Abstract
The ADAMTSs (a disintegrin and metalloproteinase with thrombospondin motifs) are a group of proteases that are found both in mammals and invertebrates. Since the prototype ADAMTS-1 was first described in 1997, there has been a rapidly expanding body of literature describing this gene family and the proteins they encode. The complete human family has 19 ADAMTS genes, together with three members of a newly identified subgroup, the ADAMTSL (ADAMTS-like) proteins, which have several domains in common with the ADAMTSs. The ADAMTSs are extracellular, multidomain enzymes whose known functions include: (i) collagen processing as procollagen N-proteinase; (ii) cleavage of the matrix proteoglycans aggrecan, versican and brevican; (iii) inhibition of angiogenesis; and (iv) blood coagulation homoeostasis as the von Willebrand factor cleaving protease. Roles in organogenesis, inflammation and fertility are also apparent. Recently, some ADAMTS genes have been found to show altered expression in arthritis and various cancers. This review highlights progress in understanding the structural organization and functional roles of the ADAMTSs in normal and pathological conditions.
Keywords: aggrecanase, angiogenesis, extracellular matrix, metalloproteinase, proteoglycan, tissue inhibitor of metalloproteinase (TIMP)
Abbreviations: ADAM, adisintegrin and metalloproteinase-like (or, alternatively, adamalysin); ADAMTS, adisintegrin and metalloproteinase with thrombospondin motifs (or, alternatively, adamalysin–thrombospondin); ADAMTSL, ADAMTS-like; CUB, complement subcomponent C1r/C1s/embryonic sea urchin protein Uegf/bone morphogenic protein 1; ECM, extracellular matrix; EDS, Ehlers–Danlos syndrome; GAG, glycosaminoglycan; IGD, interglobular domain; IL, interleukin; MMP(I), matrix metalloproteinase (inhibitor); PLAC domain, protease and lacunin domain; SPC, subtilisin-like pro-protein convertase; TIMP, tissue inhibitor of metalloproteinase; TS, thrombospondin type I-like; TSP1/2, thrombospondin 1 and 2 respectively; TTP, thrombotic thrombocytopaenic purpura; VEGF, vascular endothelial growth factor; vWF(CP), von Willebrand factor (cleaving protease)
INTRODUCTION
Kuno et al. [1] devised the name ADAMTS-1 (a disintegrin and metalloproteinase with thrombospondin motifs) to describe the founding member of a group of secreted proteases that now incorporates 19 genes in humans (Table 1). The ADAMTSs form a branch of the M12B ADAM (a disintegrin and metalloproteinase-like, or adamalysin) subfamily of metalloendopeptidases, on the basis of the similarity of their metalloproteinase domain to that of snake venom metalloproteinases (reprolysins). The adamalysins in turn are one of the constituent groups of metzincins (zinc-dependent proteases), others including the matrixins (MMPs; matrix metalloproteinases), astacins and serralysins [2]. The metzincins are so named because, apart from the zinc ion they contain, they also have a conserved methionine residue downstream from their catalytic site which forms a right-handed turn, or ‘Met-turn’ [3]. The ADAMTSs are closely related to the ADAM proteinases that are involved in ectodomain shedding or activation of diverse cell surface molecules, including growth factors and adhesion receptors [4]. However, unlike the mammalian ADAMs, which are, with few exceptions (e.g. variant forms of ADAM-12 and -28), transmembrane proteins, the ADAMTSs are secreted molecules, some of which bind to the ECM (extracellular matrix) [5,6]. As we discuss in this review, we have detailed knowledge of the functions of only a few family members, namely the aggrecanases, procollagen N-proteinases and vWFCP (von Willebrand factor-cleaving protease), with many orphan ADAMTSs still having no known substrate or activity.
Table 1. The human ADAMTS names, loci and known substrates.
The nomenclature for the ADAMTS genes and the proteins they encode has been standardized such that the protein names contain a hyphen before their respective number and the gene names do not (http://www.lerner.ccf.org/bme/apte/adamts). Abbreviations: PCINP, pro-collagen I N-proteinase.
| Gene name | Protein name | Alternative names | Chromosome location | Known substrates |
|---|---|---|---|---|
| ADAMTS1 | ADAMTS-1 | METH-1; aggrecanase-3 | 21q21 | Aggrecan; versican V1 |
| ADAMTS2 | ADAMTS-2 | PCINP | 5q35 | Procollagen I, II and III N-propeptides |
| ADAMTS3 | ADAMTS-3 | KIAA0366* | 4q21 | Procollagen II N-propeptide |
| ADAMTS4 | ADAMTS-4 | aggrecanase-1; KIAA0688* | 1q23 | Aggrecan; brevican; versican V1; fibromodulin; decorin; carboxymethylated transferrin |
| ADAMTS5 | ADAMTS-5 | aggrecanase-2; ADAMTS11 | 21q21 | Aggrecan |
| ADAMTS6 | ADAMTS-6 | 5q12 | ||
| ADAMTS7 | ADAMTS-7 | 15q24 | ||
| ADAMTS8 | ADAMTS-8 | METH-2 | 11q25 | |
| ADAMTS9 | ADAMTS-9 | KIAA1312 | 3p14 | Aggrecan; versican |
| ADAMTS10 | ADAMTS-10 | 19p13 | ||
| ADAMTS12 | ADAMTS-12 | 5q35 | ||
| ADAMTS13 | ADAMTS-13 | vWFCP | 9q34 | von Willebrand factor |
| ADAMTS14 | ADAMTS-14 | 10q21 | Procollagen I N-propeptide | |
| ADAMTS15 | ADAMTS-15 | 11q25 | Aggrecan | |
| ADAMTS16 | ADAMTS-16 | 5p15 | ||
| ADAMTS17 | ADAMTS-17 | 15q24 | ||
| ADAMTS18 | ADAMTS-18 | 16q23 | ||
| ADAMTS19 | ADAMTS-19 | 5q31 | ||
| ADAMTS20 | ADAMTS-20 | 12q12 |
* The KIAA protein database may be accessed at: http://www.kazusa.or.jp/huge (KIAA0366 is ADAMTS3; KIAA0688 is ADAMTS4 and KIAA1312 is ADAMTS9, as shown in the Table).
The ADAMTS nomenclature was initially in some confusion. Particular ADAMTSs have appeared under different names: for instance, ADAMTS11 [7] and ADAMTS5 [8] are names for the same gene encoding the protein aggrecanase-2, but the consensus is that ADAMTS5 is the correct name. The trivial name implantin appears to be specific to mouse Adamts5 [8]. METH-1 and METH-2 [9] are now more usually called ADAMTS1 and ADAMTS8 respectively. ADAMTS-2 has become the accepted name for PCINP (pro-collagen I N-proteinase); vWFCP is better known as ADAMTS-13. ADAMTS-1 has also been referred to unofficially as aggrecanase-3. With regard to the naming of the family as a whole, since the original definition is somewhat bulky, we suggest that the term ‘adamalysin–thrombospondins’ provides an accurate and convenient descriptor.
THE DOMAIN STRUCTURE OF THE ADAMTS PROTEINS
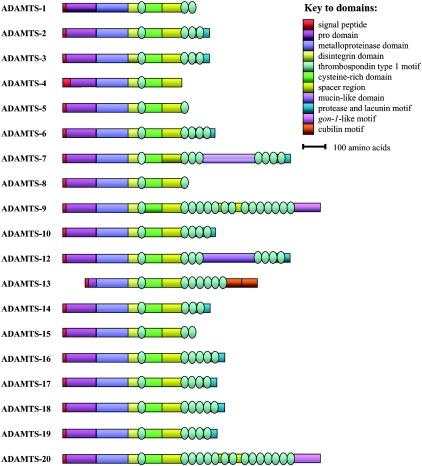
The domain structure of the ADAMTSs is illustrated in Figure 1. The molecular structure of the ADAMTS proteins can be subcategorized into domains, modules and motifs (courtesy of Dr Suneel S. Apte at The Lerner Research Institute, The Cleveland Clinic Foundation, Cleveland, OH, U.S.A.; http://www.lerner.ccf.org/bme/apte/adamts). There is considerable potential for the generation of multiple isoforms of ADAMTSs via alternative splicing. Overall, there is more variability between the different proteins at the C-terminus than at the N-terminus. The ADAMTSs are all synthesized initially as inactive pre-proenzymes: from the N- to the C-terminus, they each comprise: (i) a signal peptide; (ii) a pro-domain, the presence of which may preserve enzymatic latency; (iii) a metalloproteinase catalytic domain with a reprolysin-type zinc-binding motif, HEXXHXXG/N/SXXHD [where ‘X’ represents any amino acid residue and the conserved aspartic acid residue (highlighted in bold) distinguishes the ADAMs and ADAMTSs from other metalloproteinases], and a methionine residue within the sequence V/IMA/S, or ‘Met-turn’, downstream of the third zinc-binding histidine; (iv) a disintegrin-like domain, which shares sequence similarity to the soluble snake venom disintegrins, a family of polypeptides of which some members contain an arginine/glycine/aspartic acid (RGD) integrin recognition sequence [10] (however, no ADAMTS has an RGD sequence in its disintegrin domain, and indeed there is no evidence to date that ADAMTSs associate with integrins); (v) a central TS (thrombospondin type I-like) repeat seen in TSP1 and TSP2 (thrombospondins 1 and 2 respectively); (vi) a cysteine-rich domain with high sequence homology among the ADAMTSs, containing ten conserved cysteine residues; (vii) a spacer region of variable length with no distinguishing structural features; and (viii) a variable number of C-terminal TS repeats: these range from 14 C-terminal repeats in the case of ADAMTS-20 and the long isoform of ADAMTS-9 [11,12] to none in the case of ADAMTS-4 [13].
Figure 1. Domain structure of the ADAMTS proteins.
In the case of proteins with varying isoforms, only the longest isoform is represented here. The thrombospondin type 1 domains are highlighted by the oval shapes.
Some of the ADAMTSs have further C-terminal modules. ADAMTS-7 and ADAMTS-12 both have a mucin domain between the third and fourth of their seven C-terminal TS repeats, likened to these glycoproteins because the region is similarly rich in serine, proline and threonine residues and predicted to be heavily O-glycosylated [14]. ADAMTS-20 and the long isoform of ADAMTS-9 both have a GON domain, first described in gon-1, an ADAMTS involved in gonadal development in Caenorhabditis elegans [15]. ADAMTS-13 is unique in having two CUB [complement subcomponent C1r/C1s/embryonic sea urchin protein Uegf (urchin epidermal growth factor)/bone morphogenic protein 1] domains [16], seen in the astacin family of metzincins [17]. The CUB domain is a functionally independent module found in a diverse range of extracellular proteins. Most of these CUB domain-containing proteins are thought to be involved in developmental processes, such as embryogenesis or organogenesis [18]. ADAMTS-2, -3, -10, -12, -14, -17 and -19 all have a PLAC (protease and lacunin) domain [19–21], first described as a C-terminal domain of unknown function of the ECM protein lacunin, associated with epithelial remodelling in embryos and developing wings of the moth Manduca sexta [19]. The genomic sequence of ADAMTS16 predicts the possibility of an mRNA encoding a longer protein isoform (containing a PLAC domain) than that currently documented (I. M. Clark, unpublished work).
Phylogenetic analysis of the ADAMTSs suggests that the family members can be categorized into seven subgroups on the basis of their sequence similarities and, where known, their targeted substrates [12]. These seven subgroups are: (a) ADAMTS-1, -4, -5, -8 and -15; (b) ADAMTS-2, -3 and -14; (c) ADAMTS-9 and -20; (d) ADAMTS-7 and -12, plus ADAMTS-6 and -10; (e) ADAMTS-16 and -18; (f) ADAMTS-17 and -19; and (g) ADAMTS-13, which is least like any other ADAMTS.
All members of the subgroup consisting of ADAMTS-1, -4, -5, -8 and -15 have all been demonstrated to cleave (though with various efficiencies) the major cartilage proteoglycan aggrecan, and have thus been termed ‘aggrecanases’. Gao et al. [22] have proposed the more general term ‘hyalectanase’ to indicate an enzyme that can cleave aggrecan, brevican or versican. However, both names may ultimately prove to be inadequate, as we do not know whether these proteoglycans are the most important physiological substrates. Moreover, though ADAMTS-9 is classified as a GON-ADAMTS with ADAMTS-20, it too is able to cleave aggrecan and versican [11].
An important difference between ADAMTSs and both MMPs and ADAMs is the ability of many ADAMTSs, e.g. ADAMTS-1, ADAMTS-4 and ADAMTS-9 [6,11,23], to bind to the ECM. Though the basis of this property has not been defined in every case, there is support for the involvement of the TS repeats. Using a series of ADAMTS-1 deletion mutants, Kuno and Matsushima [6] demonstrated that ECM binding was mediated through the central and C-terminal TS repeats and the spacer region, and that sulphated GAGs (glycosaminoglycans) were probably responsible, since ADAMTS-4 cannot cleave GAG-free aggrecan. ADAMTS-4 binds to aggrecan through its sole TS motif; a peptide consisting of the putative GAG- and CD36-binding motifs of the TS domain of ADAMTS-4 can block its binding to aggrecan, and deletion mutants lacking this domain cannot cleave aggrecan [24]. The ECM binding of ADAMTS-4 has been attributed to interaction of its C-terminal spacer region with the C-terminal domain of fibronectin [25].
The ADAMTSs undergo N-terminal processing, firstly by a signal peptidase during translation and transit of the protein through the endoplasmic reticulum membrane, and subsequently by removal of the pro-domain. The pro-domain is generally considered to preserve enzyme latency, but it is also potentially important for correct protein folding and secretion [26,27]. All currently known ADAMTSs contain a SPC (subtilisin-like pro-protein convertase) cleavage site, and, with the exception of ADAMTS-10 and ADAMTS-12, they are all furin [also named SPC1 or PACE (paired basic amino acid converting enzyme)] recognition sequences, following the consensus RXR/KR [28]. Pro-protein processing has been examined for ADAMTS-1 [29], ADAMTS-7 [14], ADAMTS-9 [11] and ADAMTS-12 [20] using furin-deficient cells, and in each case, removal of the pro-domain was either completely blocked or severely inhibited, but could be restored on transfection with furin. For pro-ADAMTS-4, intracellular co-localization of the enzyme with furin has been observed [30]. Processing probably occurs in the trans-Golgi network, since it is blocked by treatment with brefeldin A. However, furin may not be the only enzyme involved, since pro-ADAMTS-4 can also be properly processed in the furin-deficient cell line RPE40 [30]. Several ADAMTSs have one or more additional SPC cleavage sites upstream of the primary site. Initial cleavage at this secondary cleavage site within the pro-domain of ADAMTS-2 has been observed [31]. It is possible that sequential processing of the N-termini of ADAMTS proteins may regulate their intracellular distribution or release.
The pro-domain of the ADAMTS proteins may not function in the same fashion as that of the MMPs, where a cysteine residue in the conserved motif PRCGVPD co-ordinates with the catalytic zinc atom to maintain enzyme latency, cleavage of the pro-region resulting in activation of the ‘cysteine switch’ [32]. Only six of the ADAMTSs (ADAMTS-1, -6, -7, -10, -12 and -15) contain a cysteine residue in their pro-domain within a XXCGVXD motif that loosely resembles that of the cysteine switch in the MMPs [33]. Moreover, it appears that some ADAMTS, e.g. ADAMTS-13 and ADAMTS-7, can be catalytically active with their pro-domains still attached [14,34].
C-TERMINAL PROCESSING AFFECTS SUBSTRATE SPECIFICITY AND ECM BINDING
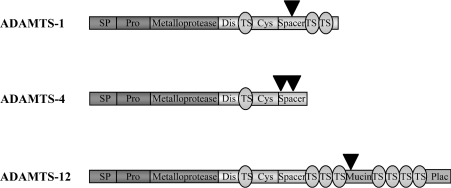
The ancillary C-terminal domains of the ADAMTSs have a profound impact on both substrate specificity and localization of the enzymes. C-terminal processing has been described for ADAMTS-1 [9,29], ADAMTS-4 [22], ADAMTS-8 [9], ADAMTS-9 [11] and ADAMTS-12 [20], and best studied in ADAMTS-1 and ADAMTS-4. Cleavage events occur within the spacer region [9,22,29,35–37] and in the case of ADAMTS-12, within the mucin domain [20]. So far, one C-terminal processing event has been described for ADAMTS-1, -8 and -12, and two for ADAMTS-4 (Figure 2).
Figure 2. Diagram to show the approximate C-terminal cleavage points (indicated by arrowheads) of ADAMTS-1, ADAMTS-4 and ADAMTS-12.
ADAMTS-8 and ADAMTS-9 are also reported to undergo C-terminal cleavage, but details are not known.
In the case of ADAMTS-4, which is the best characterized of the aggrecanases, C-terminal processing of the 75 kDa full-length active form produces isoforms of 60 and 50 kDa (note that slight variances in these molecular masses have been reported by different research groups) [35,36], and this results in release of the enzyme from the ECM and alteration of its activity profile [22]. Aggrecan is cleaved at several sites by both MMPs and aggrecanases (described in detail below in the subsection ‘The aggrecanases: ADAMTS-1, -4, -5, -8, -9 and -15’); however, the signature aggrecanase cleavage site is at Glu373–Ala374 [13]. The full-length 75 kDa ADAMTS-4 is the most potent aggrecanase of the three isoforms, but it cleaves at Glu1480–Gly1481 [23]. In contrast, cleavage at Glu373–Ala374 is a property of the 60 and 50 kDa isoforms [22] and recombinant truncated proteins lacking the C-terminal spacer region [23]. Thus the C-terminal spacer region can act to inhibit the Glu373–Ala374 aggrecanase activity of full-length ADAMTS-4. Moreover, the full-length form is ECM-associated, but deletion of the spacer domain releases the enzyme [22,23,25,36]. ECM association has been shown to involve binding to fibronectin [25]. Exogenous fibronectin blocked the aggrecanase activity of the full-length ADAMTS-4, but not a mutant lacking the spacer region [25], indicating that ECM binding could be the basis for the ability of the C-terminal spacer region to modulate substrate specificity. This is reinforced further by the observation that deletion mutants comparable with the 60 kDa truncated isoform can cleave additional substrates, including carboxymethylated transferrin, fibromodulin and decorin [23]. However, the TS repeat of ADAMTS-4 is required for binding of the enzyme to sulphated GAGs linked to aggrecan [38], and deletion of this motif reduces aggrecanase activity to 1% of normal [23], showing that aggrecan binding and ECM association may be separable functions in distinct domains in ADAMTS-4, although these domains are likely to work in concert to control function of the enzyme in vivo.
Another example of C-terminal processing affecting localization and bioactivity is provided by ADAMTS-1, which has profound anti-angiogenic properties (see the ‘Anti-angiogenesis: ADAMTS-1 and -8’ subsection below). The C-terminally processed 65 kDa form of ADAMTS-1 lacking part of the spacer region and the two C-terminal TS repeats is found mainly in the culture medium, whereas the full-length 87 kDa form is located in pericellular ECM of endothelial cell cultures [29]. The 65 kDa isoform of ADAMTS-1 is less anti-angiogenic than the full-length form. ADAMTS-1 binds to VEGF-165, the heparin-binding isoform of VEGF (vascular endothelial growth factor), inhibiting its binding to its receptor, VEGFR2 [37]. Using recombinant full-length ADAMTS-1 and a series of deletion mutants, it has been shown that it is the C-terminal TS repeats of ADAMTS-1 which mediate its binding to VEGF [37].
There is the opportunity for additional levels of regulation of cellular ADAMTS activities by the involvement of specific proteases in C-terminal processing. Using recombinant proteins in vitro, Flannery et al. [36] observed autocatalytic processing of ADAMTS-4 to generate the two C-terminally cleaved isoforms. However, an inactive Glu362→Gln active-site ADAMTS-4 mutant is properly processed when transfected into human chondrosarcoma cells [35]. Under these circumstances, C-terminal processing of ADAMTS-4 occurs on the cell surface, and involves a TIMP-1 (tissue inhibitor of metalloproteinase 1)-sensitive metalloproteinase, recently identified as MMP-17 (membrane type-4 MMP; [22,35]). Likewise, MMP-2, MMP-8 and (especially) MMP-15 (membrane type-2 MMP) are all able to mediate the C-terminal processing of ADAMTS-1 in vitro [29]. C-terminal processing of ADAMTS-12 is also dependent on an additional metalloproteinase, since it is blocked by the broad spectrum hydroxamate inhibitor BB-94, but not by mutation of the catalytic site [20].
REGULATION AND EXPRESSION OF THE ADAMTSs
Inhibition of the ADAMTSs
Although the TIMPs are, with a few exceptions, broadly effective inhibitors of the MMPs (reviewed in [39]), it is clear that they display much greater selectivity towards both the ADAMs and ADAMTSs. The aggrecanases ADAMTS-4 and ADAMTS-5 are both potently inhibited by TIMP-3, with Ki values in the sub-nanomolar range, although they are essentially insensitive to TIMP-1, -2 and -4 [40–42]. This is supported by data from an ex vivo bovine nasal cartilage explant model of cartilage destruction, where the N-terminal domain of TIMP-3 potently inhibited GAG release at 0.1 μM, whereas neither N-TIMP-1 nor N-TIMP-2 inhibited GAG release, even at 1 μM. Other ADAMTSs may have a different inhibition profile by TIMPs: ADAMTS-1 is partially inhibited by TIMP-2 and TIMP-3 at 500 nM, but TIMP-1 and TIMP-4 have no inhibitory effect at the same concentration [43]. TIMP-3 is thus likely to be the major natural inhibitor of aggrecanase activity in cartilage [44]. Both ADAMTS-4 and -5 cleave α2-macroglobulin at a novel cleavage site within the bait region, at what is also a novel site of cleavage for ADAMTS-4 and -5 [45].
The aggrecanolytic activity of ADAMTS-1, -4 and -5 is effectively inhibited by catechin gallate esters found in green tea: both epigallocatechin gallate and epicatechin gallate potently inhibited all three ADAMTSs, with approximate IC50 values of 100–150 nM for ADAMTS-4 and ADAMTS-5, and 200–250 nM for ADAMTS-1 [46]. Limited studies have been undertaken on the effect of synthetic inhibitors on the ADAMTSs: ADAMTS-1 is inhibited by EDTA, 1,10-phenanthroline [47], BB-94 [43] and MMP inhibitor 2 (from Calbiochem; [29]); ADAMTS-12 is inhibited by BB-94 [20].
It has been suggested that papilin, an essential component of the ECM of C. elegans, may modulate ADAMTSs during organogenesis [48]. Papilin, which exists in mammals as well as in invertebrates, is an ECM glycoprotein containing a conserved sequence (‘papilin cassette’) of non-enzymatic domains that it shares with the ADAMTSs, namely a signal peptide, a TS repeat, a cysteine-rich domain and several more TS repeats, and has been shown to inhibit ADAMTS-2 in vitro [48]. In this respect, papilin is close in structure to the ADAMTSL (ADAMTS-like) proteins, and may suggest a function for the latter. However, unlike either the ADAMTSs or the ADAMTSLs, papilin contains additional domains further towards the C-terminal end: cysteine-rich repeats, a species-dependent variable number of Kunitz domains [homologues of the BPTI (bovine pancreatic trypsin inhibitor)], Ig–C2 loop domains consisting of a unique cysteine-rich sequence, and a C-terminal peptide, and the functions of these domains are uncertain [48]. Despite the sequence homology with BPTI, the Kunitz domains do not inhibit trypsin, chymotrypsin or plasminogen.
Regulation of ADAMTS gene expression
Expression of ADAMTS mRNAs appears to be distributed across a wide range of normal adult tissues. Expression in fetal tissue is generally more limited. A summary of ADAMTS expression analysis is given in Table 2.
Table 2. ADAMTS expression analysis in human tissues.
nd, not detected in tissues examined; nt, not tested. Ac, adrenal cortex; Am, adrenal medulla; Ao, aorta; Bl, bladder; Bo, bone; Br, brain; Bs, breast; Co, colon; Cx, cervix; En, endothelium; Ht, heart; In, intestine; Ki, kidney; Li, liver; Lu, lung; Mu, skeletal muscle; Oe, oesophagus; Ov, ovary; Pc, pancreas; Pl, placenta; Pr, prostate; Re, retina; Sk, skin; Sl, spleen; Sm, submaxillary gland; Sp, spinal cord; St, stomach; Te, tendon; Th, thyroid; Ty, thymus; Ts, testis; Ut, uterus.
| Gene name | Fetal tissue | Normal adult tissue | Malignant adult tissue | References |
|---|---|---|---|---|
| ADAMTS1 | Ki, Lu | Ht, Pl, Li, Mu, Ki, Th, Am, Ac, St, Ut, Bl, Cx, Ao, Co, Oe, Ov, Pr, Sp | Cell lines only | [9] |
| ADAMTS2 | nt | Ao, Bo, Sk, Te, Bl, Re, Ki, Lu, In, Li, Mu | nt | [83] |
| ADAMTS3 | nt | Pl, Lu, Br, Ht, Sk | nt | [84] |
| ADAMTS4 | nt | Bl, Br, Ov, Ht, Mu, Ut, St, Sp | nt | [7] |
| ADAMTS5 | nt | Bl, Cx, Oe, Pl, Ut | nt | [7] |
| ADAMTS6 | nt | nd | nt | [8] |
| ADAMTS7 | nt | Ht, Mu, Ki, Pc, Br, Li | nt | [8] |
| ADAMTS8 | Br, Lu, Ki | Lu, Ht, Pl, Br | Cell lines only | [9] |
| ADAMTS9 | Br, Ht, Ki, Li, Lu, Mu, Sl, Ty | Ht, Pl, Lu, Mu, Ki, Pc, Ov, Co | nt | [11] |
| [88] | ||||
| ADAMTS10 | nt | Pc, Ht, Br, Lu, Pl, Li, Ki | Cell lines only | [125] |
| ADAMTS12 | Lu | nd | St carcinoma | [20] |
| ADAMTS13 | Li | Pr, Br, Li, Pl, Ht, Mu | nd | [16] |
| [110] | ||||
| ADAMTS14 | Lu | Pr, Br, Li, Re, Lu, Pl | Ki, Bs carcinoma | [110] |
| [126] | ||||
| ADAMTS15 | Li, Ki | nd | nd | [110] |
| ADAMTS16 | Lu, Ki | Br, Ov | nd | [110] |
| ADAMTS17 | Lu | Ov | nd | [110] |
| ADAMTS18 | Lu, Ki | Pr, Br, En, Sm | nd | [110] |
| ADAMTS19 | Lu | nd | Osteosarcoma | [110] |
| ADAMTS20 | nt | Ts, Pr, Ov, Ht, Pl, Lu, Pc | Br, Co, Bs carcinoma | [12] |
Our own expression profiling studies of the 19 ADAMTS genes in breast and prostate cancer using real-time PCR have demonstrated statistically significant dysregulation of several ADAMTSs under these pathological conditions. Eleven of the ADAMTS genes are dysregulated in invasive breast carcinoma: ADAMTS1, 3, 5, 8, 9, 10 and 18 mRNAs are down-regulated by 40–90%, and those for ADAMTS4, 6, 14 and 20 are up-regulated by 80–2000% with respect to normal mammary tissue; in normal breast, expression appears to be predominantly in myoepithelial cells or stroma, and rarely in luminal epithelial cells [49]. Though there was no significant difference in expression of ADAMTS15 between normal breast and mammary cancer, those patients whose tumours had low ADAMTS15 levels showed poorer event-free survival [49]. In prostate cancer, ADAMTS13 and 20 are down-regulated with respect both to samples of benign prostatic hyperplasia and matched normal prostate tissue (S. Porter, A. C. P. Riddick, K. K. Sethia, R. Y. Ball and D. R. Edwards, unpublished work).
Down-regulation of expression of particular ADAMTSs in cancer compared with normal tissue suggests that these proteases may negatively regulate tumour growth, and this would be consistent with the anti-angiogenic properties of ADAMTS-1 and -8 (see the ‘Anti-angiogenesis: ADAMTS-1 and -8’ subsection). Masui et al. [50] have also observed down-regulation of ADAMTS1 mRNA in pancreatic cancer and hepatocellular carcinoma with respect to non-neoplastic tissue. However, ADAMTS1 expression did not correlate with tumour vascularity in either hepatocellular carcinoma or pancreatic cancer. Also, metastatic pancreatic cancer was associated with significantly higher expression of ADAMTS1, and these patients had a poorer prognosis after curative surgery, which suggests that ADAMTS1 may in fact have a positive role in promoting the progression of pancreatic cancer through lymph node metastasis and local invasion. Thus, as with the MMPs, the ADAMTSs may have multiple, perhaps opposing, actions at particular stages during tumour progression.
In cartilage samples obtained from patients with osteoarthritis, eight of the ADAMTS genes showed dysregulation: ADAMTS1, 5, 9 and 15 (all aggrecanases) are down-regulated, and ADAMTS2, 12, 14 and 16 are up-regulated in osteoarthritis cartilage with respect to phenotypically normal cartilage obtained from fracture patients [51]. Although the consistent down-regulation of aggrecanase mRNAs in osteoarthritis cartilage is somewhat surprising, given the catabolic role of the enzymes, it is important to note that the tissues used in this profiling study represent end-stage disease, and therefore it will be important to analyse expression in earlier disease stages and in animal models of arthritis.
Regulation of the ADAMTS genes is still poorly understood, though there is evidence that several are regulated by growth factors, hormones and inflammatory cytokines. Transforming growth factor-β has been shown to induce expression of ADAMTS2 mRNA in MG-63 osteosarcoma cells [52], ADAMTS12 in KMST human fetal fibroblasts [20] and ADAMTS4 (but not ADAMTS5) in fibroblast-like synoviocytes [53]. Consistent with its role as a mediator of cartilage matrix breakdown, IL (interleukin)-1 stimulation of chondrocytes or bovine nasal or articular cartilage led to an increase in detectable aggrecanase activity, without increasing measured ADAMTS-4 protein [54]; however, it is possible that other aggrecanase ADAMTSs may account for the increased activity in this system. In IL-1-treated pig articular cartilage explants, increased ADAMTS-4 protein levels were apparent [23]. In the immortalized chondrocytic cell line TC28a4, ADAMTS1, ADAMTS4 and ADAMTS5 mRNAs were all regulated by a combination of IL-1α and oncostatin M, though with differing magnitude and kinetics [55]. IL-17, a major pro-inflammatory cytokine produced mainly by synovial membranes in rheumatoid arthritis, also up-regulates ADAMTS4 in bovine articular chondrocytes via phosphorylation of ERK (extracellular-signal-regulated kinase), p38 and JNK (c-Jun N-terminal kinase) mitogen-activated protein kinases [56]. ADAMTS1 mRNA is also induced in response to IL-1 in N1E-115 rat cells following hypoglossal nerve injury [57]. In human THP-1 monocytic cells, PMA strongly induced ADAMTS4 expression, and this induction was suppressed by both the peroxisome proliferator-activated receptor γ agonist GW7845 and 9-cis-retinoic acid [58], suggesting that ADAMTS4 may be important in activated macrophages in inflammation and various pathologies.
The regulation of some ADAMTS genes is controlled by hormonal stimuli: the thyroid hormone tri-iodothyronine (T3) up-regulates mRNA expression of ADAMTS5, but not ADAMTS4, with subsequent aggrecan degradation, in growth plate cartilage during endochondral ossification [59]. Parathyroid hormone up-regulates ADAMTS1 mRNA expression in bone and osteoblasts [60]. ADAMTS-1 plays a critical role in follicular rupture, and its expression in granulosa cells of pre-ovulatory follicles is induced both by luteinizing hormone and human chorionic gonadotropin, and regulated by progesterone [61,62].
ADAMTS FUNCTION
Anti-angiogenesis: ADAMTS-1 and -8
Two ADAMTS proteins that have been proven to be anti-angiogenic are ADAMTS-1 and ADAMTS-8 [9]. Both can inhibit VEGF-induced angiogenesis in a chick chorioallantoic membrane assay and suppress fibroblast-growth-factor-2-induced vascularization in a corneal pocket assay, and they both mediate a greater anti-angiogenic response than either TSP-1 or endostatin on a molar basis, with ADAMTS-1 showing a greater inhibitory capacity than ADAMTS-8.
The anti-angiogenic activity of ADAMTS-1 and -8 is thought to be mediated through their TS motifs. These repeats are a feature of TSP1 and TSP2, and out of the five TSP family members, these are the only two to have anti-angiogenic activity (for a review, see [63]). Of possible relevance to the anti-angiogenic actions of ADAMTS-1 is that TSP1 can interact with CD36, a membrane glycoprotein receptor on endothelial cells [64,65]. This involves the sequence CSVTCG, present in two of the three TS repeats in TSP1, and there is a similar conserved motif, CSRT/SCG, in the central TS repeat of all ADAMTSs. This suggests that ADAMTS-1 might potentially bind to CD36 on the surface of endothelial cells via its central TS motif, as indeed might other ADAMTS proteins, although this has not been demonstrated. However, as mentioned in the ‘C-terminal processing affects substrate specificity and ECM binding’ section above, recent data have shown that it is the region of ADAMTS-1 which contains the two C-terminal TS repeats that is responsible for its anti-angiogenic action, through which the protein can bind VEGF-165 [37]. It is possible that a GWQRRL/TVECRD motif that is common to the first C-terminal TS repeat of both ADAMTS-1 and -8, but absent from any other ADAMTS, may play an important role.
The aggrecanases: ADAMTS-1, -4, -5, -8, -9 and -15
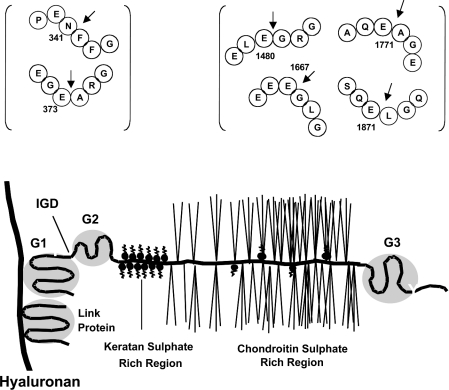
Aggrecan is the major proteoglycan of cartilage and is responsible for the ability of this tissue to resist compression by hydrating and swelling against the type II collagen scaffold [66]. It contains two N-terminal globular domains, G1 and G2, separated by an IGD (interglobular domain), followed by a GAG-attachment region and a C-terminal globular domain, G3 (see Figure 3) [7]. The G1 domain interacts with hyaluronic acid and link protein to form large aggregates that are trapped within the cartilage collagen matrix. Aggrecan protects cartilage collagen from degradation [67]. The major cleavage sites that cause aggrecan depletion from cartilage are in the IGD domain, since aggrecan which lacks the G1 domain is free to exit the matrix and so no longer contributes to cartilage function [68]. Two major proteolytic cleavage sites have been identified in this domain: one at Asn341–Phe342, at which all MMPs present in cartilage primarily act, and one at Glu373–Ala374, which is an aggrecanase cleavage site. There are also additional aggrecanase cleavage sites, but these occur within the GAG-attachment region between the G2 and G3 globular domains: Glu1480–Gly1481, Glu1667–Gly1668, Glu1771–Ala1772 and Glu1871–Leu1872 [38,69].
Figure 3. A diagrammatic representation of aggrecan and its known cleavage sites.
Aggrecan has two cleavage sites situated within the G1/G2 IGD domain. The Asn341–Phe342 is the main MMP cleavage site, whereas the Glu373–Ala374 is cleaved by aggrecanases. There are also four cleavage sites within the chondroitin sulphate-rich region of aggrecan which at least ADAMTS4 and ADAMTS5 are known to cleave. The black arrows on the diagram represent the site at which cleavage takes place.
The most studied aggrecanases are ADAMTS-4 and ADAMTS-5. These enzymes cleave aggrecan at the Glu373–Ala374 bond, and at the four other sites within the GAG-attachment region mentioned above. Cleavage of these other sites in vitro appears to be more efficient than cleavage within the IGD at the Glu373–Ala374 bond [38,69]. Aggrecan fragments generated by cleavage at all of these sites by ADAMTS-4 and ADAMTS-5 have been identified in bovine articular cartilage explants treated with IL-1 and tumour necrosis factor to stimulate aggrecan release [70]. ADAMTS-4 and ADAMTS-5 can also cleave the chondroitin sulphate proteoglycans brevican, predominantly expressed in the central nervous system [71], and versican, present in blood vessels [72]. When processed at the C-terminus, ADAMTS-4 is also active against carboxymethylated transferrin, fibromodulin and decorin (discussed above) [23]. ADAMTS-1 is also an aggrecanase [43,46,73], and cleaves versican at the same bond as does ADAMTS-4, although ADAMTS-4 can cleave both versican and aggrecan more efficiently [72]. Recently, ADAMTS-9 was also identified as an aggrecanase and versicanase, cleaving aggrecan at the Glu1771–Ala1772 bond and versican at the Glu441–Ala442 bond [11]. According to a patent application filed by the Yamanouchi company in Japan, ADAMTS-15 is also an aggrecanase [74]. Recombinant ADAMTS-8 was also recently demonstrated to possess weak (approx. 3000-fold lower than ADAMTS-4) aggrecanase activity at the Glu373–Ala374 bond [75].
Outside of a possible role in cartilage metabolism, the aggrecanases may have other functions in physiology or pathology. Adamts1-null mice have growth retardation, malformation of adipose tissue, decreased fertility with changes in histology of uterus and ovaries, and a renal phenotype similar to the human congenital disease ureteropelvic junction obstruction [76–78]. ADAMTS1 is also a transcriptional target of the progesterone receptor during ovulation [61], and is up-regulated in bone and osteoblasts by parathyroid hormone and related agents [60]. ADAMTS1 was down-regulated in endothelial cells derived from the livers of cirrhotic animals [79], and is also reduced in the subcutaneous adipose tissue of obese mice [80]. The ability of ADAMTS-4 to degrade brevican may implicate it in central nervous system physiology and pathology [81]: for example, increased aggrecanase-mediated brevican cleavage is a feature of invasive gliomas [71].
The procollagen N-proteinases: ADAMTS-2, -3 and -14
ADAMTS-2, -3 and -14 are procollagen N-proteinases, involved in the processing of procollagens to collagen by removal of the N-terminus propeptides [31,82–84]. ADAMTS-2 acts on procollagen I, II and III, whereas ADAMTS-3 and ADAMTS-14 have been linked only with procollagen II and procollagen I processing, respectively. Mutations in the ADAMTS2 gene cause dermatosparaxis in sheep and cattle and EDS (Ehlers–Danlos syndrome) type VIIc in humans [85], an autosomal recessive disorder characterized by severe skin fragility, joint laxity (hyperextensibility) and characteristic facies (short stature, micrognathia, epicanthic folds and depressed nasal bridge) [86]. In six individuals with EDS type VIIc so far described, all were homozygous for point mutations in the ADAMTS2 gene, resulting in a premature stop codon. A calf with dermatosparaxis had a 17 bp deletion, which resulted in a frame-shift mutation [85]. Adamts2-null mice, although grossly normal at birth, developed severe skin fragility after 1–2 months. It was noted that males are sterile, although Adamts2-null females have normal fertility [87]. The progressive skin fragility appeared to be caused not only by the abnormal morphology of type I collagen fibrils, distorted by a large proportion of uncleaved N-propeptides, but also by the failure of these fibrils to increase in diameter, a feature essential for their tensile strength. These results suggest that ADAMTS-2 is essential for maturation of type I collagen fibrils in skin, and that neither ADAMTS-3 nor ADAMTS-14 compensate adequately for ADAMTS-2 deficiency in this tissue.
The GON-ADAMTSs: ADAMTS-9 and -20
ADAMTS-9 was first described by Clark et al. [88] in 2000, and was found to share a high degree of sequence homology with the C. elegans ADAMTS gene gon-1 (discussed in the ‘ADAMTS in non-mammalian species’ section below). The gon-1 gene derives its name from its essential role in gonadal development in C. elegans [89]. It was later confirmed that the initially identified human ADAMTS-9 molecule, with three C-terminal TS repeats and no gon domain, was a short isoform when a longer molecule, containing 14 C-terminal TS repeats and a gon domain (a C-terminal region containing ten cysteine residues, similar to the C-terminal domain of gon-1) was identified [11,12], suggesting that the gene is alternatively spliced. Whether or not ADAMTS-9 has a role in mammalian gonadal development is not yet known.
Human ADAMTS9 has been mapped to chromosome 3 at band p14.2–p14.3, a common aphidicolin fragile site (a point on the chromosome that is liable to show gaps and breaks) [90], which is both a translocation breakpoint in hereditary renal cell carcinomas [91,92], and is frequently deleted in various other human cancers, including breast carcinoma [93]. Clark et al. [88] state that this locus is thought to contain one or more tumour suppressor genes, and that ADAMTS9 may be a possible candidate, especially as they describe abrogation of ADAMTS9 expression in ovarian and renal tumours. In our own studies of ADAMTS gene dysregulation in human breast carcinoma, we saw a down-regulation of ADAMTS9 by 83% with respect to non-neoplastic mammary tissue [49].
ADAMTS-20 shares its modular arrangement with that of the long isoform of ADAMTS-9, having 14 C-terminal TS repeats and a gon domain [11,12]. A shorter isoform of ADAMTS-20 has been identified in both humans [12] and the mouse [94], with ten and eight C-terminal TS repeats respectively. Point mutations in the mouse Adamts20 gene cause the pigment defect in belted (bt) mutant mice, a mostly pigmented phenotype with a dorsal non-pigmented region proximal to the hind limbs that appears as a white belt [94].
vWFCP/ADAMTS-13
vWFCP has been identified as ADAMTS-13 [16,95–97]. The existence of vWFCP was known for some years before its identity as an ADAMTS was elucidated [98,99]. Its substrate is vWF, a large multimeric glycoprotein present in plasma, platelets and vascular endothelial cells. vWF is a carrier protein for clotting factor VIII, supports platelet aggregation, and also mediates platelet adhesion to areas of vascular damage by binding both to glycoproteins on the surface of platelets and to exposed ECM components [97,100]. The shear stress to which vWF is subjected in the circulation leads to its enhanced proteolytic susceptibility [101]. Deficiency of vWFCP/ADAMTS-13 is responsible for TTP (thrombotic thrombocytopenic purpura), a pathological condition characterized by the formation of microvascular vWF- and platelet-rich thrombi, and associated with anaemia, renal failure and neurological dysfunction [96]. On the basis of present evidence, there are two basic mechanisms for ADAMTS-13 deficiency: (a) homozygous or compound heterozygous gene mutations; and (b) anti-(ADAMTS-13) autoantibody formation [102].
ADAMTS-13 shows the least homology with any other member of the ADAMTS family (notably, it has a much smaller pro-domain than any other), and is the only human ADAMTS so far described to contain a CUB domain [16]. ADAMTS13 is a large gene comprising 29 exons and encoding a product of 1427 amino acids, with evidence of additional variants due to alternative splicing [96].
Analysis of C-terminal deletion mutants of ADAMTS-13 has shown that the ability to cleave vWF in vitro requires the presence of the spacer region, though the more C-terminal TS and CUB domains were dispensable [103]. However, it seems likely that the C-terminal TS and CUB domains are relevant for the physiological activities of ADAMTS-13 in vivo, since TTP individuals with mutations in these regions of the gene have severe ADAMTS-13 deficiency [96,104]. Another curious feature of ADAMTS-13 is that the unusually short propeptide region seems not to be necessary for secretion, folding or maintenance of enzyme latency [34]. Mutation of the furin cleavage site or deletion of the entire pro-region led to secretion of pro-ADAMTS-13, which was fully functionally active. It is thus not clear at present what function the pro-region of ADAMTS-13 may perform, though it would seem there are no TTP-inherited mutations yet described that affect this region [96,104–106].
To date, 20 mutations (including frame-shifts, splice site mutations and amino acid substitutions) have been identified in the ADAMTS13 gene that lead to a deficient level of protein, thereby causing TTP in affected individuals [96,104,106]. These were spread across many exons, and no clustering of mutations was seen in any particular exon. A study by Kokame et al. [105] investigated four ADAMTS13 mutations in two Japanese families with a history of Upshaw–Schulman syndrome, the congenital form of TTP, and found that one of the four mutations did not completely abrogate enzyme activity. Furthermore, this mutation was found to be present in nearly 10% of the Japanese population. Two of the mutations prevented secretion of ADAMTS-13.
The development of autoantibodies to ADAMTS-13 occurs sporadically, although concordance in monozygotic twins has been reported, which implicates a genetic predisposing factor [107]. A recent study of sera obtained from 25 patients with anti-(ADAMTS-13) autoantibodies showed that, in the majority of cases, autoantibody reaction is directed against several epitopes along the entire ADAMTS-13 molecule, and that the cysteine-rich/spacer region is the only domain to be consistently involved in antibody reactivity [108]. This underlines the functional importance of this region for ADAMTS-13 activity [96,104]. An additional point of interest is that autoantibody reaction was directed against the pro-domain of ADAMTS-13 in 20% of cases, reinforcing the study by Majerus et al. [34] discussed above.
The orphan ADAMTSs
The unofficial term ‘orphan ADAMTS’ has been used by Dr S. S. Apte (http://www.lerner.ccf.org/bme/apte/adamts) to refer to those ADAMTS proteins with no known function or substrate to date. These are currently ADAMTS-6, -7, -10, -12 and -16–19 inclusive. Our studies have shown that ADAMTS6 and ADAMTS18 are dysregulated in breast carcinoma [49], and ADAMTS12 and ADAMTS16 are dysregulated in osteoarthritis [51].
A new study on ADAMTS-7 shows that the large spacer region between TS repeats 4 and 5 of the previously described long isoform, ADAMTS-7B, is a mucin domain that undergoes extensive post-translational O-glycosylation and within which is attached one or more chondroitin sulphate chains. This suggests that ADAMTS-7B may function as a proteoglycan in some tissues and cells, which has implications for its substrate recognition and cellular localization [14]. The sequence of ADAMTS-12 also predicts a mucin domain, and these two proteins therefore constitute a unique subgroup within the ADAMTS family. No substrate has been identified for ADAMTS-7, although aggrecanase activity has been ruled out.
A new paper has described ADAMTS10 mutations in three families with autosomal recessive WMS (Weill-Marchesani syndrome), predicting premature truncation of the protein within the metalloproteinase domain [109]. The clinical features of WMS, which include short stature, brachydactyly, eye and cardiac anomalies and progressive joint stiffness, suggest a role for ADAMTS-10 both in growth and in skin, lens and cardiac development in humans.
ADAMTSs IN NON-MAMMALIAN SPECIES
The ADAMTSs are much less common in non-mammalian metazoans. Apart from the five C. elegans ADAMTS genes, there are three ADAMTS-like protein sequences in Drosophila melanogaster [5,110]. Llamazares et al. [12] state that gon motifs are present in proteins predicted from the sequence analysis of D. melanogaster, Anopheles gambiae and Fugu rubripes, and propose that, together with ADAMTS-9, ADAMTS-20 and gon-1, they all define a subset of GON-ADAMTSs.
Several ADAMTS genes have been found to play a crucial role in gonadal morphogenesis in C. elegans. In this species, the forming gonad acquires U-shaped arms (two in the hermaphrodite gonad, one in the male gonad), determined by the directed migration of ‘leader’ cells in the arm tips. The C. elegans gene gon-1 is expressed both in leader cells and muscle, potentially permitting and directing expansion of the gonad by remodelling of the basement membrane [111]. Another C. elegans ADAMTS-like gene, mig-17, is required for the correct migration of leader cells: it influences the route of migration rather than permitting migration in itself, and also acts by remodelling the basement membrane [112]. mig-17 is most closely related to mouse Adamts1, comprising a signal peptide and pro-, catalytic, disintegrin-like and cysteine-rich domains, although it has no TS repeats and is not a true ADAMTS [112]. Another C. elegans ADAMTS gene, adt-1, is required for morphogenesis of the rays, which, with the fan, comprise the male-specific copulatory organs [113]. Ray morphogenesis requires the rapid remodelling of the ECM. The authors demonstrate that deletion mutants show impaired mating ability and structural ray abnormalities.
Very little information is available on ADAMTSs in non-metazoans. There are no reported ADAMTS genes in either Saccharomyces cerevisiae or Arabidopsis thaliana [110]. However, Varney et al. [114] have described an adamalysin-like protein, AmpA, in the cellular slime mould amoeba Dictyostelium discoideum. It consists of a signal peptide, two disintegrin-like domains and four repeats similar to the TS motifs of the ADAMTSs. Although it does not have all the hallmark domains of either an ADAM or ADAMTS, it carries out similar functions to them in cell-fate specification and modulates cell–cell and cell–substrate adhesion during the migration of single amoebae into aggregation centres.
ADAMTSL MOLECULES
Hirohata et al. [115] have described a novel ADAMTSL molecule, named ADAMTSL-1, which is a secreted glycoprotein thought to function in the ECM. Expression of ADAMTSL-1 in COS-1 cells results in deposition of the protein in the cell substratum in a punctate fashion; this has led to the protein being given the trivial name ‘punctin’. ADAMTSL-1 lacks the pro-domain, metalloproteinase and disintegrin domains seen in the ADAMTSs; from the N-terminus, it contains a signal peptide, a TS repeat, a cysteine-rich domain, a spacer region and three further TS repeats.
Two further members of the ADAMTSL family have been cloned to date: these are ADAMTSL2 [116] and ADAMTSL3, also called punctin-2 [117]. ADAMTSL2 and 3 differ from ADAMTSL1 in their greater length, greater number of TS repeats (6 and 13 respectively) and the fact that they are more widely expressed [117]. In addition, ADAMTSL3 is distinguished from the ADAMTS genes in its arrangement of TS repeats, which are arranged in two arrays separated by a region containing three immunoglobulin-like repeats [117], also seen in mammalian papilin [48] and a long isoform of ADAMTSL-1 [117]. The functions of the ADAMTS-like molecules are currently unknown, but as they are composed only of ADAMTS ancillary, non-enzymatic domains, it is interesting to speculate that they may have functions in cell–cell or cell–ECM adhesion, or they may regulate ADAMTS protease functions [115].
FUTURE CHALLENGES, UNANSWERED QUESTIONS
The ADAMTSs are involved in a bewilderingly diverse range of biological processes, from ECM assembly to ECM degradation, from regulation of angiogenesis to blood coagulation, and with clear links to organogenesis and fertility. We are just beginning to unravel the details of their participation in these areas. In addition to ongoing biochemical studies of function in vitro, one of the key avenues for future investigation will be the analysis of gene-knockout mice. Somewhat surprisingly, data have thus far been published on only three members, ADAMTS1 [76], ADAMTS2 [87] and ADAMTS4 [118]. The complex phenotype of Adamts1-null mice confirms a key role for this gene in formation of adipose tissue, in kidney morphogenesis and female fertility. None of these phenotypes suggest an indispensable role for ADAMTS-1 in regulation of angiogenesis, where it has been studied extensively in vitro [9,37,119]. However, ADAMTSs probably have both overlapping and unique functions, and the phenotypes of knockout mice will depend on the spatiotemporal patterns of expression of the family members. It is possible that the functions of at least some of the ADAMTSs may ultimately be found to be linked to specific aspects of organogenesis by facilitating cell migration and branching morphogenesis by cleavage of regulatory ECM molecules, which reflects the functions of the ancestral C. elegans gon-1 and mig-17 genes. It is interesting also to note that TIMP-3, which inhibits a number of ADAMTS genes, has been linked with epithelial branching morphogenesis in the developing mouse lung [120]. It will be revealing to determine whether ADAMTSs participate in alveo-bronchiolar development, since many of them are highly expressed in the fetal lung.
The discovery of the ADAMs and ADAMTSs is owed in part to the effort to develop MMPIs (MMP inhibitors) as anticancer and anti-arthritis therapies [121]. Early studies showed the MMPs were pivotal to cancer metastasis, so broad-spectrum MMPIs were developed to abrogate proteolytic activity by binding to the zinc atom in their catalytic domain. Given the encouraging results seen in early mouse models (e.g. [122]), MMPIs moved rapidly into clinical trials which, with hindsight, proved to be inappropriately hasty. We have begun to understand more fully the functions of MMPs in normal and pathological situations, and together with the discovery of the ADAMs and ADAMTSs, which share a related metalloproteinase domain, it is clear that indiscriminate targeting of the metalloproteinases was inappropriate. Targeting the catalytic domain of MMPs most likely affected the ADAMTSs, either directly (through sufficient homology with ADAMTS catalytic domains) or indirectly (because some MMPs are implicated in ADAMTS processing, as discussed above), and could have interfered with the anti-angiogenic function of which some ADAMTSs are demonstrably capable. In fact, there is some evidence for this, since some MMPIs caused musculoskeletal pain in clinical trials, which has been attributed to inhibition of sheddase activity by targeting non-MMPs such as the ADAMs [123]. The future challenge will be selective targeting, possibly requiring individually designed inhibitors for each enzyme. In the case of the ADAMTSs, our emerging knowledge of these proteins suggests that inhibitors might be effectively directed at their spacer regions. A recent paper points the way towards understanding, and capitalizing upon, the unique ability of TIMP-3 to inhibit not only MMPs, but several ADAMs and ADAMTSs, which will be likely to pave the way for selective TIMP variants to be designed [124].
In conclusion, the ADAMTSs are a complex family of enzymes, and their complete characterization will be challenging. An important factor to recognize is that several have different isoforms, generated via alternative splicing or post-translational processing, leading to differences in substrate profiles and biodistribution and, potentially at least, the unmasking of cryptic properties. This fascinating and complex story will continue to challenge us for some time to come.
Acknowledgments
We gratefully acknowledge support from the Breast Cancer Campaign, the Arthritis Research Campaign, BBSRC (Biotechnology and Biological Sciences Research Council), AstraZeneca, the Norfolk and Norwich Big C Appeal, and the European Union Framework Programmes 5 (QLG1-CT-2000–01131) and 6 (LSHC-CT-2003–503297).
References
- 1.Kuno K., Kanada N., Nakashima E., Fujiki F., Ichimura F., Matsushima K. Molecular cloning of a gene encoding a new type of metalloproteinase-disintegrin family protein with thrombospondin motifs as an inflammation associated gene. J. Biol. Chem. 1997;272:556–562. doi: 10.1074/jbc.272.1.556. [DOI] [PubMed] [Google Scholar]
- 2.Kaushal G. P., Shah S. V. The new kids on the block: ADAMTSs, potentially multifunctional metalloproteinases of the ADAM family. J. Clin. Invest. 2000;105:1335–1337. doi: 10.1172/JCI10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stocker W., Grams F., Baumann U., Reinemer P., Gomis-Rüth F. X., McKay D. B., Bode W. The metzincins — topological and sequential relations between the astacins, adamalysins, serralysins, and matrixins (collagenases) define a superfamily of zinc-peptidases. Protein Sci. 1995;4:823–840. doi: 10.1002/pro.5560040502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seals D. F., Courtneidge S. A. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17:7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- 5.Tang B. L. ADAMTS: a novel family of extracellular matrix proteases. Int. J. Biochem. Cell. Biol. 2001;33:33–44. doi: 10.1016/s1357-2725(00)00061-3. [DOI] [PubMed] [Google Scholar]
- 6.Kuno K., Matsushima K. ADAMTS-1 protein anchors at the extracellular matrix through the thrombospondin type I motifs and its spacing region. J. Biol. Chem. 1998;273:13912–13917. doi: 10.1074/jbc.273.22.13912. [DOI] [PubMed] [Google Scholar]
- 7.Abbaszade I., Liu R. Q., Yang F., Rosenfeld S. A., Ross O. H., Link J. R., Ellis D. M., Tortorella M. D., Pratta M. A., Hollis J. M., et al. Cloning and characterization of ADAMTS11, an aggrecanase from the ADAMTS family. J. Biol. Chem. 1999;274:23443–23450. doi: 10.1074/jbc.274.33.23443. [DOI] [PubMed] [Google Scholar]
- 8.Hurskainen T. L., Hirohata S., Seldin M. F., Apte S. S. ADAM-TS5, ADAM-TS6, and ADAM-TS7, novel members of a new family of zinc metalloproteases. General features and genomic distribution of the ADAM-TS family. J. Biol. Chem. 1999;274:25555–25563. doi: 10.1074/jbc.274.36.25555. [DOI] [PubMed] [Google Scholar]
- 9.Vázquez F., Hastings G., Ortega M. A., Lane T. F., Oikemus S., Lombardo M., Iruela-Arispe M. L. METH-1, a human ortholog of ADAMTS-1, and METH-2 are members of a new family of proteins with angio-inhibitory activity. J. Biol. Chem. 1999;274:23349–23357. doi: 10.1074/jbc.274.33.23349. [DOI] [PubMed] [Google Scholar]
- 10.Perutelli P. Disintegrins: potent inhibitors of platelet aggregation. Recenti Prog. Med. 1995;86:168–174. [PubMed] [Google Scholar]
- 11.Somerville R. P., Longpre J. M., Jungers K. A., Engle J. M., Ross M., Evanko S., Wight T. N., Leduc R., Apte S. S. Characterization of ADAMTS-9 and ADAMTS-20 as a distinct ADAMTS subfamily related to Caenorhabditis elegans GON-1. J. Biol. Chem. 2003;278:9503–9513. doi: 10.1074/jbc.M211009200. [DOI] [PubMed] [Google Scholar]
- 12.Llamazares M., Cal S., Quesada V., López-Otín C. Identification and characterization of ADAMTS-20 defines a novel subfamily of metalloproteinases-disintegrins with multiple thrombospondin-1 repeats and a unique GON domain. J. Biol. Chem. 2003;278:13382–13389. doi: 10.1074/jbc.M211900200. [DOI] [PubMed] [Google Scholar]
- 13.Tortorella M. D., Burn T. C., Pratta M. A., Abbaszade I., Hollis J. M., Liu R., Rosenfeld S. A., Copeland R. A., Decicco C. P., Wynn R., et al. Purification and cloning of aggrecanase-1: a member of the ADAMTS family of proteins. Science. 1999;284:1664–1666. doi: 10.1126/science.284.5420.1664. [DOI] [PubMed] [Google Scholar]
- 14.Somerville R. P., Longpre J. M., Apel E. D., Lewis R. M., Wang L. W., Sanes J. R., Leduc R., Apte S. S. ADAMTS7B, the full-length product of the ADAMTS7 gene, is a chondroitin sulfate proteoglycan containing a mucin domain. J. Biol. Chem. 2004;279:35159–35175. doi: 10.1074/jbc.M402380200. [DOI] [PubMed] [Google Scholar]
- 15.Blelloch R., Kimble J. Control of organ shape by a secreted metalloprotease in the nematode Caenorhabditis elegans. Nature (London) 1999;399:586–590. doi: 10.1038/21196. [DOI] [PubMed] [Google Scholar]
- 16.Zheng X., Chung D., Takayama T. K., Majerus E. M., Sadler J. E., Fujikawa K. Structure of von Willebrand factor-cleaving protease (ADAMTS13), a metalloprotease involved in thrombotic thrombocytopenic purpura. J. Biol. Chem. 2001;276:41059–41063. doi: 10.1074/jbc.C100515200. [DOI] [PubMed] [Google Scholar]
- 17.Geier G., Zwilling R. Cloning and characterization of a cDNA coding for Astacus embryonic astacin, a member of the astacin family of metalloproteases from the crayfish Astacus astacus. Eur. J. Biochem. 1998;253:796–803. doi: 10.1046/j.1432-1327.1998.2530796.x. [DOI] [PubMed] [Google Scholar]
- 18.Bork P., Beckmann G. The CUB domain. A widespread module in developmentally regulated proteins. J. Mol. Biol. 1993;231:539–545. doi: 10.1006/jmbi.1993.1305. [DOI] [PubMed] [Google Scholar]
- 19.Nardi J. B., Martos R., Walden K. K., Lampe D. J., Robertson H. M. Expression of lacunin, a large multidomain extracellular matrix protein, accompanies morphogenesis of epithelial monolayers in Manduca sexta. Insect Biochem. Mol. Biol. 1999;29:883–897. doi: 10.1016/s0965-1748(99)00064-8. [DOI] [PubMed] [Google Scholar]
- 20.Cal S., Argüelles J. M., Fernández P. L., López-Otín C. Identification, characterization, and intracellular processing of ADAM-TS12, a novel human disintegrin with a complex structural organization involving multiple thrombospondin-1 repeats. J. Biol. Chem. 2001;276:17932–17940. doi: 10.1074/jbc.M100534200. [DOI] [PubMed] [Google Scholar]
- 21.Engle J. M., Tam L., Goldring M., Apte S. S. 47th Annual Meeting. San Francisco, California: Orthopaedic Research Society; 2001. ADAMTS-9 and ADAMTS-10, two novel, unusual ADAMTS proteases and regulation of the ADAMTS family in three human chondrocyte cell lines. [Google Scholar]
- 22.Gao G., Westling J., Thompson V. P., Howell T. D., Gottschall P. E., Sandy J. D. Activation of the proteolytic activity of ADAMTS4 (aggrecanase-1) by C-terminal truncation. J. Biol. Chem. 2002;277:11034–11041. doi: 10.1074/jbc.M107443200. [DOI] [PubMed] [Google Scholar]
- 23.Kashiwagi M., Enghild J. J., Gendron C., Hughes C., Caterson B., Itoh Y., Nagase H. Altered proteolytic activities of ADAMTS-4 expressed by C-terminal processing. J. Biol. Chem. 2004;279:10109–10119. doi: 10.1074/jbc.M312123200. [DOI] [PubMed] [Google Scholar]
- 24.Tortorella M., Pratta M., Liu R.-Q., Abbaszade I., Ross H., Burn T., Arner E. The thombospondin motif of aggrecanase-1 (ADAMTS-4) Is critical for aggrecan substrate recognition and cleavage. J. Biol. Chem. 2000;275:25791–25797. doi: 10.1074/jbc.M001065200. [DOI] [PubMed] [Google Scholar]
- 25.Hashimoto G., Shimoda M., Okada Y. ADAMTS4 (aggrecanase-1) interaction with the C-terminal domain of fibronectin inhibits proteolysis of aggrecan. J. Biol. Chem. 2004;279:32483–32491. doi: 10.1074/jbc.M314216200. [DOI] [PubMed] [Google Scholar]
- 26.Cao J., Hymowitz M., Conner C., Bahou W. F., Zucker S. The propeptide domain of membrane type 1-matrix metalloproteinase acts as an intramolecular chaperone when expressed in trans with the mature sequence in COS-1 cells. J. Biol. Chem. 2000;275:29648–29653. doi: 10.1074/jbc.M001920200. [DOI] [PubMed] [Google Scholar]
- 27.Milla M. E., Leesnitzer M. A., Moss M. L., Clay W. C., Carter H. L., Miller A. B., Su J. L., Lambert M. H., Willard D. H., Sheeley D. M., et al. Specific sequence elements are required for the expression of functional tumor necrosis factor-alpha-converting enzyme (TACE) J. Biol. Chem. 1999;274:30563–30570. doi: 10.1074/jbc.274.43.30563. [DOI] [PubMed] [Google Scholar]
- 28.Bergeron F., Leduc R., Day R. Subtilase-like pro-protein convertases: from molecular specificity to therapeutic applications. J. Mol. Endocrinol. 2000;24:1–22. doi: 10.1677/jme.0.0240001. [DOI] [PubMed] [Google Scholar]
- 29.Rodríguez-Manzaneque J. C., Milchanowski A. B., Dufour E. K., Leduc R., Iruela-Arispe M. L. Characterization of METH-1/ADAMTS1 processing reveals two distinct active forms. J. Biol. Chem. 2000;275:33471–33479. doi: 10.1074/jbc.M002599200. [DOI] [PubMed] [Google Scholar]
- 30.Wang P., Tortorella M., England K., Malfait A. M., Thomas G., Arner E. C., Pei D. Proprotein convertase furin interacts with and cleaves pro-ADAMTS4 (aggrecanase-1) in the trans-Golgi network. J. Biol. Chem. 2004;279:15434–15440. doi: 10.1074/jbc.M312797200. [DOI] [PubMed] [Google Scholar]
- 31.Wang W. M., Lee S., Steiglitz B. M., Scott I. C., Lebares C. C., Allen M. L., Brenner M. C., Takahara K., Greenspan D. S. Transforming growth factor-beta induces secretion of activated ADAMTS-2. A procollagen III N-proteinase. J. Biol. Chem. 2003;278:19549–19557. doi: 10.1074/jbc.M300767200. [DOI] [PubMed] [Google Scholar]
- 32.Nagase H., Woessner J. F., Jr Matrix metalloproteinases. J. Biol. Chem. 1999;274:21491–21494. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- 33.Van Wart H. E., Birkedal-Hansen H. The cysteine switch: a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proc. Natl. Acad. Sci. U.S.A. 1990;87:5578–5582. doi: 10.1073/pnas.87.14.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majerus E. M., Zheng X., Tuley E. A., Sadler J. E. Cleavage of the ADAMTS13 propeptide is not required for protease activity. J. Biol. Chem. 2003;278:46643–46648. doi: 10.1074/jbc.M309872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao G., Plaas A., Thompson V. P., Jin S., Zuo F., Sandy J. D. ADAMTS4 (aggrecanase-1) activation on the cell surface involves C-terminal cleavage by glycosylphosphatidyl inositol-anchored membrane type 4-matrix metalloproteinase and binding of the activated proteinase to chondroitin sulfate and heparan sulfate on syndecan-1. J. Biol. Chem. 2004;279:10042–10051. doi: 10.1074/jbc.M312100200. [DOI] [PubMed] [Google Scholar]
- 36.Flannery C. R., Zeng W., Corcoran C., Collins-Racie L. A., Chockalingam P. S., Hebert T., Mackie S. A., McDonagh T., Crawford T. K., Tomkinson K. N., et al. Autocatalytic cleavage of ADAMTS-4 (aggrecanase-1) reveals multiple glycosaminoglycan-binding sites. J. Biol. Chem. 2002;277:42775–42780. doi: 10.1074/jbc.M205309200. [DOI] [PubMed] [Google Scholar]
- 37.Luque A., Carpizo D. R., Iruela-Arispe M. L. ADAMTS1/METH1 inhibits endothelial cell proliferation by direct binding and sequestration of VEGF 165. J. Biol. Chem. 2003;278:23656–23665. doi: 10.1074/jbc.M212964200. [DOI] [PubMed] [Google Scholar]
- 38.Tortorella M. D., Pratta M., Liu R. Q., Austin J., Ross O. H., Abbaszade I., Burn T., Arner E. Sites of aggrecan cleavage by recombinant human aggrecanase-1 (ADAMTS-4) J. Biol. Chem. 2000;275:18566–18573. doi: 10.1074/jbc.M909383199. [DOI] [PubMed] [Google Scholar]
- 39.Baker A. H., Edwards D. R., Murphy G. Metalloproteinase inhibitors: biological actions and therapeutic opportunities. J. Cell Sci. 2002;115:3719–3727. doi: 10.1242/jcs.00063. [DOI] [PubMed] [Google Scholar]
- 40.Hashimoto T., Wen G., Lawton M. T., Boudreau N. J., Bollen A. W., Yang G. Y., Barbaro N. M., Higashida R. T., Dowd C. F., Halbach V. V., Young W. L. Abnormal expression of matrix metalloproteinases and tissue inhibitors of metalloproteinases in brain arteriovenous malformations. Stroke. 2003;34:925–931. doi: 10.1161/01.STR.0000061888.71524.DF. [DOI] [PubMed] [Google Scholar]
- 41.Kashiwagi M., Tortorella M., Nagase H., Brew K. TIMP-3 is a potent inhibitor of aggrecanase 1 (ADAM-TS4) and aggrecanase 2 (ADAM-TS5) J. Biol. Chem. 2001;276:12501–12504. doi: 10.1074/jbc.C000848200. [DOI] [PubMed] [Google Scholar]
- 42.Arner E. C., Pratta M. A., Trzaskos J. M., Decicco C. P., Tortorella M. D. Generation and characterization of aggrecanase. A soluble, cartilage-derived aggrecan-degrading activity. J. Biol. Chem. 1999;274:6594–6601. doi: 10.1074/jbc.274.10.6594. [DOI] [PubMed] [Google Scholar]
- 43.Rodríguez-Manzaneque J. C., Westling J., Thai S. N., Luque A., Knauper V., Murphy G., Sandy J. D., Iruela-Arispe M. L. ADAMTS1 cleaves aggrecan at multiple sites and is differentially inhibited by metalloproteinase inhibitors. Biochem. Biophys. Res. Commun. 2002;293:501–508. doi: 10.1016/S0006-291X(02)00254-1. [DOI] [PubMed] [Google Scholar]
- 44.Gendron C., Kashiwagi M., Hughes C., Caterson B., Nagase H. TIMP-3 inhibits aggrecanase-mediated glycosaminoglycan release from cartilage explants stimulated by catabolic factors. FEBS Lett. 2003;555:431–436. doi: 10.1016/s0014-5793(03)01295-x. [DOI] [PubMed] [Google Scholar]
- 45.Tortorella M. D., Arner E. C., Hills R., Easton A., Korte-Sarfaty J., Fok K., Wittwer A. J., Liu R. Q., Malfait A. M. Alpha2-macroglobulin is a novel substrate for ADAMTS-4 and ADAMTS-5 and represents an endogenous inhibitor of these enzymes. J. Biol. Chem. 2004;279:17554–17561. doi: 10.1074/jbc.M313041200. [DOI] [PubMed] [Google Scholar]
- 46.Vankemmelbeke M. N., Jones G. C., Fowles C., Ilic M. Z., Handley C. J., Day A. J., Knight C. G., Mort J. S., Buttle D. J. Selective inhibition of ADAMTS-1, -4 and -5 by catechin gallate esters. Eur. J. Biochem. 2003;270:2394–2403. doi: 10.1046/j.1432-1033.2003.03607.x. [DOI] [PubMed] [Google Scholar]
- 47.Kuno K., Terashima Y., Matsushima K. ADAMTS-1 is an active metalloproteinase associated with the extracellular matrix. J. Biol. Chem. 1999;274:18821–18826. doi: 10.1074/jbc.274.26.18821. [DOI] [PubMed] [Google Scholar]
- 48.Kramerova I. A., Kawaguchi N., Fessler L. I., Nelson R. E., Chen Y., Kramerov A. A., Kusche-Gullberg M., Kramer J. M., Ackley B. D., Sieron A. L., et al. Papilin in development; a pericellular protein with a homology to the ADAMTS metalloproteinases. Development. 2000;127:5475–5485. doi: 10.1242/dev.127.24.5475. [DOI] [PubMed] [Google Scholar]
- 49.Porter S., Scott S. D., Sassoon E. M., Williams M. R., Jones J. L., Girling A. C., Ball R. Y., Edwards D. R. Dysregulated expression of adamalysin-thrombospondin genes in human breast carcinoma. Clin. Cancer Res. 2004;10:2429–2440. doi: 10.1158/1078-0432.ccr-0398-3. [DOI] [PubMed] [Google Scholar]
- 50.Masui T., Hosotani R., Tsuji S., Miyamoto Y., Yasuda S., Ida J., Nakajima S., Kawaguchi M., Kobayashi H., Koizumi M., et al. Expression of METH-1 and METH-2 in pancreatic cancer. Clin. Cancer Res. 2001;7:3437–3443. [PubMed] [Google Scholar]
- 51.Kevorkian L., Young D. A., Darrah C., Donell S. T., Shepstone L., Porter S., Brockbank S. M., Edwards D. R., Parker A. E., Clark I. M. Expression profiling of metalloproteinases and their inhibitors in cartilage. Arthritis Rheum. 2004;50:131–141. doi: 10.1002/art.11433. [DOI] [PubMed] [Google Scholar]
- 52.Wang W. M., Lee S., Steiglitz B. M., Scott I. C., Lebares C. C., Allen M. L., Brenner M. C., Takahara K., Greenspan D. S. Transforming growth factor-beta induces secretion of activated ADAMTS-2. A procollagen III N-proteinase. J. Biol. Chem. 2003;278:19549–19557. doi: 10.1074/jbc.M300767200. [DOI] [PubMed] [Google Scholar]
- 53.Yamanishi Y., Boyle D. L., Clark M., Maki R. A., Tortorella M. D., Arner E. C., Firestein G. S. Expression and regulation of aggrecanase in arthritis: the role of TGF-beta. J. Immunol. 2002;168:1405–1412. doi: 10.4049/jimmunol.168.3.1405. [DOI] [PubMed] [Google Scholar]
- 54.Pratta M. A., Scherle P. A., Yang G., Liu R. Q., Newton R. C. Induction of aggrecanase 1 (ADAM-TS4) by interleukin-1 occurs through activation of constitutively produced protein. Arthritis Rheum. 2003;48:119–133. doi: 10.1002/art.10726. [DOI] [PubMed] [Google Scholar]
- 55.Koshy P. J., Lundy C. J., Rowan A. D., Porter S., Edwards D. R., Hogan A., Clark I. M., Cawston T. E. The modulation of matrix metalloproteinase and ADAM gene expression in human chondrocytes by interleukin-1 and oncostatin M: a time-course study using real-time quantitative reverse transcription-polymerase chain reaction. Arthritis Rheum. 2002;46:961–967. doi: 10.1002/art.10212. [DOI] [PubMed] [Google Scholar]
- 56.Sylvester J., Liacini A., Li W. Q., Zafarullah M. Interleukin-17 signal transduction pathways implicated in inducing matrix metalloproteinase-3, -13 and aggrecanase-1 genes in articular chondrocytes. Cell Signal. 2004;16:469–476. doi: 10.1016/j.cellsig.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 57.Sasaki M., Seo-Kiryu S., Kato R., Kita S., Kiyama H. A disintegrin and metalloprotease with thrombospondin type1 motifs (ADAMTS-1) and IL-1 receptor type 1 mRNAs are simultaneously induced in nerve injured motor neurons. Brain Res. Mol. Brain Res. 2001;89:158–163. doi: 10.1016/s0169-328x(01)00046-8. [DOI] [PubMed] [Google Scholar]
- 58.Worley J. R., Baugh M. D., Hughes D. A., Edwards D. R., Hogan A., Sampson M. J., Gavrilovic J. Metalloproteinase expression in PMA-stimulated THP-1 cells. Effects of peroxisome proliferator-activated receptor-gamma (PPAR gamma) agonists and 9-cis-retinoic acid. J. Biol. Chem. 2003;278:51340–51346. doi: 10.1074/jbc.M310865200. [DOI] [PubMed] [Google Scholar]
- 59.Makihira S., Yan W., Murakami H., Furukawa M., Kawai T., Nikawa H., Yoshida E., Hamada T., Okada Y., Kato Y. Thyroid hormone enhances aggrecanase-2/ADAM-TS5 expression and proteoglycan degradation in growth plate cartilage. Endocrinology. 2003;144:2480–2488. doi: 10.1210/en.2002-220746. [DOI] [PubMed] [Google Scholar]
- 60.Miles R. R., Sluka J. P., Halladay D. L., Santerre R. F., Hale L. V., Bloem L., Thirunavukkarasu K., Galvin R. J., Hock J. M., Onyia J. E. ADAMTS-1: a cellular disintegrin and metalloprotease with thrombospondin motifs is a target for parathyroid hormone in bone. Endocrinology. 2000;141:4533–4542. doi: 10.1210/endo.141.12.7817. [DOI] [PubMed] [Google Scholar]
- 61.Robker R. L., Russell D. L., Espey L. L., Lydon J. P., O'Malley B. W., Richards J. S. Progesterone-regulated genes in the ovulation process: ADAMTS-1 and cathepsin L proteases. Proc. Natl. Acad. Sci. U.S.A. 2000;97:4689–4694. doi: 10.1073/pnas.080073497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Espey L. L., Yoshioka S., Russell D. L., Robker R. L., Fujii S., Richards J. S. Ovarian expression of a disintegrin and metalloproteinase with thrombospondin motifs during ovulation in the gonadotropin-primed immature rat. Biol. Reprod. 2000;62:1090–1095. doi: 10.1095/biolreprod62.4.1090. [DOI] [PubMed] [Google Scholar]
- 63.de Fraipont F., Nicholson A. C., Feige J. J., Van Meir E. G. Thrombospondins and tumor angiogenesis. Trends Mol. Med. 2001;7:401–407. doi: 10.1016/s1471-4914(01)02102-5. [DOI] [PubMed] [Google Scholar]
- 64.Iruela-Arispe M. L., Lombardo M., Krutzsch H. C., Lawler J., Roberts D. D. Inhibition of angiogenesis by thrombospondin-1 is mediated by 2 independent regions within the type 1 repeats. Circulation. 1999;100:1423–1431. doi: 10.1161/01.cir.100.13.1423. [DOI] [PubMed] [Google Scholar]
- 65.Lawler J. The functions of thrombospondin-1 and-2. Curr. Opin. Cell Biol. 2000;12:634–640. doi: 10.1016/s0955-0674(00)00143-5. [DOI] [PubMed] [Google Scholar]
- 66.Roughley P. J. Articular cartilage and changes in arthritis: noncollagenous proteins and proteoglycans in the extracellular matrix of cartilage. Arthritis Res. 2001;3:342–347. doi: 10.1186/ar326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pratta M. A., Yao W., Decicco C., Tortorella M. D., Liu R. Q., Copeland R. A., Magolda R., Newton R. C., Trzaskos J. M., Arner E. C. Aggrecan protects cartilage collagen from proteolytic cleavage. J. Biol. Chem. 2003;278:45539–45545. doi: 10.1074/jbc.M303737200. [DOI] [PubMed] [Google Scholar]
- 68.Sandy J. D., Neame P. J., Boynton R. E., Flannery C. R. Catabolism of aggrecan in cartilage explants. Identification of a major cleavage site within the interglobular domain. J. Biol. Chem. 1991;266:8683–8685. [PubMed] [Google Scholar]
- 69.Sugimoto K., Takahashi M., Yamamoto Y., Shimada K., Tanzawa K. Identification of aggrecanase activity in medium of cartilage culture. J. Biochem. (Tokyo) 1999;126:449–455. doi: 10.1093/oxfordjournals.jbchem.a022471. [DOI] [PubMed] [Google Scholar]
- 70.Tortorella M. D., Malfait A. M., Deccico C., Arner E. The role of ADAM-TS4 (aggrecanase-1) and ADAM-TS5 (aggrecanase-2) in a model of cartilage degradation. Osteoarthritis Cartilage. 2001;9:539–552. doi: 10.1053/joca.2001.0427. [DOI] [PubMed] [Google Scholar]
- 71.Matthews R. T., Gary S. C., Zerillo C., Pratta M., Solomon K., Arner E. C., Hockfield S. Brain-enriched hyaluronan binding (BEHAB)/brevican cleavage in a glioma cell line is mediated by a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) family member. J. Biol. Chem. 2000;275:22695–22703. doi: 10.1074/jbc.M909764199. [DOI] [PubMed] [Google Scholar]
- 72.Sandy J. D., Westling J., Kenagy R. D., Iruela-Arispe M. L., Verscharen C., Rodríguez-Manzaneque J. C., Zimmermann D. R., Lemire J. M., Fischer J. W., Wight T. N., Clowes A. W. Versican V1 proteolysis in human aorta in vivo occurs at the Glu441–Ala442 bond, a site that is cleaved by recombinant ADAMTS-1 and ADAMTS-4. J. Biol. Chem. 2001;276:13372–13378. doi: 10.1074/jbc.M009737200. [DOI] [PubMed] [Google Scholar]
- 73.Kuno K., Okada Y., Kawashima H., Nakamura H., Miyasaka M., Ohno H., Matsushima K. ADAMTS-1 cleaves a cartilage proteoglycan, aggrecan. FEBS Lett. 2000;478:241–245. doi: 10.1016/s0014-5793(00)01854-8. [DOI] [PubMed] [Google Scholar]
- 74.Yamaji N., Nishimura K., Abe K., Ohara O., Nagase T., Nomura N. Pat. Japan: Yamanouchi Pharmaceutical Co. Ltd; 2001. Novel metalloprotease having aggrecanase activity. [Google Scholar]
- 75.Collins-Racie L. A., Flannery C. R., Zeng W., Corcoran C., Annis-Freeman B., Agostino M. J., Arai M., DiBlasio-Smith E., Dorner A. J., Georgiadis K. E., et al. ADAMTS-8 exhibits aggrecanase activity and is expressed in human articular cartilage. Matrix Biol. 2004;23:219–230. doi: 10.1016/j.matbio.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 76.Shindo T., Kurihara H., Kuno K., Yokoyama H., Wada T., Kurihara Y., Imai T., Wang Y., Ogata M., Nishimatsu H., et al. ADAMTS-1: a metalloproteinase-disintegrin essential for normal growth, fertility, and organ morphology and function. J. Clin. Invest. 2000;105:1345–1352. doi: 10.1172/JCI8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yokoyama H., Wada T., Kobayashi K., Kuno K., Kurihara H., Shindo T., Matsushima K. A disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS)-1 null mutant mice develop renal lesions mimicking obstructive nephropathy. Nephrol. Dial. Transplant 17. 2002;9(suppl.):39–41. doi: 10.1093/ndt/17.suppl_9.39. [DOI] [PubMed] [Google Scholar]
- 78.Thai S. N., Iruela-Arispe M. L. Expression of ADAMTS1 during murine development. Mech. Dev. 2002;115:181–185. doi: 10.1016/s0925-4773(02)00115-6. [DOI] [PubMed] [Google Scholar]
- 79.Diamantis I., Luthi M., Hosli M., Reichen J. Cloning of the rat ADAMTS-1 gene and its down regulation in endothelial cells in cirrhotic rats. Liver. 2000;20:165–172. doi: 10.1034/j.1600-0676.2000.020002165.x. [DOI] [PubMed] [Google Scholar]
- 80.Voros G., Maquoi E., Collen D., Lijnen H. R. Differential expression of plasminogen activator inhibitor-1, tumor necrosis factor-alpha, TNF-alpha converting enzyme and ADAMTS family members in murine fat territories. Biochim. Biophys. Acta. 2003;1625:36–42. doi: 10.1016/s0167-4781(02)00589-4. [DOI] [PubMed] [Google Scholar]
- 81.Nakamura H., Fujii Y., Inoki I., Sugimoto K., Tanzawa K., Matsuki H., Miura R., Yamaguchi Y., Okada Y. Brevican is degraded by matrix metalloproteinases and aggrecanase-1 (ADAMTS4) at different sites. J. Biol. Chem. 2000;275:38885–38890. doi: 10.1074/jbc.M003875200. [DOI] [PubMed] [Google Scholar]
- 82.Colige A., Beschin A., Samyn B., Goebels Y., Van Beeumen J., Nusgens B. V., Lapiere C. M. Characterization and partial amino acid sequencing of a 107-kDa procollagen I N-proteinase purified by affinity chromatography on immobilized type XIV collagen. J. Biol. Chem. 1995;270:16724–16730. doi: 10.1074/jbc.270.28.16724. [DOI] [PubMed] [Google Scholar]
- 83.Colige A., Li S. W., Sieron A. L., Nusgens B. V., Prockop D. J., Lapiere C. M. cDNA cloning and expression of bovine procollagen I N-proteinase: a new member of the superfamily of zinc-metalloproteinases with binding sites for cells and other matrix components. Proc. Natl. Acad. Sci. U.S.A. 1997;94:2374–2379. doi: 10.1073/pnas.94.6.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fernandes R. J., Hirohata S., Engle J. M., Colige A., Cohn D. H., Eyre D. R., Apte S. S. Procollagen II amino propeptide processing by ADAMTS-3. Insights on dermatosparaxis. J. Biol. Chem. 2001;276:31502–31509. doi: 10.1074/jbc.M103466200. [DOI] [PubMed] [Google Scholar]
- 85.Colige A., Sieron A. L., Li S. W., Schwarze U., Petty E., Wertelecki W., Wilcox W., Krakow D., Cohn D. H., Reardon W., et al. Human Ehlers-Danlos syndrome type VII C and bovine dermatosparaxis are caused by mutations in the procollagen I N-proteinase gene. Am. J. Hum. Genet. 1999;65:308–317. doi: 10.1086/302504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McKusick V. A. 10th edn. Baltimore: The Johns Hopkins University Press; 1992. Mendelian Inheritance in Man. [Google Scholar]
- 87.Li S. W., Arita M., Fertala A., Bao Y., Kopen G. C., Langsjo T. K., Hyttinen M. M., Helminen H. J., Prockop D. J. Transgenic mice with inactive alleles for procollagen N-proteinase (ADAMTS-2) develop fragile skin and male sterility. Biochem. J. 2001;355:271–278. doi: 10.1042/0264-6021:3550271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Clark M. E., Kelner G. S., Turbeville L. A., Boyer A., Arden K. C., Maki R. A. ADAMTS9, a novel member of the ADAM-TS/metallospondin gene family. Genomics. 2000;67:343–350. doi: 10.1006/geno.2000.6246. [DOI] [PubMed] [Google Scholar]
- 89.Blelloch R., Anna-Arriola S. S., Gao D., Li Y., Hodgkin J., Kimble J. The gon-1 gene is required for gonadal morphogenesis in Caenorhabditis elegans. Dev. Biol. 1999;216:382–393. doi: 10.1006/dbio.1999.9491. [DOI] [PubMed] [Google Scholar]
- 90.Hecht F., Sutherland G. R. Detection of fragile sites on human chromosomes. Clin. Genet. 1985;28:95–96. doi: 10.1111/j.1399-0004.1985.tb01227.x. [DOI] [PubMed] [Google Scholar]
- 91.Lott S. T., Lovell M., Naylor S. L., Killary A. M. Physical and functional mapping of a tumor suppressor locus for renal cell carcinoma within chromosome 3p12. Cancer Res. 1998;58:3533–3537. [PubMed] [Google Scholar]
- 92.van den Berg A., Buys C. H. Involvement of multiple loci on chromosome 3 in renal cell cancer development. Genes Chromosomes Cancer. 1997;19:59–76. doi: 10.1002/(sici)1098-2264(199706)19:2<59::aid-gcc1>3.3.co;2-o. [DOI] [PubMed] [Google Scholar]
- 93.Matsumoto S., Kasumi F., Sakamoto G., Onda M., Nakamura Y., Emi M. Detailed deletion mapping of chromosome arm 3p in breast cancers: a 2-cM region on 3p14.3–21.1 and a 5-cM region on 3p24.3–25.1 commonly deleted in tumors. Genes Chromosomes Cancer. 1997;20:268–274. [PubMed] [Google Scholar]
- 94.Rao C., Foernzler D., Loftus S. K., Liu S., McPherson J. D., Jungers K. A., Apte S. S., Pavan W. J., Beier D. R. A defect in a novel ADAMTS family member is the cause of the belted white-spotting mutation. Development. 2003;130:4665–4672. doi: 10.1242/dev.00668. [DOI] [PubMed] [Google Scholar]
- 95.Fujikawa K., Suzuki H., McMullen B., Chung D. Purification of human von Willebrand factor-cleaving protease and its identification as a new member of the metalloproteinase family. Blood. 2001;98:1662–1666. doi: 10.1182/blood.v98.6.1662. [DOI] [PubMed] [Google Scholar]
- 96.Levy G. G., Nichols W. C., Lian E. C., Foroud T., McClintick J. N., McGee B. M., Yang A. Y., Siemieniak D. R., Stark K. R., Gruppo R., et al. Mutations in a member of the ADAMTS gene family cause thrombotic thrombocytopenic purpura. Nature (London) 2001;413:488–494. doi: 10.1038/35097008. [DOI] [PubMed] [Google Scholar]
- 97.Soejima K., Mimura N., Hirashima M., Maeda H., Hamamoto T., Nakagaki T., Nozaki C. A novel human metalloprotease synthesized in the liver and secreted into the blood: possibly, the von Willebrand factor-cleaving protease? J. Biochem. (Tokyo) 2001;130:475–480. doi: 10.1093/oxfordjournals.jbchem.a003009. [DOI] [PubMed] [Google Scholar]
- 98.Furlan M., Robles R., Lamie B. Partial purification and characterization of a protease from human plasma cleaving von Willebrand factor to fragments produced by in vivo proteolysis. Blood. 1996;87:4223–4234. [PubMed] [Google Scholar]
- 99.Tsai H. M. Physiologic cleavage of von Willebrand factor by a plasma protease is dependent on its conformation and requires calcium ion. Blood. 1996;87:4235–4244. [PubMed] [Google Scholar]
- 100.Sadler J. E. Biochemistry and genetics of von Willebrand factor. Annu. Rev. Biochem. 1998;67:395–424. doi: 10.1146/annurev.biochem.67.1.395. [DOI] [PubMed] [Google Scholar]
- 101.Tsai H. M., Sussman I. I., Nagel R. L. Shear stress enhances the proteolysis of von Willebrand factor in normal plasma. Blood. 1994;83:2171–2179. [PubMed] [Google Scholar]
- 102.Furlan M., Robles R., Solenthaler M., Lammle B. Acquired deficiency of von Willebrand factor-cleaving protease in a patient with thrombotic thrombocytopenic purpura. Blood. 1998;91:2839–2846. [PubMed] [Google Scholar]
- 103.Zheng X., Nishio K., Majerus E. M., Sadler J. E. Cleavage of von Willebrand factor requires the spacer domain of the metalloprotease ADAMTS13. J. Biol. Chem. 2003;278:30136–30141. doi: 10.1074/jbc.M305331200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schneppenheim R., Budde U., Oyen F., Angerhaus D., Aumann V., Drewke E., Hassenpflug W., Haberle J., Kentouche K., Kohne E., et al. von Willebrand factor cleaving protease and ADAMTS13 mutations in childhood TTP. Blood. 2003;101:1845–1850. doi: 10.1182/blood-2002-08-2399. [DOI] [PubMed] [Google Scholar]
- 105.Kokame K., Matsumoto M., Soejima K., Yagi H., Ishizashi H., Funato M., Tamai H., Konno M., Kamide K., Kawano Y., et al. Mutations and common polymorphisms in ADAMTS13 gene responsible for von Willebrand factor-cleaving protease activity. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11902–11907. doi: 10.1073/pnas.172277399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Assink K., Schiphorst R., Allford S., Karpman D., Etzioni A., Brichard B., van de Kar N., Monnens L., van den Heuvel L. Mutation analysis and clinical implications of von Willebrand factor-cleaving protease deficiency. Kidney Int. 2003;63:1995–1999. doi: 10.1046/j.1523-1755.63.6s.1.x. [DOI] [PubMed] [Google Scholar]
- 107.Studt J. D., Hovinga J. A., Radonic R., Gasparovic V., Ivanovic D., Merkler M., Wirthmueller U., Dahinden C., Furlan M., Lammle B. Familial acquired thrombotic thrombocytopenic purpura: ADAMTS13 inhibitory autoantibodies in identical twins. Blood. 2004;103:4195–4197. doi: 10.1182/blood-2003-11-3888. [DOI] [PubMed] [Google Scholar]
- 108.Klaus C., Plaimauer B., Studt J. D., Dorner F., Lammle B., Mannucci P. M., Scheiflinger F. Epitope mapping of ADAMTS13 autoantibodies in acquired thrombotic thrombocytopenic purpura. Blood. 2004;103:4514–4519. doi: 10.1182/blood-2003-12-4165. [DOI] [PubMed] [Google Scholar]
- 109.Dagoneau N., Benoist-Lasselin C., Huber C., Faivre L., Megarbane A., Alswaid A., Dollfus H., Alembik Y., Munnich A., Legeai-Mallet L., Cormier-Daire V. ADAMTS10 mutations in autosomal recessive Weill-Marchesani syndrome. Am. J. Hum. Genet. 2004;75:801–806. doi: 10.1086/425231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cal S., Obaya A. J., Llamazares M., Garabaya C., Quesada V., López-Otín C. Cloning, expression analysis, and structural characterization of seven novel human ADAMTSs, a family of metalloproteinases with disintegrin and thrombospondin-1 domains. Gene. 2002;283:49–62. doi: 10.1016/s0378-1119(01)00861-7. [DOI] [PubMed] [Google Scholar]
- 111.Blelloch R., Newman C., Kimble J. Control of cell migration during Caenorhabditis elegans development. Curr. Opin. Cell Biol. 1999;11:608–613. doi: 10.1016/s0955-0674(99)00028-9. [DOI] [PubMed] [Google Scholar]
- 112.Nishiwaki K., Hisamoto N., Matsumoto K. A metalloprotease disintegrin that controls cell migration in Caenorhabditis elegans. Science. 2000;288:2205–2208. doi: 10.1126/science.288.5474.2205. [DOI] [PubMed] [Google Scholar]
- 113.Kuno K., Baba C., Asaka A., Matsushima C., Matsushima K., Hosono R. The Caenorhabditis elegans ADAMTS family gene adt-1 is necessary for morphogenesis of the male copulatory organs. J. Biol. Chem. 2002;277:12228–12236. doi: 10.1074/jbc.M200144200. [DOI] [PubMed] [Google Scholar]
- 114.Varney T. R., Ho H., Petty C., Blumberg D. D. A novel disintegrin domain protein affects early cell type specification and pattern formation in Dictyostelium. Development. 2002;129:2381–2389. doi: 10.1242/dev.129.10.2381. [DOI] [PubMed] [Google Scholar]
- 115.Hirohata S., Wang L. W., Miyagi M., Yan L., Seldin M. F., Keene D. R., Crabb J. W., Apte S. S. Punctin, a novel ADAMTS-like molecule, ADAMTSL-1, in extracellular matrix. J. Biol. Chem. 2002;277:12182–12189. doi: 10.1074/jbc.M109665200. [DOI] [PubMed] [Google Scholar]
- 116.Nagase T., Ishikawa K., Miyajima N., Tanaka A., Kotani H., Nomura N., Ohara O. Prediction of the coding sequences of unidentified human genes. IX. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 1998;5:31–39. doi: 10.1093/dnares/5.1.31. [DOI] [PubMed] [Google Scholar]
- 117.Hall N. G., Klenotic P., Anand-Apte B., Apte S. S. ADAMTSL-3/punctin-2, a novel glycoprotein in extracellular matrix related to the ADAMTS family of metalloproteases. Matrix Biol. 2003;22:501–510. doi: 10.1016/s0945-053x(03)00075-1. [DOI] [PubMed] [Google Scholar]
- 118.Glasson S. S., Askew R., Sheppard B., Carito B. A., Blanchet T., Ma H. L., Flannery C. R., Kanki K., Wang E., Peluso D., et al. Characterization of and osteoarthritis susceptibility in ADAMTS-4-knockout mice. Arthritis Rheum. 2004;50:2547–2558. doi: 10.1002/art.20558. [DOI] [PubMed] [Google Scholar]
- 119.Iruela-Arispe M. L., Carpizo D., Luque A. ADAMTS1: a matrix metalloprotease with angioinhibitory properties. Ann. N. Y. Acad. Sci. 2003;995:183–190. doi: 10.1111/j.1749-6632.2003.tb03221.x. [DOI] [PubMed] [Google Scholar]
- 120.Gill S. E., Pape M. C., Khokha R., Watson A. J., Leco K. J. A null mutation for tissue inhibitor of metalloproteinases-3 (Timp-3) impairs murine bronchiole branching morphogenesis. Dev. Biol. 2003;261:313–323. doi: 10.1016/s0012-1606(03)00318-x. [DOI] [PubMed] [Google Scholar]
- 121.Coussens L. M., Fingleton B., Matrisian L. M. Matrix metalloproteinase inhibitors and cancer, trials and tribulations. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 122.Reich R., Thompson E. W., Iwamoto Y., Martin G. R., Deason J. R., Fuller G. C., Miskin R. Effects of inhibitors of plasminogen activator, serine proteinases, and collagenase IV on the invasion of basement membranes by metastatic cells. Cancer Res. 1988;48:3307–3312. [PubMed] [Google Scholar]
- 123.Baxter A. D., Bhogal R., Bird J., Keily J. F., Manallack D. T., Montana J. G., Owen D. A., Pitt W. R., Watson R. J., Wills R. E. Arylsulphonyl hydroxamic acids: potent and selective matrix metalloproteinase inhibitors. Bioorg. Med. Chem. Lett. 2001;11:1465–1468. doi: 10.1016/s0960-894x(01)00259-1. [DOI] [PubMed] [Google Scholar]
- 124.Nagase H., Brew K. Designing TIMP (tissue inhibitor of metalloproteinases) variants that are selective metalloproteinase inhibitors. Biochem. Soc. Symp. 2003;70:201–212. doi: 10.1042/bss0700201. [DOI] [PubMed] [Google Scholar]
- 125.Somerville R. P., Jungers K. A., Apte S. S. ADAMTS10: discovery and characterization of a novel, widely expressed metalloprotease and its proteolytic activation. J. Biol. Chem. 2004;279:51208–51217. doi: 10.1074/jbc.M409036200. [DOI] [PubMed] [Google Scholar]
- 126.Bolz H., Ramirez A., von Brederlow B., Kubisch C. Characterization of ADAMTS14, a novel member of the ADAMTS metalloproteinase family. Biochim. Biophys. Acta. 2001;1522:221–225. doi: 10.1016/s0167-4781(01)00329-3. [DOI] [PubMed] [Google Scholar]