Abstract
Cell‐free plasma DNA is elevated in cancer patients and decreases in response to effective treatments. Consequently, these nucleic acids have potential as new tumor markers. In our current study, we investigated whether the plasma DNA concentrations in patients with cancer are altered during the course of radiation therapy. To first determine the origin of cell‐free plasma DNA, plasma samples from mice bearing transplanted human tumors were analyzed for human‐specific and mouse‐specific cell‐free DNA. Human‐specific DNA was detectable only in plasma from tumor‐bearing mice. However, mouse‐specific plasma DNA was significantly higher in tumor‐bearing mice than in normal mice, suggesting that cell‐free plasma DNA originated from both tumor and normal cells. We measured the total cell‐free plasma DNA levels by quantitative polymerase chain reaction in 15 cancer patients undergoing radiation therapy and compared these values with healthy control subjects. The cancer patients showed higher pretreatment plasma DNA concentrations than the healthy controls. Eleven of these patients showed a transient increase of up to eightfold in their cell‐free plasma DNA concentrations during the first or second week of radiation therapy, followed by decreasing concentrations toward the end of treatment. In two other cancer patients, the cell‐free plasma DNA concentrations only decreased over the course of the treatment. The total cell‐free plasma DNA levels in cancer patients thus show dynamic changes associated with the progression of radiation therapy. Additional prospective studies will be required to elucidate the potential clinical utility and biological implications of dynamic changes in cell‐free plasma DNA during radiation therapy. (Cancer Sci 2009; 100: 303–309)
The response of malignant tumors to radiation therapy varies, even if they show similar histopathological origins and clinical staging. An accurate assessment of the radiation sensitivity of individual tumors during the early phase of radiation therapy could provide valuable guidance for selecting the most appropriate treatment plan for individual patients, such as patient‐specific modifications of the dose and field of radiation therapy, or additional or alternative therapeutic modalities earlier in the course of treatment. This in turn might contribute to better patient outcomes. However, conventional methods, including imaging diagnoses using computed tomography and magnetic resonance imaging or histopathological examination of biopsy samples, are limited in their ability to evaluate the radiation sensitivity of individual tumors during or immediately following radiation therapy. Hence, alternative assessment methods are required.
Previous studies have shown that cell‐free DNA is present in the bloodstream of cancer patients at higher concentrations than in healthy controls.( 1 , 2 , 3 , 4 ) Although the cell origins and the mechanisms underlying cell‐free DNA generation in cancer patients are not yet fully understood, it has been suggested that tumor cells might actively secrete DNA into the bloodstream, or that DNA could be released following tumor cell lysis.( 1 , 5 ) In addition, an elevated cell‐free DNA integrity in the plasma of cancer patients has been reported.( 6 , 7 , 8 ) Cell death in normal tissues of healthy individuals occurs mainly through apoptosis, producing small and uniform DNA fragments.( 8 ) In contrast, cell death in tumor tissues occurs mainly through necrosis and generates DNA fragments with varying strand lengths resulting from the random and incomplete digestion of genomic DNA by a variety of deoxyribonucleases. Consistent with these findings, cell‐free plasma DNA integrity, as measured by the ratio of longer DNA fragments (300~400‐bp <) to the total DNA content, is increased in cancer patients compared with healthy controls.( 9 , 10 , 11 , 12 , 13 ) The plasma concentrations of other types of nucleic acids, such as mRNA and mitochondrial DNA, are also elevated in cancer patients.( 14 , 15 ) These data suggest that quantification of the absolute levels of nucleic acids or of the integrity of the DNA in plasma may be useful for the diagnosis of certain types of cancer.( 2 , 9 )
Other studies have shown that changes in the levels of circulating cell‐free DNA correlates with the clinical status of a tumor. Sozzi et al. and Gautschi et al. have reported that the cell‐free plasma DNA concentration in patients with cancer decreased following effective treatment and increased in association with tumor recurrence.( 2 , 16 ) These data suggest that quantification of plasma DNA might represent a novel approach to the monitoring of tumor status and the response to treatment.( 4 , 17 , 18 ) However, the kinetics of plasma DNA concentrations either during or after radiation therapy remains unknown. To address this issue in our current study, we first analyzed the plasma concentrations of tumor‐ and mouse‐specific cell‐free DNA in mice implanted with human tumor cells to determine its origin in these samples. We then conducted serial quantitative analyses of cell‐free plasma DNA in patients with cancer undergoing radiation therapy. This is the first report to show the dynamic changes that occur for cell‐free plasma DNA in cancer patients during radiation therapy.
Materials and Methods
Cell culture and tumor‐bearing mice. The human tumor cell lines RPMI 1788 (lymphoblastoid cell), KM12C (colon adenocarcinoma), SQ5 (lung squamous cell carcinoma), DLD‐1 (colon adenocarcinoma), A431 (epidermoid carcinoma), and SR‐OV‐3 (lung small cell carcinoma) were used. All cell lines were maintained as described previously.( 19 ) Cell aliquots (1.0 × 107 in 100 µL medium) from individual cell lines were subcutaneously implanted into the backs of 7–9‐week‐old pathogen‐free female BALB/c nude mice (Clea Japan, Tokyo, Japan). After approximately 15 days, when tumors were at least 5 mm in diameter, blood samples were collected. Blood samples from nude mice with no human tumor xenografts were used as controls.
Patient and control sample collection. Fifteen patients with localized cancers, all without distant metastases and undergoing radiation therapy at the Yokohama City University Hospital, were enrolled in the present study. The demographics and treatment information for this patient group are listed in Table 1. In accordance with our Institutional Research Ethics Committee approval, informed consent was obtained from all patients. Blood samples were collected prior to radiation therapy, on the third day of treatment, and then once every 1 or 2 weeks during the course of treatment. Radiation therapy was delivered using a 4–15 MV linear accelerator. Standard fractions of 1.8–2.0 Gy per fraction on 5 days per week were applied. With the exception of patients 2 and 5, no patient had received any treatment prior to radiation therapy. Patient 2 had received a previous course of chemotherapy. Patient 5 had undergone a mastectomy followed by chemotherapy 2 years previously and the present course of radiation therapy was administered for a recurrent disease consisting of a solitary parasternal lymph node metastasis. Patient 8 had an additional 50 Gy of external radiation therapy to the mediastinum lymph node metasis following on from the dose administered to the primary tumor. Eight of the 15 cancer patients in our treatment group had concurrent chemotherapy. Blood samples taken from 20 healthy volunteers (13 men and seven women, mean age 45.6 ± 15.1 years) were used as controls. There were no significant age or sex differences between the cancer patients and individuals in the healthy control group (Mann–Whitney's U‐test and χ2‐test, respectively).
Table 1.
patient characteristics and treatment information
| Patient no. | Cancer location | Sex | Age (years) | Tumor stage | Pathology | Radiation total dose (Gy) | Fraction | Concurrent chemotherapy | Operation after radiation therapy |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Lung | F | 81 | T2N0M0 | Unknown | 70.2 | 39 | – | – |
| 2 | Gallbladder | M | 74 | Local advanced | Adenocarcinoma | 66 | 33 | – | – |
| 3 | Skin | M | 83 | T4N0M0 | SCC | 70 | 35 | – | – |
| 4 | Lymphoma | F | 62 | StageIE | Follicular | 40.6 | 22 | – | – |
| 5 | Breast | F | 40 | LN recurrence | Adenocarcinoma | 66 | 33 | – | – |
| 6 | Hypopharynx | F | 67 | T4N2cM0 | SCC | 70.2 | 39 | CBDCA, 5FU | Laryngectomy |
| 7 | Esophagus | M | 75 | T1N1–2M0 | SCC | 60 | 30 | – | – |
| 8 | Hypopharynx | M | 86 | T3N2cM0 | SCC | 70.2 | 39 | CBDCA, UFT | – |
| 9 | Ethmoid sinus | F | 63 | T4N0M0 | SCC | 66 | 33 | CDDP, 5FU, MTX, LV | – |
| 10 | Tongue | F | 36 | T2N2bM0 | SCC | 70.2 | 39 | CDDP, 5FU, MTX, LV | – |
| 11 | Tongue | M | 86 | T3N2bM0 | SCC | 36 | 20 | – | – |
| 12 | Larynx | M | 77 | T4N0M0 | SCC | 30.6 | 17 | CBDCA, UFT | Laryngectomy |
| 13 | Maxillar sinus | M | 58 | T4N0M0 | SCC | 66 | 33 | CDDP, 5FU, TXT | – |
| 14 | Larynx | M | 62 | T4N2bM0 | SCC | 70.2 | 39 | CDDP, 5FU, MTX, LV | – |
| 15 | Hypopharynx | M | 75 | T4N2cM0 | SCC | 70.2 | 39 | CDDP, 5FU, MTX, LV | – |
LN, lymph node; unknow, unknow pathological type; SCC, Squamous Cell Carcinoma; follicular, follicular lymphoma; RT, Radiation Therapy; CBDCA, Carboplatin; 5FU, Fluorouracil; UFT, Tegafur‐Uracil; CDDP, Cisplatin; MTX, Methotrexate; LV, Leucovorin; TXT, Paclitaxel.
DNA extraction from plasma samples. All blood samples were centrifuged at 1500 g for 10 min. Supernatants were then carefully collected from the top portion of the plasma to eliminate the possibility of contamination by cells, and stored at –80°C until further use. Patient and mouse DNA were purified from 200 µL plasma using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). Patient mRNA was extracted from 1000 µL plasma using the RNeasy Mini Kit (Qiagen). Patient cDNA was synthesized from mRNA extracts using the SuperScript™ III First‐Strand synthesis system for qRT‐PCR (Invitrogen, Carlsbad, CA, USA).
Quantification of human and mouse nucleic acids in plasma samples using real‐time polymerase chain reaction. Nucleic acid concentrations in all plasma samples were measured by quantitative polymerase chain reaction (PCR) using the ABI PRISM 7700 Sequence Detector (Applied Biosystems, Foster City, CA, USA) and TaqMan Universal PCR Master Mix (Applied Biosystems) according to the manufacturer's protocol. TaqMan Gene Expression Assay mouse β‐actin Mm00607939_s1 (Applied Biosystems) was used for amplification of mouse genomic DNA. For quantification of human β‐actin genomic DNA in mouse plasma samples, the following custom primers and probe sets were used: forward primer, 5′‐ATCCTAAAAGCCACCCCACT‐3′; reverse primer, 5′‐CTCAAGTTGGGGGACAAAAA‐3′; and probe, 5′‐FAM‐CACAGGGGAGGTGATAGCAT‐TAMURA‐3′. Similarly, the levels of human 100‐bp and 400‐bp β‐actin genomic DNA fragments in human plasma samples were analyzed using custom primers and probes. Both 100‐bp and 400‐bp human PCR fragments were amplified using the same forward primer 5′‐ATCGCTCACCGCAAATGC‐3′. Reverse primers were 5′‐CATCTTGTTTTCTGCGCAAGTT‐3′, and 5′‐AATGCTATCACCTCCCCTGTGT‐3′ for the 100‐bp and 400‐bp products, respectively. The probe used for both fragments was 5′‐FAM‐CTAGGCGGACTATGACTTAGTTGCGTTACACCC‐TAMRA‐3′. The 100‐bp primer successfully amplified products from most DNA fractions and these fragments reflected the total quantity of cell‐free DNA resulting from both apoptosis and necrosis. In contrast, the amplified 400‐bp products were representative of concentrations of longer DNA fragments released from non‐apoptotic cells. DNA integrity was calculated as the ratio of these concentrations (i.e. 400‐bp fragments/100‐bp fragments) in each assay. Human β‐actin cDNA was quantified using Pre‐Developed TaqMan Assay Reagents 4310881E (Applied Biosystems). Mitochondrial DNA was quantified using the primers and probe previously described by Chiu et al.( 20 )
Statistical analysis. All statistical calculations to determine the significance of differences between the plasma DNA levels in human and animal samples were carried out using the Mann–Whitney U‐test.
Results
Origin of cell‐free genomic DNA in the plasma of mice bearing human tumors and their control counterparts. To determine the origin of cell‐free DNA in plasma samples, tumor‐specific DNA (human‐specific DNA) and mouse‐specific DNA were analyzed separately in blood samples from mice implanted with human tumor cells. Preliminary quantitative PCR experiments showed no cross‐reaction between human‐ and mouse‐specific DNA amplification systems. We analyzed 24 plasma samples from mice bearing human tumors arising from different implanted cell lines (two of RPMI 1788, four of SQ5‐SLK, three of KM12C, seven of DLD‐1, two of SROV‐3, six of A431) and 11 plasma samples from control mice for the presence of human‐specific DNA. As shown in Figure 1(a), 16 of 24 plasma samples from tumor‐bearing mice (66.7%) showed amplification of various levels of human‐specific DNA, whereas no human‐specific DNA was detected in the samples from control mice (P < 0.001). The concentration of human‐specific DNA in the plasma varied according to the implanted tumor cell type.
Figure 1.
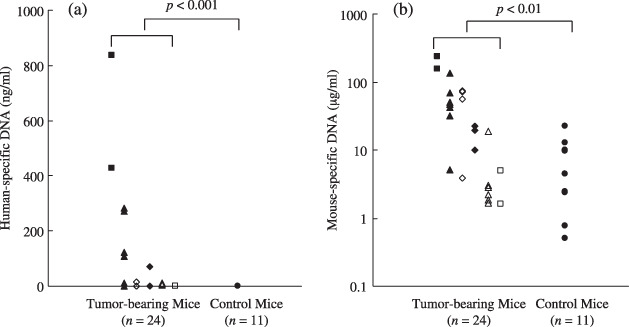
Concentrations of (a) human‐specific DNA and (b) mouse‐specific DNA in the plasma of mice bearing various types of human tumors ( , SROV‐3;
, SROV‐3;  , DLD‐1;
, DLD‐1;  , SQ5‐SLK;
, SQ5‐SLK;  , KM12C;
, KM12C;  , A431;
, A431;  , RPMI 1788) and in control mice. (a) Sixteen of 24 plasma samples from mice bearing human tumors, but none of the 11 samples from control mice, contained human‐specific genomic DNA (P < 0.001). The concentration of human‐specific DNA in plasma varied according to the implanted tumor cell type. (b) Samples from mice bearing human tumors (n = 24) contained significantly greater concentrations of cell‐free mouse DNA than samples from control mice (n = 11) (P < 0.01). When the concentration of mouse‐specific DNA of each tumor type were compared with the control group, the difference depended on the tumor type implanted.
, RPMI 1788) and in control mice. (a) Sixteen of 24 plasma samples from mice bearing human tumors, but none of the 11 samples from control mice, contained human‐specific genomic DNA (P < 0.001). The concentration of human‐specific DNA in plasma varied according to the implanted tumor cell type. (b) Samples from mice bearing human tumors (n = 24) contained significantly greater concentrations of cell‐free mouse DNA than samples from control mice (n = 11) (P < 0.01). When the concentration of mouse‐specific DNA of each tumor type were compared with the control group, the difference depended on the tumor type implanted.
Mouse‐specific cell‐free DNA was detected in plasma samples from both mice bearing human tumors and control mice. As shown in Figure 1(b), plasma samples of 24 mice bearing human tumors showed significantly higher concentrations of mouse‐specific DNA when compared with the control group (P < 0.01). When the concentrations of mouse‐specific DNA in each tumor type were compared with those of the control group, the difference was found to be dependent on the implanted cell type. ( , SROV‐3, P < 0.05;
, SROV‐3, P < 0.05;  , DLD‐1, P < 0.01;
, DLD‐1, P < 0.01;  , SQ5‐SLK, P < 0.05;
, SQ5‐SLK, P < 0.05;  , KM12C, P < 0.05;
, KM12C, P < 0.05;  , A431, P > 0.1; and
, A431, P > 0.1; and  , RPMI 1788, P > 0.1).
, RPMI 1788, P > 0.1).
Cell‐free nucleic acids in plasma samples from cancer patients and healthy controls. The concentrations of 100‐bp or 400‐bp fragments of genomic DNA, mitochondrial DNA, and mRNA were measured in plasma samples from our cancer patient group prior to radiation therapy and from healthy control subjects. The cancer patients had significantly higher plasma concentrations of the genomic DNA fragments (Fig. 2a,b), and marginal increases in plasma mitochondrial DNA (Fig. 2c) when compared with healthy control subjects. These patients also showed significantly elevated mRNA concentrations (Fig. 2d). As shown in Figure 2(e), DNA integrity (the ratio of longer DNA fragments to the total DNA, 400‐bp fragments/100‐bp fragments) was also significantly greater in plasma from the cancer patients compared with the healthy control subjects.
Figure 2.
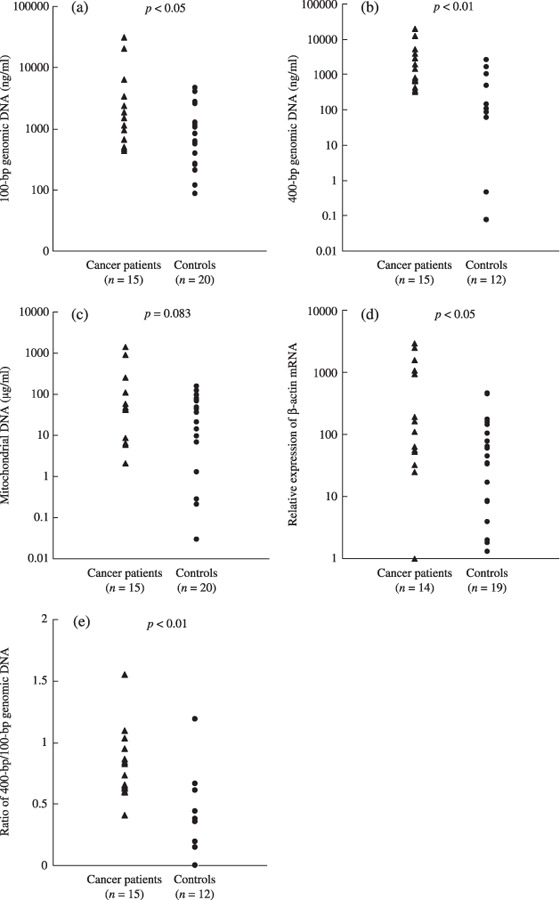
The concentrations of cell‐free (a) 100‐bp genomic DNA, (b) 400‐bp genomic DNA, (c) mitochondrial DNA, and (d) the relative expression of β‐actin mRNA in plasma samples from cancer patients (n = 15) and healthy controls (n = 20). An mRNA sample from patient 15 was not available. Control plasma samples for the measurement of 400‐bp genomic DNA and mRNA were not available from all 20 healthy subjects (n = 12, and n = 19, respectively). cDNA reverse‐transcribed from 2 µg mRNA from exponentially growing HeLa cells was used as a standard. All types of DNA except for mitochondrial DNA were significantly elevated in plasma samples from cancer patients compared with healthy controls. (a) 100 bp genomic DNA, P < 0.05; (b) 400‐bp genomic DNA, P < 0.01; (c) mitochondrial DNA, P = 0.083. (d) mRNA expression levels were higher in cancer patients than in healthy controls (P < 0.05). (e) DNA integrity (the ratio of 400‐bp fragments/100‐bp fragments) was higher in samples from cancer patients than in healthy controls (P < 0.01).
Kinetics of cell‐free DNA in plasma samples from cancer patients during radiation therapy. Serial plasma samples from cancer patients undergoing radiation therapy were analyzed for changes in their concentrations of cell‐free DNA. These nucleic acid concentrations during radiation therapy and the associated patient outcomes are presented in Table 2. Changes in the plasma DNA concentrations over time for representative patients are shown in Figure 3. The initial plasma concentrations before treatment were normalized to 100% and the data are reported in relation to this initial value. As shown in Table 2, the plasma concentrations of 100‐bp and 400‐bp cell‐free DNA fragments in 13 of 15 patients decreased during radiation therapy (indicated as ‘ ’ in Table 2, patients 1, 5 and 9 in Figure 3a). Of these 13 patients, a total of 10 and 11 individuals had a transient increase in their 100‐bp and 400‐bp DNA fragment levels, respectively, during the first or second week of therapy. These increases were followed by a steady decrease in the plasma DNA concentration by the end of the treatment period (peak + in Table 2, patients 1 and 5 in Figure 3a). The peak percentage, which is the ratio of the DNA concentration at the transient peak to that before treatment, increased from 106% up to 869% (Table 2). In the remaining patients, plasma DNA concentrations continued to increase (indicated by ‘
’ in Table 2, patients 1, 5 and 9 in Figure 3a). Of these 13 patients, a total of 10 and 11 individuals had a transient increase in their 100‐bp and 400‐bp DNA fragment levels, respectively, during the first or second week of therapy. These increases were followed by a steady decrease in the plasma DNA concentration by the end of the treatment period (peak + in Table 2, patients 1 and 5 in Figure 3a). The peak percentage, which is the ratio of the DNA concentration at the transient peak to that before treatment, increased from 106% up to 869% (Table 2). In the remaining patients, plasma DNA concentrations continued to increase (indicated by ‘ ’ in Table 2, patient 3 in Figure 3a) or showed unclassified patterns of change (‘unclassified’ in Table 2). Changes in 100‐bp and 400‐bp cell‐free plasma DNA concentrations were similar in most of the patients. Although the mitochondrial DNA concentrations also decreased (with or without a transient increase) during radiation therapy in 9 of 15 patients (Table 2, patients 12 and 2 in Figure 3b), this pattern of change was independent of changes in 100‐bp and 400‐bp DNA concentrations (Table 2; Fig. 3b).
’ in Table 2, patient 3 in Figure 3a) or showed unclassified patterns of change (‘unclassified’ in Table 2). Changes in 100‐bp and 400‐bp cell‐free plasma DNA concentrations were similar in most of the patients. Although the mitochondrial DNA concentrations also decreased (with or without a transient increase) during radiation therapy in 9 of 15 patients (Table 2, patients 12 and 2 in Figure 3b), this pattern of change was independent of changes in 100‐bp and 400‐bp DNA concentrations (Table 2; Fig. 3b).
Table 2.
Plasma DNA concentrations during radiation therapy and outcomes
| Patient no. | 100‐bp DNA | 400‐bp DNA | Mitochondrial DNA | Disease status | Days after RT | Follow up period after RT (days) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Change | Peak | Day | Peak % | Change | Peak | Day | Peak % | Change | Peak | Day | Peak % | ||||
| 1 |

|
+ | 6 | 279 |

|
+ | 15 | 249 |

|
Local | 115 | 423 | |||
| 2 |

|

|

|
Local | 180 | 365 | |||||||||
| 3 |

|

|

|
CR | 1532 | ||||||||||
| 4 |

|
+ | 2 | 378 |

|
+ | 2 | 452 | Unclassified | CR | 1465 | ||||
| 5 |

|
+ | 3 | 869 |

|
+ | 3 | 692 |

|
Dis met (lung) | 31 | 366 | |||
| 6 |

|
+ | 12 | 267 |

|
+ | 12 | 292 | Unclassified | CR | 1274 | ||||
| 7 |

|
+ | 7 | 287 |

|
+ | 7 | 424 |

|
Dis met (brain) | 74 | 134 | |||
| 8 |

|
+ | 19 | 343 |

|
+ | 8 | 126 |

|
+ | 19 | 250 | CR | 767 | |
| 9 |

|

|

|
CR | 286 | ||||||||||
| 10 |

|
+ | 6 | 106 |

|
+ | 10 | 109 |

|
CR | 1150 | ||||
| 11 |

|
+ | 7 | 132 |

|
+ | 7 | 101 | Unclassified | CR | 128 | ||||
| 12 |

|
+ | 3 | 140 |
 |

|
+ | 3 | 108 | Dis met (lung) | 321 | 712 | |||
| 13 |

|
+ | 10 | 432 |

|
+ | 10 | 240 |

|
CR | 191 | ||||
| 14 | Unclassified |

|
+ | 11 | 188 |

|
Resi, dis met (lung) | 252 | 786 | ||||||
| 15 |

|
+ | 20 | 311 |

|

|
CR | 1090 | |||||||
Plasma DNA concentration showed decreasing ( ) or increasing (
) or increasing ( ) tendency during RT period; unclassified, changing without any tendency; peak +, DNA concentration increasing with a transient peak followed by decreasing; day, day of transient peak; peak %, ratio of DNA concentration at transient peak to that before treatment; local, local recurrence; CR, complete remission; dis met, distant metastasis; resi, residual tumor; A/D, alive with disease; A, alive without disease; D, dead of disease; d, dead of other cause.
) tendency during RT period; unclassified, changing without any tendency; peak +, DNA concentration increasing with a transient peak followed by decreasing; day, day of transient peak; peak %, ratio of DNA concentration at transient peak to that before treatment; local, local recurrence; CR, complete remission; dis met, distant metastasis; resi, residual tumor; A/D, alive with disease; A, alive without disease; D, dead of disease; d, dead of other cause.
Figure 3.
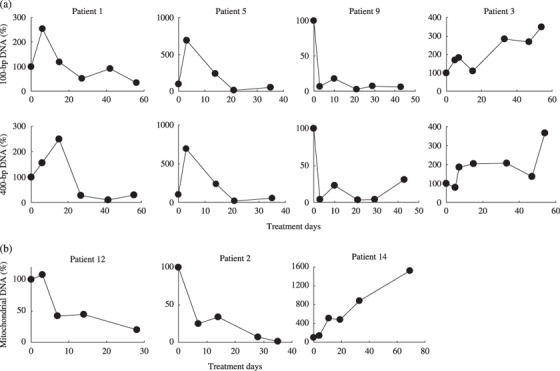
Dynamic changes with time in the plasma concentrations of (a) 100‐bp and 400‐bp genomic DNA and (b) mitochondrial DNA in samples from cancer patients undergoing radiation therapy. Plasma samples were obtained before radiation therapy (day 0), on the third day of treatment, and then every 1 or 2 weeks during the treatment period. Initial plasma concentrations prior to treatment were normalized to 100% and the data are reported in relation to this initial value. (a) Top panels, 100‐bp DNA; bottom panels, 400‐bp DNA. (b) Mitochondrial DNA.
The follow‐ups indicated that 3 of 15 patients had developed a local recurrent disease and four patients developed distant metastases after treatment (Table 2). There was no obvious association found between patient outcome and either the concentrations of the DNA before treatment or the pattern of change during the course of treatment.
Discussion
In our present study, we first confirmed that plasma samples from mice bearing human tumors contained human‐specific DNA that must have originated from implanted human tumor cells. More importantly, we found that plasma from tumor‐bearing mice had higher concentrations of mouse‐specific DNA than plasma from control mice. These results suggest that the increased plasma DNA concentrations observed in cancer patients may consist of DNA originating from both tumor and non‐tumor cells. The mechanism of release of DNA from non‐tumor cells into the plasma remains unclear. Increasing concentrations of cell‐free DNA have been reported in disorders other than cancer that are associated with tissue and cell injury, including systemic lupus erythematodes, rheumatoid arthritis, pulmonary or brain embolism, and myocardial infarction.( 3 , 5 ) Therefore, non‐tumor, cell‐free DNA in the plasma of patients with cancer may be associated with increased tissue damage or the disintegration of non‐tumor interstitial cells inside tumor tissue. Localized tumor growth and invasion may also damage cells in adjacent normal tissue, resulting in the release of cell‐free DNA into the circulation.
We also report that the plasma concentrations of cell‐free genomic DNA were higher in patients with cancer than in healthy control subjects, consistent with previous reports.( 2 , 3 , 17 , 18 ) In addition, serial analyses of plasma DNA concentrations in samples from patients with cancer undergoing radiation therapy showed dynamic changes over time associated with the progression of this treatment. In the majority of patients, we observed a transient rise in the plasma DNA concentrations during the first or second week of treatment, followed by a trend toward a decrease through the remaining treatment period.
There are few reports of changes in the cell‐free plasma DNA concentrations in response to radiation therapy or chemotherapy.( 3 ) In one study by Lo et al. serial analyses of plasma Epstein–Barr virus DNA levels in patients with nasopharyngeal cancer showed an initial rise in these concentrations, followed by a rapid decrease in most cases during the first week of therapy.( 21 ) According to the findings of Kamat et al. and Rago et al. in mice bearing human tumors, a marked, transient rise occurs in the plasma concentrations of cell‐free human‐specific DNA (tumor‐specific DNA) immediately after chemotherapy or surgery, followed by a rapid decrease.( 22 , 23 ) These observations indicated that the dynamics of tumor‐specific DNA in plasma may reflect tumor burden, that is, the treatments may have caused rapid cell death, resulting in the release of a bolus of tumor cell DNA into the circulation, which decreased with tumor regression. Hence, such analyses may have a potential role in monitoring the efficacy of cancer therapies during the early phases of these treatments. In our present study, we analyzed the total cell‐free plasma DNA, which lacks tumor specificity. However, we found that the total amount of cell‐free DNA showed dynamic changes during radiation therapy with a transient rise followed by a rapid decrease. These observations are similar to those reported for tumor‐specific DNA by Lo et al.( 21 ) or Kamat et al.( 22 ) mentioned above. Furthermore, it is possible that a decrease in cell‐free plasma DNA during radiation is due to not only the decrease of tumor cells themselves but also a reduction of the whole tumor lesion. It is a point of some interest as to whether the pattern of change in plasma DNA concentrations during the early phase of radiation therapy correlates with treatment efficacy. To clarify this point, future clinical studies of larger patient populations will be very valuable.
There have been some analyses of the tumor‐specific plasma DNA in cancer patients, such as the presence of mutated or hypermethylated genes or gene microinstability, that are associated with the tumor itself.( 7 ) Tumor‐specific DNA originates from tumor cells that have degenerated either spontaneously or in response to therapeutic intervention, whereas total DNA originates from both tumor and non‐tumor tissue. Hence, tumor‐specific DNA may be ideal for the diagnosis or monitoring of tumors. However, there are some noteworthy limitations to the clinical applications of tumor‐specific DNA. First, tumor‐specific genetic alterations vary among patients, with the exception of tumor‐specific viral DNA such as Epstein–Barr virus. To identify tumor‐specific plasma DNA in a cancer patient, genetic analysis of the primary tumor is first required.( 12 ) In addition, the amount of tumor‐specific DNA in plasma is reported to vary between patients and according to cancer type, with the proportion ranging between less than 1% and greater than 90% of the total cell‐free plasma DNA.( 4 , 18 , 24 ) We also found that the amount of human‐specific DNA (tumor‐specific DNA) in the plasma of tumor‐bearing mice varied among tumor types. Analyses of tumor‐specific plasma DNA would be challenging and less sensitive when these amounts are at very low levels, thus limiting their clinical utility. However, development of new techniques could negate some of these disadvantages of assaying tumor‐specific DNA. For example, Diehl et al. recently described a highly sensitive assay to quantify circulating tumor‐specific mutated DNA in plasma.( 25 ) These authors have found that these DNA molecules closely reflect the total systemic tumor burden even in the presence of microscopic lesion recurrence. Developments that will enable the practical use of this assay for a large number of patients would be of great interest.
We find from our analyses that the DNA integrity (i.e. the ratio of longer DNA fragments to total DNA in plasma samples) was significantly higher in patients with cancer than in the healthy controls, consistent with previously published data.( 6 , 7 , 8 , 9 , 10 , 11 , 12 , 13 ) If the tumor volume decreases in response to a therapeutic intervention, the DNA integrity in the plasma may also decrease because longer DNA in the plasma is considered to mainly originate from tumor cells. In this regard, we have found that the DNA integrity, as in the case of the absolute DNA amount, showed a tendency to decrease until the end of treatment in 10 of 15 patients (data not shown). Chan et al. have also reported previously that 70% of nasopharyngeal cancer patients showed a reduction in plasma DNA integrity after radiotherapy.( 6 ) Furthermore these authors have shown that a persistent (i.e. no reduction) high plasma DNA integrity following treatment could be associated with a lower disease‐free survival for these patients. We thus speculated whether the measurement of plasma DNA integrity would serve as a useful marker of malignant disease. In addition, a detailed assessment of different fragment lengths of cell‐free plasma DNA may provide more precise information on the biological mechanisms underlying tumor cell death in response to anticancer treatments.
There are few published reports on the monitoring of cell‐free mitochondrial DNA and mRNA in the plasma of patients with cancer. We found in our present study that concentrations of cell‐free mitochondrial plasma DNA and mRNA in patients with cancer were generally higher than in healthy control subjects, consistent with previous reports.( 14 , 15 ) We also found that changes in mitochondrial DNA concentrations showed a pattern independent of 100‐bp and 400‐bp genomic DNA concentrations during radiation therapy. These results suggest that mitochondrial DNA in plasma is influenced by factors other than genomic DNA. The importance of cell‐free plasma mitochondrial DNA and mRNA in patients with cancer requires further investigation.
In summary, we herein report that the cell‐free plasma DNA levels are elevated in cancer patients and show dynamic changes associated with progressive radiation therapy. Serial plasma DNA analyses during treatment could thus be a potentially useful tool for monitoring the tumor response to radiation therapy. Additional prospective studies including larger groups of patients and longer follow‐up periods are required in order to fully evaluate the clinical usefulness of this assay for estimating the radiation sensitivity of an individual tumor.
Acknowledgments
This research was supported by the Ministry of Education, Science, Sports, and Culture, Grant‐in‐Aid for Young Scientists (B) 15790678 (2003–04), and by a grant under the 2003 Strategic Research Project of Yokohama City University, Japan. We thank Ms M. Nagano at Applied Biosystems Japan for advice on some of the experimental aspects of this study and the support of the medical staff at the Radiation Oncology Department of Yokohama City University Hospital.
References
- 1. Anker P, Mulcahy H, Chen XQ, Stroun M. Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer Metastasis Rev 1999; 18: 65–73. [DOI] [PubMed] [Google Scholar]
- 2. Sozzi G, Conte D, Leon M et al . Quantification of free circulating DNA as a diagnostic marker in lung cancer. J Clin Oncol 2003; 21: 3902–8. [DOI] [PubMed] [Google Scholar]
- 3. Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 1977; 37: 646–50. [PubMed] [Google Scholar]
- 4. Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer – a survey. Biochim Biophys Acta 2007; 1775: 181–232. [DOI] [PubMed] [Google Scholar]
- 5. Fournie GJ, Courtin JP, Laval F et al . Plasma DNA as a marker of cancerous cell death. Investigations in patients suffering from lung cancer and in nude mice bearing human tumours. Cancer Lett 1995; 91: 221–7. [DOI] [PubMed] [Google Scholar]
- 6. Chan KC, Leung SF, Yeung SW, Chan AT, Lo YM. Persistent aberrations in circulating DNA integrity after radiotherapy are associated with poor prognosis in nasopharyngeal carcinoma patients. Clin Cancer Res 2008; 14: 4141–5. [DOI] [PubMed] [Google Scholar]
- 7. Deligezer U, Eralp Y, Akisik EE et al . Size distribution of circulating cell‐free DNA in sera of breast cancer patients in the course of adjuvant chemotherapy. Clin Chem Laboratory Med 2008; 46: 311–17. [DOI] [PubMed] [Google Scholar]
- 8. Suzuki N, Kamataki A, Yamaki J, Homma Y. Characterization of circulating DNA in healthy human plasma. Clin Chim Acta 2008; 387: 55–8. [DOI] [PubMed] [Google Scholar]
- 9. Wang BG, Huang HY, Chen YC et al . Increased plasma DNA integrity in cancer patients. Cancer Res 2003; 63: 3966–8. [PubMed] [Google Scholar]
- 10. Umetani N, Giuliano AE, Hiramatsu SH et al . Prediction of breast tumor progression by integrity of free circulating DNA in serum. J Clin Oncol 2006; 24: 4270–6. [DOI] [PubMed] [Google Scholar]
- 11. Jiang WW, Zahurak M, Goldenberg D et al . Increased plasma DNA integrity index in head and neck cancer patients. Int J Cancer 2006; 119: 2673–6. [DOI] [PubMed] [Google Scholar]
- 12. Sai S, Ichikawa D, Tomita H et al . Quantification of plasma cell‐free DNA in patients with gastric cancer. Anticancer Res 2007; 27: 2747–51. [PubMed] [Google Scholar]
- 13. Tomita H, Ichikawa D, Ikoma D et al . Quantification of circulating plasma DNA fragments as tumor markers in patients with esophageal cancer. Anticancer Res 2007; 27: 2737–41. [PubMed] [Google Scholar]
- 14. Ng EK, Tsui NB, Lam NY et al . Presence of filterable and nonfilterable mRNA in the plasma of cancer patients and healthy individuals. Clin Chem 2002; 48: 1212–17. [PubMed] [Google Scholar]
- 15. Mehra N, Penning M, Maas J, Van Daal N, Giles RH, Voest EE. Circulating mitochondrial nucleic acids have prognostic value for survival in patients with advanced prostate cancer. Clin Cancer Res 2007; 13: 421–6. [DOI] [PubMed] [Google Scholar]
- 16. Gautschi O, Bigosch C, Huegli B et al . Circulating deoxyribonucleic acid as a prognostic marker in non‐small‐cell lung cancer patients undergoing chemotherapy. J Clin Oncol 2004; 22: 4157–64. [DOI] [PubMed] [Google Scholar]
- 17. Pathak AK, Bhutani M, Kumar S, Mohan A, Guleria R. Circulating cell‐free DNA in plasma/serum of lung cancer patients as a potential screening and prognostic tool. Clin Chem 2006; 52: 1833–42. [DOI] [PubMed] [Google Scholar]
- 18. Gormally E, Caboux E, Vineis P, Hainaut P. Circulating free DNA in plasma or serum as biomarker of carcinogenesis: practical aspects and biological significance. Mutat Res 2007; 635: 105–17. [DOI] [PubMed] [Google Scholar]
- 19. Hara T, Omura‐Minamisawa M, Chao C, Nakagami Y, Ito M, Inoue T. Bcl‐2 inhibitors potentiate the cytotoxic effects of radiation in Bcl‐2 overexpressing radioresistant tumor cells. Int J Radiat Oncol Biol Phys 2005; 61: 517–28. [DOI] [PubMed] [Google Scholar]
- 20. Chiu RW, Chan LY, Lam NY et al . Quantitative analysis of circulating mitochondrial DNA in plasma. Clin Chem 2003; 49: 719–26. [DOI] [PubMed] [Google Scholar]
- 21. Lo YM, Leung SF, Chan LY et al . Kinetics of plasma Epstein–Barr virus DNA during radiation therapy for nasopharyngeal carcinoma. Cancer Res 2000; 60: 2351–5. [PubMed] [Google Scholar]
- 22. Kamat AA, Bischoff FZ, Dang D et al . Circulating cell‐free DNA. A novel biomarker for response to therapy in ovarian carcinoma. Cancer Biol Ther 2006; 5: 1369–74. [DOI] [PubMed] [Google Scholar]
- 23. Rago C, Huso DL, Diehl F et al . Serial assessment of human tumor burdens in mice by the analysis of circulating DNA. Cancer Res 2007; 67: 9364–70. [DOI] [PubMed] [Google Scholar]
- 24. Jahr S, Hentze H, Englisch S et al . DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001; 61: 1659–65. [PubMed] [Google Scholar]
- 25. Diehl F, Schmidt K, Choti MA et al . Circulating mutant DNA to assess tumor dynamics. Nat Med 2008; 14: 985–90. [DOI] [PMC free article] [PubMed] [Google Scholar]


