Genome Research 17: 1146–1160 (2007)
The authors would like to correct an error in Figure 1, which may have inadvertently displayed duplicated and mislabeled figure panels. The corrected Figure 1 is provided with the removal of panels D and F. The text referring to these panels has also been modified in the Results section (pages 1147–1149) and in the figure legend (see below). These corrections do not alter the conclusions of the article. The authors apologize for any confusion.
Figure 1.
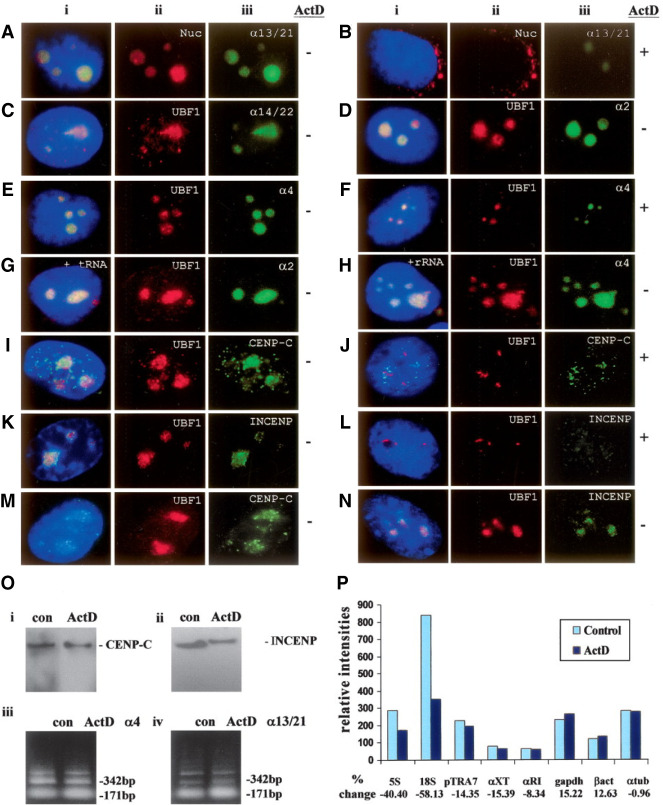
(A–H) Subnuclear distribution of centromeric α-satellite RNA and proteins in HeLa cells. Unless otherwise stated, images were captured on a conventional fluorescence microscope. Simultaneous RNA-FISH detection of α-satellite RNA specific for chromosomes 13/21 (Aiii; green), 14/22 (Ciii; green), 2 (Diii; green) and 4 (Eiii; green); and immunofluorescence detection of nucleolar proteins nucleolin (Nuc) (Aii; red) and UBF1 (Cii, Dii, Eii; red) in human HeLa cells. ActD treatment at 0.05 μg/mL resulted in the delocalization of α13/21- (Biii; green) and α4- (Fiii; green) satellite RNA, nucleolin (Bii; red), and UBF1 (Fii; red) from the nucleoli. UBF1 displayed a typical relocalization toward the nucleolar periphery to give a concentrated cap-like signal at the tip of the nucleoli (Fii; red). Competition with tRNA and rRNA (10× excess amount) did not affect the detection of α-satellite RNA at the nucleoli as exemplified by the chromosome 2- and 4-specific α-satellite RNA signals (Giii, Hiii; green). (I–L) Nucleolar accumulation of CENPC1 (Iiii; green) and INCENP (Kiii; green), as indicated by coimmunostaining with UBF1 (Iii, Kii, respectively; red). CENPC1 and INCENP both delocalized from the nucleoli following ActD treatment (Jiii, Liii, respectively; green). (M) Confocal laser scanning microscopy analysis showing nucleolar accumulation of CENPC1 by differential interference contrast images (iii; green) and coimmunostaining with UBF1 (ii; red). (N) Confocal microscopy analysis showing nucleolar accumulation of INCENP (iii; green) and coimmunostaining with UBF1 (ii; red). (i) Merged images of ii and iii. Antibodies used were as described in the Methods, except for the anti-CENPC1 antibody used in Miii, which was kindly provided by Dr. Bill Earnshaw. (O) Effect of ActD treatment on the expression level of CENPC1, INCENP, and α-satellite RNA. Total cell lysates were prepared from HeLa cells with (ActD) and without (con) prior treatment with 0.05 μg/mL ActD for 6 h, and subjected to Western blotting analysis. The expression levels of both CENPC1 (i) and INCENP (ii) remained unchanged following ActD treatment. (iii) Total RNA was prepared from untreated and ActD-treated HeLa cells. RT-PCR was performed on the RNA using primer sets corresponding to the α4- and α13/21-specific α-satellite sequences. A 171-bp ladder of monomeric α-satellite sequences was seen. No significant difference in the intensities of PCR products was detected following 6 h ActD treatment. (P) Nuclear run-on transcription assay to assess the effect of ActD treatment on α-satellite transcription. Equal numbers of nuclei were used for each assay, as determined by the DNA concentration. The effect of ActD on the transcription of α-satellite was measured by comparing the hybridization signals of in vitro-synthesized 32P-labeled run-on transcripts with immobilized DNA. The radioactive signals for the ribosomal genes 5S and 18S were decreased following ActD treatment, but the signals for the transcription of RNA polymerase II-driven genes (such as GAPDH, beta-actin, and alpha-tubulin) and those of α-satellite DNA (pTRA7 [chromosomes 13/14/21-specific], αXT [chromosomes 14/22-specific], and αRI [chromosomes 13/21-specific]) (Lo et al. 1999) were not significantly affected.
Centromere RNA accumulates in the transcriptionally active nucleolus
“The results with these antisera confirmed that α-satellite RNA was enriched in the nucleolus (Fig. 1A,C,D,E). Competition with tRNA and rRNA did not affect the detection of the α-satellite RNA signals at the nucleoli (examples of chromosome 2- and 4-specific α-satellite RNA are shown in Fig. 1G,H).”
“As expected, nucleolin displaced to the nuclear periphery following ActD treatment, whereas UBF1 displayed a typical relocalization toward the nucleolar periphery to give a concentrated cap-like signal at the tip of the nucleoli (Zhang et al. 2004) (Fig. 1Fii). In the presence of the ActD level used, α-satellite RNA also significantly, but not completely, delocalized from the nucleoli, indicating that the nucleolar localization of α-satellite RNA is sensitive to RNA polymerase I inhibition (Fig. 1Biii, Fiii).”
“As controls, parallel experiments were performed to determine the effect of ActD treatment on the expression levels of both centromere proteins and α-satellite RNA by Western blotting and RT-PCR analysis, respectively (Fig. 1O). The levels of expression of the centromere proteins CENPC1 and INCENP (Fig. 1O) were not affected with 6 h of ActD treatment at 0.05 μg/mL, suggesting that the observed reduction in nucleolar signals was not due to inhibition of general transcription (Fig. 1O). Likewise, the α-satellite transcription was not noticeably reduced in cells subjected to treatment with 3–6 h of ActD, as indicated by RT-PCR performed using primer sets corresponding to the chromosome 4- and 13/21-specific α-satellite (Fig. 1O).”
“As controls, we showed that the expression levels of the ribosomal genes 5S and 18S were greatly reduced by the ActD treatment, but not those of the RNA polymerase II-dependent genes such as GAPDH, beta-actin, and alpha-tubulin (Fig. 1P).”
Nucleolar localization of centromere proteins CENPC1 and INCENP is sensitive to actinomycin D treatment and RNA depletion
“By immunofluorescence analysis with conventional and confocal laser scanning microscopy (Fig. 1I,M, respectively), we have demonstrated a clear subcellular localization of CENPC1 at the nucleolus. The high specificity of the antibody used in immunofluorescence was evident from the detection of a single CENPC1 protein band in Western blot analysis (Fig. 1O).”
“In order to identify other centromere proteins that may localize to the nucleolus, we performed immunofluorescence analysis with a panel of antisera against centromere proteins. We found that INCENP (Fig. 1K,N), but not survivin, CENPA, CENPB, and CENPE (data not shown), were enriched at the nucleolus.”
“We have also found that treatment with 0.05 μg/mL ActD for 6 h resulted in the delocalization of CENPC1 and INCENP from the nucleolus (Fig. 1Jiii,Liii, respectively), suggesting that the nucleolar localization of these proteins, as was the case with the α-satellite RNA, was also dependent on RNA polymerase I activity.”
Identification of nucleolus localization sequences (NoLS) of CENPC1 and INCENP
“It is unclear why the previous study (Pluta and Earnshaw 1996) detected only limited localization of CENPC1 at the nucleoli, as we have shown in our own experiments that the anti-CENPC1 antibodies from both laboratories gave similar strong localization within the nucleoli (see Fig. 1I,M).”
doi: 10.1101/gr.279693.124