Abstract
oriP is a 1.7-kb region of the Epstein-Barr virus (EBV) chromosome that supports the replication and stable maintenance of plasmids in human cells. oriP contains two essential components, called the DS and the FR, both of which contain multiple binding sites for the EBV-encoded protein, EBNA-1. The DS appears to function as the replicator of oriP, while the FR acts in conjunction with EBNA-1 to prevent the loss of plasmids from proliferating cells. Because of EBNA-1's role in stabilizing plasmids through the FR, it has not been entirely clear to what extent EBNA-1 might be required for replication from oriP per se, and a recent study has questioned whether EBNA-1 has any direct role in replication. In the present study we found that plasmids carrying oriP required EBNA-1 to replicate efficiently even when assayed only 2 days after plasmids were introduced into the cell lines 143B and 293. Significantly, using 293 cells it was demonstrated that the plasmid-retention function of EBNA-1 and the FR did not contribute significantly to the accumulation of replicated plasmids, and the DS supported efficient EBNA-1-dependent replication in the absence of the FR. The DS contains two pairs of closely spaced EBNA-1 binding sites, and a previous study had shown that both sites within either pair are required for activity. However, it was unclear from previous work what additional sequences within the DS might be required. We found that each “half” of the DS, including a pair of closely spaced EBNA-1 binding sites, had significant replicator activity when the other half had been deleted. The only significant DNA sequences that the two halves of the DS share in common, other than EBNA-1 binding sites, is a 9-bp sequence that is present twice in the “left half” and once in the “right half.” These nonamer repeats, while not essential for activity, contributed significantly to the activity of each half of the DS. Two thymines occur at unique positions within EBNA-1 binding sites 1 and 4 at the DS and become sensitive to oxidation by permanganate when EBNA-1 binds, but mutation of each to the consensus base, adenine, actually improved the activity of each half of the DS slightly. In conclusion, the DS of oriP is an EBNA-1-dependent replicator, and its minimal active core appears to be simply two properly spaced EBNA-1 binding sites.
The circularized, 165-kb chromosome of Epstein-Barr virus (EBV) is maintained autonomously in proliferating cells that are latently infected (23, 32). A 1.7-kb region of the EBV chromosome, termed oriP, can support the replication and maintenance of recombinant plasmids in human cells (43) if a single EBV-encoded protein, the nuclear protein EBNA-1, is present (25, 47). Initiation of replication at oriP can occur no more than once per cell cycle (46) and thus appears to be controlled by the cellular regulatory mechanism known as licensing (22). EBNA-1 is not a DNA helicase (8, 29), so the initial unwinding of DNA at the origin must also be performed by the cell. Because oriP can be studied easily and the protein that activates it is known, investigations of oriP offer the potential to reveal important aspects of mammalian DNA replication.
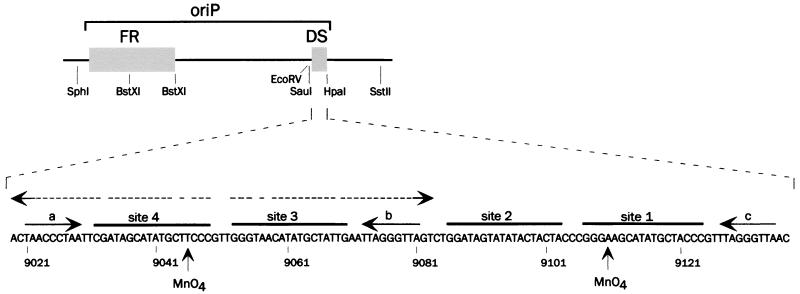
When oriP was discovered, it was not yet appreciated that a replication origin alone would be insufficient to support autonomous maintenance of plasmids in human cells (4). In fact, oriP contains two separate components (34) that perform distinct, essential functions (Fig. 1). A 120-bp region, called the DS, contains four EBNA-1 binding sites (33) and is the functional replicator (15, 42), i.e., it contains the cis-acting elements that lead DNA replication to initiate. Bidirectional DNA replication begins at the approximate location of the DS (11). The other component of oriP is the FR, named for a 20-member family of 30-bp repeats in which each repeat contains an EBNA-1 binding site (33). The essential function of the FR is to prevent the loss of plasmids from mitotically active cells (21), which it appears to accomplish by tethering plasmids via EBNA-1 to condensed human chromosomes as they segregate during mitosis (28, 37).
FIG. 1.
Structure of oriP and relevant restriction sites (above) and its DS component (expanded below). The DS is considered to extend from nucleotide 9019 at the left edge of the dyad symmetry to nucleotide 9137, at an HpaI site, based on previous studies (5, 34). EBNA-1 binding sites 1 to 4 are numbered according to the method of Rawlins et al. (33). The three copies of the nonamer repeat, labeled a, b, and c as described by Vogel et al. (41), are indicated by solid arrows to indicate their relative orientations. The positions of two thymines, in the upper strand of site 4 and the lower strand of site 1, that become reactive to permanganate when EBNA-1 binds are indicated by “MnO4.” The 65-bp hyphenated dyad symmetry is indicated by the dotted lines with divergent arrows.
Because EBNA-1 acts (mainly through the FR) to prevent the loss of plasmids carrying oriP, a degree of uncertainty has lingered over the extent to which EBNA-1 is directly required for replication from oriP. In a recent study, oriP was seen to support plasmid replication independently of EBNA-1 when replication was measured 48 h after plasmids were introduced into cells; after 72 or 96 h, intervals that had been used in most of the previous studies, EBNA-1 became essential, leading to the conclusion that EBNA-1 is needed only to prevent replicated plasmids from being lost from cells (1). This would seem to conflict with other studies in which the DS component alone, in the absence of the FR and its plasmid-retention function, was found to support plasmid replication in a transient (96 h) transfection assay only if EBNA-1 was provided (15, 36). However, the four EBNA-1 binding sites at the DS might themselves provide protection against plasmid loss for a few days, since as few as two high-affinity EBNA-1 binding sites have been reported to show some plasmid-retention activity (30). On the other hand, strong circumstantial evidence indicates that EBNA-1 is involved in the function of the DS. First, double point mutations that prevented EBNA-1 binding to certain combinations of sites at the DS were found to abolish oriP-dependent plasmid replication (15). Second, in vivo footprints made by treating cells or nuclei with three different agents, DNase I, dimethyl sulfate, and permanganate, revealed near-perfect signatures of EBNA-1 binding to its DS sites, indicating that these sites are occupied by EBNA-1 throughout most of the cell cycle (20).
Experiments described here should resolve this issue. The results show that oriP-specific plasmid replication requires EBNA-1 even during the first 48 h after transfection. Importantly, this was demonstrated under conditions in which the FR did not contribute appreciably to the accumulation of replicated plasmids, and the DS supported efficient EBNA-1-dependent replication in the absence of the FR. Some EBNA-1-independent replication could be attributed to oriP after transient transfection, but it was relatively inefficient, was not dependent on the DS, and may not be very sequence specific.
Insight into how the DS replicator functions should come from determining what nucleotide sequences determine its function. The functional boundaries of the DS were found to include approximately the 120-bp region shown in Fig. 1, based on the effects of deletions (5, 34, 36, 43). This region includes four EBNA-1 binding sites that were shown to function in pairs. Using double point mutations to inactivate individual EBNA-1 binding sites, Harrison et al. found that the DS was active so long as both sites within either functional pair were intact (sites 1 and 2 or sites 3 and 4 in Fig. 1) (15). The EBNA-1 sites within each functional pair have the same spacing, and inserting 5 or 10 bp between the sites of either pair abolished its activity (15).
Previous studies have not resolved whether sequences at the DS in addition to the EBNA-1 binding sites are required for the replicator to function. EBNA-1 sites 3 and 4 are part of a hyphenated inverted repeat (dyad symmetry) that is 65 bp long. Chittenden et al. studied the effects of deletions entering the DS from either direction using an assay for long-term plasmid maintenance under selection and concluded that the sequences within the dyad symmetry itself, and presumably EBNA-1 sites 3 and 4 within it, were needed for the plasmids to be maintained efficiently and without rearrangements, while EBNA-1 sites 1 and 2 were essential for activity in their assay (5). However, in the original report on oriP, it was shown that a deletion from the right that removed all of EBNA-1 site 1 allowed long-term plasmid maintenance with only a moderate reduction in efficiency (43). The results of Harrison et al., described above, appeared to be inconsistent as to whether inactivating individual EBNA-1 binding sites or both sites in one functional pair caused a decrease in the efficiency of plasmid maintenance (15). It is thus unclear to what extent the effects of some of the deletions studied by Chittenden et al. resulted from the removal of EBNA-1 binding sites or neighboring sequences. There are three copies of a 9-bp (nonamer) sequence within the DS, and the simultaneous substitution of all three copies was shown to increase moderately the rate at which oriP-dependent plasmids were lost from cells (41).
Here we report a study of a large number of deletions and substitution mutations within the DS. The results lead us to the conclusion that a minimal DS replicator is likely to require no more than two EBNA-1 binding sites with proper spacing. However, every EBNA-1 binding site and each nonamer repeat at the DS was found to contribute to replicator function under certain circumstances.
MATERIALS AND METHODS
Plasmids.
pHyg is a 4.8-kb plasmid that carries the hph (hygromycin B phosphotransferase) gene linked to the HSV tk promoter, and pHEBo (7.0 kb) is pHyg carrying oriP and flanking EBV sequences extending to the SphI and SstII sites indicated in Fig. 1 (39). The deletion removing the FR, ΔFR, was made by deleting between the BstXI site to the right of the FR and a SalI site flanking oriP in pHEBo (42). p367 is pHEBo carrying the EBNA-1 gene expressed from the Rous sarcoma virus long terminal repeat (RSV LTR) (45). p396 is a similar plasmid with a deletion (dl46) removing codons 540 to 580 within the DNA binding-dimerization domain-encoding part of the EBNA-1 gene, which abolishes EBNA-1 activity (45). p367 (9.9 kb) is smaller than p396 (10.6 kb) because it lacks most of the triplet repeats which encode the Gly-Gly-Ala repetitive part of EBNA-1.
The mutations at the DS of pHEBo are described in Table 1. For all mutations generated using mutagenesis in M13 or PCR, the nucleotide sequence across the DS was determined for both DNA strands. The sequences of the oligonucleotides that were used for mutagenesis will be provided on request. To generate the “Bst” substitution mutations, the DS was inserted as an EcoRV-HpaI fragment into the HincII site of M13 mp11 and used in site-directed mutagenesis with the T7-Gen kit (U.S. Biochemicals). The DS carrying each mutation was excised by using SauI and HincII and inserted between the SauI and HpaI sites of pHEBo. The Δ1 deletion, 9107 (SmaI site) to 9134 (HpaI site), was introduced into pHEBo and its mutant derivatives by amplifying the DS by PCR using the primers ODJ4 (5′-GGAATCCTGACCCCATGT) and ODJ11 (5′-AACGTCAATCAGAGGGGC), which have 5′ ends at positions 8943 and 9220, respectively, of the B95-8 sequence. From the resulting 278-bp product, the region between the SauI and SmaI sites was excised and inserted between the SauI and HpaI sites of pHEBo. Δ9009–9066 was made by deleting between the SauI site and the Bst1107I site of the 3-Bst mutation. Δ9036–9066 was made by deleting between the Bst1101I sites of the double mutant, 3-Bst 4-Bst.
TABLE 1.
Plasmids carrying mutations at oriP
| Plasmid | Mutation(s) | Description of mutation(s) |
|---|---|---|
| p571a | Δ1-4 | Deletion of EBNA-1 sites 1 to 4, positions 8995 to 9134 (EcoRV to HpaI) |
| p568a | Δ1 | Deletion of EBNA-1 site 1, positions 9107 to 9134 (SmaI to HpaI) |
| p653a | 1-Bst | 9107-GGGAAGCAGTATACACCC (mutant EBNA-1 site 1, substitution underlined) |
| p654a | 2-Bst | 9086-GGATGTATACTACTACTA (mutant EBNA-1 site 2) |
| p655a | 3-Bst | 9053-GGGTAACAGTATACATTG (mutant EBNA-1 site 3) |
| p656a | 4-Bst | 9032-CGATGTATACTGCTTCCC (mutant EBNA-1 site 4) |
| p657a | Δ1, 2-Bst | |
| p658a | Δ1, 3-Bst | |
| p659a | Δ9009–9066 | Positions 9009 to 9066 replaced by TAC |
| p688b | 3-Bst, 4-Bst | |
| p689a | Δ9036–9066 | Positions 9036 to 9066 replaced by GTATAC (Bst1107 I site) |
| p727b | Δ8994–9018 | Positions 8994 to 9018 replaced by GCTC (created SstI site) |
| p715b | S, X, Δ3-4 | S = TA to GC at positions 8994 8995 (made SstI site); X = G to A at position 9086 (made XbaI site); Δ3-4 = deletion of positions 8996 to 9082, including EBNA-1 sites 3 and 4 |
| p716b | S, X, Δ3-4, 2con | 2con = position 9091 T to consensus C; position 9097 A to consensus G |
| p717b | S, X, Δ3-4, 1con | 1con = position 9110 A to consensus T |
| p718b | S, X, Δ3-4, 1con, 2con | |
| p738b | S, X, 4con | 4con = position 9046 T to consensus A |
| p739b | S, X, 4con, 1con | |
| p740b | S, X, 4con, Δ1-2 | Δ1-2 = deletion of EBNA-1 sites 1 and 2, positions 9088 to 9134 (XbaI to HpaI) |
| p751bc | S, X | |
| p752bc | S, X, Δ1-2 | |
| p753b | S, X, a-Tth | a-Tth = 9020-CTAACCCTAATT to AGACTTTGTCAA |
| p754b | S, X, a-Tth, Δ1 | |
| p755b | S, X, a-Tth, b-Tth | b-Tth = 9071-AATTAGGGTTAG to AGACTTTGTCAA |
| p756b | S, X, a-Tth, b-Tth, Δ1 | |
| p758b | S, X, Δ1-2, a-Tth | |
| p766b | S, X, Δ1-2, a-Tth, b-Tth | |
| p784b | S, X, a-Tth, b-Tth, c-Stu | c-Stu = 9127-TTAGGGTTA to ACTTCAGGA |
| p785bc | S, X, c-Stu | |
| p785Δ | X, Δ3-4′, c-Stu | Δ3-4′ = deletion of positions 8993 to 9082 |
Constructed in pHEBo (p152) (39).
Constructed in pHEBoΔRV and identical to pHEBo except deleted for 349 bp between ClaI and BamHI sites to remove an EcoRV site.
Also contains an inadvertent and apparently innocuous mutation, T to A at position 9079.
p715 was made by first inserting an XbaI linker at the EcoRV site of pHEBoΔRV (Table 1) to create pHEBoΔX. In the process, the T at 8994 was lost, and this, together with the juxtaposition of the linker DNA, led to the creation of an SstI site to the left of the XbaI site. Next, a 29-nucleotide primer beginning several positions before EBNA-1 site 2 and containing a G-to-A mutation at 9086 (mutation “X,” creating an XbaI site) was used with ODJ10 to amplify the DNA extending to the right, from which the right half of the DS was excised using XbaI and HpaI and inserted between these sites of pHEBoΔX. This created the Δ3-4 deletion. The deleted region was restored to p715 by using a 103-bp synthetic DNA ending at the SstI and XbaI sites to create p751 in which the DS is wild type except for the harmless point mutation, “X,” providing a useful XbaI site between the two “halves” of the DS. The mutation 4con was introduced in the same manner. The remaining mutations were incorporated into specific primers, extending from one side of either the XbaI site or the HpaI site to beyond the site of mutation, and used for PCR in combination with outside flanking primers, ODJ4 or ODJ10. The mutated regions were then introduced into either p715 or p751 using the SstI, XbaI, and HpaI sites, as appropriate.
Transfections.
143B cells, 293 cells, and Raji cells are all available from the American Type Culture Collection. 143/98.2 cells were described previously as 143/SVoB-H2.9, clone 4 (47). Adherent cell lines 143B, 143/98.2, and 293 were cultured in Iscove's modified Dulbecco medium (IMDM) supplemented with 9% fetal bovine serum, penicillin, and streptomycin, and Raji cells were grown in suspension in RPMI 1640 medium with the same supplements. The adherent cell lines were transfected in 6-cm dishes, while the cells were 50 to 70% confluent using the calcium phosphate coprecipitation method (13). Generally, 2.5 μg of plasmid DNA in 225 μl of HEPES buffered saline was mixed with 25 μl of 1.25 M CaCl2 and then added to a dish of cells containing 2.5 ml of complete medium. For the cotransfections for Fig. 3, 2 μg of each plasmid was used. After incubation under normal culture conditions for 4.5 to 5.5 h, cells were shocked for 2.5 min with 20% glycerol in IMDM at room temperature (10). The transient transfections were considered to begin during the glycerol shock since most of the uptake of DNA occurs in response to this step (10). To remove plasmid DNA not taken into the cells, in the case of the transient transfections, the cells were detached from the plates the day after transfection using trypsin (Gibco-BRL) at 1/5 the recommended concentration in phosphate-buffered saline (PBS)–1 mM EDTA. The cells were added to 4 ml of complete medium, collected by centrifugation, resuspended in 4 ml of PBS-EDTA, pelleted again, washed a third time in PBS-EDTA, and then returned to culture in 10-cm dishes.
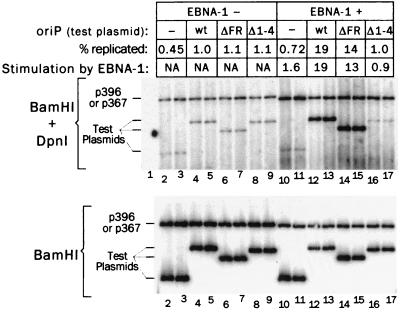
FIG. 3.
The DS component of oriP is an EBNA-1-dependent replicator. Duplicate plates of 293 cells were transfected with the oriP test plasmids as indicated. Cells were also cotransfected with either p367, a derivative of pHEBo that produces EBNA-1 (lanes 10 to 17), or with p396, a similar plasmid that produces mutant EBNA-1 that cannot bind to DNA (lanes 2 to 9). Plasmids were extracted from cells 46 h after transfection and were analyzed for DpnI-resistant plasmids (40% of each sample, upper blot) and total plasmids (2% of each sample, lower blot). The blots were processed in parallel using the same labeled probe, and signals from beta-image analysis were compared directly to calculate the percentages of replicated DNA. Stimulation by EBNA-1 is the ratio of the average amount of replication of each test plasmid in the presence of EBNA-1 to that in the absence of EBNA-1. NA, not applicable. A control for DpnI digestion was included in lane 1, upper blot, as was done for Fig. 2. The lower part of the gel containing the products of DpnI digestion is not shown.
For assays of plasmid maintenance under selection, 143/98.2 cells were removed from their dishes the day after transfection, and 1/10th of the cells were returned to culture in 6-cm dishes. Medium containing 275 μg of hygromycin B (Calbiochem) per ml was added the next day, and resistant cultures were selected and passaged as described in the text and figure legends. Raji cells were electroporated at room temperature with a Gibco-BRL Cell-Porator set at 300 V and 330 μF using 5 × 106 cells and 2 μg of plasmid DNA in 0.6 ml of growth medium in chambers spaced 4 mm between electrodes. After 2 days in culture, viable cells were counted and placed into multiple wells of 24-well Falcon culture dishes at different dilutions in medium containing 300 μg of hygromycin B per ml. The numbers of positive wells were counted after 3 weeks, and resistant clones were expanded under selection until 3 × 107 to 4 × 107 cells could be harvested at 30 to 39 days after electroporation.
Assays for plasmid replication and maintenance.
For long-term maintenance assays, plasmids were extracted from one confluent 10-cm plate of 143/98.2 cells (∼2 × 107 cells) or from 3 × 107 to 4 × 107 Raji cells using the method of Hirt (19) and purified as previously described (42). DNA samples were electrophoresed through 0.7% agarose gels in the absence of ethidium bromide and transferred to nylon membranes, which were probed with radiolabeled pHyg. Radiographic images were made using a Molecular Dynamics PhosphorImager.
For the transient transfections, plasmids were extracted using the alkaline lysis procedure that is commonly used to isolate plasmids from bacteria (35). Each 10-cm plate of cells was rinsed with 2 ml of PBS-EDTA, and then 2.4 ml of premixed GET (glucose-EDTA-Tris) and NaOH-sodium dodecyl sulfate solutions was added. The lysates were scraped into polypropylene tubes, placed on ice, and neutralized with 1.2 ml of potassium acetate-acetic acid solution between 15 and 20 min after lysis. After removal of the precipitated protein and chromosomal DNA by centrifugation, the supernatants were deproteinized with chloroform-phenol (1:1). Nucleic acids were precipitated with an equal volume of isopropanol, resuspended in 0.4 ml of TE (10 mM Tris-HCl, 0.5 mM EDTA; pH 8.0), treated with phenol, and precipitated using 2 volumes of ethanol after adding NaCl to 200 mM. The nucleic acid pellets were dissolved in 30 to 50 μl of TE. To test for resistance to DpnI, a portion (up to 40%) was digested with 4 U of DpnI, 5 U of BamHI, and 1 μg of RNase A for 4 h.
Electrophoretic mobility shift assay (EMSA).
DS DNA probes were made by PCR using pHEBo or its mutant derivatives as the template with primers ODJ4 and ODJ11 (see above), each end labeled with [γ-32P]ATP and polynucleotide kinase to 2 to 3 μCi/pmol. The 278-bp DNAs were centered between nucleotides 9081 and 9082, located between EBNA-1 sites 2 and 3. Then, 15 fmol of end-labeled DS DNA (adjusted to 2,000 dpm per fmol using unlabeled DNA), 2 μg of salmon sperm DNA, 60 or 240 fmol of EBNA-1 NΔ407 dimers (26), and 50 μg of protein from COS7 cells were combined in a volume of 15 μl containing 128 mM KCl, 6 mM (NH4)2 SO4, 5 mM MgCl2, 1.3 mM dithiothreitol, 10 mM Tris-HCl (pH 7.4), and 7.6% glycerol. After 10 min at room temperature, the samples were analyzed by electrophoresis through a 4% polyacrylamide gel as described earlier (6) except that 50 mM Tris-HCl–190 mM glycine (pH 8.5) was used as the electrophoresis buffer. Purified NΔ407 protein, produced in Escherichia coli, was a generous gift of D. Mackey and B. Sugden (26). The COS7 protein, a ribosome-free soluble fraction (S100) of a whole-cell extract made by using 0.5 M KCl, had no effect on the number of complexes seen or their mobilities but had a “carrier” effect, reducing by severalfold the amount of EBNA-1 required.
RESULTS
Efficient oriP-specific replication requires EBNA-1.
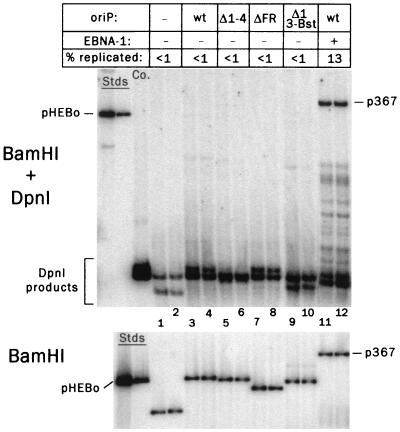
To test whether oriP would support plasmid replication in the absence of EBNA-1, the following plasmids were transfected into 143 cells: a 4.8-kb plasmid vector, pHyg (39); a 7.0-kb derivative of pHyg carrying oriP, pHEBo (39); three derivatives of pHEBo carrying different mutations in oriP; and p367, a 9.7-kb derivative of pHEBo carrying the EBNA-1 gene expressed from the RSV LTR (45). At 48 h after transfection, plasmids were extracted from the cells and digested with BamHI to linearize the plasmids and with DpnI to digest unreplicated plasmids, which retain the dam methylation pattern that the plasmids acquired during propagation in E. coli. As shown in Fig. 2, full-length DpnI-resistant plasmid was detected readily by Southern analysis only in the case of p367, which carries both oriP and the EBNA-1 gene, indicating that the plasmid had replicated in the transfected cells (upper image, lanes 11 and 12). The replication of pHEBo and its mutant derivatives was near the limit of detection, with bands representing DpnI-resistant molecules barely visible above background in some of the lanes in the original image.
FIG. 2.
Dependence of oriP-specific plasmid replication on EBNA-1 in 143B cells during the first 48 h after transfection. Duplicate dishes of 143B cells were transfected with test plasmids, pHyg (vector, “−”), pHEBo (pHyg carrying oriP, “wt”), or pHEBo carrying different mutations in oriP, as indicated for samples 1 to 10. For samples 11 and 12, cells were transfected with p367, which is pHEBo carrying the EBNA-1 gene. Plasmids were harvested from cells 48 h after transfection. For Southern analysis, 40% of each sample was digested with BamHI plus DpnI (upper blot), and 10% was digested with BamHI alone (lower blot). The percentages of total plasmid (lower blot) that replicated (full-length, DpnI-resistant plasmid, upper blot) were calculated by comparison to the quantitative standards of each blot. To test for the completeness of DpnI cutting, 2 ng of pHEBo was combined with DNA extracted from mock-transfected cells and digested with DpnI plus BamHI (lane marked “Co.,” upper blot). The standards were linearized pHEBo: 125 and 32 pg for the upper gel and 500 and 125 pg for the lower gel.
To control for variations in the efficiency of transfection, a portion of the extracted DNA from each sample was digested with BamHI (without DpnI) and analyzed similarly (Fig. 2, lower blot). To remove plasmid DNA associated with the outer surfaces of cells, the cells were removed from their dishes the day after transfection and washed extensively with PBS containing EDTA before being returned to culture in new dishes. When cells so treated were harvested 48 h after transfection and nuclear and cytoplasmic fractions were prepared, essentially all of the transfected plasmid was found in the nuclear fraction, indicating that the washing procedure was effective (data not shown). In comparing the upper and lower images of Fig. 2, note that four times more extracted DNA was analyzed for DpnI resistance than for total plasmid DNA, that fivefold-higher amounts of plasmid standards were loaded for the measurement of total plasmid, and that the images have been displayed at different settings. It was determined by quantative image analysis that 14 and 12% of the p367 DNA in the cells of the duplicate transfections had been replicated at least once. The amounts of replicated pHEBo and its derivatives were too low to be measured reliably but were estimated to be <1% of the plasmid in the cells.
This experiment indicated that EBNA-1 was needed to allow detection of oriP-specific replication in 143B cells as early as 48 h after transfection. The experiment was repeated several times using 143B cells and 293 cells, and in all tests the presence of the EBNA-1 gene increased the accumulation of replicated plasmid by at least 10-fold. However, it could not be determined from such experiments how much, if any, of this increase resulted from a stimulation of replication, per se, by EBNA-1. Conceivably, much or all of the increase might have resulted from EBNA-1 acting in conjunction with the FR of oriP to prevent the loss of plasmids before or after replication. In the following experiment, a strong stimulation of oriP-specific replication by EBNA-1 was demonstrated under conditions in which the effect of EBNA-1 on plasmid stability was found to be minimal.
The DS is the EBNA-1-dependent replicator of oriP and does not require the FR.
The 293 cell line, which was derived from human embryonic kidney cells by transformation with adenovirus DNA (12), can be transfected very efficiently, at least in part because of the stabilizing effect that the antiapoptotic 19-kDa E1B protein of adenovirus has on introduced plasmids (18). Using 293 cells, we could reliably detect the relatively inefficient replication of pHyg and pHEBo in the absence of EBNA-1 as early as 48 h following transfection. For the experiment shown in Fig. 3, these plasmids were each mixed before transfection with either p367, to supply functional EBNA-1, or with p396, a derivative of p367 in which the EBNA-1 gene is nonfunctional because of a deletion affecting the DNA-binding domain, dl46 (45). In the absence of EBNA-1, just under 0.5% of the vector was replicated (lanes 2 and 3), and the presence of oriP on the plasmid increased this level to 1% (lanes 4 and 5). The plasmids from which either the DS or the FR of oriP were deleted (Δ1-4 or ΔFR) replicated similarly, indicating that neither of the components of oriP was required for this low level of replication. The replication of p396, which includes oriP, the RSV LTR, and a nonfunctional EBNA-1 gene, was somewhat more efficient, at 2.7% of the plasmid in the cells, on average (lanes 2 to 9). The replication of these plasmids in the absence of EBNA-1 may not be very specific in nature. It has been noted previously that plasmids with nonspecific DNA inserts replicate in 293 cells with efficiencies that increase as a function of the size of the plasmid (17).
When EBNA-1 was provided by cotransfection with p367, DpnI-resistant pHEBo accumulated to 19% of the retained plasmid DNA, a 19-fold increase over accumulation in the absence of EBNA-1 (Fig. 3, compare lanes 12 and 13 with lanes 4 and 5). EBNA-1 had little effect on the levels of replicated pHyg that accumulated (lanes 10 to 11 compared with lanes 2 to 3). On average, 14.3% of the EBNA-1-expressing plasmid, p367, replicated (lanes 10 to 17), which was fivefold higher than with p396 (lanes 2 to 9), which produces a nonfunctional EBNA-1 (both plasmids also carry oriP). The stimulation by EBNA-1 was lower in this case than with pHEBo, primarily because p396 had higher levels of background replication in the absence of EBNA-1.
The results with the derivatives of pHEBo lacking either component of oriP indicated that the DS alone was responsible for the EBNA-1-dependent replication of pHEBo. The derivative of pHEBo lacking the FR replicated nearly as well as pHEBo in the presence of EBNA-1 (lanes 14 and 15), reaching 14% of the plasmid in the cells versus 19% for pHEBo. This means that under the conditions of this assay, the plasmid retention function of the FR and EBNA-1 had very little effect, if any, on the amounts of replicated plasmids that accumulated. In sharp contrast, deletion of the DS (Δ1-4) entirely eliminated EBNA-1-dependent replication (lanes 16 to 17). It is significant that pHEBo lacking the DS replicated at a very low level and that this level was not increased by the presence of EBNA-1. This plasmid carries the FR of oriP, so if replicated plasmids were lost rapidly during the course of the assay and if the FR and EBNA-1 stabilized them, then EBNA-1 would have made a difference in this case.
From these results we can conclude that oriP requires EBNA-1 for essentially all of its replication activity and that the DS component is its EBNA-1-dependent replicator. Under the conditions of transient transfection of 293 cells, the FR is not needed to protect plasmids against loss and, perhaps for this reason, the FR contributes very little, if at all, to oriP-specific, EBNA-1-dependent plasmid replication.
All four EBNA-1 binding sites at the DS are needed for full activity in Raji cells.
To investigate the sequence determinants of the oriP replicator, numerous deletions and sequence substitutions were introduced between the EcoRV site at nucleotide 8995 and the HpaI site at nucleotide 9134 (Fig. 1 and Table 1), the region originally defined as the DS component of oriP. First, each of the four EBNA-1 sites was individually inactivated by introducing a six-base substitution near the center of each site. The substituting sequence, GTATAC, a Bst1101I recognition site, resulted in a transversion at each position from the consensus EBNA-1 binding site and would be expected to abolish binding by EBNA-1 (2).
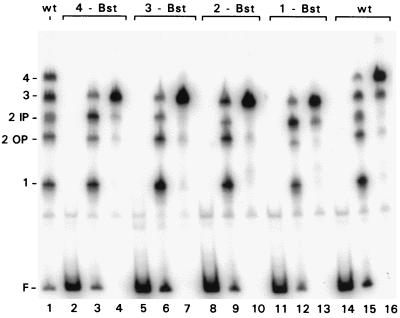
We examined the ability of EBNA-1 to bind to the DS when each of these mutations was present, using an EMSA as shown in Fig. 4. To allow resolution of individual bands in the assay, we used a truncated EBNA-1 protein, NΔ407, which lacks the linking domains of the amino-terminal half of the protein (26). When DS DNA is bound by submolar amounts of EBNA-1 that are insufficient to occupy all four sites on each molecule, it might be expected that four distinct complexes would result with EMSA, corresponding to one, two, three, and four sites simultaneously bound. The result, however, was that five shifted bands were present (Fig. 4, lanes 1, 15, and 16), as was found previously (48). The reason for this is that two different overall DNA conformations are possible when two sites are bound. (EBNA-1 bends DNA when it binds [J.B. and J.Y., unpublished data], and the pairs of sites 1 and 2 and sites 3 and 4 are in the same helical phase, while all other combinations of two sites, such as sites 1 and 4, are not.) With any one of the EBNA-1 binding sites mutant, only four shifted bands were present when limiting amounts of EBNA-1 were added (lanes 3, 6, 9, and 12). The mobility of the largest complex with each mutant DS (the predominant complex when EBNA-1 was in excess) was consistent with the loss of one binding site (lanes 4, 7, 10, and 13). Therefore, the substitution mutations, which are called Bst-1, Bst-2, Bst-3, and Bst-4, inactivated EBNA-1 binding at sites 1, 2, 3, and 4, respectively, as expected.
FIG. 4.
Analysis of EBNA-1 binding to DS mutants lacking individual binding sites, as determined by EMSA. An end-labeled, 278-bp DNA including the DS was prepared by PCR from wild-type oriP or from mutants lacking individual EBNA-1 binding sites, as indicated. EBNA-1 NΔ407 (26) was mixed with each DNA, using a molar ratio of EBNA-1 dimers to DNA of either 4 (for lanes 1, 3, 6, 9, 12, and 15) or 16 (for lanes 4, 7, 10, 13, and 16). EBNA-1 was omitted from lanes 2, 5, 8, 11, and 14. Complexes were electrophoresed through a 4% polyacrylamide gel. At the left, the positions of free probe (F) and complexes with EBNA-1 bound to one site, two sites, three sites, or all four sites are indicated. Complexes with two sites bound have different mobilities, depending primarily on whether the sites are in helical phase (2 IP) or out of helical phase (2 OP) (see the text).
To test the effects of the mutations on oriP function, derivatives of pHEBo carrying each of the four mutations were tested for their ability to replicate stably under selection in Raji cells, an EBV-positive Burkitt's lymphoma cell line. Raji cells that had been electroporated with each of the plasmids were diluted serially and cultured in multiwell dishes in medium containing hygromycin B. As shown in Table 2, pHEBo (wild-type oriP) conferred stable resistance to hygromycin B efficiently enough that resistant clones emerged in all 16 wells into which 100 cells were plated in duplicate electroporations. From this it could be estimated using Poisson statistics that 3% or more of the cells were able to form clones under selection. With each of the plasmids in which any one of the EBNA-1 binding sites was mutant, drug-resistant clones emerged at a noticeably lower frequency of between 2 × 10−3 and 8 × 10−3. A 27-bp deletion removing site 1, the Δ1 mutation, had the same effect. Deleting site 1 and inactivating site 2 simultaneously (Δ1 and 2-Bst) reduced the efficiency further, down to ca. 4 × 10−4. In this case, with only EBNA-1 sites 3 and 4 remaining at the DS, the efficiency was reduced to roughly 1% that of wild type, but the activity remaining was still significant because it was at least 100-fold higher than that observed when the entire DS was deleted (Δ1-4). A deletion of nearly half of the DS, Δ9009–9066, removing most of site 3, all of site 4, and flanking DNA to the left, reduced activity to a marginal level of around 2 × 10−5, indicating that EBNA-1 sites 1 and 2 function poorly if at all in the context of this deletion. Simultaneously deleting site 1 and inactivating site 3 (Δ1, 3-Bst), leaving intact neither of the pairs of sites (sites 1 and 2 or sites 3 and 4) that function together reduced the frequency to a level that was undetectable by this assay, i.e., to less than ∼3 × 10−6.
TABLE 2.
Frequencies of establishment of hygromycin B-resistant clones of Raji cells after electroporation with plasmids carrying wild-type or mutant oriP
| Wild type or oriP mutant | No. of positive wells, of 16 tested at:
|
Frequencya | ||
|---|---|---|---|---|
| 10,000 cells/well | 1,000 cells/well | 100 cells/well | ||
| Wild type | 16, 16 | 16, 16 | 16, 16 | >3 × 10−2 |
| Δ1-4 | 0, 0 | 0, 0 | 0, 0 | <6 × 10−6 |
| Δ1 | 16, 16 | 16, 15 | 2, 6 | 3 × 10−3 |
| 1-Bst | 16, 16 | 14, 15 | 2, 3 | 2 × 10−3 |
| 2-Bst | 16, 16 | 16, 16 | 3, 4 | 3 × 10−3 |
| 3-Bst | 16, 16 | 16, 16 | 6, 4 | 4 × 10−3 |
| 4-Bst | 16, 16 | 16, 16 | 8, 9 | 8 × 10−3 |
| Δ1 2-Bst | 16, 16 | 2, 7 | 0, 0 | 4 × 10−4 |
| Δ1 3-Bst | 0, 0 | 0, 0 | 0, 0 | <6 × 10−6 |
| Δ9009–9066 | 2, 3 | 0, 0 | 0, 0 | 2 × 10−5 |
Average number of G418-resistant clones per cell plated as estimated by fluctuation analysis.
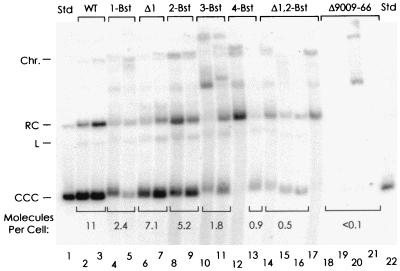
Hygromycin B-resistant clones carrying the different plasmids were taken from two or more positive wells into which the lowest possible number of cells had been plated. The cells were expanded under selection for about 24 population doublings in total, requiring 30 to 40 days from the time of transfection. Plasmids were isolated for Southern analysis, which is shown in Fig. 5. The mutant plasmids that lacked any single EBNA-1 binding site were maintained as circular monomers in all but one of the clones examined, at levels ranging from 0.9 to 8.6 copies per cell (lanes 4 to 13). Wild-type oriP supported plasmid maintenance more efficiently, with 11 monomeric copies maintained per cell on average (lanes 2 and 3). With the mutant plasmids, rearranged or multimeric forms arose in most of the clones that were analyzed, implying that replication of the monomeric forms was limiting. With one of the clones carrying the plasmid with the 4-Bst mutation, the most prominent band was consistent with it representing a supercoiled dimer of the plasmid (seen at the position of the relaxed circular monomeric form in lane 12). A form consistent with a trimer of the plasmid carrying the 3-Bst mutation was prominent in the clone analyzed in lane 10. The plasmid with Δ1 and 2-Bst mutations was maintained at approximately 0.6 monomers per cell in three of the four clones that were tested (lanes 14 to 16), while only a larger form, possibly a dimer, was present in the fourth clone (lane 17). The plasmid carrying the deletion Δ9009–9066 was clearly detectable in only one of four clones and in that case as a larger form, possibly a trimer (lanes 18 to 21). A faint band at the position of the supercoiled monomer was visible for two of the clones in the original image, suggesting that the plasmid had replicated inefficiently and had been lost over time.
FIG. 5.
Long-term maintenance of plasmids carrying mutations in oriP in Raji cells. Shown is a Southern analysis of plasmids extracted from 3 million cells of two or four individual hygromycin B-resistant Raji clones carrying pHEBo (WT) or its mutant derivatives, as indicated at the top. The positions of supercoiled (CCC), relaxed circular (RC), and linear (L) forms of pHEBo are indicated at the left. “Chr.” marks the position of residual, fragmented chromosomal DNA, visible on the stained gel. As standards, 200- and 50-pg amounts of pHEBo were loaded in lanes 1 and 22, corresponding to 9 and 2.3 molecules per cell. The numbers of supercoiled monomeric plasmids detected per cell are given as the average for the bands that are bracketed.
To summarize, these results show that there are conditions under which every EBNA-1 binding site at the DS is required in order for it to function with full efficiency. In the absence of EBNA-1 binding sites 1 and 2, the DS was still functional but much reduced in its efficiency, while deletion of sites 3 and 4 and some flanking sequences made it essentially nonfunctional for long-term plasmid maintenance. These results differ from the results of Harrison et al., who reported that oriP was fully functional when sites 1 and 2 or sites 3 and 4 were inactivated with double point mutations (15). One possible explanation for the different results is that the substitution mutations and deletions that we used could have removed functional sequences that were not affected by the point mutations used by Harrison et al. Another difference is that Harrison et al. used for their studies D98/Raji cells, a hybrid cell line that is phenotypically similar to the parent D98 carcinoma cell line, and it is possible that less of the DS is required in these cells than is required in Raji cells. We favor the latter explanation because, in the experiments that are described below using a different adherent cell line, we have found that either half of the DS can function fairly efficiently when the other half has been deleted.
Either half of the DS is a functional replicator requiring two EBNA-1 binding sites.
For the remainder of the experiments of this study, we used 143/98.2 cells, an EBNA-1-containing derivative of the fibrosarcoma cell line 143B (47). When the replication of plasmids carrying mutations at the DS was determined 2 to 3 days after transfection, replication was found to be insensitive to inactivation of individual EBNA-1 sites and even to the deletion of either half of the DS; the Δ1 3-Bst combination, however, abolished activity (data not shown). The results were thus consistent with the conclusion of Harrison et al. (15) that either functional pair of EBNA-1 sites, 1 and 2 together or 3 and 4 together, is sufficient for DS function. However, if the plasmids were assayed in 143/98.2 cells for maintenance under selection for 2 to 4 weeks, the subtle effects of inactivating individual EBNA-1 binding sites became apparent. For this reason, we used this long-term plasmid maintenance assay to investigate the sequence requirements of the DS.
Initially, we tested the effects of deleting the EBNA-1 sites individually and in certain pairs. For the assay, duplicate 6-cm dishes of cells were transfected with each plasmid, 1/10 of the cells in each dish were transferred to new 6-cm dishes the next day, and selection for hygromycin B resistance was started on the third day. Generally, by the sixth to seventh day of the experiment, over 600 drug-resistant colonies were apparent, and these usually had entirely covered the dish by the eight day if oriP was wild type. If any one of the EBNA-1 sites was mutant or if both sites of either functional pair were inactive, similar numbers of colonies appeared, but they grew more slowly and needed 1 or 2 more days to fill the dishes. With the entire DS deleted, similar numbers of colonies appeared initially, but they grew more slowly, never reached confluence, and began to die after 10 to 12 days under selection. This abortive drug resistance is due to the FR of oriP and EBNA-1, which allow the plasmids to be retained by the cells for several generations even though they cannot replicate (21, 34). The plasmid with both EBNA-1 sites 1 and 3 inactive (Δ1 3-Bst) also gave abortive drug-resistant colonies. The cultures that carried the functional plasmids were grown in selective medium with successive 1:4 and 1:3 splits before being harvested at 18 to 21 days after transfection.
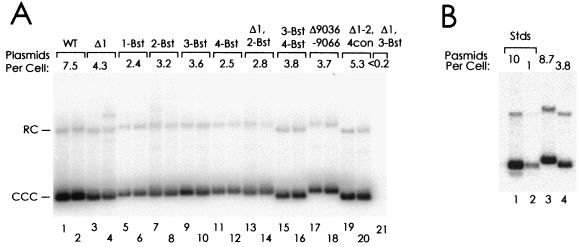
A Southern analysis of the plasmids present in the hygromycin B-resistant cells is shown in Fig. 6. pHEBo, carrying wild-type oriP, was maintained at an average of 7.5 molecules per cell (Fig. 6A, lanes 1 and 2), and the elimination of individual EBNA-1 binding sites reduced this to between 2.4 and 4.3 molecules per cell (lanes 3 to 12). The simultaneous elimination of sites 1 and 2 (lanes 13 and 14) or of sites 3 and 4 (lanes 15 to 18) caused a similar reduction in the number of plasmids that were detected. Thus, the elimination of individual EBNA-1 sites caused measureable reductions in plasmid maintenance, but there was no further reduction in activity when both sites of either functional pair of sites were eliminated. The simultaneous elimination of sites 1 and 3 abolished activity (lane 20). The results are consistent with those of Harrison et al., in which the simultaneous elimination of sites 2 and 3 or sites 1 and 4 abolished activity. It is clear that both sites of a functional pair, sites 1 and 2 or sites 3 and 4, must be intact to support DS function.
FIG. 6.
Support of long-term plasmid maintenance in 143/98.2 cells by oriP with various mutations at the DS. (A) Duplicate plates of cells were transfected with pHEBo (WT) or its derivatives carrying the indicated mutations at the DS. One-tenth of the cells were selected for hygromycin B resistance and then split successively 1:4 and 1:3 before being expanded and harvested at 18 to 21 days after transfection, except that in the case of the Δ1 3-Bst mutant, cells were not split and required 29 days for expansion. Plasmids extracted from ∼7 million cells were analyzed by Southern blot. For each plasmid tested, the average number of plasmid molecules detected per cell is indicated. The supercoiled (CCC) and relaxed circular (RC) forms of the plasmids are indicated. Note that the vector portion of the plasmids tested in lanes 15, 16, 19, and 20 is slightly smaller than the vector portion of the others (Table 1). (B) To convert the relative amounts detected in panel A to copies per cell, the samples of lanes 1 and 20 in panel A were analyzed again on this blot, in lanes 3 and 4, and compared to known amounts of pHEBo in lanes 1 and 2.
From similar experiments we found that deletions removing either half of the DS caused moderate reductions in activity that were similar to the effects of deleting individual EBNA-1 sites. The deletion Δ3-4 (positions 8994 to 9082) removed EBNA-1 sites 3 and 4 and the overlapping long dyad symmetry element (for which the DS component of oriP was named), along with some flanking DNA. The effect of Δ3-4 was to reduce the number of plasmids maintained in the cells to less than half the number maintained with wild-type oriP (Fig. 7B, lanes 3 and 4 compared to lanes 1 and 2; Fig. 8, lane 13 compared to lanes 1 and 2). The smaller deletion, Δ9009–9066 was reproducibly somewhat more deleterious (Fig. 8, lanes 11 and 12). The other half of the DS was removed by the deletion Δ1-2 (positions 9088 to 9134), and the effect was similarly moderate (Fig. 7A, lanes 18 and 19 compared to lanes 2 and 3).
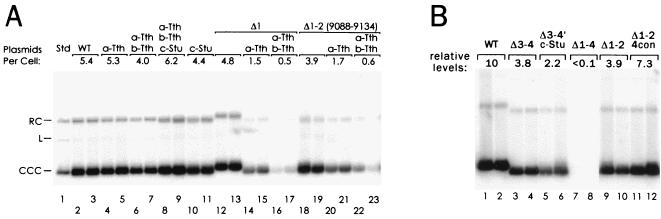
FIG. 7.
Activities of oriP mutants with substitutions of the nonamer repeats. (A) Duplicate plates of 143/98.2 cells were transfected with pHEBo or its derivatives carrying the indicated mutations. Cultures were split 1:10 before selection for hygromycin resistance, split 1:6 and 1:10 after selection, and harvested 20 to 21 days (lanes 2 to 11) or 24 days (lanes 12 to 23) after transfection. Plasmids extracted from approximately 5 million cells were analyzed by Southern blot. In lane 1, 50 pg of pHEBoΔRV was loaded as a standard corresponding to 1.4 copies per cell. The average calculated numbers of plasmid copies per cell are indicated. (B) Similar experiment to that shown in panel A except that cultures were grown without hygromycin B beyond the 13th day following transfection and the cultures were harvested sooner. After selection, the resistant cultures were split successively 1:10 and then 1:2, except as noted below, and harvested at 15 to 16 days following transfection. With Δ3-4, c-Stu, the cultures could only be split once at 1:5 after selection and before harvesting. With Δ1-4, the cultures were not split after selection and could not be harvested until 24 days posttransfection. The relative average numbers of plasmids detected have been indicated, with pHEBo (WT) set arbitrarily at 10.
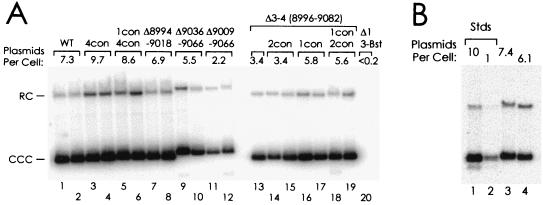
FIG. 8.
Activity of oriP when the EBNA-1 binding sites at the DS were made to resemble FR consensus binding sites. (A) 143B/98.2 cells were transfected with plasmids carrying the indicated mutations, selected for hygromycin resistance, and propagated as described for Fig. 6. Cultures were harvested 20 to 21 days (lanes 1 to 8) or 25 to 26 days (lanes 9 to 20) after transfection. Average number of plasmid copies per cell are indicated. (B) The samples from lanes 1 and 19 were analyzed again on this blot (lanes 3 and 4) in comparison to known amounts of plasmid (lanes 1 and 2) to provide references for quantitating the signals of the blot in panel A.
The deletions Δ1-2 (positions 9088 to 9134) and Δ3-4 (positions 8994 to 9082) divide the DS into functional “halves,” separable just to the left of site 2 (Fig. 1). It is clear from these experiments that either half of the DS can function fairly well without the other half. The experiments that are described next were performed to investigate what sequences allow each half to function.
An ancillary role for the nonamer repeats.
The left half of the DS includes EBNA-1 sites 3 and 4 and the dyad symmetry element, which is centered over the EBNA-1 sites and spans 65 bp. The right functional half lacks such a degree of internal symmetry. Other than the EBNA-1 binding sites, the only obvious similarity between the two functional halves is a 9-bp sequence, TAACCCTAA, which is present twice in the left half, in opposite orientations near the ends of the dyad symmetry, and once in the right half (Fig. 1). Previously, Vogel et al. replaced all three copies of the nonamer sequence with a G+C-rich sequence and noted a moderate decrease in the ability of oriP to support plasmid maintenance measured over a period of weeks (41). We wondered whether the nonamer sequence might appear to be less important if replaced with a sequence that preserved its high A+T content or whether it might be more important for the function of either isolated half of the DS than it is for the DS as a whole.
To test these possibilities, the “a” and “b” copies of the nonamer at each end of the dyad symmetry were replaced as part of a 12-bp substitution that included a TthI site (Table 1). The “a” and “b” copies of the nonamer are in opposite orientations, while the substituting sequence was placed in the same orientation at both sites. We were unable to introduce the substitutions in opposite orientations, which would have preserved the dyad symmetry. The substitution at the left end of the dyad, “a-Tth,” changed 8 of 9 bp of nonamer a, and the substitution “b-Tth” at the other end changed 7 of 9 bp of nonamer b. The A+T-rich composition of the sequence at each end of the dyad symmetry was not changed. Finally, nonamer c was replaced with a 9-bp substitution, called c-Stu, that changed 8 of 9 bp of the nonamer.
One, two, or all three substitutions of the nonamer together had no noticeable effect on plasmid maintenance under selection in 143/98.2 cells in a 3-week assay (Fig. 7A, lanes 1 to 10). However, the substitution mutations had marked effects on the activities of either half of the DS. The a-Tth mutation caused plasmid copy numbers to decrease by a factor of about 3 when tested in the context of either the Δ1 mutation or the Δ1-2 mutation (Fig. 7A, lanes 14 and 15 compared to lanes 12 and 13 and lanes 20 and 21 compared to lanes 18 and 19). The b-Tth and a-Tth mutations together in these contexts reduced plasmid levels by another threefold, to close to 0.5 copies per cell (lanes 16, 17, 22, and 23). Placing the b-Tth substitution in the context of a-Tth did not reduce the length of the dyad symmetry, since a-Tth had already eliminated the symmetry extending outside of the two EBNA-1 binding sites. Since b-Tth caused a further decrease in activity, the substitution itself, rather than a loss of symmetry, was responsible for the loss of function in this case. In a separate experiment using a shorter plasmid maintenance assay, the c-Stu substitution was seen to reduce plasmid maintenance by more than a factor of 2 in the context of the Δ3-4 deletion (Fig. 7B, lanes 7 and 8 compared to lanes 3 and 4).
The results suggest that the nonamer repeats contribute to the replicator function of the DS, particularly to the capacity of either half of the DS to function alone. Each half of the DS can function in the absence of the nonamer sequences, however, so the contribution of the nonamer sequence is ancillary.
The permanganate-sensitive, nonconsensus T that is unique to sites 1 and 4 is unimportant.
The four EBNA-1 binding sites that are present at the DS differ from the high-affinity binding site consensus sequence at several positions (2). Of particular interest were the nonconsensus thymines that are unique to sites 1 and 4 and become permanganate-sensitive when EBNA-1 binds (see above). These occur at the −6 position of binding site 1 (relative to the center of the binding site) and at the symmetrical +6 position on the lower strand of binding site 4 (Fig. 1). Mutations were made at these positions to convert the nonconsensus thymine at nucleotide 9046 in site 4 and at nucleotide 9110 (lower strand) in site 1 to the consensus adenine, and the mutations were called 4con and 1con, respectively.
It was also of interest that among the 24 EBNA-1 binding sites at oriP, sites 2 and 3 have the lowest G+C content, with only 5 of 18 and 7 of 18 bp, respectively, being GC pairs. The central 6 bp in site 2 are TATATA, instead of the consensus CATATG, helping to give this site the lowest G+C content. The T and A at positions −3 and +3 of site 2 were converted to the consensus C and G, respectively, and the double point mutation was called 2con. Hearing et al. found that converting these positions to the consensus C and G had no measurable effect on the function of the DS as a whole (16), but it remained to be tested whether these mutations would reduce the ability of the right half of the DS to function alone.
Neither the 4con mutation alone nor the combination of 4con and 1con mutations significantly affected the ability of oriP to support plasmid maintenance under selection (Fig. 8A, lanes 1 to 6). In the context of the Δ3-4 deletion, 1con led to an increase in the number of plasmids that were maintained per cell to a level midway between the levels of the wild type and the Δ3-4 mutant (lanes 16 and 17). The combination of the 1con and 2con mutations was associated with an identical increase in plasmid maintenance (lanes 18 and 19), and this appeared to be due to 1con alone, since 2con by itself had no effect (lanes 14 and 15 compared to lane 13). The 4con mutation also led to a reproducible increase in activity when tested in the context of the deletion of sites 1 and 2 (Fig. 7B, lanes 3 to 7). In the experiment shown in Fig. 6A, the plasmid carrying 4con and Δ1-2 mutations was maintained at higher levels than were any of the plasmids carrying mutations that inactivated one or two EBNA-1 binding sites within a functional pair.
The mutations 1con and 4con might have increased the replicator function of each DS half by increasing the binding affinity for EBNA-1 (2). Whether or not this is the explanation, these experiments ruled out the possibility that some of the unique aspects of EBNA-1 sites 1, 2, and 4 contribute in any critical way to replicator function. All of the results point to the conclusion that the only features of the DS that are likely to be essential for replicator activity are its EBNA-1 binding sites and the spacing between them.
DISCUSSION
Initiation of DNA replication at oriP requires EBNA-1.
Not long ago, the view that EBNA-1 directs replication to initiate at the DS of oriP seemed compelling because of (i) genetic evidence that the EBNA-1 binding sites are essential (15), (ii) physical evidence for the association of EBNA-1 with the DS in living cells (20), and (iii) direct demonstrations that EBNA-1 is required for DS-dependent replication of plasmids after transient transfection of cells (15, 36). However, the latter, most direct evidence was compromised by the fact that EBNA-1 also confers stability to plasmids carrying EBNA-1 binding sites (21, 30). This problem was pointed out in a recent study by Aiyar et al., who showed that introduced plasmids are unstable after replication in most cell lines (1).
This study reaffirms that EBNA-1 is required for efficient oriP-specific replication of plasmids. The results of Fig. 2 and 3 show that even after early times after the transfection of cells with plasmids (<48 h), EBNA-1 was necessary for more than 90% of the replication of plasmids containing oriP or its DS component. The DS component was shown to be the replicator of oriP, confirming previous studies (15, 36, 42). Most importantly, DS replicator activity was shown to require EBNA-1 in transfected 293 cells, where the FR components of oriP and EBNA-1 were unimportant for the accumulation of replicated plasmids, indicating that EBNA-1 contributed to the replication and not the maintenance of the plasmids over the brief course (46 h) of the assay. Some residual replication activity could be attributed to oriP in the absence of EBNA-1, but it did not require the DS component and may not be of a very specific nature. Finally, a study of many deletion and substitution mutations at the DS led to the conclusion that the DS depends on its EBNA-1 binding sites and that, although other sequences appear to contribute to activity, little if anything other than two EBNA-1 binding sites is essential for activity.
These results appear to conflict with the findings of Aiyar et al., who reported that oriP could support the replication of plasmids during the first 48 h following the transfection of cells in the absence of EBNA-1 and that EBNA-1 stimulated replication by no more than twofold (1). It is worth noting the two methodological differences between our study and the study of Aiyar et al. First, for most of the measurements, Aiyar et al. used rather harsh conditions for digestion of DNA with DpnI, which resulted in the degradation of plasmids unless both DNA strands lacked adenine methylation. This required the plasmids to have been replicated two or more times (i.e., during at least two successive cell cycles) within 48 h; so, for EBNA-1 to stimulate replication detectably, it needed to accumulate to adequate levels very soon after transfection. Under the more normal conditions that we used for DpnI digestion, plasmids that have been replicated once resist cleavage (46). Second, while we determined the amounts of replicated plasmids by the rather direct Southern blot assay, Aiyar et al. used competitive PCR, which is less direct and has more potential for error.
The relatively inefficient replication that we could attribute to oriP in the absence of EBNA-1 was unaffected by deletion of the DS (Fig. 3). Aiyar et al. found that a region of roughly 800 bp that included the DS supported replication in the absence of EBNA-1, but the relevance of the DS to this activity was not determined, and a fragment containing mainly the FR was also active (1). Without knowing what regions of oriP or flanking DNA are responsible for the EBNA-1-independent activity, it would be premature to speculate whether this activity has any functional significance. Mammalian cells have the ability to initiate replication inefficiently with relatively low specificity within cloned DNA segments on plasmids that have been introduced into cells (17), so such activity by itself does not signify a normal role in replication.
Does the FR activate the DS replicator under any circumstances?
The DS supports EBNA-1-dependent replication in the absence of the FR when tested in HeLa cells and in D98/Raji cells transfected for 96 h (15, 36), as well as in 293 cells (this study). In Raji cells, however, the DS requires the FR to support plasmid replication measured 96 h after transfection (34, 45). In preliminary tests with 143B cells, the FR appears to stimulate the level of replication that can be detected after 48 h following transfection and to be essential for replication measured after 96 h (data not shown). One possible explanation for this difference among cell lines is that plasmids are lost more quickly from some cell lines (Raji and 143B) than from others (HeLa, D98/Raji, and 293) and that the FR is merely preventing the loss of plasmids. In support of this, Aiyar et al. found that expression of luciferase from an introduced plasmid was lost much more quickly with 143B cells than with 293 cells (1). On the other hand, EBNA-1 molecules interact with each other while bound at the FR and the DS to form a DNA loop, and the interaction can stabilize the binding of EBNA-1 to the lower-affinity sites at the DS in vitro (9, 27, 38). It is conceivable that this interaction is important under some circumstances and that it might contribute to the dependence of replication on the FR that is observed with some cell lines.
A minimal DS replicator.
Our studies of deletion and substitution mutations at the DS yielded two immediate conclusions. First, all four EBNA-1 binding sites at the DS are needed in order for it to function with optimal efficiency. Second, either half of the DS, having only two EBNA-1 binding sites, works fairly effectively under certain conditions, providing a glimpse of the minimal DS replicator.
The importance of each of the four EBNA-1 binding sites had not been clearly documented previously. It was particularly evident in the assay for plasmid maintenance under selection in Raji cells, where the absence any single EBNA-1 binding site reduced the efficiency of colony outgrowth by roughly an order of magnitude (Table 2). The importance of individual EBNA-1 binding sites was more subtle, although still apparent, in EBNA-1-positive 143B cells. Given that the choice of the cell line, the length of time under selection, and the choice of a vector (e.g., transcriptional control elements can interfere with oriP function [14]) can each affect the stringency of an assay for oriP activity, it is understandable why some results of previous studies (5, 15, 43) appear to be contradictory.
Previously, it had been shown that the DS remained functional when either the left functional pair or the right functional pair of EBNA-1 binding sites were inactivated by double point mutations (15), but it was unclear how well each “half” of the DS could function with the other half deleted. The fact that either half of the DS can function fairly well in the absence of the other half, as demonstrated in this study, means that each half contains the essential elements of a minimal replicator. We know that both EBNA-1 binding sites are required for the activity of each half of the DS (reference 15; this study). We also know that the two EBNA-1 binding sites must be properly spaced (15), with exactly 21 bp between their centers (unpublished data). Other than the EBNA-1 binding sites and their spacing, the only obvious sequences that are shared by the two halves of the DS are within the repeated nonamer sequence.
Nonamer repeats and sequences flanking the EBNA-1 binding sites.
The 9-bp sequence, 5′-TTAGGGTTA, flanks EBNA-1 binding sites 3 and 4 in opposite orientations in the left half of the DS, and it flanks site 1 in the right half. It was suggested, on the basis of the differential sensitivity to dimethyl sulfate in cells arrested at different points of the cell cycle, that a cellular protein interacts with the nonamer sequences (31). The substitution of all three nonamer repeats with a G+C-rich sequence only moderately reduced the maintenance of an oriP-dependent plasmid in a previous study (41). In the present study, the simultaneous mutation of all three copies had no noticeable effect on the activity of the DS as a whole, but these mutations noticeably reduced plasmid maintenance when each half of the DS was tested independently. The nonamer sequence, while not essential, thus appears to play an ancillary role. It should be pointed out that more tests would be required to establish a role for the nonamer sequence itself. Instead, sequences that overlap it may contribute to this process, or perhaps a property of the DNA, such as ease of unwinding, is involved. The DS of a close relative of EBV, herpesvirus papio, does not appear to contain a sequence resembling the nonamer sequence (24).
Unique, oxidation-sensitive thymines in EBNA-1 binding sites 1 and 4.
Two thymines at symmetric positions in EBNA-1 binding sites 1 and 4 at the DS become sensitive to oxidation by permanganate when EBNA-1 binds to the sites and distorts the helical structure of the DNA (3, 7, 20, 40). These two thymines represent transversions from the adenine that occupies this position in the consensus EBNA-1 binding site and are unique to these two sites. The nonconsensus thymine is not present in any of the 50 remaining half sites within the 26 EBNA-1 binding sites that are present in the EBV genome of strain B95-8. Since the unique thymines occur at equivalent positions within each functional pair of EBNA-1 binding sites and at sites of helical distortion, it was reasonable to test for their functional significance. The simultaneous conversion of these thymines to the consensus base, adenine (1con and 4con mutations), did not noticeably affect the function of the DS, and individually these mutations actually improved the ability of each half of the DS to function. Thymine at these positions of an EBNA-1 binding site (−6, lower strand; +6, upper strand) in place of the consensus adenine is known to decrease the affinity for EBNA-1 (2). Presumably, the affinity of sites 1 and 4 for EBNA-1 is less than what is optimal for each half of the DS to function independently as a replicator, and so 1con and 4con stimulate activity by increasing the affinity for EBNA-1. While the significance of these unique thymines remains unclear, the results point to the importance of EBNA-1 binding sites per se, as the determinants of this replicator.
Conclusion.
The results of this study reaffirm that the DS component of oriP is dependent on EBNA-1 for replicator activity. The core determinant of the DS replicator appears to be simply two properly spaced EBNA-1 binding sites. The only marked resemblance between the DS of herpesvirus papio and the DS of EBV is the presence of multiple EBNA-1 binding sites and a spacing of 21 bp (two exact turns of the DNA helix) between the sites, center to center (44). In related studies, we have found that changing the distance between a functional pair of EBNA-1 binding sites by just 1 or 2 bp abolishes replicator activity. This suggests that a very specific structure of EBNA-1 proteins and DS DNA is required for replication, which is consistent with a direct role for EBNA-1 in an initiation step.
ACKNOWLEDGMENTS
We thank David Mackey and Bill Sugden for the generous gift of EBNA-1 NΔ407 protein, Angela Ying for constructing the Bst mutants and for performing the Southern analysis of Fig. 5, and Prasad Kularni for comments on the manuscript.
This work was supported by grant CA4312212 from the National Institutes of Health.
REFERENCES
- 1.Aiyar A, Tyree C, Sugden B. The plasmid replicon of EBV consists of multiple cis-acting elements that facilitate DNA synthesis by the cell and a viral maintenance element. EMBO J. 1998;17:6394–6403. doi: 10.1093/emboj/17.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambinder R F, Shah W A, Rawlins D R, Hayward G S, Hayward S D. Definition of the sequence requirements for binding of the EBNA-1 protein to its palindromic target sites in Epstein-Barr virus DNA. J Virol. 1990;64:2369–2379. doi: 10.1128/jvi.64.5.2369-2379.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bochkarev A, Bochkareva E, Frappier L, Edwards A M. The 2.2 Å structure of a permanganate-sensitive DNA site bound by the Epstein-Barr virus origin binding protein, EBNA1. J Mol Biol. 1998;284:1273–1278. doi: 10.1006/jmbi.1998.2247. [DOI] [PubMed] [Google Scholar]
- 4.Calos M P. Stability without a centromere. Proc Natl Acad Sci USA. 1998;95:4084–4085. doi: 10.1073/pnas.95.8.4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chittenden T, Lupton S, Levine A J. Functional limits of oriP, the Epstein-Barr virus plasmid origin of replication. J Virol. 1989;63:3016–3025. doi: 10.1128/jvi.63.7.3016-3025.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chodosh L A. Mobility shift DNA-binding assay using gel electrophoresis. In: Ausubel F M, Brent R, Kingston R E, Moore D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons, Inc.; 1988. pp. 12.2.1–12.2.10. [Google Scholar]
- 7.Frappier L, M. O D. EBNA1 distorts oriP, the Epstein-Barr virus latent replication origin. J Virol. 1992;66:1786–1790. doi: 10.1128/jvi.66.3.1786-1790.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frappier L, M. O D. Overproduction, purification, and characterization of EBNA1, the origin binding protein of Epstein-Barr virus. J Biol Chem. 1991;266:7819–7826. [PubMed] [Google Scholar]
- 9.Frappier L, O'Donnell M. Epstein-Barr nuclear antigen 1 mediates a DNA loop within the latent replication origin of Epstein-Barr virus. Proc Natl Acad Sci USA. 1991;88:10875–10879. doi: 10.1073/pnas.88.23.10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frost E, Williams J. Mapping temperature-sensitive and host-range mutations of adenovirus type 5 by marker rescue. Virology. 1978;91:39–50. doi: 10.1016/0042-6822(78)90353-7. [DOI] [PubMed] [Google Scholar]
- 11.Gahn T A, Schildkraut C L. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989;58:527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- 12.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 13.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 14.Haase S B, Heinzel S S, Calos M P. Transcription inhibits the replication of autonomously replicating plasmids in human cells. Mol Cell Biol. 1994;14:2516–2524. doi: 10.1128/mcb.14.4.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison S, Fisenne K, Hearing J. Sequence requirements of the Epstein-Barr virus latent origin of DNA replication. J Virol. 1994;68:1913–1925. doi: 10.1128/jvi.68.3.1913-1925.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hearing J, Mulhaupt Y, Harper S. Interaction of Epstein-Barr virus nuclear antigen 1 with the viral latent origin of replication. J Virol. 1992;66:694–705. doi: 10.1128/jvi.66.2.694-705.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinzel S S, Krysan P J, Tran C T, Calos M P. Autonomous DNA replication in human cells is affected by the size and the source of the DNA. Mol Cell Biol. 1991;11:2263–2272. doi: 10.1128/mcb.11.4.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrmann C H, Mathews M B. The adenovirus E1B 19-kilodalton protein stimulates gene expression by increasing DNA levels. Mol Cell Biol. 1989;9:5412–5423. doi: 10.1128/mcb.9.12.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 20.Hsieh D J, Camiolo S M, Yates J L. Constitutive binding of EBNA1 protein to the Epstein-Barr virus replication origin, oriP, with distortion of DNA structure during latent infection. EMBO J. 1993;12:4933–4944. doi: 10.1002/j.1460-2075.1993.tb06187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krysan P J, Haase S B, Calos M P. Isolation of human sequences that replicate autonomously in human cells. Mol Cell Biol. 1989;9:1026–1033. doi: 10.1128/mcb.9.3.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laskey R, Madine M. Roles of nuclear structure in DNA replication. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 119–130. [Google Scholar]
- 23.Lindahl T, Adams A, Bjursell G, Bornkamm G W, Kaschka-Dierich C, Jehn U. Covalently closed circular duplex DNA of Epstein-Barr virus in a human lymphoid cell line. J Mol Biol. 1976;102:511–530. doi: 10.1016/0022-2836(76)90331-4. [DOI] [PubMed] [Google Scholar]
- 24.Loeb D D, Sung N S, Pesano R L, Sexton C J, Hutchison C D, Pagano J S. Plasmid origin of replication of herpesvirus papio: DNA sequence and enhancer function. J Virol. 1990;64:2876–2883. doi: 10.1128/jvi.64.6.2876-2883.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lupton S, Levine A J. Mapping genetic elements of Epstein-Barr virus that facilitate extrachromosomal persistence of Epstein-Barr virus-derived plasmids in human cells. Mol Cell Biol. 1985;5:2533–2542. doi: 10.1128/mcb.5.10.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mackey D, Middleton T, Sugden B. Multiple regions within EBNA1 can link DNAs. J Virol. 1995;69:6199–6208. doi: 10.1128/jvi.69.10.6199-6208.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mackey D, Sugden B. Studies on the mechanism of DNA linking by Epstein-Barr virus nuclear antigen 1. J Biol Chem. 1997;272:29873–29879. doi: 10.1074/jbc.272.47.29873. [DOI] [PubMed] [Google Scholar]
- 28.Marechal V, Dehee A, Chikhi-Brachet R, Piolot T, Coppey-Moisan M, Nicolas J C. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J Virol. 1999;73:4385–4392. doi: 10.1128/jvi.73.5.4385-4392.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Middleton T, Sugden B. EBNA1 can link the enhancer element to the initiator element of the Epstein-Barr virus plasmid origin of DNA replication. J Virol. 1992;66:489–495. doi: 10.1128/jvi.66.1.489-495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Middleton T, Sugden B. Retention of plasmid DNA in mammalian cells is enhanced by binding of the Epstein-Barr virus replication protein EBNA1. J Virol. 1994;68:4067–4071. doi: 10.1128/jvi.68.6.4067-4071.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niller H H, Glaser G, Knuchel R, Wolf H. Nucleoprotein complexes and DNA 5′-ends at oriP of Epstein-Barr virus. J Biol Chem. 1995;270:12864–12868. doi: 10.1074/jbc.270.21.12864. [DOI] [PubMed] [Google Scholar]
- 32.Nonoyama M, Pagano J S. Separation of Epstein-Barr virus DNA from large chromosomal DNA in non-virus-producing cells. Nat New Biol. 1972;238:169–171. doi: 10.1038/newbio238169a0. [DOI] [PubMed] [Google Scholar]
- 33.Rawlins D R, Milman G, Hayward S D, Hayward G S. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985;42:859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- 34.Reisman D, Yates J, Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985;5:1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Shirakata M, Hirai K. Identification of minimal oriP of Epstein-Barr virus required for DNA replication. J Biochem. 1998;123:175–181. doi: 10.1093/oxfordjournals.jbchem.a021907. [DOI] [PubMed] [Google Scholar]
- 37.Simpson K, McGuigan A, Huxley C. Stable episomal maintenance of yeast artificial chromosomes in human cells. Mol Cell Biol. 1996;16:5117–5126. doi: 10.1128/mcb.16.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su W, Middleton T, Sugden B, Echols H. DNA looping between the origin of replication of Epstein-Barr virus and its enhancer site: stabilization of an origin complex with Epstein-Barr nuclear antigen 1. Proc Natl Acad Sci USA. 1991;88:10870–10874. doi: 10.1073/pnas.88.23.10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugden B, Marsh K, Yates J. A vector that replicates as a plasmid and can be efficiently selected in B-lymphoblasts transformed by Epstein-Barr virus. Mol Cell Biol. 1985;5:410–413. doi: 10.1128/mcb.5.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Summers H, Fleming A, Frappier L. Requirements for Epstein-Barr nuclear antigen 1 (EBNA1)-induced permanganate sensitivity of the Epstein-Barr virus latent origin of DNA replication. J Biol Chem. 1997;272:26434–26440. doi: 10.1074/jbc.272.42.26434. [DOI] [PubMed] [Google Scholar]
- 41.Vogel M, Wittmann K, Endl E, Glaser G, Knuchel R, Wolf H, Niller H H. Plasmid maintenance assay based on green fluorescent protein and FACS of mammalian cells. BioTechniques. 1998;24:540–544. [PubMed] [Google Scholar]
- 42.Wysokenski D A, Yates J L. Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein-Barr virus. J Virol. 1989;63:2657–2666. doi: 10.1128/jvi.63.6.2657-2666.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yates J, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci USA. 1984;81:3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yates J L. Epstein-Barr virus DNA replication. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 751–774. [Google Scholar]
- 45.Yates J L, Camiolo S M. Dissection of DNA replication and enhancer activation functions of Epstein-Barr virus nuclear antigen 1. In: Kelley B S A T, editor. Cancer cells. Vol. 6. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. pp. 197–205. [Google Scholar]
- 46.Yates J L, Guan N. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol. 1991;65:483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yates J L, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 48.Zhang D, Frappier L, Gibbs E, Hurwitz J, M. O D. Human RPA (hSSB) interacts with EBNA1, the latent origin binding protein of Epstein-Barr virus. Nucleic Acids Res. 1998;26:631–637. doi: 10.1093/nar/26.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]