D. Experimental transmission of Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) to SCID-hu Thy/Liv mice. J Exp Med. 1999;190:1857–1868. doi: 10.1084/jem.190.12.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foreman K E, Friborg J, Jr, Kong W P, Woffendin C, Polverini P J, Nickoloff B J, Nabel G J. Propagation of a human herpesvirus from AIDS-associated Kaposi's sarcoma. N Engl J Med. 1997;336:163–171. doi: 10.1056/NEJM199701163360302. [DOI] [PubMed] [Google Scholar]
- 12.Friborg J, Jr, Kong W, Hottiger M O, Nabel G J. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 1999;402:889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- 13.Gelfand E W, Mills G B, Cheung R K, Lee J W, Grinstein S. Transmembrane ion fluxes during activation of human T lymphocytes: role of Ca2+, Na+/H+ exchange and phospholipid turnover. Immunol Rev. 1987;95:59–87. doi: 10.1111/j.1600-065x.1987.tb00500.x. [DOI] [PubMed] [Google Scholar]
- 14.Godden-Kent D, Talbot S J, Boshoff C, Chang Y, Moore P, Weiss R A, Mittnacht S. The cyclin encoded by Kaposi's sarcoma-associated herpesvirus stimulates cdk6 to phosphorylate the retinoblastoma protein and histone H1. J Virol. 1997;71:4193–4198. doi: 10.1128/jvi.71.6.4193-4198.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gradoville L, Gerlach J, Grogan E, Shedd D, Nikiforow S, Metroka C, Miller G. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J Virol. 2000;74:6207–6212. doi: 10.1128/jvi.74.13.6207-6212.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harvey D M, Levine A J. p53 alteration is a common event in the spontaneous immortalization of primary BALB/c murine embryo fibroblasts. Genes Dev. 1991;5:2375–2385. doi: 10.1101/gad.5.12b.2375. [DOI] [PubMed] [Google Scholar]
- 17.Hayward G S. KSHV strains: the origins and global spread of the virus. Semin Cancer Biol. 1999;9:187–199. doi: 10.1006/scbi.1998.0116. [DOI] [PubMed] [Google Scholar]
- 18.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 19.Kirshner J R, Staskus K, Haase A, Lagunoff M, Ganem D. Expression of the open reading frame 74 (G-protein-coupled receptor) gene of Kaposi's sarcoma (KS)-associated herpesvirus: implications for KS pathogenesis. J Virol. 1999;73:6006–6014. doi: 10.1128/jvi.73.7.6006-6014.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kliche S, Kremmer E, Hammerschmidt W, Koszinowski U, Haas J. Persistent infection of Epstein-Barr virus-positive B lymphocytes by human herpesvirus 8. J Virol. 1998;72:8143–8149. doi: 10.1128/jvi.72.10.8143-8149.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M, Lee H, Guo J, Neipel F, Fleckenstein B, Ozato K, Jung J U. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor. J Virol. 1998;72:5433–5440. doi: 10.1128/jvi.72.7.5433-5440.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, Lee H, Yoon D W, Albrecht J C, Fleckenstein B, Neipel F, Jung J U. Kaposi's sarcoma-associated herpesvirus encodes a functional cyclin. J Virol. 1997;71:1984–1991. doi: 10.1128/jvi.71.3.1984-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Berk A J. Reversal of in vitro p53 squelching by both TFIIB and TFIID. Mol Cell Biol. 1995;15:6474–6478. doi: 10.1128/mcb.15.11.6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Longo M C, Berninger M S, Hartley J L. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene. 1990;93:125–128. doi: 10.1016/0378-1119(90)90145-h. [DOI] [PubMed] [Google Scholar]
- 25.Lukac D M, Kirshner J R, Ganem D. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J Virol. 1999;73:9348–9361. doi: 10.1128/jvi.73.11.9348-9361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lukac D M, Renne R, Kirshner J R, Ganem D. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology. 1998;252:304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- 27.Mack D H, Vartikar J, Pipas J M, Laimins L A. Specific repression of TATA-mediated but not initiator-mediated transcription by wild-type p53. Nature. 1993;363:281–283. doi: 10.1038/363281a0. [DOI] [PubMed] [Google Scholar]
- 28.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 29.Mesri E A, Cesarman E, Arvanitakis L, Rafii S, Moore M A, Posnett D N, Knowles D M, Asch A S. Human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus is a new transmissible virus that infects B cells. J Exp Med. 1996;183:2385–2390. doi: 10.1084/jem.183.5.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov V M, Grossberg S, Chang Y. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol. 1997;71:314–324. doi: 10.1128/jvi.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monini P, Colombini S, Sturzl M, Goletti D, Cafaro A, Sgadari C, Butto S, Franco M, Leone P, Fais S, Melucci-Vigo G, Chiozzini C, Carlini F, Ascherl G, Cornali E, Zietz C, Ramazzotti E, Ensoli F, Andreoni M, Pezzotti P, Rezza G, Yarchoan R, Gallo R C, Ensoli B. Reactivation and persistence of human herpesvirus-8 infection in B cells and monocytes by Th-1 cytokines increased in Kaposi's sarcoma. Blood. 1999;93:4044–4058. [PubMed] [Google Scholar]
- 32.Moore P S, Chang Y. Kaposi's sarcoma-associated herpesvirus-encoded oncogenes and oncogenesis. J Natl Cancer Inst Monogr. 1998;1998(23):65–71. doi: 10.1093/oxfordjournals.jncimonographs.a024176. [DOI] [PubMed] [Google Scholar]
- 33.Moses A V, Fish K N, Ruhl R, Smith P P, Strussenberg J G, Zhu L, Chandran B, Nelson J A. Long-term infection and transformation of dermal microvascular endothelial cells by human herpesvirus 8. J Virol. 1999;73:6892–6902. doi: 10.1128/jvi.73.8.6892-6902.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muralidhar S, Pumfery A M, Hassani M, Sadaie M R, Kishishita M, Brady J N, Doniger J, Medveczky P, Rosenthal L J. Identification of kaposin (open reading frame K12) as a human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) transforming gene. J Virol. 1998;72:4980–4988. doi: 10.1128/jvi.72.6.4980-4988.1998. . (Erratum, 73:2568, 1999.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neipel F, Albrecht J C, Ensser A, Huang Y Q, Li J J, Friedman-Kien A E, Fleckenstein B. Human herpesvirus 8 encodes a homolog of interleukin-6. J Virol. 1997;71:839–842. doi: 10.1128/jvi.71.1.839-842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neipel F, Albrecht J C, Fleckenstein B. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nonkwelo C, Skinner J, Bell A, Rickinson A, Sample J. Transcription start sites downstream of the Epstein-Barr virus (EBV) Fp promoter in early-passage Burkitt lymphoma cells define a fourth promoter for expression of the EBV EBNA-1 protein. J Virol. 1996;70:623–627. doi: 10.1128/jvi.70.1.623-627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poole L J, Zong J C, Ciufo D M, Alcendor D J, Cannon J S, Ambinder R, Orenstein J M, Reitz M S, Hayward G S. Comparison of genetic variability at multiple loci across the genomes of the major subtypes of Kaposi's sarcoma-associated herpesvirus reveals evidence for recombination and for two distinct types of open reading frame K15 alleles at the right-hand end. J Virol. 1999;73:6646–6660. doi: 10.1128/jvi.73.8.6646-6660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ragoczy T, Miller G. Role of the Epstein-Barr virus RTA protein in activation of distinct classes of viral lytic cycle genes. J Virol. 1999;73:9858–9866. doi: 10.1128/jvi.73.12.9858-9866.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rainbow L, Platt G M, Simpson G R, Sarid R, Gao S-J, Stoiber H, Herrington C S, Moore P S, Schulz T F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40a.Renne R, Barry C, Dittmer D, Compitello N, Brown P O, Ganem D. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J Virol. 2001;75:458–468. doi: 10.1128/JVI.75.1.458-468.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renne R, Lagunoff M, Zhong W, Ganem D. The size and conformation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J Virol. 1996;70:8151–8154. doi: 10.1128/jvi.70.11.8151-8154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 43.Rickenson A B, Kief E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Virology. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2397–2446. [Google Scholar]
- 44.Roizman B. Herpesviriddae. In: Fields B N, Knipe D M, Howley P M, editors. Virology. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2221–2230. [Google Scholar]
- 45.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sadler R, Wu L, Forghani B, Renne R, Zhong W, Herndier B, Ganem D. A complex translational program generates multiple novel proteins from the latently expressed kaposin (K12) locus of Kaposi's sarcoma-associated herpesvirus. J Virol. 1999;73:5722–5730. doi: 10.1128/jvi.73.7.5722-5730.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarid R, Flore O, Bohenzky R A, Chang Y, Moore P S. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol. 1998;72:1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarid R, Wiezorek J S, Moore P S, Chang Y. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J Virol. 1999;73:1438–1446. doi: 10.1128/jvi.73.2.1438-1446.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaefer B C, Strominger J L, Speck S H. Redefining the Epstein-Barr virus-encoded nuclear antigen EBNA-1 gene promoter and transcription initiation site in group I Burkitt lymphoma cell lines. Proc Natl Acad Sci USA. 1995;92:10565–10569. doi: 10.1073/pnas.92.23.10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schulz T F. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) J Gen Virol. 1998;79:1573–1591. doi: 10.1099/0022-1317-79-7-1573. [DOI] [PubMed] [Google Scholar]
- 51.Seaman W T, Ye D, Wang R X, Hale E E, Weisse M, Quinlivan E B. Gene expression from the ORF50/K8 region of Kaposi's sarcoma-associated herpesvirus. Virology. 1999;263:436–449. doi: 10.1006/viro.1999.9963. [DOI] [PubMed] [Google Scholar]
- 52.Seto E, Usheva A, Zambetti G P, Momand J, Horikoshi N, Weinmann R, Levine A J, Shenk T. Wild-type p53 binds to the TATA-binding protein and represses transcription. Proc Natl Acad Sci USA. 1992;89:12028–12032. doi: 10.1073/pnas.89.24.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 54.Staskus K A, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson D J, Ganem D, Haase A T. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun R, Lin S F, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J Virol. 1999;73:2232–2242. doi: 10.1128/jvi.73.3.2232-2242.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Talbot S J, Weiss R A, Kellam P, Boshoff C. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology. 1999;257:84–94. doi: 10.1006/viro.1999.9672. [DOI] [PubMed] [Google Scholar]
- 57.Todaro G J, Green H. 3T3 cells. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu T-T, Usherwood E J, Stewart J P, Nash A A, Sun R. Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J Virol. 2000;74:3659–3667. doi: 10.1128/jvi.74.8.3659-3667.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang T Y, Chen S C, Leach M W, Manfra D, Homey B, Wiekowski M, Sullivan L, Jenh C H, Narula S K, Chensue S W, Lira S A. Transgenic expression of the chemokine receptor encoded by human herpesvirus 8 induces an angioproliferative disease resembling Kaposi's sarcoma. J Exp Med. 2000;191:445–454. doi: 10.1084/jem.191.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu Y, Black J B, Goldsmith C S, Browning P J, Bhalla K, Offermann M K. Induction of human herpesvirus-8 DNA replication and transcription by butyrate and TPA in BCBL-1 cells. J Gen Virol. 1999;80:83–90. doi: 10.1099/0022-1317-80-1-83. [DOI] [PubMed] [Google Scholar]
- 61.Zambetti G P, Quartin R S, Martinez J, Georgoff I, Momand J, Dittmer D, Finlay C A, Levine A J. Regulation of transformation and the cell cycle by p53. Cold Spring Harbor Symp Quant Biol. 1991;56:219–225. doi: 10.1101/sqb.1991.056.01.027. [DOI] [PubMed] [Google Scholar]
- 62.Zhang L, Chiu J, Lin J C. Activation of human herpesvirus 8 (HHV-8) thymidine kinase (TK) TATAA-less promoter by HHV-8 ORF50 gene product is SP1 dependent. DNA Cell Biol. 1998;17:735–742. doi: 10.1089/dna.1998.17.735. [DOI] [PubMed] [Google Scholar]
- 63.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci USA. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu F X, Cusano T, Yuan Y. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J Virol. 1999;73:5556–5567. doi: 10.1128/jvi.73.7.5556-5567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]