Abstract
The parts of the RNA genome of infectious bronchitis virus (IBV) required for replication and packaging of the RNA were investigated using deletion mutagenesis of a defective RNA (D-RNA) CD-61 (6.1 kb) containing a chloramphenicol acetyltransferase reporter gene. A D-RNA with the first 544, but not as few as 338, nucleotides (nt) of the 5′ terminus was replicated; the 5′ untranslated region (UTR) comprises 528 nt. Region I of the 3′ UTR, adjacent to the nucleocapsid protein gene, comprised 212 nt and could be removed without impairment of replication or packaging of D-RNAs. A D-RNA with the final 338 nt, including the 293 nt in the highly conserved region II of the 3′ UTR, was replicated. Thus, the 5′-terminal 544 nt and 3′-terminal 338 nt contained the necessary signals for RNA replication. Phylogenetic analysis of 19 strains of IBV and 3 strains of turkey coronavirus predicted a conserved stem-loop structure at the 5′ end of region II of the 3′ UTR. Removal of the predicted stem-loop structure abolished replication of the D-RNAs. D-RNAs in which replicase gene 1b-derived sequences had been removed or replaced with all the downstream genes were replicated well but were rescued poorly, suggesting inefficient packaging. However, no specific part of the 1b gene was required for efficient packaging.
Infectious bronchitis virus (IBV) belongs to the genus Coronavirus of the family Coronaviridae in the order Nidovirales (5). Coronaviruses have a single-stranded, nonsegmented, positive-sense RNA genome of between 27.4 and 31 kb, that of IBV being 27.6 kb (16). Defective RNAs (D-RNAs) are being used to identify the cis-acting sequences required for coronavirus replication, transcription, and packaging.
The 3′-most 55 nucleotides (nt) of the virus genome have been shown previously to be sufficient for negative-strand synthesis of mouse hepatitis coronavirus (MHV) D-RNAs (18). However, larger regions of both the 5′ and 3′ termini of the D-RNAs were required for synthesis of positive-stranded RNA. The numbers of nucleotides identified as being required at the 5′ end were approximately 460 for MHV strain JHM, reviewed by Lai and Cavanagh (16), and up to 1,348 for transmissible gastroenteritis virus (TGEV) (13). At the 3′ terminus, between 436 and 461 nt were required for MHV D-RNAs (14, 17, 32); most or all of the 3′ untranslated region (UTR) (288 nt), including a pseudoknot, was required for the related bovine coronavirus (BCoV) (35); and 492 nt were required for TGEV (13). Hsue and Masters (12) showed that a sequence located at the 5′ end of the 3′ UTR of a D-RNA of MHV was predicted to fold into a stable stem-loop structure and was essential for replication. The BCoV D-RNA pDrep1 required the complete N gene sequence in addition to the 3′ UTR; deletions within the N gene prevented replication (6).
Comparisons of naturally occurring MHV D-RNA sequences accompanied by deletion mutagenesis studies have identified a region considered to be essential for RNA packaging. This is a 61-nt stem-loop structure present in the 1b region of gene 1, the replicase gene (10, 22, 31). Cologna and Hogue have identified a similar sequence in BCoV, a group II coronavirus like MHV (8). Notwithstanding, MHV-JHM D-RNA DIssE does not have this 61-nt stem-loop but was replicated and packaged, although to a lesser extent than were the D-RNAs with this stem-loop (20). The analogous region is also absent from D-RNA pDrep1 of BCoV, which contains no replicase 1b sequence. This indicated that the packaging signal for the BCoV D-RNA is in one or more of the regions that comprise the D-RNA; the 5′ UTR, part of the 1a sequence of gene 1, the N gene, or the 3′ UTR (4). No single region for packaging has been identified in TGEV D-RNAs. Rather, two regions, designated F1 and F2, from the replicase 1b region have been implicated, although neither is absolutely required for packaging (13).
The starting point for this work was D-RNA CD-61, derived from a naturally occurring 9.1-kb IBV D-RNA, CD-91 (24), by deletion of 3.0 kb (Fig. 1) (25). We have made an additional 21 deletion mutant D-RNAs, using a chloramphenicol acetyltransferase (CAT) reporter gene to distinguish the processes of replication and packaging. Previously, we relied upon several passages of the rescued D-RNAs to produce sufficient RNA to be detectable by Northern blot analysis. The incorporation of a CAT reporter gene into the D-RNAs (28) has allowed us to detect replication of IBV D-RNA constructs in transfected cells, without reliance on packaging to indicate that replication has occurred. Thus, we have been able to distinguish the processes of replication and packaging.
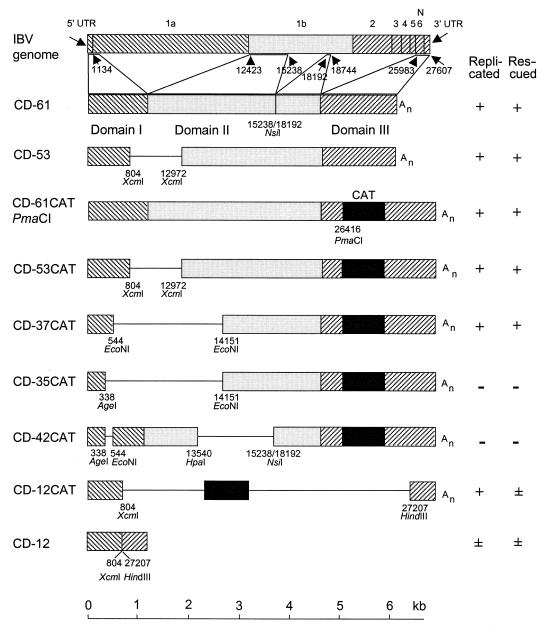
FIG. 1.
D-RNAs made to investigate the sequences derived from the 5′ end of the IBV genome necessary for replication and rescue (rescue indicates replication and packaging of the RNA into virus particles). The top diagram shows the genome of IBV (not to the same scale as that of the other diagrams), with the six genes marked, and shows the parts which have been retained in D-RNA CD-61. Thin horizontal lines indicate deletions relative to CD-61. Numbers under the diagrams are restriction site positions relative to the IBV Beaudette genome (27,607 nt; data bank accession no. M95169). Replication and rescue were determined by quantification of CAT protein by ELISA or, for D-RNAs without a CAT gene, by Northern blot analysis. A “+” for replication and for rescue indicates that the amount of CAT protein expressed from a D-RNA was very similar to the amount of CAT protein expressed by D-RNA CD-61CATPmaCI in passage P0 (Vero cells; replication) and after serial passage in CK cells (rescue), respectively. The “±” for CD-12 indicates that it was detected only by RT-PCR. The “±” for CD-12CAT indicates that only very low levels of CAT were expressed during passage. Restriction sites are indicated by their positions in the IBV Beaudette genome. An, poly(A).
MATERIALS AND METHODS
Virus and cells.
IBV Beaudette-US was grown in embryonated eggs (29). African green monkey kidney Vero cells were used for the electroporation step (passage P0) in most experiments, and primary chick kidney (CK) cells were used in all experiments for passage of the D-RNAs for up to five or six passages (P1 to P6) to amplify the D-RNAs, as previously described (24, 25, 28, 29).
Construction of cDNAs encoding D-RNAs.
A plasmid, pCD-91, containing the cDNA of the IBV D-RNA CD-91, derived from IBV Beaudette (24), was used as the starting point for the production of pCD-61, which contained D-RNA CD-61 cDNA (25). Other D-RNAs were produced by deletion mutagenesis of pCD-61 using the restriction enzymes indicated in Fig. 1, 2, 4, and 6. CAT was inserted into pCD-61 at two positions, the SnaBI (nt 13045) (Fig. 2) and PmaCI (nt 26416) (Fig. 1) sites, under the control of IBV transcription-associated sequence 5 (TAS5) (28). These constructs were used as templates for the construction of smaller D-RNAs, as indicated in Fig. 1, 2, 4, and 6.
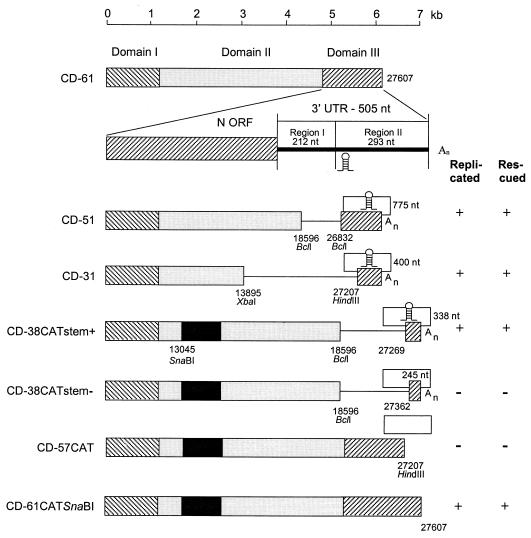
FIG. 2.
D-RNAs made to investigate the sequences derived from the 3′ end of the IBV genome necessary for replication and rescue. The 507-nt 3′ UTR has been expanded to show the variable and conserved regions I and II, respectively. The open boxes on the right-hand side indicate regions I and II of the 3′ UTR and the poly(A) tail, and the numbers indicate how many of these nucleotides remain. Other details are explained in the legend to Fig. 1.
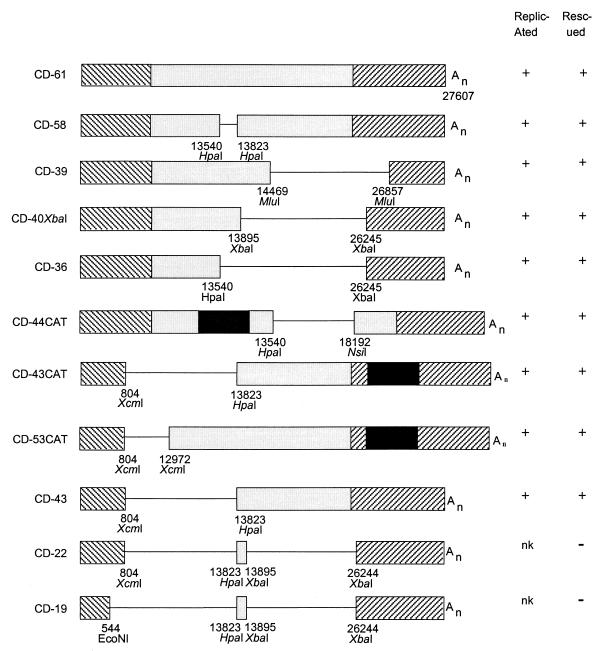
FIG. 4.
D-RNAs made to investigate sequences necessary for packaging of the RNA into viruslike particles. nk, not known. Any D-RNA without a CAT reporter gene could be detected by Northern blot analysis only after one or more passages (P1 onwards)—replication could not be assessed if rescue had not occurred. Other details are explained in the legend to Fig. 1.
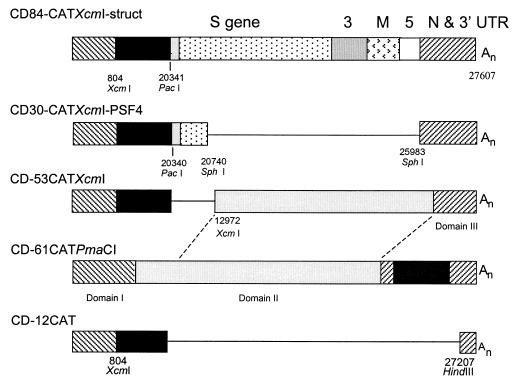
FIG. 6.
D-RNAs used to study the deletion of replicase 1b sequence and its replacement by structural protein genes. In these constructs, the CAT reporter gene was inserted at an XcmI site, 804 nt from the 5′ end.
PCR mutagenesis was used to create the D-RNA clones pCD-38CATstem+ and CD-38CATstem− (Fig. 2). The positive-sense oligonucleotides used were dom1 end (TGAT27269CATTAGTTTGCTTTATCGTAG27290) and stmout (TGATCAG27362TCTAATCTGTCTACTTAG27383), respectively; underlined nucleotides show a BclI restriction site, and the superscript numbers denote the IBV genomic position of the nucleotide to the left of the superscript numbers. The negative-sense oligonucleotide was 3′ beau (GCGGCCGCTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGCTCTAACTC), where a NotI site is underlined and GCTCTAACTC are the last 10 nt of the Beaudette genome. 3′ UTRs of the desired length were cloned into CD-61CATSnaBI digested with BclI and NotI.
The restriction enzyme sites XcmI and HindIII were joined onto the 5′ and 3′ ends, respectively, of the IBV TAS5-CAT sequence, using PCR with oligonucleotides XcmICAT+ (CCACTTCTAAGTTGGGTGTTTTACTTAACAAAAACT) and HinCAT− (AAGCTTGGGGGTTACGCCCCGCCCTGC), where the underlined nucleotides indicate the XcmI and HindIII restriction sites. This product was ligated to the 5′-terminal XcmI fragment and 3′-terminal HindIII fragment of the IBV genome, to produce CD-12CAT, modified to contain unique PacI and SacI sites between the XcmI and HindIII sites.
A D-RNA containing all the genes downstream of the replicase gene, CD-84CATXcmI-struct, was generated by insertion of a PacI-SacI fragment, comprising nt 20341 to 27608 of the IBV genome, downstream of the CAT gene in CD-12CAT (Fig. 6). Another D-RNA without any replicase 1b gene and only a small part of the S gene plus part of the N gene and all of the 3′ UTR, CD-30CATXcmI-PSF4 (Fig. 6), was generated from CD-84CATXcmI-struct by deletion mutagenesis using SphI present at position 20743 of the IBV genome and SphI introduced at nt 25983 by PCR.
Sequence analysis of cloned D-RNAs was performed to check the integrity of the D-RNAs at the positions where deletions, insertions, or other mutations had been made.
RNA electroporation of Vero cells.
T7-derived RNA transcripts corresponding to the various D-RNAs were synthesized in vitro from 2 μg of the corresponding NotI-linearized D-RNA-containing plasmids (25). Vero cells (P0) were grown to 80 to 90% confluence in 25-cm2 tissue culture flasks (Falcon) and infected with 0.5 ml of Beaudette helper virus in allantoic fluid. At 8 h postinfection (p.i.), the cells were electroporated with the transcription reaction mixtures (29). Following incubation of the electroporated cells for 16 h, virus (V1) in 1 ml of culture medium was used to infect CK cells (P1) and after 20 to 24 h p.i. virus (V2) in culture medium was passaged on CK cells (P2) for up to P5 or P6. Experiments showed that optimal rescue was obtained when the electroporation stage (P0) was done with Vero cells and subsequent passages (P1 to P6) were done with CK cells.
Analysis of IBV-derived RNAs.
Northern blot analysis on total cellular RNA was performed as described in reference 29. Two probes were used: (i) a 590-bp IBV 3′ probe, to detect all IBV-derived RNAs, corresponding to nt 27017 to 27607 at the 3′ end of the IBV genome, and (ii) an IBV 5′ probe, minus the leader sequence, to detect IBV genomic RNA and D-RNAs, consisting of an 1,120-bp AgeI-SphI fragment (nt 340 to 1460). All the probes were labeled with [32P]dCTP (29).
Analysis of CAT reporter gene.
The CAT reporter gene has been described previously (28). Briefly, cells were lysed, serial dilutions were made, and CAT protein was detected by enzyme-linked immunosorbent assay (ELISA) (Boehringer Mannheim; product no. 1363727), the amount of CAT protein present being determined by comparison to standard amounts of CAT protein.
Nucleotide sequence accession numbers.
The sequences of regions I and II of the 3′ UTRs of five strains of IBV sequenced for this report have been submitted to the EMBL database and have been assigned accession numbers as follows: strain H120, AJ278336; D207, AJ278335; HV10, AJ278337; HVI-140, AJ278338; and UK/918/68, AJ278334. The accession number for the complete sequence of the IBV Beaudette genome is M95169.
RESULTS
Penzes et al. (25) had shown that the number of cells both infected with helper IBV and electroporated with D-RNAs was low; several passages of the D-RNAs with helper virus were usually required to detect the D-RNAs by Northern blot analysis. If a D-RNA was not detected after several passages (not rescued), it could have been because (i) the D-RNA had not been replicated or (ii) it had been replicated but not packaged. Replication of CD-61CAT could be detected at P0 by a CAT ELISA when a CAT reporter gene, under the control of an IBV TAS, had been inserted at the PmaCI or SnaBI site to produce CD-61CATPmaCI (Fig. 1) and CD-61CATSnaBI (Fig. 2), respectively (28). Transcription of a unique CAT mRNA would occur only if the D-RNA was replication competent. Rescue was detected by production of CAT protein following serial passage of CAT gene-containing D-RNAs. This confirmed that the D-RNAs had been replicated and were capable of being packaged. At least two experiments were performed with each CAT-containing D-RNA. Absolute CAT values varied between experiments with a given D-RNA, probably due to differences in the adverse effects of electroporation on cells at P0 and the use of primary CK cells with batch-to-batch variation.
Packaging was also studied using D-RNAs without CAT, in which case Northern blot analysis was used after several passages.
Extent of domain I (5′ end) required for D-RNA replication.
Domain I of CD-61 comprises the 528 nt of the 5′ UTR plus the first 608 nt of open reading frame (ORF) 1a. A number of deletion mutants were created with progressively fewer nucleotides at the 3′ end of domain I (Fig. 1). D-RNAs CD-61CATPmaCI and CD-61, with and without CAT, respectively, were used as positive controls. Examples of CAT ELISA values are shown in Table 1. The number in the name of each D-RNA refers to the size, in kilobases, of the D-RNA moiety, excluding the reporter gene, when present.
TABLE 1.
Replication and rescue of D-RNAs containing structural protein genes and the CAT reporter gene at the position 804 XcmI site
| Expt. no. and D-RNA | Amt of CAT (ng/106 cells) in passage:
|
|||||
|---|---|---|---|---|---|---|
| P0 | P1 | P2 | P3 | P4 | P5 | |
| Expt. 1 | ||||||
| CD-61CATPmaCI | 3.12 | 40.74 | 73.26 | 70.39 | NDa | ND |
| CD-84CATXcmI-struct | 4.1 | 0.61 | 0.65 | 0.15 | ND | ND |
| CD-30CATXcmI-PSF4 | 3.10 | 2.08 | 0.26 | 0.00 | ND | ND |
| Expt. 2 | ||||||
| CD-61CATPmaCI | 14.94 | 162.14 | 1,748.63 | 526.93 | 862.03 | 388.58 |
| CD-53CATXcmI | 35.57 | 156.64 | 84.56 | 390.76 | 37.06 | 7.9 |
| CD-12CATXcmI | 5.36 | 0.09 | 0.04 | 0.19 | 0.01 | 0.01 |
| CD-84CATXcmI-struct | 1.21 | 0.8 | 0.15 | 0.04 | 0.01 | 0 |
| CD-30CATXcmI-PSF4 | 49.61 | 34.17 | 3.12 | 0.64 | 0.03 | 0 |
ND, not done.
Constructs CD-53CAT and CD-37CAT (Fig. 1) produced CAT protein, showing that the 5′ first 804 and 544 nt, respectively, were sufficient for replication. CD-53 was also detected in infected cells by Northern blot analysis after serial passage. Constructs CD-35CAT (comprising 338 nt of the 5′ UTR) and CD-42CAT (missing nt 338 to 544) (Fig. 1) did not produce any CAT in P0. This indicated that the 5′-terminal 339 or more nt, up to 544 nt, were required for replication.
No part of domain II (replicase gene) is specifically required for D-RNA replication.
Domain II of CD-61 comprised discontinuous parts of ORF 1b (6,322 nt) (Fig. 1). A construct, CD-12CAT (Fig. 1), consisting of the 5′ 804 nt and 3′ 400 nt from CD-61 plus a CAT gene was created. CD-12CAT produced CAT protein at P0, with amounts of CAT similar to those for CD-61CAT, showing that the 5′-terminal 804 nt and 3′-terminal 400 nt were sufficient for replication; no part of ORF 1b of the replicase gene or of the N gene was required.
Extent of domain III (3′ end) required for D-RNA replication.
Domain III (1,626 nt in total) of CD-61 comprised part of the N gene and the whole of the 3′ UTR. A series of deletion mutants were created within domains II and III of CD-61. Defective RNAs CD-51 and CD-31 retained the 3′-terminal 775 and 400 nt, respectively, and lacked the remainder of domain III and part of domain II (Fig. 2). They were rescued, as shown by detection following Northern blot analysis after serial passage in CK cells, confirming that no part of the N gene was required in cis for replication or packaging.
Williams et al. (33) compared the sequences of the 3′ UTRs of six IBV strains (Beaudette, M41, Gray, Ark99, KB8523, and H52), isolated over a period of several decades and showed that they could be divided into two regions. Region I, adjacent to the N gene (Fig. 2), was hypervariable (53.2 to 92.8% nucleotide identity), including large deletions. In contrast, the 3′-most region II (Fig. 2) was highly conserved (94.3 to 97.8% identity). We have sequenced the 3′ UTR of the H120 strain (closely related to strain H52) and of four additional European isolates (D207, HV10, HVI-140, and 918/68). Sapats et al. (27) and Breslin et al. (3) sequenced the 3′ UTRs of eight Australian IBV and three turkey coronavirus (TCoV) isolates, respectively. Taken together, the data confirm that region I is highly variable (comprising 212 nt for strain Beaudette) and that region II is relatively conserved (comprising 293 nt for strain Beaudette).
CD-38CATstem+ was designed to lack most of region I of the UTR and to retain the last 338 nt of the genome, i.e., it retained region II (Fig. 2). This D-RNA was replicated and packaged, as observed by detection of CAT protein from P0 to P5.
Construct CD-38CATstem− was similar to CD-38CATstem+ except for the deletion of a further 93 nt from the 3′ UTR, corresponding to the rest of region I and the 5′ end of region II (Fig. 2). This construct was not replicated by helper virus; P0 and subsequent passages were negative for CAT protein, and the D-RNA was not detectable by reverse transcription-PCR (RT-PCR) using oligonucleotides 93/117 and Beau3′. Thus, the first 57 nt of region II of the 3′ UTR were essential for replication.
Predicted stem-loop.
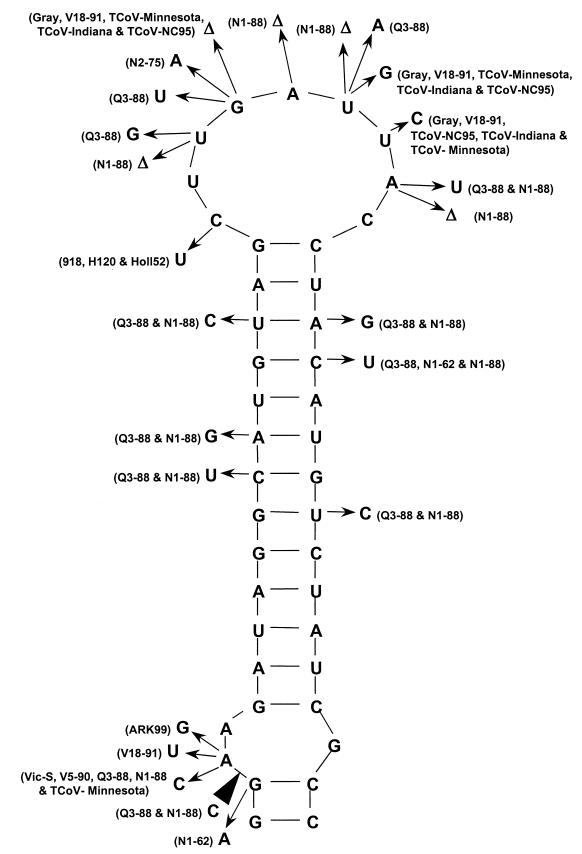
Secondary structure analysis of the entire 3′ UTRs of the 19 U.S., European, and Australian IBV strains referred to above, plus 3 strains of TCoV (2, 11), using the software package RNAdraw (23), predicted a conserved stem-loop structure of 42 nt situated from nt 27312 to 27353 in the Beaudette genome. Figure 3 shows the predicted stem-loop structure for IBV Beaudette and the nucleotide substitutions identified for the U.S., European, and Australian IBV strains. The nucleotide differences were predicted not to affect the stem-loop structure. Either the base changes in one side of the stem were covariant, or a single base change did not lead to loss of base pairing and alteration of the predicted structure. These changes strengthened the likelihood that the predicted stem-loop structure did exist. The Australian N1-88 and V18-91 strains and the American Gray strain showed the most sequence differences, including transitions, transversions, and deletions, from the Beaudette-U.S. sequence. The deletions occurred exclusively in the predicted loop region (N1-88, Gray, and V18-91). The pseudoknot predicted by Williams et al. (35) for BCoV commenced 9 nt downstream from the 3′ end of the predicted stem-loop structure and was retained in CD-38CATstem−. Thus, if the pseudoknot is present in IBV at the analogous location it alone is not sufficient for replication. Other predicted structures in the 3′ UTRs differed from strain to strain. CD-38CATstem+, which retained the nucleotides comprising the predicted stem-loop, was replicated, and CD-38CATstem−, which lacked the stem-loop, was not replicated, as reported above (Fig. 2). This indicated that nt 183 to 276 of the 3′ UTR (genome nt 27291 to 27384), inclusive of the predicted stem-loop structure, were essential for replication of the D-RNA.
FIG. 3.
Schematic representation of the predicted stem-loop structure present in region II of the 3′ UTR of IBV; 19 IBV and 3 TCoV isolates were compared. The sequence of the predicted structure corresponds to nt 27312 to 27353 of the genome of the Beaudette strain. The arrows show the positions of nucleotide differences of the strains indicated alongside. Those nucleotides not marked by arrows did not vary between the strains. Deletions are indicated by “▵,” and an insertion is indicated by a black arrowhead. The 3′ UTR sequences were established by Boursnell et al. (1) (IBV Beaudette and M41), Williams et al. (34) (IBV Gray, Arkansas 99, and Holland 52), Sutou et al. (30) (IBV KB8523), Sapats et al. (27) (IBV Vic S, V5/90, N1/62, N9/74, N2/75, N1/88, Q3/88, and V18/91), and Breslin et al. (3) (TCoV isolates Minnesota, Indiana, and NC95) and by us for the remaining isolates. Strains M41, D207, HV10, HV140, KB8523, and N9-74 had the same sequence and potential structure as IBV Beaudette.
Sequences required for packaging of IBV D-RNA.
Several constructs which each lacked part of the ORF 1b replicase sequence were made (Fig. 4). All of them were replicated and packaged, including CD-44CAT. A previous version of CD-44 without CAT (25) had not been rescued; we assume that a mutation during the production of CD-44 was responsible for that. The rescue of several D-RNAs that did not contain the CAT gene is shown in Fig. 5. The results showed that no specific region of 1b was required for replication or packaging.
FIG. 5.
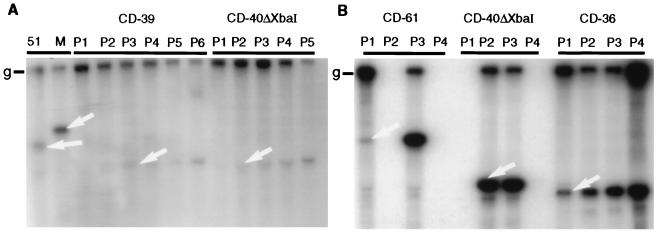
Detection of D-RNAs by Northern blot analysis. D-RNAs CD-61, CD-36, CD-39, and CD-40ΔXbaI without a CAT reporter gene were electroporated into helper virus-infected CK cells (A) or Vero cells (B) at 8 h p.i. Resultant particles were serially passaged in CK cells (P1 to P6). Total cellular RNA was extracted, electrophoresed in an agarose gel, and blotted onto nitrocellulose filters that were then probed with a 5′ genomic probe. The arrows show the position of the expected D-RNA band. Lane 51 shows CD-51 at P0, and lane M is a marker RNA that contains CD-61 P4 RNA. Blank lanes were a consequence of unsuccessful recovery of total RNA. The replication and rescue of these D-RNAs are shown in Fig. 4. g, IBV genomic RNA.
CD-22 and CD-19 (Fig. 4) and CD-12 and CD-12CAT (same deletions as each other) (Fig. 1) contained the 5′- and 3′-terminal regions of the genome that had been shown to be the only specific sequences required for replication of IBV D-RNA. However, none of these four D-RNAs were detected by Northern blot analysis after rescue attempts, following several passages in CK cells, although CD-12 was detectable by RT-PCR in P2 and P3 extracts. CD-12CAT was replicated in P0 cells, as evidenced by CAT production, and on subsequent passage (P1 to P3) cells were still positive for CAT though to a lesser extent than in P0 and less than control CD-61CAT in P1 to P3 cells. Thus, although the small D-RNAs were replicated, the rescue was poor.
D-RNAs containing structural protein genes were rescued poorly.
The small size of CD-12CAT might have contributed to its poor rescue compared with CD-61CAT. To investigate this, CD-84CATXcmI-struct was constructed. This comprised the 5′ end of CD-12CAT, corresponding to the first 804 nt of CD-61 with the CAT gene inserted at the XcmI site; the 3′ part of the replicase gene, starting at position 20341 (a PacI site); and all the remainder of the genome, i.e., including all the structural protein genes, total length, including the CAT gene, being 9,140 nt (Fig. 6). The amount of CAT expressed at P0 and at P1 to P5 produced from this construct was compared with that from CD-61CATPmaCI in two experiments (Table 1). Although the absolute CAT values varied between the two experiments, the trends in the two experiments were the same. Thus, whereas passage of CD-61CATPmaCI resulted in increasing amounts of CAT, the amount of CAT protein produced by CD-84CATXcmI-struct declined after P0.
D-RNA CD-30CATXcmI-PSF4 (3.7 kb) was derived from CD-84CATXcmI-struct by removal of the sequence corresponding to genome positions 20740 to 25983 (Fig. 6). Thus, this D-RNA had approximately 400 nt corresponding to the beginning of the S protein gene and most of the N gene. This construct produced amounts of CAT protein similar to those for CD-61CATPmaCI in P0, after which CAT expression declined rapidly on serial passage, as it did for CD-84CATXcmI-struct (Table 1). The small D-RNA CD-12CAT behaved similarly to CD-84CATXcmI-struct and CD-30CATXcmI-PSF4 (Table 1; in other experiments, the amounts of CAT protein expressed by CD-12CAT in P0 were the same as or higher than those expressed by CD-61CATPmaCI). In case the expression of CAT was affected by the position of the CAT reporter gene, i.e., at the XcmI site at position 804 rather than further downstream, as in CD-61CATSnaBI and CD-61CATPmaCI, D-RNA CD-53CATXcmI was made. In this D-RNA, the CAT gene was at position 804 (Fig. 6) but most of domain II from CD-61 was conserved. The amounts of CAT produced by CD-53CATXcmI were similar to those of CD-61CATPmaCI (Table 1, experiment 2). Thus, the results showed that D-RNAs lacking most of domain II (replicase 1b gene) were rescued very inefficiently and that domain II could not be successfully replaced by structural gene sequences.
DISCUSSION
Sequences required for RNA replication.
Our data show that the only sequences required for the replication of IBV D-RNAs are within the 5′ 544 nt and 3′ 388 nt, i.e., essentially within the 5′ and 3′ UTRs of the IBV genome. The first 544 nt of the genome include the first 15 nt of ORF 1a; it is possible that these ORF 1a nucleotides are part of the sequence required for replication. Our results are in contrast to those for MHV D-RNA DIssF, which requires a 58-nt region, approximately 3 kb from the 5′ end of ORF 1a, that folds into a stem-loop secondary structure (15, 17, 26). Our findings are similar to those with the D-RNAs of MHV A-59 (19) and BCoV (7), which do not require internal regions from the replicase 1a gene for replication.
A small (2.1 kb) D-RNA of TGEV was efficiently replicated (13). The first 5′ continuous part of this D-RNA was 1.3 kb in length, the first 315 nt being the 5′ UTR (9). It is possible that only a part of the first 1.3 kb is essential; smaller fragments were not investigated. No sequences corresponding to replicase 1b sequence were required for the replication of the IBV D-RNAs, in keeping with the findings for small D-RNAs of TGEV (13), MHV-JHM (21), and BCoV (7).
The BCoV pDrep1 D-RNA required almost the entire N gene for replication (6). Our D-RNAs CD-12, CD-12CAT, CD-31, CD-51, and CD-38CATstem+ did not contain any N gene sequence nor did the 9.7-kb TGEV DI-C or the small (3.3-kb) derivative DI-M33 which was replicated by helper TGEV (13). Defective RNAs of MHV contain part of the N gene, much of which can be removed without impairment of replication. Thus, some 378 to 463 nt of the 3′-terminal end of the genome are required for replication, depending on the strain of MHV and the particular D-RNA, most being derived from the 3′ UTR (14, 17, 32).
Predicted stem-loop in the 3′ UTR.
Comparison of the 3′ UTRs of 19 isolates of IBV and 3 of TCoV has confirmed that the UTR comprises a highly variable region I, adjacent to the N gene, and a much more conserved region II, proximal to the poly(A) tract. Structure analysis predicted that a stem-loop structure exists at the 5′ end of region II (Fig. 3), and deletion analysis indicated that this structure is essential for replication of the IBV D-RNAs (Fig. 2). It is also possible that the fusion of the specific sequences in CD-38CATstem−, which lacked the predicted stem-loop, affected replication, as has been seen elsewhere for MHV A-59 D-RNAs (19). However, the phylogenetic data presented in Fig. 3 strongly suggest that there is a stem-loop in this region, and we propose that this potential stem-loop is essential for IBV replication. This potential stem-loop is analogous in position to the stem-loops found in MHV and BCoV D-RNAs (12).
Situated 9 nt downstream of the predicted IBV stem-loop structure is a potential pseudoknot identified by Williams et al. (35), analogous to a pseudoknot structure demonstrated to be present in BCoV and essential for RNA replication. CD-38CATstem−, which lacked the predicted stem-loop structure and retained the sequence corresponding to the predicted IBV pseudoknot, was not replicated. It is possible that the predicted pseudoknot might have been impaired in CD-38CATstem− as a consequence of removal of the sequence ordinarily adjacent to it. The rescue of CD-38CATstem+, which lacked region I of the 3′ UTR, showed that this region was not essential for replication or packaging of this IBV D-RNA. No other strong candidate secondary structures were identified at the 3′ end of the genome.
We have previously identified three potential stem-loop structures within the 5′ UTR (29), one of which (nt 7 to 30) would appear to be analogous to the stem-loop identified by Chang et al. (7) in BCoV and concluded to be essential for the cis-acting replication signal associated with the leader sequence.
Sequences required for packaging of the D-RNAs.
The results of our experiments investigating packaging led us to the conclusion that only the sequences in the 5′ UTR and/or region II of the 3′ UTR were specifically required for packaging. This is similar to the findings for BCoV (6) and TGEV (13) D-RNAs but in contrast to other findings with MHV (10, 22, 31), in which a small part of gene 1b was found to act as an RNA packaging signal. However, the D-RNA DIssE of MHV-JHM lacks this 1b packaging signal but is incorporated into defective-interfering particles, at low efficiency, and is detectable by Northern blot analysis after multiple passages (21).
IBV D-RNAs CD-12 and CD-12CAT, which contain only 5′- and 3′-terminal sequences, totaling 1.2 kb of IBV sequence, were replicated, but overall rescue was poor. This might have been attributed in part to the small size of these D-RNAs, which at 1.2 and 1.9 kb (inclusive of the CAT gene), respectively, are smaller than any coronavirus D-RNA so far successfully rescued. However, our results showed that increasing the size of CD-12 with IBV sequence corresponding to the structural protein genes did not improve rescue. A D-RNA of TGEV, M21 (2.1 kb), was rescued but with a lower efficiency than that of those D-RNAs that contained one or the other of two fragments (F1 and F2) of the ORF 1b sequence (13). Izeta and colleagues (13) concluded that either both F1 and F2 fragments contain a packaging signal or their presence is required for the proper folding of a packaging signal in one or the other terminal region. Our results were similar to those of Izeta et al. (13) in that parts of the replicase 1b sequence—but not any specific part—were required for efficient rescue. Replacement of region II of CD-61 by sequence downstream of gene 1 (CD-84CATXcmI-struct) (Fig. 6) did not result in efficient rescue (Table 1). Thus, a large size alone was not sufficient for efficient packaging. The results suggest that either some D-RNAs folded in a way that was not conducive to packaging or the D-RNAs were highly unstable.
ACKNOWLEDGMENTS
This work was supported by the Ministry of Agriculture, Fisheries and Food, UK (project code OD1904), and by grant no. CT950064 of the Fourth RTD Framework Programme of the European Commission (EC). Kevin Dalton and K. Stirrups were the holders of Research Studentships from the Biotechnology and Biological Sciences Research Council (BBSRC). Sharon Evans was supported by a BBSRC Realising Our Potential Award. Rosa Casais was the recipient of an EC TMR Marie Curie Research Training Grant.
REFERENCES
- 1.Boursnell M E G, Binns M M, Foulds I J, Brown T D K. Sequences of the nucleocapsid genes from two strains of avian infectious bronchitis virus. J Gen Virol. 1985;66:573–580. doi: 10.1099/0022-1317-66-3-573. [DOI] [PubMed] [Google Scholar]
- 2.Breslin J J, Smith L G, Fuller F J, Guy J S. Sequence analysis of the matrix/nucleocapsid gene region of turkey coronavirus. Intervirology. 1999;42:22–29. doi: 10.1159/000024956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breslin J J, Smith L G, Fuller F J, Guy J S. Sequence analysis of the turkey coronavirus nucleocapsid protein gene and 3′ untranslated region identifies the virus as a close relative of infectious bronchitis virus. Virus Res. 1999;65:187–193. doi: 10.1016/S0168-1702(99)00117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brian D A, Chang R-Y, Hofmann M A, Sethna P B. Role of subgenomic minus-strand RNA in coronavirus replication. Arch Virol. 1994;9(Suppl.):173–180. doi: 10.1007/978-3-7091-9326-6_17. [DOI] [PubMed] [Google Scholar]
- 5.Cavanagh D, Brian D A, Brinton M A, Enjuanes L, Holmes K V, Horzinek M C, Lai M M C, Laude H, Plagemann P G W, Siddell S G, Spaan W, Taguchi F, Talbot P J. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- 6.Chang R Y, Brian D A. cis requirement for N-specific protein sequence in bovine coronavirus defective interfering RNA replication. J Virol. 1996;70:2201–2207. doi: 10.1128/jvi.70.4.2201-2207.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang R Y, Hofmann M A, Sethna P B, Brian D A. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J Virol. 1994;68:8223–8231. doi: 10.1128/jvi.68.12.8223-8231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cologna R, Hogue B G. Identification of a bovine coronavirus packaging signal. J Virol. 2000;74:580–583. doi: 10.1128/jvi.74.1.580-583.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eleouet J F, Rasschaert D, Lambert P, Levy L, Vende P, Laude H. Complete sequence (20 kilobases) of the polyprotein-encoding gene 1 of transmissible gastroenteritis virus. Virology. 1995;206:817–822. doi: 10.1006/viro.1995.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fosmire J A, Hwang K, Makino S. Identification and characterization of a coronavirus packaging signal. J Virol. 1992;66:3522–3530. doi: 10.1128/jvi.66.6.3522-3530.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guy J S. Turkey coronavirus is more closely related to avian infectious bronchitis virus than to mammalian coronaviruses—a review. Avian Pathol. 2000;29:207–212. doi: 10.1080/03079450050045459. [DOI] [PubMed] [Google Scholar]
- 12.Hsue B, Masters P S. A bulged stem-loop structure in the 3′ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J Virol. 1997;71:7567–7578. doi: 10.1128/jvi.71.10.7567-7578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Izeta A, Smerdou C, Alonso S, Penzes Z, Mendez A, Plana-Durán J, Enjuanes L. Replication and packaging of transmissible gastroenteritis coronavirus-derived synthetic minigenomes. J Virol. 1999;73:1535–1545. doi: 10.1128/jvi.73.2.1535-1545.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim Y-N, Jeong Y S, Makino S. Analysis of cis-acting sequences essential for coronavirus defective interfering RNA replication. Virology. 1993;197:53–63. doi: 10.1006/viro.1993.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim Y N, Makino S. Characterization of a murine coronavirus defective interfering RNA internal cis-acting replication signal. J Virol. 1995;69:4963–4971. doi: 10.1128/jvi.69.8.4963-4971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai M M, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin Y J, Lai M M C. Deletion mapping of a mouse hepatitis virus defective interfering RNA reveals the requirement of an internal and discontiguous sequence for replication. J Virol. 1993;67:6110–6118. doi: 10.1128/jvi.67.10.6110-6118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin Y J, Liao C L, Lai M M. Identification of the cis-acting signal for minus-strand RNA synthesis of a murine coronavirus: implications for the role of minus-strand RNA in RNA replication and transcription. J Virol. 1994;68:8131–8140. doi: 10.1128/jvi.68.12.8131-8140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luytjes W, Gerritsma H, Spaan W J. Replication of synthetic defective interfering RNAs derived from coronavirus mouse hepatitis virus-A59. Virology. 1996;216:174–183. doi: 10.1006/viro.1996.0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makino S, Shieh C, Keck J G, Lai M M C. Defective-interfering particles of murine coronavirus: mechanism of synthesis of defective viral RNAs. Virology. 1988;163:104–111. doi: 10.1016/0042-6822(88)90237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makino S, Shieh C-K, Soe L H, Baker S C, Lai M M C. Primary structure and translation of a defective interfering RNA of murine coronavirus. Virology. 1988;166:550–560. doi: 10.1016/0042-6822(88)90526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makino S, Yokomori K, Lai M M C. Analysis of efficiently packaged defective interfering RNAs of murine coronavirus: localization of a possible RNA-packaging signal. J Virol. 1990;64:6045–6053. doi: 10.1128/jvi.64.12.6045-6053.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matzura O, Wennborg A. RNAdraw: an integrated program for RNA secondary structure calculation and analysis under 32-bit Microsoft Windows. CABIOS. 1996;12:247–249. doi: 10.1093/bioinformatics/12.3.247. [DOI] [PubMed] [Google Scholar]
- 24.Penzes Z, Tibbles K, Shaw K, Britton P, Brown T D K, Cavanagh D. Characterization of a replicating and packaged defective RNA of avian coronavirus infectious bronchitis virus. Virology. 1994;203:286–293. doi: 10.1006/viro.1994.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penzes Z, Wroe C, Brown T D K, Britton P, Cavanagh D. Replication and packaging of coronavirus infectious bronchitis virus defective RNAs lacking a long open reading frame. J Virol. 1996;70:8660–8668. doi: 10.1128/jvi.70.12.8660-8668.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Repass J F, Makino S. Importance of the positive-strand RNA secondary structure of a murine coronavirus defective interfering RNA internal replication signal in positive-strand RNA synthesis. J Virol. 1998;72:7926–7933. doi: 10.1128/jvi.72.10.7926-7933.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sapats S I, Ashton F, Wright P J, Ignjatovic J. Novel variation in the N protein of avian infectious bronchitis virus. Virology. 1996;226:412–417. doi: 10.1006/viro.1996.0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stirrups K, Shaw K, Evans S, Dalton K, Casais R, Cavanagh D, Britton P. Expression of reporter genes from the defective RNA CD-61 of the coronavirus infectious bronchitis virus. J Gen Virol. 2000;81:1687–1698. doi: 10.1099/0022-1317-81-7-1687. [DOI] [PubMed] [Google Scholar]
- 29.Stirrups K, Shaw K, Evans S, Dalton K, Cavanagh D, Britton P. Leader switching occurs during the rescue of defective RNAs by heterologous strains of the coronavirus infectious bronchitis virus. J Gen Virol. 2000;81:791–801. doi: 10.1099/0022-1317-81-3-791. [DOI] [PubMed] [Google Scholar]
- 30.Sutou S, Sato S, Okabe T, Nakai M, Sasaki N. Cloning and sequencing of genes encoding structural proteins of avian infectious bronchitis virus. Virology. 1988;165:589–595. doi: 10.1016/0042-6822(88)90603-4. [DOI] [PubMed] [Google Scholar]
- 31.van der Most R G, Bredenbeek P J, Spaan W J M. A domain at the 3′ end of the polymerase gene is essential for encapsidation of coronavirus defective interfering RNAs. J Virol. 1991;65:3219–3226. doi: 10.1128/jvi.65.6.3219-3226.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Most R G, Luytjes W, Rutjes S, Spaan W J M. Translation but not the encoded sequence is essential for the efficient propagation of the defective interfering RNAs of the coronavirus mouse hepatitis virus. J Virol. 1995;69:3744–3751. doi: 10.1128/jvi.69.6.3744-3751.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams A K, Wang L, Sneed L W, Collisson E W. Analysis of a hypervariable region in the 3′ non-coding end of the infectious bronchitis virus genome. Virus Res. 1993;28:19–27. doi: 10.1016/0168-1702(93)90086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams A K, Wang L, Sneed L W, Collisson E W. Comparative analyses of the nucleocapsid genes of several strains of infectious bronchitis virus and other coronaviruses. Virus Res. 1992;25:213–222. doi: 10.1016/0168-1702(92)90135-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams G D, Chang R Y, Brian D A. A phylogenetically conserved hairpin-type 3′ untranslated region pseudoknot functions in coronavirus RNA replication. J Virol. 1999;73:8349–8355. doi: 10.1128/jvi.73.10.8349-8355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]