Abstract
Six predicted Kaposi's sarcoma virus herpesvirus (KSHV) proteins have homology with other well-characterized herpesvirus core DNA replication proteins and are expected to be essential for viral DNA synthesis. Intact Flag-tagged protein products from all six were produced from genomic expression vectors, although the ORF40/41 transcript encoding a primase-helicase component proved to be spliced with a 127-bp intron. The intracellular localization of these six KSHV replication proteins and the mechanism of their nuclear translocation were investigated. SSB (single-stranded DNA binding protein, ORF6) and PPF (polymerase processivity factor, ORF59) were found to be intrinsic nuclear proteins, whereas POL (polymerase, ORF9), which localized in the cytoplasm on its own, was translocated to the nucleus when cotransfected with PPF. PAF (primase-associated factor, ORF40/41), a component of the primase-helicase tripartite subcomplex together with PRI (primase, ORF56) and HEL (helicase, ORF44), required the presence of all five other replication proteins for efficient nuclear translocation. Surprisingly, even in the absence of a lytic cycle replication origin (ori-Lyt) and any known initiator or origin binding protein, the protein products of all six KSHV core replication genes cooperated in a transient cotransfection assay to form large globular shaped pseudo-replication compartments (pseudo-RC), which excluded cellular DNA. These pseudo-RC structures were confirmed to include POL, SSB, PRI, and PAF but did not contain any newly synthesized DNA. Similar to the human cytomegalovirus system, the peripheries of these KSHV pre-RC were also found to be surrounded by punctate PML oncogenic domains (PODs). Furthermore, by transient cotransfection, the six KSHV core replication machinery proteins successfully replicated a plasmid containing EBV ori-Lyt in the presence of the Epstein-Barr virus-encoded DNA binding initiator protein, ZTA. The KSHV-encoded K8 (ORF-K8) protein, which is a distant evolutionary homologue to ZTA, was incorporated into pseudo-RC structures formed by transient cotransfection with the six core KSHV replication genes. However, unlike ZTA, K8 displayed a punctate nuclear pattern both in transfected cells and at early stages of lytic infection and colocalized with the cellular PML proteins in PODs. Finally, K8 was also found to accumulate in functional viral RC, detected by incorporation of pulse-labeled bromodeoxyuridine into newly synthesized DNA in both tetradecanoyl phorbol acetate-induced JSC-1 primary effusion lymphoblasts and in KSHV lytically infected endothelial cells.
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV), also called human herpesvirus 8, is a gamma-2 class herpesvirus that is distantly related to herpesvirus saimiri (HVS) and Epstein-Barr virus (EBV) but contains several novel loci (9, 12, 50, 58). KSHV DNA is present in virtually all tumor samples of classical, endemic, and AIDS-associated forms of KS (12), as well as in peripheral blood mononuclear cells in up to 50% of homosexual AIDS patients with KS (72). KSHV is also present in a limited subset of AIDS-associated lymphoproliferative disorders referred to as primary effusion lymphomas (PELs), as well as in a large proportion of cases of multicentric Castleman's disease, but is rare in normal blood donors except in Africa (7, 8, 65).
Gammaherpesviruses characteristically establish latent infections in lymphoid cells. The initiation of the viral lytic cycle of KSHV in PEL cells can be induced by various chemicals, such as tetradecanoyl phorbol acetate (TPA) and n-butyrate (48, 57). In EBV, a critical event in the reactivation of the lytic cycle is the expression of both the ZTA (BZLF1) and RTA (BRLF1) proteins, both known DNA binding transcriptional activators, whose mRNAs are among the first viral mRNAs expressed after reactivation in latently infected B cells (17, 29, 36, 39, 41, 75). Reactivation of the whole EBV lytic cycle process including the expression from the delayed-early EBV promoters required for viral DNA replication can be activated by either ZTA or RTA (15, 30, 33, 55, 75).
In EBV, lytic cycle DNA replication depends on EBV-encoded core replication proteins, whereas latent state DNA replication requires the cellular DNA replication machinery (74). Unlike controlled plasmid-state EBV latent replication, which depends on EBNA-1 binding to multiple binding sites in ori-P (56), in the lytic cycle, multiple rounds of DNA replication are initiated from within a different origin, ori-Lyt (origin of lytic DNA replication), and require many viral gene products (28). In a cotransfection-replication assay first developed for herpes simplex virus (HSV) (10, 73), EBV ori-Lyt was used to demonstrate the functional requirement for six EBV-encoded core lytic replication proteins, POL (polymerase; BALF5), PPF (polymerase processivity factor; BMRF1), SSB (single-stranded DNA binding protein; BALF2), PRI (primase; BSLF1), HEL (helicase; BBLF4), and PAF (primase-associated factor; BBLF2/3) (22), as well as ZTA (21). The EBV-encoded MTA (BMLF1) and RTA (BRLF1) proteins also acted as nonessential accessory proteins in these assays. ZTA binds as a homodimer to both AP-1 sites and related sequences called ZTA response elements (ZREs) that are present in both ori-Lyt and key lytic cycle promoters (6, 13, 20, 39, 41, 68) and stabilizes the formation of a DNA-bound complex containing the basal transcription factors TFIID and TFIIA (16, 40). In addition to transcriptional regulation, ZTA also has a role in lytic cycle reactivation, serving as an essential initiatior protein for replication of ori-Lyt (21, 60, 62). In fact, ZTA is implicated as an origin binding protein (OBP) because it is indispensable for the ori-Lyt-dependent replication in transient cotransfection-replication assays (21), and deletion of specific ZRE motifs within ori-Lyt abolishes its ability to replicate (62, 63).
Except for ZTA, the other essential EBV replication protein genes have sequence and positional homology with the known core DNA replication protein genes of HSV and human cytomegalovirus (HCMV) (14, 18, 47, 53). Their functions are sufficiently conserved that the six core proteins of HSV can replicate EBV ori-Lyt in the presence of ZTA (21) and those of EBV can replicate HCMV ori-Lyt in the presence of the UL84 initiator protein (61).
In all herpesviruses, infected cells at late stages of the lytic cycle display large globular or kidney-shaped subnuclear domains which exclude the nucleolus and are filled with a specific set of viral replication proteins and display active viral progeny genome DNA synthesis. These herpesvirus subnuclear domains are known as viral DNA replication compartments (RC). In HSV, the assembly of complete and functionally active viral RC was detected in cells receiving all six HSV core DNA replication genes (UL5, UL8, UL42, UL52, UL30, and UL29) as well as HSV ori-S DNA and the specific ori-S and ori-L DNA binding protein OBP (UL9) in transient DNA cotransfection assays with bromodeoxyuridine (BrdU) incorporation (45, 76). The isolated genes for these same seven proteins had already been shown to be essential and sufficient for the specific amplification of cotransfected bacterial plasmid DNA containing the HSV origin (ori-S) as assayed by DpnI resistance and Southern blot hybridization (10, 73). Similar cotransfection studies have also been carried out to detect phosphonoacetic acid (PAA)-sensitive HCMV ori-Lyt-dependent DNA replication and assembly of RC in the presence of the six HCMV core replication proteins and a set of auxiliary proteins (53, 61).
KSHV also encodes a set of six genes which have various levels of homology to the equivalent EBV, HSV, and HCMV replication genes. These include ORF9 (POL), ORF59 (PPF), ORF6 (SSB), ORF56 (PRI), ORF40/41 (PAF), and ORF44 (HEL), which are expected to represent the six core replication proteins (Table 1). However, an OBP and the ori-Lyt of KSHV are still unidentified. In the immediate-early (IE) phase of the KSHV lytic cycle, a 3.6-kb mRNA encompassing both RTA (ORF50) and K8 (ORF-K8) is transcribed (42, 77). A separate 1.0-kb spliced K8 mRNA encodes a 237-amino-acid (aa) protein with a leucine zipper domain near its C terminus, which is distantly related to the ZTA (BZLF1) protein of EBV and Jun/Fos family proteins (42). Although K8 exhibits certain properties suitable for an OBP because of analogous genome location and similar splicing pattern with respect to EBV ZTA, there is no evidence that K8 itself is either a DNA binding protein or a transcription factor. Furthermore, RTA but not K8 can reactivate the KSHV lytic cycle from latency (26, 66).
TABLE 1.
KSHV replication genes and expression plasmids used in this study
| KSHV gene | Protein | Function | Expression vectora
|
Nucleotide positionb
|
EBV gened | ||
|---|---|---|---|---|---|---|---|
| pSG5 | pSG5-Flag | ATG | Stop | ||||
| Core machinery | |||||||
| ORF6 | SSB | Single-stranded DNA binding | pJX3 | 3210 | 6611 | BALF2 | |
| ORF9 | POL | DNA polymerase | pJX1 | pJX8 | 11363 | 14401 | BMRF1 |
| ORF40/41 | PAF | Primase associated | pJX5 | pJX12 | 60308 | 62444 | BBLF2/3 |
| ORF44 | HEL | Helicase subunit | pJX7 | pJX14 | 64892 | 67258 | BBLF4 |
| ORF56 | PRI | Primase subunit | pJX4 | pJX11 | 79436 | 81967 | BSLF1 |
| ORF59 | PPF | Polymerase processivity | pJX2 | pJX9 | 96739 | 95549 | BMRF1 |
| Associated components | |||||||
| ORF57 | MTA | Posttranscriptional | pJX6 | pJX13 | 82717 | 83544 | BMLF1 |
| ORF-K8 | K8 | DNA replication (?) | pCJC581 | 74850 | 75569 | BZLF1 | |
| pFYW1c | |||||||
pJX8 to pJX14 are Flag-tagged pSG5 expression plasmids, and pJX1 to pJX7 are nontagged versions.
Genomic locations of the translated start and stop sites for the individual open reading frames of the KSHV replication loci were subcloned into an expression vector (pSG5) containing SV40 enhancer, β-globin intron, and SV40 polyadenylation signals as described in Materials and Methods.
pSG5-Myc expression plasmid for ORF-K8.
KSHV homologue.
Recent studies have revealed that the cellular promyelocytic leukemia protein (PML)-associated nuclear bodies known as PML oncogenic domains (PODs) or nuclear domain 10 are the sites for input viral DNA accumulation in adenovirus, simian virus 40 (SV40), HSV, and HCMV infections, as well as for IE transcription in HCMV (31, 32). PODs consist of 20 to 30 spherical 0.3- to 0.5-μm structures that are present in most cells and are thought to be associated with the nuclear matrix. Herpesvirus DNA replication proteins seem to target PODs for immediate access to viral genomes deposited there; this was further implied when HCMV RC were seen growing from the PODs into kidney-shaped RC structures surrounded by PODs (2). The PML domains are targeted by several known herpesvirus regulatory proteins such as HSV IE110 (ICP0) and HCMV IE1 and IE2, which colocalize with or adjacent to PODs, and in the case of IE2 later also associate with viral DNA RC (2). In addition, other key proteins involved in herpesvirus DNA replication that are not known to target to PODs, including HSV IE175 (ICP4), HSV OBP (UL9), and EBV ZTA, also efficiently colocalize with their homologous functionally active RC.
Very little is known about the lytic DNA replication of KSHV. In this study, we cloned the six putative KSHV core replication genes based on homology to EBV and HSV and confirmed that they expressed protein products of the expected sizes in transfected Vero cells. We also showed that the six functional KSHV core replication proteins could substitute for their EBV counterparts in the replication of EBV ori-Lyt. We then investigated (i) the intracellular localization of these proteins, as well as their complex interactive requirements for nuclear translocation; (ii) the requirements for formation of subnuclear RC-like viral domains in a transient transfection system when all six core genes were cotransfected together; and (iii) the association of ORF-K8 and cellular PODs with the transiently assembled RC-like structures. Finally, we confirmed that functional KSHV RC in lytic cycle-induced PEL cells and KSHV-infected dermal microvascular endothelial cells (DMVEC) also colocalized with K8 and PODs.
MATERIALS AND METHODS
Cells and virus.
Vero cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (FBS) in humidified 5% CO2 in a 37°C incubator. Vero cells were seeded at 8 × 104 cells per well in two-well slide chambers for transfection. JSC-1 PEL cells (5a) were grown in RPMI medium containing 5% FBS in a humidified 5% CO2 incubator. Harvesting of the KSHV supernatant virons from PELs and infection of human primary dermal microvascular endothelial cells (DMVEC) were all performed as described elsewhere (D. J. Ciufo, J. Cannon, L. Poole, F. Y. Wu, J. Orenstein, R. Ambinder, and G. S. Hayward, submitted for publication).
Expression plasmids.
Plasmid pJX1 carries the unspliced 3.0-kb KSHV ORF9 (POL) gene driven by the SV40 enhancer-promoter inserted at the BamHI site downstream from the SV40 promoter in the pSG5 eukaryotic expression vector (Stratagene). Using KSHV(BCBL-R) phage lambda clones as templates (50), a 3.0-kb intact ORF9 gene fragment with BamHI cohesive ends was obtained by PCR with primers LGH2481 and LGH2482. Plasmid pJX2 contains a 1.19-kb ORF59 (PPF) gene fragment with BamHI and BglII cohesive ends obtained with primers LGH2483 and LGH2484 inserted at the same BamHI site of pSG5. Plasmid pJX3 contains a 3.4-kb ORF6 (SSB) gene fragment with EcoRI cohesive ends obtained with primers LGH2485 and LGH2486 inserted at the EcoRI site in pSG5. Plasmid pJX4 contains a 2.5-kb ORF56 (PRI) gene fragment with BglII cohesive ends obtained with primers LGH2487 and LGH2488 inserted at the BglII site of pSG5. Plasmid pJX5 contains a 2.13-kb ORF40/41 (PAF) gene genomic fragment with BglII cohesive ends obtained with primers LGH2489 and LGH2490 inserted at the BglII site in pSG5. Plasmid pJX6 contains a 1.58-kb ORF57 (MTA) genomic DNA fragment with BamHI cohesive ends obtained with primers LGH2491 and LGH2492 inserted at the BglII sites in pSG5. Plasmid pJX 7 contains a 2.36-kb ORF44 gene fragment with BglII cohesive ends obtained with primers LGH2493 and LGH2446 inserted at the BamHI and BglII sites in pSG5. The oligonucleotide PCR primers used to clone the pJX plasmid series are listed in Table 2.
TABLE 2.
Oligonucleotide primers designed for KSHV replication and replication-associated genes
| Primer | Genomic position | Sequence (5′-3′) | Target gene (product) |
|---|---|---|---|
| LGH2485 | 3210–3240 | CAG TGG ATC CAT GGC GCT AAA GGG ACC | 5′ ORF6 (SSB) |
| LGH2486 | 6585–6611 | TGA CAG ATC TCT ACA AAT CCA GGT CAG | 3′ ORF6 |
| LGH2481 | 11363–11389 | CAG TGG ATC CAT GGA TTT TTT CAA TCC | 5′ ORF9 (POL) |
| LGH2482 | 14375–14401 | TGC AGG ATC CCT AGG GCG TGG CAA AAG | 3′ ORF9 |
| LGH2489 | 60308–60333 | CAG TAG ATC TAT GGC AAC GAG CGA AG | 5′ ORF40/41 (PAF) |
| LGH2490 | 62412–62444 | TGA CAG ATC TTC AAA ATA AAG ATA AAA GCC TGG | 3′ ORF40/41 |
| LGH2493 | 64892–64918 | CAG TAG ATC TAT GGA CAG CTC GGA AGG | 5′ ORF44 (HEL) |
| LGH2446 | 67228–67258 | CAG TAG ATC TTC AGT AGA TCA GAG TAG TCT T | 3′ ORF44 |
| LGH2487 | 79436–79462 | CAG TAG ATC TAT GGA GAC GAC ATA CCG | 5′ ORF56 (PRI) |
| LGH2488 | 81942–81967 | TGA CAG ATC TTT AAC TGG CCA GTC CC | 3′ ORF56 |
| LGH2491 | 82717–82745 | CAG TGG ATC CAT GGT ACA AGC AAT GAT AG | 5′ ORF57 (MTA) |
| LGH2492 | 83516–83544 | TGA CAG ATC TTT AAG AAA GTG GAT AAA AG | 3′ ORF57 |
| LGH2483 | 96739–96712 | CAG TGG ATC CAT GCC TGT GGA TTT TCA | 5′ ORF59 (PPF) |
| LGH2484 | 95578–95549 | TGA CAG ATC TTC AAA TCA GGG GGT TAA ATG | 3′ ORF59 |
Empty vector plasmid pYW51 carries an oligonucleotide Flag epitope tag bounded by BglII and BamHI cohesive ends and inserted at the BamHI site of the pSG5 vector. Plasmid pJX8 carries the gene encoding POL (ORF9) from pJX1 with an added in-frame 5′ Flag epitope from plasmid pYW51. Similarly, plasmid pJX9 carries the PPF (ORF59) gene from pJX2 with an in-frame 5′ Flag epitope, plasmid pJX11 contains the PRI (ORF56) gene from JX4 with an added in-frame Flag epitope, plasmid pJX12 contains the PAF (ORF40/41) genomic DNA fragment from pJX5 with an added 5′ Flag epitope, plasmid pJX13 carries the MTA (ORF57) genomic DNA fragment with an added in-frame 5′ Flag epitope, and plasmid pJX14 carries the HEL (ORF44) gene with an added in-frame 5′ Flag epitope.
Plasmid pDY048 contains a polylinker insert bounded by EcoRI and BamHI sites added to pSG5 with an oligonucleotide sequence encoding a Myc epitope tag with NotI and SrfI cohesive ends inserted within the polylinker. Plasmid pCJC565 contains an intact multiply spliced 714-bp KSHV K8 cDNA clone flanked by BamHI sites in a pUC18 background (C. J. Chiou, F. Y. Wu, D. M. Ciufo, D. Alcendor, S. J. Kim, J. C. Zong, and G. S. Hayward, submitted for publication). To construct a Myc-K8 expression plasmid, the K8 cDNA was inserted into the BamHI site of pDY048 in frame with the 5′ Myc epitope tag. Plasmid pYNC100 carries an intact 735-bp EBV transactivator ZTA cDNA clone placed behind the leader plus ATG region from a black beetle virus for efficient in vitro translation (13). Plasmid pCJC514 carries the K8 cDNA clone with both BamHI cohesive ends inserted into the BglII site of plasmid pGH255, which contains the same black beetle virus leader region as pYNC100. Plasmid pEF52 is a pBR322 vector that carries a 5.4-kb BamHI fragment of EBV ori-Lyt from genomic coordinates 48848 to 54858 (21, 22). Plasmid pRTS21 is an pSG5 expression vector for EBV ZTA (60). pSG5 plasmids containing the six EBV core lytic replication genes, pRTS13, -14, -11, -28, -25, and -12, expressing EBV POL, PPF, PRI, HEL, PAF, and SSB, respectively, were also used (23).
Transient DNA transfection.
Transient DNA transfection assays for immunofluorescence assay (IFA) were carried out with 8 × 104 Vero cells in two-well slide chambers. Different combinations of up to eight DNA plasmids (0.3 μg of each) carrying tagged and/or nontagged versions of the gene encoding KSHV ORF9 (POL), ORF59 (PPF), ORF56 (PRI), ORF40/41 (PAF), ORF44 (HEL), ORF6 (SSB), ORF57 (MTA), or K8 driven by the SV40 enhancer region were transfected into each well to study the intracellular localization of the protein products into RC-like structures. The CsC1-purified plasmid DNAs were cotransfected by the calcium phosphate precipitation procedure in BBS buffer (49). Empty vector plasmid pSG5 DNA was used as a carrier to normalize the total amount of transfected DNA. Transfected cells were incubated in Dulbecco modified Eagle medium supplemented with 10% FBS in a 3% CO2 incubator at 35°C overnight. The medium was changed 18 h after transfection, and the slides were placed into a 5% CO2 incubator at 37°C. Cells were fixed 48 h after transfection for IFA. BrdU was added to the culture medium at a final concentration of 10 μM for 30 min before fixation when appropriate.
Transient transfection replication assay.
Vero cells were seeded at 106 cells per 100-mm-diameter dish and were transfected by the procedures described above. Approximately 10 μg of total plasmid DNA containing either the EBV replication genes including ZTA or the KSHV replication genes plus ZTA were cotransfected with 5.0 μg of EBV viral origin DNA (pEF52). For harvesting at 80 h after the posttransfection medium change, the cell monolayer was washed twice with phosphate-buffered saline (PBS) and subsequently scraped into 4 ml of 40 mM Tris-hydrochloride (pH 7.5)–1 mM EDTA–150 mM NaCl. The cells were then pelleted and lysed in 2 ml of lysis buffer (10 mM Tris-HCl [pH 8.0], 10 mM EDTA, 2% sodium dodecyl sulfate [SDS], 100 μg of proteinase K per ml). Following overnight incubation at 37°C, the samples were diluted to 4 ml with Tris-EDTA (pH 8.0), extracted with phenol, phenol-chloroform, and chloroform, and ethanol precipitated after the addition of sodium acetate (pH 5.2) to a final concentration of 0.3 M. The DNA pellets were resuspended in 450 μl of distilled H2O (dH2O), treated with 100 μg of RNase A per ml, ethanol precipitated, and resuspended in 300 μl of dH2O. Approximately 10 μg of extracted cellular DNA was digested with 30 U of BamHI in a 100-μl reaction volume for 5 at 37°C. The reaction digest containing BamHI was then inactivated at 80°C for 20 min and cooled on ice for 30 min. The cooled reaction sample was then incubated overnight with 30 U of DpnI at 37°C. To monitor DpnI activity, 5 μl of the DpnI reaction digest was removed and incubated simultaneously with 500 ng of pUC19 DNA overnight at 37°C. Complete cleavage of the pUC19 DNA indicated that the experimental DNA was also completely digested. The cellular DNA was resolved by electrophoresis on a 0.8% agarose gel at 35 V for 20 h, transferred to a Nytran membrane (Schleicher & Schuell) after treatment of the gel at 20°C for 10 min in 200 mM HCl, and then incubated in 0.4 M NaOH–0.6 M NaCl for 20 min. The agarose gel was then vacuum transferred to the nylon membrane in the presence of 10× SSC (1.5 M NaCl, 0.15 M sodium citrate) for 30 min. After the membrane was dried completely at 20°C, the DNA was irreversibly cross-linked by UV radiation (Stratalinker; Stratagene). For hybridization, the membrane was incubated overnight at 60°C in 25 ml of buffer consisting of 1% SDS, 0.5 mg of heparin per ml, and 5× SSC (750 mM NaCl, 50 mM Na2HPO4, 5 mM Na2EDTA). Approximately 100 ng of a gel-purified BamHI ori-Lyt fragment was radiolabeled with [α-32P]dCTP by random priming to a specific activity of 108 cpm/μg (Boehringer Mannheim-Roche kit). The membrane was then incubated at 60°C with 106 cpm of denatured radiolabeled ori-Lyt probe DNA per ml and fresh hybridization buffer. Following hybridization for 8 h to 12 h, the membrane was washed twice in 0.1× SSC–0.1% SDS at 65°C for 45 min and exposed to Kodak XAR5 film for 24 h at −80°C using an intensifying screen. The resulting autoradiograms were quantified with a Kontes fiber optic scanner.
TPA induction and BrdU incorporation in PEL cells.
Starting with an 80% viable suspension culture of the cell line JSC-1, cells were pelleted at 3,000 rpm and 5 × 105 were seeded into each well of a standard six-well tissue culture dish. After initial pelleting, half of the original cell medium was replaced with fresh RPMI medium with 5% FBS warmed to 37°C to a final volume of 2 ml. Cells were pelleted and transferred to the each of the six wells. Then cells were incubated at 37°C with 5% CO2 for 10 min. TPA was added to each well at a final concentration of 20 ng/ml. After 12, 48, or 72 h of incubation, the viability of the suspension culture was determined again, and 106 cells were pelleted and resuspended in 1 ml of fresh prewarmed medium containing 10 μM BrdU. The caps of the Eppendorf tubes were punctured with sterile needles to allow air diffusion. The cells were incubated at 37°C in a 5% CO2 incubator for 45 min. The cells were pelleted again and washed once with fresh 1 ml 1× PBS. The cell pellet was suspended in 200 μl of fresh PBS and plated it onto the polylysine-loaded glass slides for 20 min or until the cells become fully adherent to the slides. Extra liquid was drained, and the slides were dried completely. The slides were either stored at −20°C or used for immunofluorescence staining immediately.
IFA.
Infected, induced, or transfected cells were washed in 1× Tris-saline (100 mM NaCl, 10 mM Tris-HCl [pH 7.5]), fixed with 2% paraformaldehyde in PBS for 10 min at room temperature, and then permeabilized in 0.2% Triton X-100 in PBS for 20 min on ice. To expose incorporated BrdU residues, pulse-labeled cells were incubated with 4 N HCl for 10 min at room temperature and then washed in PBS for three times with 5-min intervals. The primary mouse monoclonal antibody (MAb) and rabbit polyclonal antibody (PAb) were diluted together in PBS with 2% goat serum for double-label assays or diluted separately for single-label assays. Primary antibodies were incubated for 1 h at 37°C followed by incubation with the appropriate combination of fluorescein isothiocyanate (FITC)-conjugated and rhodamine-conjugated anti-mouse and anti-rabbit secondary antibodies at 1:100 dilution for 30 min at 37°C for double-label assays. Rhodamine-conjugated anti-mouse secondary antibody was diluted at 1:100 for single labeling. To visualize cellular chromatin, a drop (20 μl) of 4′, 6-diamidino-2-phenylindole (DAPI)-containing antifade slide mounting solution (Vector Shield) was added to the slide prior to microscopy. Antibodies used included mouse anti-BrdU MAb (Becton Dickinson), rabbit anti-ORF6 PAb (SSB), mouse anti-ORF59 MAb (PPF), rabbit polyclonal anti-K8, rabbit polyclonal anti-PML(C), directed against amino acid positions 484 to 498 of the human 90-kDa PML isoform (1), mouse MAb and rabbit PAb anti-Flag (Sigma), and mouse MAb and rabbit PAb anti-Myc (Santa Cruz). Rabbit anti-ORF K8 peptide PAb was generated by immunization with the N-terminal peptide (16-DNSEKDEAVIEED-28) bounded by N-Y and C-CS residues. Rabbit anti-ORF6 (SSB) peptide PAb was generated by immunization with C-terminal peptide (1116-GKKRKIASLLSDL-1128). Anti-ORF59 (PPF) MAb was a gift from Bala Chandran (University of Kansas) (11).
Slides were screened and photographed with 40×, 63×, or 100× oil immersion objectives on a Leitz Dialux 20EB epifluorescence microscope with Image-Pro software (Media Cybernetics, Silver Spring, Md.) and appropriate narrow-band FITC or rhodamine filters. For confocal microscopy, a Noran OZ CLSM confocal microscope system with Intervision software (Noran Inc., Madison, Wis.) was used.
Western blot analysis.
Vero cells were washed with PBS while still attached to 100-mm-diameter tissue culture plates and harvested with 10 ml of fresh PBS using a cell scraper. Cells were then pelleted at 6,000 rpm for 5 min and washed twice with 5 ml of PBS. The cell pellet was lysed with 0.4 ml of ice-cold lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 1.5 mM Na3VO4, 50 mM NaF, 10 mM sodium pyrophosphate, 10 mM glycerol phosphate, 1 mM phenylmethylsulfonyl fluoride). Clarified cell extracts from the equivalent of 104 cells were separated by electrophoresis on SDS–10% polyacrylamide gels followed by electroblotting onto nitrocellulose. The filter sheets were blocked by incubation for 1 h at 20°C in PBS plus 0.1% Tween 20 containing 5% nonfat dry milk, then washed twice with PBS-Tween 20 for 15 min, and incubated with appropriate MAb at a dilution of 1:3,000 for 1 h at 20°C. After three 10-min washes with PBS-Tween 20, the filter sheet were incubated with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG; Bio-Rad) for 1 h at 20°C and then washed three times, and the reacting protein bands were detected with an enhanced chemiluminescence (ECL) system (Amersham ECL RP2106) using Kodak XAR film.
RT-PCR and sequencing.
Total RNAs (DNase treated) from both TPA-induced (48 h) and uninduced BCBL-1 cells were used for reverse transcriptase-mediated PCR (RT-PCR). First, 3 μg of total RNA suspended in 100% ethanol was centrifuged at 4°C for 30 min. The RNA pellet was air dried and resuspended in 10 μl of diethyl pyrocarbonate-treated dH2O. Second, to produce cDNA from total RNA, 10 μl of resuspended RNA was added to a reaction mixture containing 1 μl of 40 mM oligo(dT) primer; the mixture was heated at 70°C for 10 min and quickly chilled on ice. To the chilled mixture, 1 μl of RNA inhibitor (Promega), 4 μl of first-strand buffer, 2 μl of 0.1 M dithiothreitol, 1 μl of 10 mM deoxynucleoside triphosphate, and 1 μl of Superscript II reverse transcriptase (GIBCO BRL) were added. The final mixture was incubated at 50°C for 1 h to generate cDNA. Third, 2 μl of cDNA was used as template for Taq DNA polymerase, and two sets of oligonucleotides were used as PCR primers: first, LGH2489 and LGH2490 (described above), covering the 5′ and 3′ ends of the full genomic coding region encompassing both ORF40 and ORF41; and second, LGH3752 (5′-GAA GAT CTC CAT CCG GTC TGG TGG CCG TG-3′) and LGH3753 (5′-GAA GAT CTC CCC ATT TCC CTC AGT GTC TGG-3′), flanking the 127-bp putative intron of the ORF40/41 mRNA transcript. Total DNA were isolated from BCBL-1 PEL cells as described for the transient transfection replication assay, and 1 μl of total DNA was used as the PCR template. PCR amplification was conducted in a Thermal Cycler (Eppendorf) with the following program: 94°C for 3 min; 40 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C 130 s; 72°C for 3 min. PCR products were resolved by electrophoresis on a 1% agarose gel, gel purified, and cloned into TA cloning vector pCR2.1 (Invitrogen). The cloned inserts from RT-PCR were sequenced with a T7 primer from the flanking 5′ end of the TA cloning site of pCR2.1 using an automated model 310 Genetic Analyzer (ABI Prism).
RESULTS
Cloning and expression of isolated KSHV core DNA replication and replication-associated proteins.
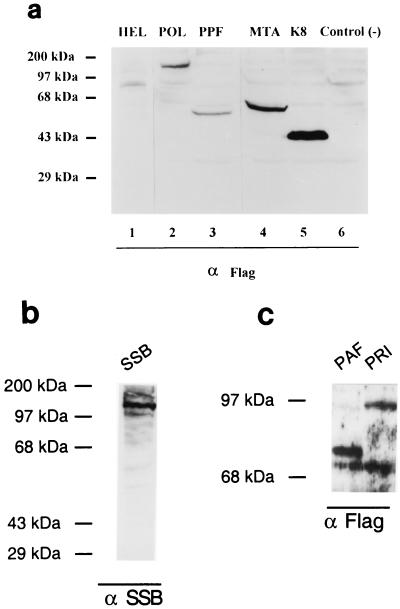
The genomic positions of KSHV ORFs encoding homologues of the six core EBV DNA replication proteins were predicted from analysis of the primary DNA sequence (Tables 1 and 2). The KSHV genes referred to as KSHV ORF9 (POL), ORF59 (PPF), ORF6 (SSB), ORF56 (PRI), ORF40/41 (PAF), and ORF44 (HEL) were predicted to encode proteins of 1,012, 396, 1,130, 843, 669, and 788 aa, respectively. Genomic DNA fragments encompassing each of these ORFs were amplified as PCR products derived from the KSHV (BCBL-R) PEL cell line DNA as template and cloned into both Flag-tagged and untagged versions of the pSG5 expression plasmid driven by the SV40 early region promoter-enhancer. The synthesis of each protein with the expected approximate molecular weight from these expression plasmids was confirmed in DNA-transfected Vero cells by Western blot analysis after incubating the membranes with either a mouse MAb directed against the Flag epitope (Fig. 1a, lanes 1 to 3, and c) or a rabbit PAb specific for KSHV SSB (Fig. 1b).
FIG. 1.
Western blot analysis of proteins expressed by eight KSHV DNA replication and replication-associated genes in transient DNA transfection assays. (a) Protein products from Flag-tagged expression plasmids encoding HEL, POL, PPF, MTA, and K8 were transfected into Vero cells. After 48 h, whole-cell protein extracts were electrophoretically fractionated on SDS– 10% polyacrylamide gels, and Western blot analysis was performed by incubating the membrane with a specific MAb or PAb followed by ECL color development. (b) Protein products of untagged pSG5 plasmid vector encoding KSHV SSB detected with rabbit anti-SSB PAb, (c) Protein products of Flag-tagged expression plasmids encoding PAF and PRI detected with mouse anti-Flag MAb.
By analogy to EBV, the KSHV ORF57 (MTA) and ORF-K8 nuclear regulatory proteins were also suspected to be either directly or indirectly associated with the viral DNA replication process (Table 1). The structures of intact cDNAs for these two genes have been analyzed previously (Chiou et al., submitted). The Flag-tagged ORF-K8 expression plasmid was generated from an isolated intact spliced cDNA clone that encodes the putative leucine zipper motif, whereas the Flag-tagged MTA version represents a genomic DNA version. ORF57 and ORF-K8 were predicted to encode proteins of 275 and 237 aa, which was confirmed by Western blot analysis with MAb Flag antibody (Fig. 1a, lanes 4 and 5).
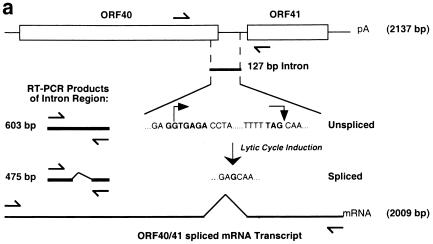
Evidence that ORF40 and ORF41 are transcribed as a single spliced mRNA species encoding PAF.
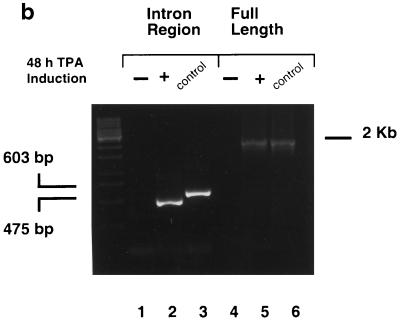
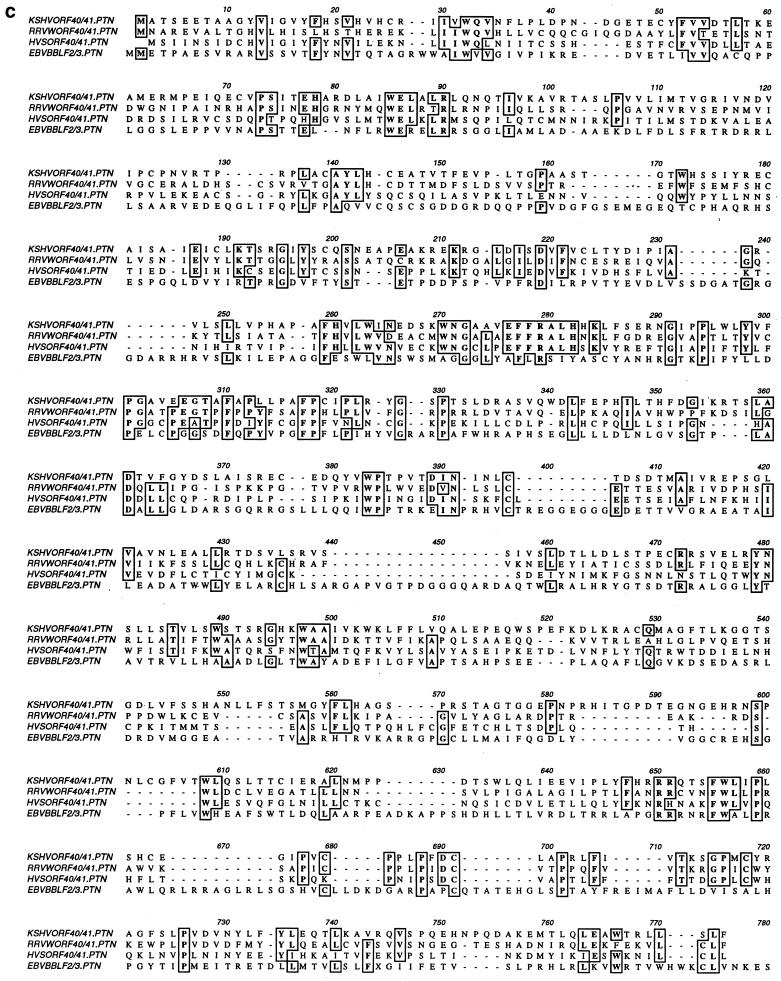
ORF40 and ORF41 are homologous to the EBV BBLF2 and BBLF3 ORFs, which are transcribed as a single mRNA transcript with an intron of 128 bp that encodes PAF (21). The KSHV PAF mRNA transcript was also believed to span two ORFs, ORF40 and ORF41. Consequently, the construction of the PAF genomic DNA expression plasmid in this case encompassed both the ORF40 and ORF41 genomic DNA regions to permit appropriate cellular splicing (pJX5). The Flag-tagged version of ORF40/41 was constructed by attaching a Flag epitope-encoding sequence adjacent to the 5′ ATG start codon of ORF40/41 (pJX12). To confirm the presence of a spliced ORF40/41 joint transcript, RT-PCR was conducted on total RNA extracted from TPA-induced BCBL-1 PEL cells. A set of primers that flanks the entire 2.0-kb ORF40/41 coding sequence, from the initiation codon of ORF40 (5′) to the termination codon of ORF41 (3′), was designed and used to confirm the total length of the ORF40/41 mRNA transcript. We designed a second set of flanking oligonucleotide primers encompassing the putative 127-nucleotide intron (Fig. 2a) and spanning 603 bp (Fig. 2b, lane 3) between genomic nucleotide positions 61409 to 62011. The results obtained from PCR with the intron flanking primers showed that a 475-bp product was obtained from the 48-h-TPA-induced total RNA (Fig. 2b, lane 2). With the full sequence primers, a 2.0-kb RT-PCR product was obtained in the induced total RNA (Fig. 2b, lane 5). Sequencing of the 475-bp RT-PCR product confirmed that the lytic cycle ORF40/41 RNA encompassed a 127-bp intron spliced out from 1,350 bp downstream from the initiation codon ATG of ORF40 (nucleotide position 60308), which also removed the termination codon for ORF40. The splice donor site located at nucleotide position 61657 in ORF40 proved to be fused in frame to the predicted acceptor site at nucleotide position 61784 in ORF41, resulting in an intact cDNA sequence of 2,009 bp. Sequence alignment showed that the KSHV PAF protein encoded by the spliced form of ORF40/41 cDNA shares over 20% amino acid identity throughout its length with EBV BBLF2/3 (PAF) protein and up to 35% amino acid identity with equivalent predicted spliced PAF proteins from the rhesus rhadinovirus (RRV) and HVS gamma-2 rhadinoviruses (Fig. 2c).
FIG. 2.
A spliced ORF40 and ORF41 transcript is induced in TPA-treated BCBL-1 PEL cells. (a) Diagram of the genomic organization of ORF40 and ORF41 between coordinates 60308 to 62444 in KSHV. The locations and orientations of the PCR primers used (→) and the sizes of PCR products from spliced and unspliced cDNA templates are shown. (b) Photograph of an ethidium bromide-stained 1% agarose gel showing the separated RT-PCR products. Lanes 1 and 4, no RT-PCR product detected from DNase-treated total RNA isolated from BCBL-1 PEL cell line before TPA induction; lanes 2 and 5, RT-PCR products from RNA isolated 48 h after TPA induction; lanes 3 and 6, PCR products from the total DNA isolated from BCBL-1 cells. For lanes 1 to 3, PCR primers covering the intron region were used; for lanes 4 to 6, PCR primers covering the entire coding region of ORF40/41 were used. (c) Amino acid sequence alignment of KSHV ORF40/41, rhesus rhadinovirus (RRV) ORF40/41, HVS ORF40/41, and EBV BBLF2/3.
Intracellular localization of each KSHV core DNA replication and replication-associated protein in transfected mammalian cells.
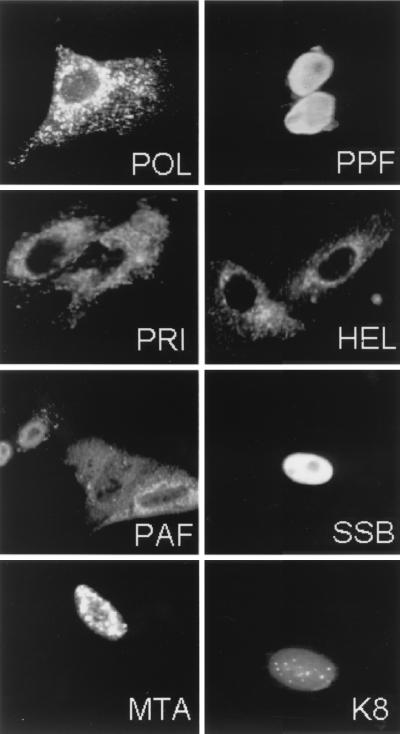
To investigate the intracellular localization patterns of the eight KSHV presumed replication-associated proteins, Vero cells were first transfected with individual expression plasmids encoding Flag-tagged POL, PPF, PRI, PAF, HEL, or MTA or the untagged SSB or K8 protein. The results of single-label IFA carried out at 48 h showed that POL, HEL, and PRI were each localized in the cytoplasm, whereas PAF gave a mixed, primarily cytoplasmic staining pattern (Fig. 3). In contrast, SSB, PPF, MTA, and K8 were all transported in the nucleus as individual isolated proteins. SSB and PPF gave a diffuse nuclear pattern, whereas most of the MTA- and K8-positive cells also formed nuclear punctate patterns within a nuclear diffuse background (Fig. 3). As shown elsewhere, these punctate domains colocalize with SC35 spliceosomes and PML nuclear bodies, respectively (Chiou et al., submitted).
FIG. 3.
Intracellular localization of single transfected KSHV DNA replication and replication-associated proteins detected with IFA. Equal samples of plasmid DNA (0.2 μg) were used for each gene in transient transfection assays in Vero cells. All except SSB and K8, were detected with mouse anti-Flag MAb and donkey FITC or rhodamine-labeled anti-mouse IgG. SSB and K8 were detected with SSB or K8-specific rabbit PAb and donkey FITC or rhodamine-labeled anti-rabbit IgG.
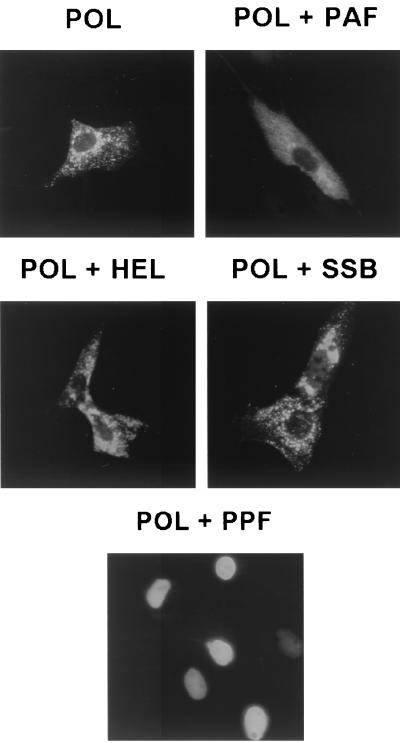
Cotransfection with PPF is sufficient to translocate POL into the nucleus.
The localization pattern of KSHV POL alone was intrinsically cytoplasmic. However, cotransfection of the POL plasmid with all five of the other core viral replication protein genes together allowed efficient nuclear translocation of POL; 71% of the transfected cells now showed a nuclear staining pattern for POL, but with 29% remaining cytoplasmic (presumably because of inefficient cotransfection) (Table 3). To determine which factor might be responsible for POL translocation into the nucleus, each core protein was omitted in turn from the cotransfection mixture described above. Interestingly, POL remained localized in the cytoplasm only when PPF was omitted, whereas sequential omission of any single viral protein other than PPF in the cotransfection mixtures still produced up to 70% nuclear translocation of POL (data not shown). To test whether PPF on its own is sufficient to translocate POL to the nucleus, the two were cotransfected in a 1:1 ratio (Fig. 4, bottom image). The results revealed that the cellular localization of POL was efficiently altered from cytoplasmic to nuclear in the presence of PPF, with 78% of the positive cells showing a nuclear diffuse POL pattern and 16% showing a predominantly nuclear pattern with some cytoplasmic background (Table 3). Again, 6% of the positive cells still showed completely cytoplasmic POL localization, which can presumably be explained by the lack of expression of PPF in those cells. As negative controls, differential pairing with each of the other four core proteins in cotransfection assays did not affect the cytoplasmic localization pattern of POL (Fig. 4, top and middle images).
TABLE 3.
KSHV ORF9 (POL) nuclear translocation in cotransfection assaysa
| Protein | Distribution (%)
|
Total no. of positive cells counted | ||||
|---|---|---|---|---|---|---|
| Nuclear | Mixed
|
Cytoplasmic | ||||
| PN | N>C | Whole-cell diffuse | ||||
| All | 71 | >1 | >1 | >1 | 29 | 88 |
| POL | >1 | >1 | >1 | >1 | <99 | 76 |
| POL + PPF | 78 | 14 | 2 | >1 | 6 | 181 |
Pairing with other replication genes other than PPF did not alter the cytoplasmic localization of POL.
FIG. 4.
Contribution of PPF but not PAF, PRI, HEL, or SSB to nuclear translocation of POL. The Flag-tagged POL expression plasmid was paired with other untagged replication genes, cotransfected into Vero cells, and then detected with mouse anti-Flag and rhodamine-labeled anti-mouse IgG. POL alone shows cytoplasmic staining but was efficiently translocated into the nucleus in the presence of PPF.
All six core replication proteins are required for nuclear translocation of the KSHV PAF (ORF40/41).
In EBV, PRI, HEL, and PAF are all intrinsically cytoplasmic on their own but can form a tripartite subcomplex after cotransfection, with HEL (BBLF4) and PRI (BSLF1) being sufficient to completely translocate PAF (BBLF2/3) into the nucleus (23). However, the KSHV requirements for PAF (ORF40/41) nuclear translocation proved to be more complex than the EBV requirements.
Our initial IFA showed that the KSHV PAF (ORF40/41) protein on its own also produced a mixed pattern with predominantly cytoplasmic staining (85%) in transfected Vero cells (Table 4; Fig. 5k and m). To further differentiate between partial nuclear states and cytoplasmic localization of PAF, three additional subclasses of staining were defined under the categories of mixed nuclear staining (Table 4): predominantly nuclear (PN) (Fig. 5c and d), nuclear more than cytoplasmic (N>C) (Fig. 5e and g), and whole-cell diffuse (Fig. 5i and j).
TABLE 4.
KSHV ORF40/41 (PAF) nuclear translocation in cotransfection assaysa
| Protein | Distribution (%)
|
Total no. of positive cells counted | ||||
|---|---|---|---|---|---|---|
| Nuclear | Mixed
|
Cytoplasmic | ||||
| PN | N>C | Whole-cell diffuse | ||||
| All | 55 | 10 | 4 | 7 | 22 | 92 |
| All − POL | <1 | 56 | 24 | <1 | 20 | 34 |
| All − PPF, POL | <1 | 43 | 24 | 2 | 31 | 42 |
| All − SSB, PPF, POL | <1 | 12 | 51 | 9 | 26 | 136 |
| PAF + SSB | <1 | 64 | 32 | 1 | 3 | 31 |
| PAF + PPF | <1 | 24 | 23 | 3 | 50 | 94 |
| PAF + PRI | <1 | 4 | 29 | 21 | 46 | 24 |
| PAF | <1 | 3 | 10 | 2 | 85 | 296 |
| PAF + MTA | <1 | <1 | 40 | 15 | 45 | 20 |
| PAF + K8 | <1 | 47 | 39 | 5 | 9 | 64 |
ORF40/41 was Flag tagged; Flag MAb and rhodamine-labeled anti-mouse IgG were used for IFA visualization.
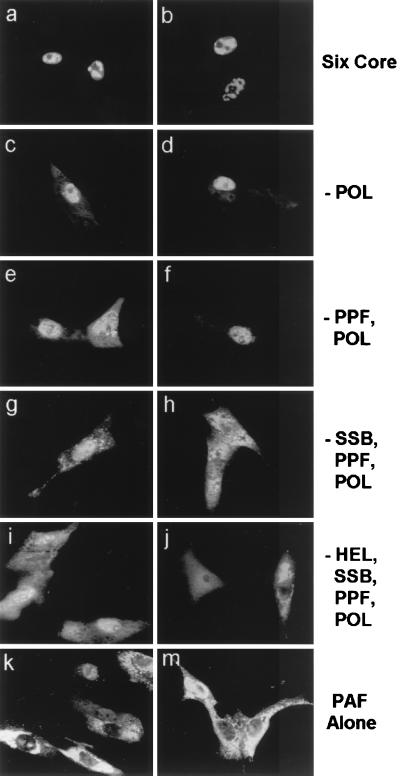
FIG. 5.
All six viral replication gene products are required for full nuclear translocation of PAF in cotransfected Vero cells. Flag-tagged PAF plasmids were used along with various combinations of the other untagged replication genes, and mouse anti-Flag and rhodamine-labeled anti-mouse IgG were used to visualize the PAF intracellular localization. (a and b) Two separate single-label frames showing cells cotransfected with the whole set of plasmids encoding POL, PPF, PRI, PAF, HEL, and SSB; (e to j) omission experiments showing cotransfection of all plasmids except those encoding POL (c and d), PPF and POL (e and f), SSB, POL, and PPF (g and h), and HEL, SSB, PPF, and POL (i and j). Sequential omission of the other replication genes reverts PAF to a cytoplasmic localization similar to that obtained by transfection with PAF alone (k and m).
Efficient nuclear localization of PAF was achieved only when all six KSHV core replication proteins (POL, PPF, PRI, PAF, HEL, and SSB) were present; 55% of the PAF-positive cells showed completely nuclear staining (Fig. 5a and b), 21% showed a mixed pattern, and 22% retained a completely cytoplasmic localization (Table 4). Complete nuclear staining patterns were never observed for PAF when one or more of the six core genes were omitted. When only POL was omitted, 56% of the positive cells showed a PN staining pattern (Fig. 5c and d); when both POL and PPF were omitted, 43% of the positive cells showed PN staining pattern (Fig. 5e and f); and when POL, PPF, and SSB were all omitted, leaving only the PRI-HEL-PAF tripartite complex present, the PN staining pattern dropped to 12%, although 51% still gave an N>C staining pattern (Fig. 5g and h).
Differential pairing of PAF with each of the other core proteins was done to determine which of them was involved in significantly altering the PAF localization pattern (Table 4). Cotransfection of PAF with SSB elevated the number of cells displaying a PN staining pattern from 3% to 64% compared to the basal localization state of PAF alone. In addition, cotransfection of PPF with PAF also raised the PN state of PAF from the basal 3% to 24%. Cotransfection of PRI and HEL with Flag-tagged PAF, the analogous components of a presumed KSHV triparte primase-helicase subcomplex, significantly elevated the number of cells showing an N>C staining pattern of PAF (from 10% to 51%). Repeating the experiment with PRI as the Flag-tagged version also increased the number of positive cells showing an N>C pattern for PRI, whereas for a negative control, cotransfection of PAF with PRI alone did not significantly increase the proportion of PN or N>C patterns for PAF (Fig. 5i and j; Table 4). Cotransfection of POL with PAF also did not alter the cytoplasmic localization of PAF (data not shown). Therefore, although formation of the tripartite helicase-primase complex contributed, more efficient translocation of PAF to the nucleus required additional interactions with SSB and PPF.
In addition, cotransfection of KSHV MTA with PAF did not significantly increase the number of PN cells, but it did dramatically elevate the N>C cell number up to 40% compared to the basal level of 10% (Table 4). Clearly, KSHV MTA is not required for full nuclear translocation of the spliced cDNA PAF, whereas the six core replication proteins themselves were sufficient for stable nuclear accumulation of the PAF protein. Furthermore, cotransfection of K8 with PAF alone significantly increased the efficiency of PAF nuclear translocation, with the PN state of PAF being elevated from the basal value of 3% up to 47% (Table 4). Therefore, in the case of KSHV, the collective effort of all six core replication proteins as well as MTA and K8 appears to be required for maximal nuclear accumulation of the primase-helicase subcomplex.
Assembly of KSHV RC-like structures in transient cotransfection assays.
When carrying out preliminary assays to attempt to identify the lytic cycle replication origin and the putative OBP or initiation protein of KSHV, we were surprised to find that DNA replication-related structures were formed in the negative control cells receiving the set of plasmids expressing just the six core KSHV replication proteins. This was also evident in the positive control panels from the PAF translocation experiment described above (Fig. 5a and b).
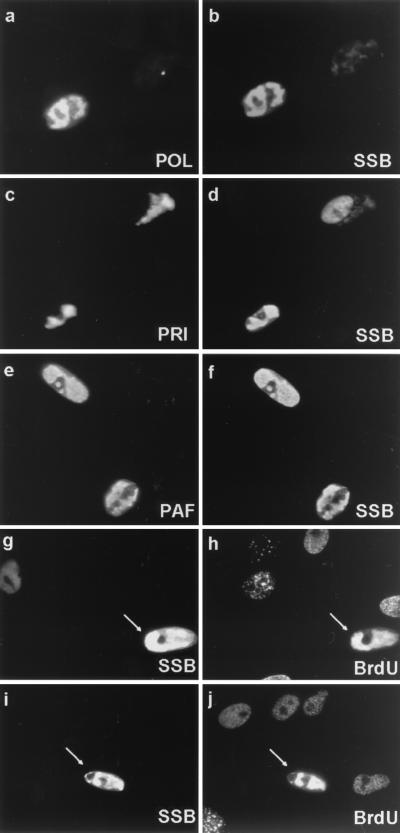
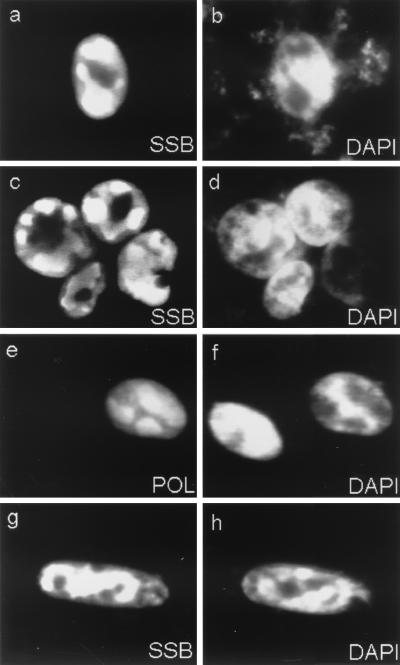
To examine whether SSB colocalized with the other core proteins in these structures, cell cultures were cotransfected with various combinations of plasmids expressing five nontagged replication genes and a sixth plasmid expressing either Flag-tagged POL, PRI, or PAF. At 48 h after transfection, the cells were fixed and examined by double-label IFA. Interestingly, 20 to 30% of the positive cells showed large irregularly shaped RC-like nuclear bodies detected with anti-SSB PAb, and these same cells also showed identical patterns for Flag-tagged POL, PRI, or PAF (Fig. 6a to f). Some cells contained small punctate SSB patterns (see below), but the rest of the SSB-positive cells generally showed a nuclear diffuse pattern for SSB, probably because of the lack of at least one of the other five proteins (Fig. 6d). As negative controls, when we included only POL, PPF, and SSB in the cotransfection assay or sequentially omitted any one of the individual core DNA replication genes, none of the cells displayed any large or small RC-like bodies in the nucleus, and only nuclear diffuse patterns were observed for both SSB and POL (data not shown). A similar result was obtained for PAF when POL was omitted from the mixture (Fig. 5c and d).
FIG. 6.
All six core KSHV DNA replication proteins are required for the assembly of complete RC-like structures in contransfected Vero cells, and active DNA synthesis occurs within KSHV RC assembled in the presence of EBV ori-Lyt and ZTA. (a to f) Double-label IFA demonstrating colocalization of POL, PRI, and PAF with SSB in large pseudo-RC in transient assembly assays. SSB was detected by IFA with FITC-labeled anti-SSB rabbit PAb, whereas POL, PRI, and PAF Flag-tagged fusion proteins were detected with rhodamine-labeled anti-Flag mouse MAb. (a and b) Flag-tagged POL cotransfected with the untagged plasmids encoding PPF, PRI, PAF, HEL, and SSB; (c and d) Flag-tagged PRI cotransfected with all five other untagged plasmids; (e and f) Flag-tagged PAF cotransfected with all five other untagged plasmids. The incompletely transfected cell at the upper right in panels c and d displays nuclear diffuse SSB and cytoplasmic PRI. Deliberate omission of any of the six replication genes also disrupted RC formation (not shown). (g to j) Evidence for newly synthesized DNA within KSHV RC formed in Vero cells cotransfected with the complete set of untagged KSHV core replication expression plasmids (POL, PPF, PRI, PAF, HEL, and SSB) plus EBV ori-Lyt and the ZTA DNA binding protein. Cells were pulse labeled for 30 min with BrdU prior to double-label screening for RC formation with an anti-SSB PAb and for active DNA synthesis with an anti-BrdU MAb. (g and i) Anti-KSHV SSB antibody to identify intranuclear RC containing core viral DNA replication proteins. (h and j) Anti-BrdU antibody to identify sites of ongoing DNA synthesis in the same cells (arrows indicate structures that resemble complete viral DNA RC). The anti-BrdU antibody also detected typical background speckled BrdU incorporation patterns found in the 25% of untransfected cells in S phase.
Evidently the formation of these nuclear structures, which closely resembled the complete mature RC seen in other herpesvirus systems, required and incorporated all six of the KSHV core DNA replication proteins. However, obviously we had to consider these as only RC-like instead of complete functionally active RC on the presumption that in the absence of ori-Lyt, they did not synthesize any viral DNA. To exclude the possibility that those structures were synthesizing viral or cellular DNA, we pulse-labeled DNA-transfected cells with BrdU prior to fixation and detected no BrdU incorporation in any viral protein-positive Vero cells, including those with the large RC-like structures (data not shown). Furthermore, by double-label techniques with the appropriate fluorescence filters, we were able to visualize the distribution of cellular chromatin by including DAPI in the mounting solution and found that nuclear DNA proved to be excluded from the large viral RC-like structures (Fig. 7). Therefore, the absence of cellular DNA from these domains appeared to confirm their existence as distinct structures or bodies that could physically displace cellular chromosomal DNA to the margins of the nucleus. Because of the apparent absence of progeny viral DNA also, we have elected to refer to those structures as pseudo-RC to avoid confusion with the authentic but much smaller viral DNA-negative pre-RC structures observed in herpesvirus-infected cells in the presence of PAA or before viral DNA synthesis initiates.
FIG. 7.
Nuclear DNA staining reveals that the RC-like structures assembled by cotransfection of the six core KSHV DNA replication genes exclude or displace cellular chromatin. (a, c, and g) RC-like structures visualized with anti-SSB PAb and FITC or rhodamine-labeled anti-rabbit IgG; (e and f) RC-like structures formed by Flag-POL fusion protein product detected with anti-Flag MAb and rhodamine-labeled anti-mouse IgG; (b, d, f, and h) nuclear DNA in the same cells detected by adding mounting solution contain the intercalating fluorescent dye DAPI.
KSHV core replication proteins replicate EBV ori-Lyt in the presence of the EBV ZTA protein.
We wished to examine whether the KSHV RC-like domains could in fact represent functional precursors to the typical mature herpesvirus RC that actively synthesize viral DNA. Because of the lack of a known KSHV ori-Lyt, we used instead EBV ori-Lyt, which can be efficiently replicated by HSV core replication proteins plus the EBV ZTA initiation protein in transient assays (21). The six core replication genes of KSHV (POL, PPF, PRI, PAF, HEL, and SSB) were cotransfected with a target pBR322 plasmid containing a 5.4-kb EBV ori-Lyt (pEF52) as well as an expression plasmid encoding the multifunctional EBV OBP ZTA (pRTS21). KSHV MTA (ORF57) was included in the cotransfection mixture to potentially enhance the expression level of the KSHV genes, based on previous evidence that the EBV and HCMV versions enhance the efficiency of such assays (21, 61). Prior to fixation, the cells were pulse-labeled with BrdU and subsequently subjected to double-label IFA with KSHV SSB-specific PAb and a BrdU-specific MAb. In a small proportion (0.5 to 1%) of cotransfected cells, the SSB protein displayed large RC-like structures similar to those described above, and these exactly colocalized with large subnuclear domains that actively incorporated BrdU, indicating the presence of ongoing new DNA synthesis (Fig. 6g to j). These compartments were characteristic of the assembled herpesvirus DNA RC seen in both the HCMV and HSV transient assembly assay systems (61, 76) and were not detected in a negative control experiment when EBV ori-Lyt and/or EBV ZTA were omitted (data not shown).
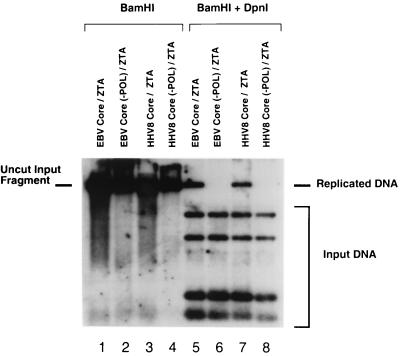
To further verify this finding, a demethylation hybridization assay for transient replication (10) was conducted by cotransfecting the six KSHV core replication genes (POL, PPF, PRI, PAF, HEL, and SSB) with KSHV MTA, EBV ZTA, and EBV ori-Lyt. Total Vero cell DNA recovered from this experiment was analyzed by Southern blot hybridization with an ori-Lyt BamHI fragment probe which revealed the presence of newly synthesized EBV ori-Lyt plasmid DNA that was resistant to DnpI digestion (Fig. 8, lane 7). Therefore, we infered that the input methylated bacterial EBV ori-Lyt plasmid was indeed replicated to produce unmethylated progeny copies by the KSHV core replication genes. When KSHV POL was omitted in the negative control experiment (Fig. 8, lane 8), no replicated EBV ori-Lyt DNA was detected. Therefore, the KSHV pseudo-RC are clearly capable of replicating herpesvirus DNA when an appropriate origin plasmid and OBP are included in the assay.
FIG. 8.
Successful replication of EBV ori-Lyt in cotransfected Vero cells receiving all six essential KSHV viral replication proteins plus EBV ori-Lyt and ZTA. Southern blotting to detect unmethylated EBV ori-Lyt progeny DNA (Challberg assay) was performed with size-fractionated total DNA from cotransfected Vero cells receiving various combinations of replication gene plasmids. Lanes 1 and 5, EBV POL, PPF, PRI, PAF, HEL, and SSB core genes plus ZTA and EBV ori-Lyt; lanes 2 and 6, all EBV expression plasmids except EBV POL; lanes 3 and 7, KSHV POL, PPF, PRI, PAF, HEL, and SSB core genes plus KSHV MTA, EBV ZTA, and EBV ori-Lyt; lanes 4 and 8, all KSHV plasmids plus ZTA and ori-Lyt except for the plasmid encoding KSHV POL. Each DNA sample (10 μg) was digested with BamHI to excise and detect all EBV ori-Lyt DNA (lanes 1 to 4) or digested with BamHI and DpnI to degrade methylated input plasmid DNA and detect only amplified unmethylated ori-Lyt-containing progeny DNA in transfected cells (lanes 5 to 8). A 32P-labeled 5.4-kb BamHI DNA fragment containing the ori-Lyt target sequences isolated from pEF52 was used as the hybridization probe.
K8 targets to PODs and accumulates in the pseudo-RC that are formed by the six core proteins in cotransfection assays.
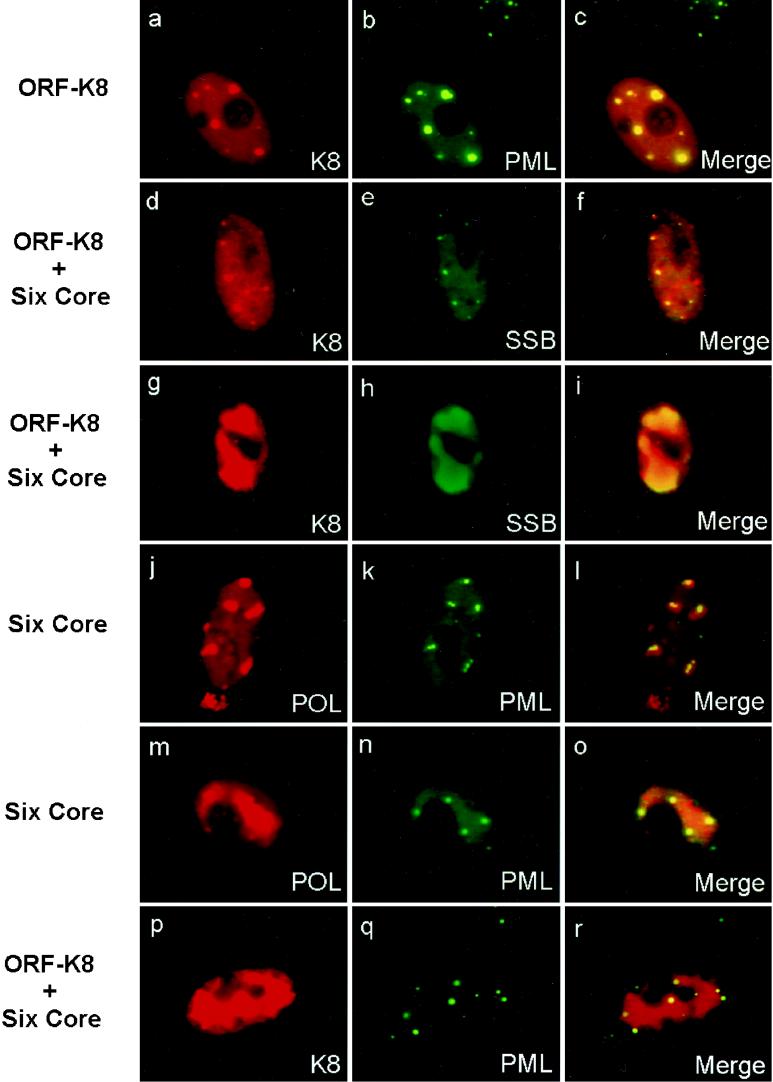
Although there is no significant residual amino acid homology, the K8 protein of KSHV is thought to be evolutionarily equivalent to the ZTA protein of EBV, based on its colinear genomic localization and similar C-terminal splicing pattern producing a leucine zipper motif (42; Chiou et al., submitted). Given the role of ZTA in EBV DNA replication, we investigated whether K8 also accumulates in the pseudo-RC formed in transient cotransfection assays. As shown elsewhere (Chiou et al., submitted), when Vero cells were transfected with a plasmid expressing K8 alone, the K8 protein produced numerous small nuclear punctate bodies, which perfectly colocalized with similar punctate nuclear bodies detected with PAb against the PML proto-oncogene in double-label IFA experiments (Fig. 9a to c). We have shown previously that in HCMV infection, two of the accessory regulatory proteins needed for efficient DNA replication, IE2 and UL112–113, initially target to the PODs and subsequently accumulate into the functionally active RC (2). Therefore, we wished to test whether K8 was also directly incorporated into the KSHV RC.
FIG. 9.
Formation of early KSHV pre-RC incorporating cotransfected ORF-K8 initiates from the periphery of cellular PODs. (a to c) Double-label IFA showing that transfected K8 targets to PODs by itself; detection with rhodamine-labeled anti-Myc MAb and FITC-labeled anti-PML PAb. (d to f) Double-label IFA showing that SSB forms nuclear punctate pre-RC foci associated with PODs when cotransfected with the six core replication genes and K8; detection with FITC-labeled anti-SSB PAb and rhodamine-labeled anti-Myc MAb (K8). (g to i) K8 accumulates in the pre-RC formed by the six core proteins; detection with rhodamine-labeled anti-Myc MAb and FITC-labeled anti-SSB PAb. (j to o) Small pre-RC initiating from the periphery of PODs (j to l), and mature pre-RC closely surrounded by PODs (m to o); detection with rhodamine-labeled anti-Flag MAb (POL) and FITC-labeled anti-PML PAb. (p to r) K8 accumulated in pre-RC surrounded by PODs; detection with rhodamine-labeled anti-Myc MAb and FITC-labeled anti-PML PAb.
To permit greater flexibility in double-label IFA transient assembly assays, a Myc epitope-tagged version of the ORF-K8 mammalian expression plasmid (pFW1) was used. To examine whether the K8 protein colocalized with POL and PAF, Flag-tagged versions of these two core replication genes (pJX8 and pJX12, respectively) were used separately in combination with the Myc-tagged K8 and the five other nontagged core replication genes. Anti-SSB PAb was used to detect SSB in the same cotransfection assays when needed. The results revealed that when cotransfected with the six core replication genes, Myc-K8 was indeed incorporated into large irregularly shaped RC-like structures similar to those previously described for the replication core proteins. Furthermore, using anti-Myc MAb and anti-SSB PAb in double-label IFA, K8 was observed to precisely colocalize with SSB in the large viral pseudo-RC domains (Fig. 9g to i). In similar experiments carried out with Flag-tagged POL and PAF (data not shown), 18, 23, and 16% of the double-positive cells contained RC-like structures for POL and K8, for PAF and K8, and for SSB and K8, respectively. Among these cells, the K8 protein colocalized with POL, PAF, and SSB in the pseudo-RC with over 80% efficiency but still occasionally remained nuclear diffuse (20%) in some cells with positive pseudo-RC. We emphasize that K8 itself was not required for the formation of those core replication structures, because the six replication genes alone were sufficient for generating the pseudo-RC. However, if any of the replication genes were intentionally omitted or not expressed in the cells, RC were not formed and K8 displayed either a nuclear diffuse or nuclear punctate pattern.
The pre-RC of KSHV initially form in association with PODs and finally become surrounded by PODs.
As previously described, the peripheries of the PODs are known to be sites of initiation of RC formation in both HSV and HCMV (31, 32). Therefore, we examined whether PODs were associated with the formation of KSHV pre-RC in a similar manner. Double-label IFA experiments with POL, SSB, and PML as markers were carried out in cotrasfection assays receiving all six KSHV replication genes and K8. We found both typical large irregularly shaped pseudo-RC formed by POL as well as less frequently occurring small globular or nuclear punctate structures. Double staining with anti-PML(C) PAb and anti-flag MAb showed that several PML punctate domains precisely bounded the periphery of each of the large pseudo-RC structures containing Flag-tagged POL (Fig. 9m to o). A similar assay carried out with Flag-tagged PAF yielded the same results (data not shown). A Myc-tagged K8 expression plasmid was also included in the transfection mixture; after double-label IFA with anti-Myc MAb and anti-PML PAb, we found that K8 also accumulated in large RC-like structures that were surrounded by punctate PML bodies (Fig. 9p to r).
In addition to the large pseudo-RC, a small number of the transfected cells contained five to eight much smaller POD-like nuclear punctate structures detected with anti-SSB PAb, which perfectly colocalized with K8 (and presumably PML) (Fig. 9d to f). As a negative control, when SSB or PPF was cotransfected with K8, SSB and PPF remained nuclear diffuse whereas K8 was still nuclear punctate. Clearly, in the absence of all six core replication proteins, SSB or PPF did not associate with K8 or PODs, which confirmed that neither SSB nor PPF intrinsically targeted PODs on their own. In contrast, the viral punctate structures were formed in association with PODs and K8 only when all six core replication proteins were present. These viral nuclear punctate bodies closely resembled the pre-replication initiation foci (pre-RF) found in HSV-infected cells in the presence of PAA to block viral DNA synthesis.
Furthermore, also in the presence of all six core replication proteins, Flag-tagged POL was sometimes found in several nuclear globular foci, which were two to three times larger than the PODs and pre-RF described above, and each globule was typically associated with two adjacent PML nuclear bodies (Fig. 9j to 1). These larger globules were interpreted to be the early or intermediate stages of pre-RC or pseudo-RC formation because they resembled similar early-stage intermediate-sized structures found subsequent to replication initiation in HCMV-infected cells, as well as in transient assembly assays (1, 2, 61). In infection with IE1-deleted HCMV, typically several PML nuclear bodies initially aggregate around pre-RC and subsequently spread out and decorate the boundaries of growing and fused larger RC (2). The finding of these KSHV apparent early-stage pre-RC associated with two or more PML bodies further suggested that the initial stages of KSHV RC formation indeed take place adjacent to PODs just as in HCMV and HSV. In addition, these SSB-positive nuclear globules were also found to colocalize with the K8 globular bodies (data not shown), showing that K8 was also efficiently incorporated into these growing globular structures stemming from the PODs as the pre-RF enlarged to become pseudo-RC.
Overall, the results of these experiments confirmed that just as in both HSV and HCMV-infected cells and transient cotransfection assembly assays, the nuclear substructures formed by KSHV core replication proteins are closely associated with PODs, providing further evidence that these punctate and globular structures are indeed valid pre-RF and pre-RC, even though they are not actually replicating viral genomes because of the absence of ori-Lyt.
Characterization of viral DNA replication compartments in TPA-induced JSC-1 PEL cells and KSHV-infected human primary DMVEC.
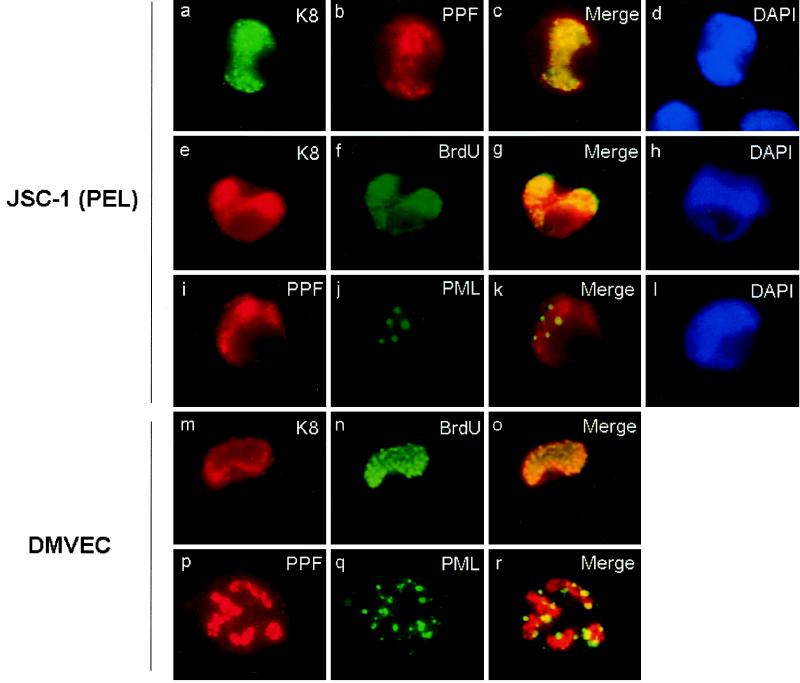
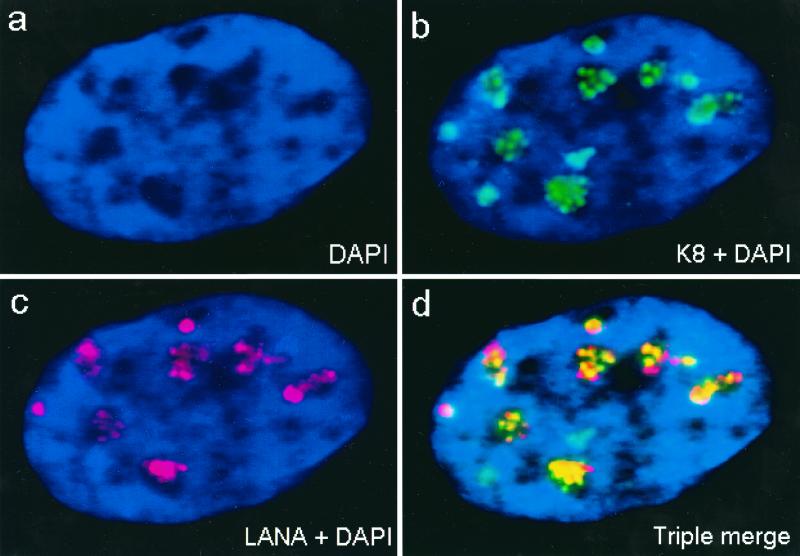
To examine RC in KSHV-infected cells, we used the JSC-1 PEL cell line (5a), which is latently infected with KSHV. After lytic cycle induction with TPA over 72 h, the cells were pulse-labeled with BrdU for 30 min before fixation. Some of the BrdU-labeled cells contained DNA synthesis initiation sites that merged to form kidney-shaped globular subnuclear structures and proved to colocalize with similar structures formed by K8 when examined by double-label IFA with anti-BrdU MAb and anti-K8 PAb (Fig. 10e to h). Furthermore, viral RC could also be detected with anti-PPF MAb, and double-label IFA revealed that K8 also accumulated into the PPF-positive RC as well (Fig. 10a to d). The in vivo finding that K8 was present in the KSHV RC in TPA-induced lymphocytes reinforced the possibility of a role for K8 in lytic cycle DNA replication.
FIG. 10.
Visualization and characterization of viral DNA RC in KSHV-infected DMVEC and TPA-induced JSC-1 PEL cells. (a to d) Double-label IFA detection of ORF-K8 accumulated in RC at 72 h after TPA treatment of JSC-1 PEL cells using FITC anti-K8 PAb and rhodamine anti-PPF MAb. (e to h) Double-label IFA detection of K8 colocalized with newly synthesized DNA in TPA-treated PEL cell RC after a 30-min BrdU pulse-label experiment using rhodamine anti-K8 PAb and FITC anti-BrdU MAb. (i to l) Double-label IFA detection of replication compartments surrounded by PODs in TPA-treated PEL cells, using rhodamine-labeled anti-PPF MAb and FITC-labeled anti-PML PAb. (m to o) Double-label IFA detection of K8 accumulated in RC formed in KSHV-infected DMVEC, using rhodamine anti-K8 PAb and FITC anti-BrdU MAb (30 min of BrdU pulse-labeling). (p to r) Double-label IFA detection of PPF in infected DMVEC RC surrounded by PODs, using rhodamine anti-PPF MAb and FITC anti-PML PAb.
Human DMVEC are permissive host cells for primary infection with supernatant KSHV virons generated from JSC-1 PEL cells after TPA induction (Ciufo et al., submitted), Addition of filter-purified pelleted KSHV virions can completely convert contact-inhibited DMVEC cultures into latently infected spindle cells that all express KSHV LANA (latency-associated nuclear antigen). In addition, the presence of up to 10% of the spindle cells undergoing spontaneous lytic cycle was confirmed by IFA for K8 and other lytic cycle proteins. These experiments also revealed typical herpesvirus RC structures formed by K8 within the nucleus of a subset of lytically infected DMVEC that showed both cytopathic rounding effects and expressed late viral proteins (Ciufo et al., submitted).
To examine RC formation and POD association in infected monolayer cells (which are morphologically superior to the PEL lymphocytes for this purpose), we infected DMVEC with KSHV, 4 days later pulse-labeled the cells with BrdU for 30 min, and then subjected them to double-label IFA with anti-BrdU MAb and anti-K8 PAb. In both virus-infected and mock-infected cultures, 2 to 5% of cells, presumably those cells in S phase, showed BrdU incorporation into randomly distributed nuclear microspeckles or networks representing typical cellular DNA initiation sites or replisomes (data not shown). In the KSHV-infected cultures, those S-phase cells that displayed microspeckled BrdU structures did not express any K8 protein. However, in some cells, the BrdU-labeled DNA synthesis sites coalesced to form distinctive globular or large irregularly shaped nuclear structures, characteristic of viral RC, and K8 was also found to accumulate in most of those BrdU-positive RC (Fig. 10m to o). In contrast, in uninfected cells, no RC-like compartments were detected and no K8 signal was observed.
We also investigated whether the pre-RC in infected cells budded from the periphery of PODs and subsequently were surrounded by PODs as in the cotransfection assembly model. Both KSHV latently infected DMVEC and JSC-1 PEL cells were treated with TPA to induce the viral lytic cycle, and viral RC were visualized by double-label IFA with PPF MAb and anti-PML PAb. Again we found that the cellular PODs indeed closely surrounded the periphery of the PPF-labeled RC in both DMVEC and JSC-1 PEL cells (Fig. 10i to l and p to r). This finding further confirmed the validity of the structures formed in the transient cotransfection assays and the observation that unlike the situation in wild-type HSV and HCMV infections, PML is not displaced from PODs before or during replication in KSHV-infected cells.
Finally, Fig. 11 shows a highly magnified DMVEC nucleus in which the transition between latent and lytic cycle infection appears to be occurring. In this cell, LANA, which associates in a punctate pattern with the multicopy KSHV episomes during latency (3) but disappears after lytic induction, is still present in a punctate pattern (red fluorescence) in interchromatic spaces identified by DAPI staining (blue fluorescence). Interestingly, double IFA for the K8 protein (green fluorescence) showed numerous punctate pre-RC (presumably POD associated) both surrounding and partially colocalized with LANA (yellow and white merge patterns), where the viral episomal genomes are presumably also located.
FIG. 11.
Close association of the KSHV ORF-K8 and LANA proteins in punctate patterns in interchromatin spaces in a KSHV-infected DMVEC spindle cell nucleus undergoing early stages of the transition from latent to lytic cycle gene expression. The four photomicrograph panels show the same nucleus with different combinations of DAPI staining plus FITC or rhodamine IFA. (a) DAPI staining alone (blue) of an infected DMVEC monolayer spindle cell showing several interchromatin gaps. (b) Combined DAPI staining plus IFA with anti-K8 rabbit PAb (FITC, green) showing multiple small K8-positive punctate domains presumably representing PODs that are predominantly localized within some of the interchromatin spaces. (c) Combined DAPI staining plus IFA with anti-LANA mouse MAb (rhodamine, red) showing LANA punctate domains that are also in interchromatin spaces and presumably associated with latent state plasmid KSHV genomes (3). (d) Triple merge combining DAPI, FITC IFA (K8), and rhodamine IFA (LANA), showing close juxaposition and partial colocalization of the K8 punctate domains with each residual LANA punctate domain in the interchromatin gaps.
DISCUSSION
We have shown here that even in the absence of a KSHV ori-Lyt, Vero cells cotransfected with plasmids expressing the six presumed essential KSHV core replication proteins, POL, PPF, PRI, PAF, HEL, and SSB, formed nuclear structures that were morphologically identical to the RC observed in KSHV lytically infected cells. These structures share the following properties with the typical DNA RC seen in other herpesvirus-infected cells: (i) they formed large globular nuclear structures that spared the nucleolus and excluded cellular chromatin to the margins of the nucleus, (ii) they required and incorporated all six essential KSHV core DNA replication proteins, (iii) they were sites of new BrdU-positive DNA synthesis when EBV ori-Lyt and the EBV OBP ZTA were included, and (iv) they initiated from and were found in close association with cellular PODs.
Evaluation of KSHV POL and PAF nuclear translocation.
Intrinsically nuclear proteins often contain nuclear localization signals (NLS) in their protein sequences which allow interaction with cellular nuclear transport proteins to mediate nuclear accumulation (37, 54). Cytoplasmic proteins either lack such NLS or contained masked NLS, but some proteins that have functional roles in the nucleus are transported to the nucleus by physically interacting with other NLS-containing nuclear proteins or proteins that unmask the hidden NLS domains. In KSHV, four of the six core replication proteins (POL, PAF, PRI, and HEL) were intrinsically localized in the cytoplasm. Their subsequent nuclear translocation was one subject of our investigation.
In EBV, PPF (BMRF1), which is thought to be similar to the cellular PCNA sliding clamp, is known to physically interact with POL (BALF5) and serve as an accessory protein to activate and modulate the catalytic activity of POL (34, 35, 38, 69). In HSV, the affinity of POL (UL30) for a synthetic primer-template junction is increased 10-fold by the presence of PPF (UL42), consistent with the notion that herpesvirus PPF acts as a sliding clamp for the herpesvirus POL (25). In KSHV, PPF (ORF59) is thought to be similar to both EBV and HSV PPF and is therefore likely to nonspecifically bind to DNA and interact specifically with POL (ORF9) to improve processivity during DNA replication. Since KSHV PPF is intrinsically a nuclear protein and KSHV POL (unlike EBV POL) is localized in the cytoplasm, the ability of PPF to efficiently translocate POL into the nucleus is consistent with their physical interaction as a complex.
In EBV, PRI (BSLF1), PAF (BBLF2/3), and HEL (BBLF4) are known to form a tripartite primase-helicase complex that translocates efficiently into the nucleus in transient colocalization assays (23). In KSHV, the fact that PRI and HEL must both be present to at least partially translocate PAF into the nucleus, whereas PRI alone was not sufficient, supported expectations that HEL is likely to be needed in addition to PRI and PAF to form the stable KSHV PRI-HEL-PAF tripartite complex. However, unlike the case for EBV, efficient nuclear accumulation of the primase-helicase subcomplex was achieved only in the presence of all three other core replication proteins (SSB, POL, and PPF) as well.
Furthermore, we asked if there were any individual proteins in addition to PRI and HEL that are able to play a direct role in PAF nuclear translocation. KSHV SSB alone was able to partially translocate PAF into the nucleus, which is consistent with evidence that EBV PAF (BBLF2/3) demonstrates a physical interaction with EBV SSB (BALF2) in in vitro assays (23). A similar interaction between HSV SSB (UL29 or ICP8) and the primase-helicase tripartite subcomplex containing PRI (UL52), HEL (UL5), and PAF (UL8) has been proposed (76). Our nuclear translocation results might be evidence for a physical interaction between SSB and PAF in KSHV also. Cotransfection of KSHV PAF with the intrinsically nuclear PPF protein did not significantly alter the cytoplasmic localization of PAF compared to the effect of SSB on PAF, which provides a negative control supporting the specificity of the other interactions by PAF.
In addition, we identified two more KSHV proteins that positively contributed to PAF nuclear translocation; K8 very efficiently and MTA (ORF57) less efficiently. The homologous HCMV UL69 and EBV MTA proteins act as accessory proteins in transient RC assembly and DNA replication assays. EBV MTA has properties of an RNA export or shuttle protein that specifically increases cytoplasmic accumulation of unspliced EBV viral replication gene mRNA (64). The related and positionally homologous KSHV MTA (ORF57) protein also colocalizes with spliceosomes (Chiou et al., submitted) and is likely to be homologous in function to EBV MTA. Although the equivalent protein of HSV (IE63 or ICP27) is essential for viral DNA replication (59) and also partially colocalizes in RC (76), the MTA family proteins are not known to directly interact with replication proteins in any herpesvirus system. Therefore, the exact role or mechanism of MTA, if any, as an accessory factor in KSHV DNA replication has yet to be elucidated. On the other hand, EBV ZTA appears to interact directly with the helicase-primase subunit proteins and is able to translocate them into the nucleus in transient transfection assays (23). Cotransfection with K8, the putative ZTA homologue in KSHV, increased the nuclear localization of PAF almost as efficiently as did SSB, suggesting a possible analogous physical interaction of K8 with the KSHV primase-helicase subcomplex.
Ori-Lyt independent formation of pseudo-RC and associated cellular DNA exclusion.
RC represent subnuclear domains where viral proteins concentrate and promote a favorable environment for viral DNA replication to take place. The classic replicon model for herpesviruses postulates that in order for DNA synthesis to begin, a sequence-specific recognition event by an initiator protein is typically required. In most DNA viruses studied to date, the core origin region is first bound by a virus-specified OBP such as HSV UL9 that recruits the core replication machinery and often also possesses helicase or unwinding activity itself (27, 51, 52, 71). The prevalent idea is that the core replication machinery assembles in association with and after recruitment of the OBP to the origin. In contrast, our results clearly suggest that assembly of the core protein complex can occur independently of its attachment to the origin DNA. Two other studies have hinted that the HSV ori-Lyt and an OBP may not be absolute requirements for assembly of visible replication structures at least in transient cotransfection systems (45, 76), although they certainly are required for viral DNA synthesis itself. In particular, structures resembling pre-RF, pre-RC, and even some large irregular bodies similar to the complete functional RC were formed in the absence of ori-S, although inclusion of an ori-S plasmid did increase the efficiency of the formation of the larger functional RC (76). UL9 (OBP) does appear to be required for SSB to localize in pre-RF in HSV-infected cells (5, 24, 43), but only UL5 (HEL), UL8 (PAF), and UL52 (PRI) are sufficient for the localization of SSB in S-phase-cell micropunctate replisomes in DNA-transfected cells (43, 76).
Perhaps the components of RC formed in some cells cotransfected with the six core replication proteins in the absence of ori-Lyt are overexpressed in comparison to the levels encountered during infection, and an overabundance of the replication proteins may obviate the need for an OBP and the lytic origin DNA. However, it is also plausible that the formation of structurally normal but nonfunctional pseudo-RC containing only the appropriate viral (and cellular) proteins without progeny DNA is an inherent property of the system. The fact that the KSHV pseudo-RC assembled in the nucleus excluded cellular DNA as demonstrated by DAPI staining was interesting. It has long been known that herpesvirus infection marginates cellular chromatin during infection. Therefore, our ability to generate nonfunctional pseudo-RC assembled by cotransfection of only the six core replication genes provides insight into the nuclear environment in which only the empty protein shells of the replication compartments are present but are still able to marginate the cellular chromatin.
To confirm that the endogenous functional RC in herpesvirus-infected cells do not also show DAPI-negative patterns, we infected human foreskin HF cells with HCMV for 72 h and examined the DAPI and IFA patterns after BrdU incorporation (data not shown). We detected distinct HCMV RC by both IFA with IE2-specific PAb and with BrdU-specific MAb. As expected, instead of seeing cellular chromatin exclusion as in the case of pseudo-RC assembly by transient cotransfection, cells with functional RC gave equally intense uniform DAPI staining throughout the nucleoplasm, except for a very narrow dark boundary ring around the RC itself. Furthermore, in situ hybridization with probes specific for HCMV genomic sequences confirmed that viral DNA is indeed found exclusively in the RC at 48 and 72 h after infection (data not shown). These results show unambiguously that the nuclear void observed for the pseudo-RC was now filled with newly synthesized viral DNA in the case of virus-infected cells. Perhaps herpesviruses can create a cellular DNA-free nuclear domain where their own viral replication proteins congregate and then initiate viral DNA replication to fill the nuclear void rather than letting the accumulation of progeny viral DNA itself be the driving force for chromosome margination.
We also observed that in these experiments KSHV replication proteins were not synthesized in S-phase cells in the transfected cultures, as judged by the absence of any speckled BrdU nuclear patterns, which are typical for cells undergoing S phase. Similarly, the transfected HSV and HCMV replication genes were apparently transcribed only in non-S-phase cells in our previous transient assembly assays (61, 76).
EBV ori-Lyt replication by KSHV core replication proteins.
The fact that cotransfection with the six putative core replication genes of KSHV could replicate EBV ori-Lyt in the presence of the EBV OBP (ZTA) proved that they indeed encoded and expressed functional proteins capable of replicating a herpesvirus DNA replication origin. Furthermore, this result showed that these six genes of KSHV are indeed the functional homologues of the complete set of EBV core replication genes in that they could substitute for their EBV counterparts as a whole. This result also supports our previous reports showing that different herpesviruses (HCMV and EBV or EBV and HSV) can complement each other in cotransfection DNA replication assays, provided that the entire core replication complex, rather than just single genes, is replaced (21, 61).
Association of pre-RC with cellular PODs.
PODs are spherical 0.3- to 0.5-μm structures present in most cells, with an average number of 10 to 20 per cell, in which PML and other cellular proteins surround an electron-dense core that is associated with the nuclear matrix. They are believed to be important in viral infection because they are closely associated with sites for initiation of viral IE transcription and DNA replication. For example, the input viral genomes of adenovirus, HSV, and HCMV are preferentially deposited at the periphery of the PODs (2, 4, 19, 31, 32). The idea that the periphery of PODs is also associated with viral DNA relication was demonstrated first for HSV (31), and RC have also been shown to form initially at sites coincident with or adjacent to PODs in both HSV- and HCMV-infected cells. In cotransfected cells receiving the HSV origin DNA plasmid, all seven essential HSV DNA replication proteins are required to initially form three to six spherical pre-RF structures in non-S-phase cells that can subsequently develop into large mature globular and kidney-shaped RC that are actively engaged in viral DNA synthesis as assayed by PAA-sensitive incorporation of BrdU (76). Those few initial spherical pre-RFs are thought to be associated with PODs and are presumed to be the true replication intermediates, whereas the S-phase microspeckled SSB-positive structures are not (44, 45, 70, 76). Some of the HCMV core DNA replication proteins, including SSB (UL57) and PPF (UL44), as well as the nonessential accessory proteins IE2 and UL112-113, also have been shown to be localized in nuclear RC both in infected cells and in transient assembly assays. Both the HCMV IE2 and UL112-113 accessory proteins also localize in or adjacent to PODs at very early times after infection, and they are both subsequently incorporated into large viral RC that appear to initiate and grow from the periphery of PODs (2).
Alternatively, because PML nuclear bodies are found to be situated in the interchromosomal spaces (46) and are also excluded from the large KSHV RC, they may be passively forced to the periphery of RCs by default. However, the observation that ORF-K8 initially targets to the PODs, and that the pre-RFs exactly colocalize with PODs instead of initiating near the PODs, suggests a more active role for PODs as targeting sites for KSHV core replication proteins during the lytic cycle. Because PODs are known to be targets for viral genome deposition, it is likely that the viral replication proteins target to PODs as an effective way to colocate with the template viral DNA. Even in infected DMVEC, PPF (ORF59) can be detected transiently in punctate forms that colocalize with K8 and the PODs (data not shown). Clearly the six core replication proteins, which individually do not associate with PODs, eventually target to PODs when all components of the core complex are present, and K8 (although not needed for this process) may serve as a viral adapter that enhances their association with both the PODs and origin DNA.
Interestingly, unlike HSV and HCMV, which normally displace or degrade PML and effectively disrupt the PODs before efficient viral DNA replication proceeds, KSHV did not demonstrate any signs of loss of PML or disruption of PODs prior to or during lytic DNA replication. Furthermore, unlike HSV IE110 or HCMV IE1, K8 by itself in transient assays targeted to PODs but did not displace or degrade PML. Therefore, K8 more closely resembles HCMV IE2 in this regard, including the ability to later retarget to RC. On the other hand, neither HSV ICP4 or UL9 (OBP) nor the EBV ZTA protein, which all also associate efficiently with RC, are known to target to PODs (although some mutant forms of ZTA do). In other experiments, we have found that the leucine zipper domain of K8 does not substitute for that of ZTA or c-Fos in dimerization and DNA binding assays and that it is therefore extremely unlikely that K8 itself can function as a typical bZIP family DNA binding protein (Chiou et al., submitted). Therefore, the exact functional significance of the association of K8 with both PODs and RC is still unresolved.
In the latent state, LANA associates with episomal KSHV genomes at loci associated with cellular chromosomes but not with PML nuclear bodies (3, 67). Consequently, the partial colocalization of LANA with K8 during the putative switch from latency to the lytic cycle (Fig. 11) implies the possibility of a physical transfer of the KSHV genomes from the cellular chromosome-associated LANA domains to K8-associated PODs. Examination of the mechanism of this putative switching process seems likely to be informative.
ACKNOWLEDGMENTS
These studies were funded by a National Cancer Institute research grant (R01 CA73585) to G.S.H. from the National Institutes of Health. F.Y.W. was a graduate student in Departments of Pharmacology and Molecular Sciences and of Oncology at Johns Hopkins University School of Medicine and was partially supported by the Anti-Cancer Drug Development Training Program (T32 CA09243).
We thank Gangling Liao, Dolores Ciufo, Jianchao Zong, Masahiro Fujimuro, Jae Myun Lee, and Honglin Chen for valuable technical assistance or advice. The gift of mouse anti-ORF59 MAb from Bala Chandran (University of Kansas Medical Center) is gratefully acknowledged.
REFERENCES
- 1.Ahn J H, Brignole III E J, Hayward G S. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol Cell Biol. 1998;18:4899–4913. doi: 10.1128/mcb.18.8.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn J H, Jang W J, Hayward G S. The human cytomegalovirus IE2 and UL112–113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10) J Virol. 1999;73:10458–10471. doi: 10.1128/jvi.73.12.10458-10471.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballestas M E, Chatis P A, Kaye K M. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 4.Burkham J, Coen D M, Weller S K. ND10 protein PML is recruited to herpes simplex virus type 1 prereplicative sites and replication compartments in the presence of viral DNA polymerase. J Virol. 1998;72:10100–10107. doi: 10.1128/jvi.72.12.10100-10107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bush M, Yager D R, Gao M, Weisshart K, Marcy A I, Coen D M, Knipe D M. Correct intranuclear localization of herpes simplex virus DNA polymerase requires the viral ICP8 DNA-binding protein. J Virol. 1991;65:1082–1089. doi: 10.1128/jvi.65.3.1082-1089.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Cannon J S, Ciufo D, Hawkins A L, Griffin C A, Borowitz M J, Hayward G S, Ambinder R F. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi's sarcoma herpesvirus-containing supernatant. J Virol. 2000;74:10187–10193. doi: 10.1128/jvi.74.21.10187-10193.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carey M, Kolman J, Katz D A, Gradoville L, Barberis L, Miller G. Transcriptional synergy by the Epstein-Barr virus transactivator ZEBRA. J Virol. 1992;66:4803–4813. doi: 10.1128/jvi.66.8.4803-4813.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 8.Cesarman E, Moore P S, Rao P H, Inghirami G, Knowles D M, Chang Y. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 9.Cesarman E, Nador R G, Bai F, Bohenzky R A, Russo J J, Moore P S, Chang Y, Knowles D M. Kaposi's sarcoma-associated herpesvirus contains G protein-coupled receptor and cyclin D homologs which are expressed in Kaposi's sarcoma and malignant lymphoma. J Virol. 1996;70:8218–8223. doi: 10.1128/jvi.70.11.8218-8223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Challberg M D. A method for identifying the viral genes required for herpesvirus DNA replication. Proc Natl Acad Sci USA. 1986;83:9094–9098. doi: 10.1073/pnas.83.23.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan S R, Bloomer C, Chandran B. Identification and characterization of human herpesvirus-8 lytic cycle-associated ORF 59 protein and the encoding cDNA by monoclonal antibody. Virology. 1998;240:118–126. doi: 10.1006/viro.1997.8911. [DOI] [PubMed] [Google Scholar]
- 12.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 13.Chang Y N, Dong D L, Hayward G S, Hayward S D. The Epstein-Barr virus Zta transactivator: a member of the bZIP family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J Virol. 1990;64:3358–3369. doi: 10.1128/jvi.64.7.3358-3369.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison C A d, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 15.Chevallier-Greco A, Manet E, Chavrier P, Mosnier C, Daillie J, Sergeant A. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 1986;5:3243–3249. doi: 10.1002/j.1460-2075.1986.tb04635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chi T, Lieberman P, Ellwood K, Carey M. A general mechanism for transcriptional synergy by eukaryotic activators. Nature. 1995;377:254–257. doi: 10.1038/377254a0. [DOI] [PubMed] [Google Scholar]
- 17.Countryman J, Miller G. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc Natl Acad Sci USA. 1985;82:4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davison A J, Taylor P. Genetic relations between varicella-zoster virus and Epstein-Barr virus. J Gen Virol. 1987;68:1067–1079. doi: 10.1099/0022-1317-68-4-1067. [DOI] [PubMed] [Google Scholar]
- 19.Doucas V, Ishov A M, Romo A, Juguilon H, Weitzman M D, Evans R M, Maul G G. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 20.Farrell P J, Rowe D T, Rooney C M, Kouzarides T. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 1989;8:127–132. doi: 10.1002/j.1460-2075.1989.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fixman E D, Hayward G S, Hayward S D. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J Virol. 1995;69:2998–3006. doi: 10.1128/jvi.69.5.2998-3006.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fixman E D, Hayward G S, Hayward S D. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J Virol. 1992;66:5030–5039. doi: 10.1128/jvi.66.8.5030-5039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao Z, Krithivas A, Finan J E, Semmes O J, Zhou S, Wang Y, Hayward S D. The Epstein-Barr virus lytic transactivator Zta interacts with the helicase-primase replication proteins. J Virol. 1998;72:8559–8567. doi: 10.1128/jvi.72.11.8559-8567.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodrich L D, Schaffer P A, Dorsky D I, Crumpacker C S, Parris D S. Localization of the herpes simplex virus type 1 65-kilodalton DNA-binding protein and DNA polymerase in the presence and absence of viral DNA synthesis. J Virol. 1990;64:5738–5749. doi: 10.1128/jvi.64.12.5738-5749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottlieb J, Challberg M D. Interaction of herpes simplex virus DNA polymerase and the UL42 accessory protein with a model primer template. J Virol. 1994;68:4937–4945. doi: 10.1128/jvi.68.8.4937-4945.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gradoville L, Gerlach J, Shedd D, Nikiforow S, Metroka C, Miller G. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J Virol. 2000;74:6207–6212. doi: 10.1128/jvi.74.13.6207-6212.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammarsten O, Elias P, Stow N D. Characterization of a binding site for the herpes simplex virus type 1 UL9 origin-binding protein within the UL9 gene. J Gen Virol. 1996;77:969–976. doi: 10.1099/0022-1317-77-5-969. [DOI] [PubMed] [Google Scholar]
- 28.Hammerschmidt W, Sugden B. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell. 1988;55:427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- 29.Hardwick J M, Tse L, Applegren N, Nicholas J, Veliuona M A. The Epstein-Barr virus R transactivator (Rta) contains a complex, potent activation domain with properties different from those of VP16. J Virol. 1992;66:5500–5508. doi: 10.1128/jvi.66.9.5500-5508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holley-Guthrie E A, Quinlivan E B, Mar E C, Kenney S. The Epstein-Barr virus (EBV) BMRF1 promoter for early antigen (EA-D) is regulated by the EBV transactivators, BRLF1 and BZLF1, in a cell-specific manner. J Virol. 1990;64:3753–3759. doi: 10.1128/jvi.64.8.3753-3759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishov A M, Maul G G. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishov A M, Stenberg R M, Maul G G. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J Cell Biol. 1997;138:5–16. doi: 10.1083/jcb.138.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kenney S, Holley-Guthrie E, Mar E C, Smith M. The Epstein-Barr virus BMLF1 promoter contains an enhancer element that is responsive to the BZLF1 and BRLF1 transactivators. J Virol. 1989;63:3878–3883. doi: 10.1128/jvi.63.9.3878-3883.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiehl A, Dorsky D I. Bipartite DNA-binding region of the Epstein-Barr virus BMRF1 product essential for DNA polymerase accessory function. J Virol. 1995;69:1669–1677. doi: 10.1128/jvi.69.3.1669-1677.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiehl A, Dorsky D I. Cooperation of EBV DNA Pol and EA-D (BMRF-1) in vitro and colocalization in nuclei of infected cells. Virology. 1991;184:330–340. doi: 10.1016/0042-6822(91)90849-7. [DOI] [PubMed] [Google Scholar]
- 36.Kolman J L, Taylor N, Gradoville L, Countryman J, Miller G. Comparing transcriptional activation and autostimulation by ZEBRA and ZEBRA/c-Fos chimeras. J Virol. 1996;70:1493–1504. doi: 10.1128/jvi.70.3.1493-1504.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kutay U, Bischoff F R, Kostka S, Kraft R, Gorlich D. Export of importin alpha from nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- 38.Li J S, Zhou B S, Dutschman G E, Grill S P, Tan R S, Cheng Y C. Association of Epstein-Barr virus early antigen diffuse component and virus-specified DNA polymerase activity. J Virol. 1987;61:2947–2949. doi: 10.1128/jvi.61.9.2947-2949.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lieberman P M, Berk A J. In vitro transcriptional activation, dimerization, and DNA-binding specificity of the Epstein-Barr virus Zta protein. J Virol. 1990;64:2560–2568. doi: 10.1128/jvi.64.6.2560-2568.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lieberman P M, Berk A J. A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA–promoter DNA complex formation. Genes Dev. 1994;8:995–1006. doi: 10.1101/gad.8.9.995. [DOI] [PubMed] [Google Scholar]
- 41.Lieberman P M, Hardwick J M, Sample J, Hayward G S, Hayward S D. The Zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J Virol. 1990;64:1143–1155. doi: 10.1128/jvi.64.3.1143-1155.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin S F, Robinson D R, Miller G, Kung H J. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J Virol. 1999;73:1909–1917. doi: 10.1128/jvi.73.3.1909-1917.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liptak L M, Uprichard S L, Knipe D M. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J Virol. 1996;70:1759–1767. doi: 10.1128/jvi.70.3.1759-1767.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lukonis C J, Burkham J, Weller S K. Herpes simplex virus type 1 prereplicative sites are a heterogeneous population: only a subset are likely to be precursors to replication compartments. J Virol. 1997;71:4771–4781. doi: 10.1128/jvi.71.6.4771-4781.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lukonis C J, Weller S K. Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. J Virol. 1997;71:2390–2399. doi: 10.1128/jvi.71.3.2390-2399.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maul G G, Negorev D, Bell P, Ishov A M. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J Struct Biol. 2000;129:278–287. doi: 10.1006/jsbi.2000.4239. [DOI] [PubMed] [Google Scholar]
- 47.McGeoch D J, Dalrymple M A, Dolan A, McNab D, Perry L J, Taylor P, Challberg M D. Structures of herpes simplex virus type 1 genes required for replication of virus DNA. J Virol. 1988;62:444–453. doi: 10.1128/jvi.62.2.444-453.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov V M, Grossberg S, Chang Y. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol. 1997;71:314–324. doi: 10.1128/jvi.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mullen M A, Gerstberger S, Ciufo D M, Mosca J D, Hayward G S. Evaluation of colocalization interactions between the IE110, IE175, and IE63 transactivator proteins of herpes simplex virus within subcellular punctate structures. J Virol. 1995;69:476–491. doi: 10.1128/jvi.69.1.476-491.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicholas J, Ruvolo V, Zong J, Ciufo D, Guo H G, Reitz M S, Hayward G S. A single 13-kilobase divergent locus in the Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genome contains nine open reading frames that are homologous to or related to cellular proteins. J Virol. 1997;71:1963–1974. doi: 10.1128/jvi.71.3.1963-1974.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olivo P D, Nelson N J, Challberg M D. Herpes simplex virus DNA replication: the UL9 gene encodes an origin-binding protein. Proc Natl Acad Sci USA. 1988;85:5414–5418. doi: 10.1073/pnas.85.15.5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olivo P D, Nelson N J, Challberg M D. Herpes simplex virus type 1 gene products required for DNA replication: identification and overexpression. J Virol. 1989;63:196–204. doi: 10.1128/jvi.63.1.196-204.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pari G S, Anders D G. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J Virol. 1993;67:6979–6988. doi: 10.1128/jvi.67.12.6979-6988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Powers M A, Forbes D J. Cytosolic factors in nuclear transport: what's importin? Cell. 1994;79:931–934. doi: 10.1016/0092-8674(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 55.Ragoczy T, Heston L, Miller G. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J Virol. 1998;72:7978–7984. doi: 10.1128/jvi.72.10.7978-7984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rawlins D R, Milman G, Hayward S D, Hayward G S. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985;42:859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- 57.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 58.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sacks W R, Greene C C, Aschman D P, Schaffer P A. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J Virol. 1985;55:796–805. doi: 10.1128/jvi.55.3.796-805.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sarisky R T, Gao Z, Lieberman P M, Fixman E D, Hayward G S, Hayward S D. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J Virol. 1996;70:8340–8347. doi: 10.1128/jvi.70.12.8340-8347.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sarisky R T, Hayward G S. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J Virol. 1996;70:7398–7413. doi: 10.1128/jvi.70.11.7398-7413.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schepers A, Pich D, Hammerschmidt W. A transcription factor with homology to the AP-1 family links RNA transcription and DNA replication in the lytic cycle of EBV. EMBO J. 1993;12:3921–3929. doi: 10.1002/j.1460-2075.1993.tb06070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schepers A, Pich D, Mankertz J, Hammerschmidt W. cis-acting elements in the lytic origin of DNA replication of Epstein-Barr virus. J Virol. 1993;67:4237–4245. doi: 10.1128/jvi.67.7.4237-4245.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Semmes O J, Chen L, Sarisky R T, Gao Z, Zhong L, Hayward S D. Mta has properties of an RNA export protein and increases cytoplasmic accumulation of Epstein-Barr virus replication gene mRNA. J Virol. 1998;72:9526–9534. doi: 10.1128/jvi.72.12.9526-9534.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 66.Sun R, Lin S F, Gradoville L, Yuan F, Miller G. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1998;95:10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Szekely L, Kiss C, Mattsson K, Kashuba E, Pokrovskaja K, Juhasz A, Holmvall P, Klein G. Human herpesvirus-8-encoded LNA-1 accumulates in heterochromatin-associated nuclear bodies. J Gen Virol. 1999;80:2889–2900. doi: 10.1099/0022-1317-80-11-2889. [DOI] [PubMed] [Google Scholar]
- 68.Taylor N, Flemington E, Kolman J L, Baumann R P, Speck S H, Miller G. ZEBRA and a Fos-GCN4 chimeric protein differ in their DNA-binding specificities for sites in the Epstein-Barr virus BZLF1 promoter. J Virol. 1991;65:4033–4041. doi: 10.1128/jvi.65.8.4033-4041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsurumi T, Daikoku T, Nishiyama Y. Further characterization of the interaction between the Epstein-Barr virus DNA Pol catalytic subunit and its accessory subunit with regard to the 3′-to-5′ exonucleolytic activity and stability of initiation complex at primer terminus. J Virol. 1994;68:3354–3363. doi: 10.1128/jvi.68.5.3354-3363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Uprichard S L, Knipe D M. Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology. 1997;229:113–125. doi: 10.1006/viro.1996.8430. [DOI] [PubMed] [Google Scholar]
- 71.Weir H M, Stow N D. Two binding sites for the herpes simplex virus type 1 UL9 protein are required for efficient activity of the oriS replication origin. J Gen Virol. 1990;71:1379–1385. doi: 10.1099/0022-1317-71-6-1379. [DOI] [PubMed] [Google Scholar]
- 72.Whitby D, Howard M R, Tenant-Flowers M, Brink N S, Copas A, Boshoff C, Hatzioannou T, Suggett F E, Aldam D M, Denton A S. Detection of Kaposi sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi's sarcoma. Lancet. 1995;346:799–802. doi: 10.1016/s0140-6736(95)91619-9. [DOI] [PubMed] [Google Scholar]
- 73.Wu C A, Nelson N J, McGeoch D J, Challberg M D. Identification of herpes simplex virus type 1 genes required for origin-dependent DNA synthesis. J Virol. 1988;62:435–443. doi: 10.1128/jvi.62.2.435-443.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yates J, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci USA. 1984;81:3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zalani S, Holley-Guthrie E, Kenney S. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc Natl Acad Sci USA. 1996;93:9194–9199. doi: 10.1073/pnas.93.17.9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhong L, Hayward G S. Assembly of complete, functionally active herpes simplex virus DNA replication compartments and recruitment of associated viral and cellular proteins in transient cotransfection assays. J Virol. 1997;71:3146–3160. doi: 10.1128/jvi.71.4.3146-3160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu F X, Cusano T, Yuan Y. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J Virol. 1999;73:5556–5567. doi: 10.1128/jvi.73.7.5556-5567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]