Abstract
The existence of a compensatory mechanism in response to cell wall damage has been proposed in yeast cells. The increase of chitin accumulation is part of this response. In order to study the mechanism of the stress-related chitin synthesis, we tested chitin synthase I (CSI), CSII, and CSIII in vitro activities in the cell-wall-defective mutant gas1Δ. CSI activity increased twofold with respect to the control, a finding in agreement with an increase in the expression of the CHS1 gene. However, deletion of the CHS1 gene did not affect the phenotype of the gas1Δ mutant and only slightly reduced the chitin content. Interestingly, in chs1 gas1 double mutants the lysed-bud phenotype, typical of chs1 null mutant, was suppressed, although in gas1 cells there was no reduction in chitinase activity. CHS3 expression was not affected in the gas1 mutant. Deletion of the CHS3 gene severely compromised the phenotype of gas1 cells, despite the fact that CSIII activity, assayed in membrane fractions, did not change. Furthermore, in chs3 gas1 cells the chitin level was about 10% that of gas1 cells. Thus, CSIII is the enzyme responsible for the hyperaccumulation of chitin in response to cell wall stress. However, the level of enzyme or the in vitro CSIII activity does not change. This result suggests that an interaction with a regulatory molecule or a posttranslational modification, which is not preserved during membrane fractionation, could be essential in vivo for the stress-induced synthesis of chitin.
Yeast cells are surrounded by a matrix composed of β(1,3)/(1,6)-glucans and mannoproteins as major components and chitin as a minor one (21). Chitin constitutes only 1 to 2% of the cell wall dry weight, but it plays a key role in yeast morphogenesis and is essential for the viability of yeast and fungal cells. During vegetative growth chitin is deposited at the site of bud emergence, forms a ring that surrounds the neck between the mother and daughter cells, and constitutes the primary septum. On the surface of mother cells a chitin ring is still recognizable after cell division, the so-called bud scar, and in the corresponding site on the daughter surface a birth scar is present. A tiny amount of chitin is also layered over the whole of the lateral cell wall, and this occurs in the mother cell.
Three chitin synthase (CS) activities, CSI, CSII, and CSIII, are responsible for the deposition of cell wall chitin. The three isoenzymes differ in certain properties, such as the optimum pH, metal specificity, and susceptibility to inhibitors (6). CSI and CSII activities are determined only by the product of CHS1 and CHS2 genes, respectively, which encode the polypeptides containing the catalytic domain of each chitin synthases. Chs1p is responsible for the synthesis of chitin after cell separation. It plays a repair function, since it counterbalances the acid-induced increase in the chitinase activity that hydrolyzes the chitin present in the primary septum at the end of cytokinesis (3–5, 17, 18). CSI represents about 90% of the in vitro measurable chitin synthase activity, but its contribution to the production of chitin in vivo is negligible. Chs2p is responsible for deposition of the primary septum and is thus necessary for cell division (33, 34). CSIII activity is responsible for the deposition of chitin in the ring and lateral cell walls and contributes to the synthesis of most cell wall chitin during vegetative growth (33). During cell cycle progression the Chs3p level remains constant (10, 38), but its localization changes (10, 31). A complex regulation of synthesis and transport determines the spatial and temporal control of chitin deposition by Chs3p, the catalytic component. The CHS4 to CHS7 genes are involved in this regulation (19, 32, 35, 36, 39). Additionally, CS activities exhibit in vitro zymogenic properties, suggesting that they are regulated at a posttranslational level (6, 8, 9).
In the present study we investigated the increase in chitin accumulation which appears to be part of the responses that a yeast cell activates to counteract cell wall damage. The fks1Δ mutant, which lost a subunit of the β(1,3)-glucan synthase, has a reduced level of β(1,3)-glucan and exhibits an induction of chitin accumulation (15, 28). A similar response is present in gas1 cells which lack a β(1,3)-glucosyltransferase activity (20) that is important for the correct incorporation of glucan and mannoproteins (see references 15, 24, 28, and 27 for a review). Moreover, the loss of Fks1p or Gas1p induces also the expression of Fks2p, the alternative subunit of the β(1,3)-glucan synthase, a 20-fold increase in the cross-links between cell wall mannoproteins and chitin, and a 3-fold increase in CWP1 expression (15, 24, 28). The increase in chitin deposition could be included in a compensation mechanism that mutants defective in cell wall synthesis or assembly activate to maintain cell integrity. In our study we focused on the role of CS activities in the possible mechanism of increased chitin synthesis in the gas1Δ mutant.
MATERIALS AND METHODS
Strains, growth conditions, and genetic methods.
The yeast strains used here are listed in Table 1. Standard techniques were used for diploid construction, sporulation, and tetrad dissection. Cells were grown in batches at 30°C in YNB-glucose (Difco yeast nitrogen base without amino acids at 6.7 g/liter, 2% glucose, and the required supplements) or in YEPD (1% yeast extract, 2% Bacto-Peptone, 2% dextrose). For solid media, 2% agar was added. Diploids were sporulated in New Sporulation Medium (8.2 g of sodium acetate, 1.9 g of KCl, 0.35 g of MgSO4, 1.2 g of NaCl, and 15 g of agar per liter) at 24°C. Spore germination was carried out at 24°C on YEPDAT plates (YEPD, 2% agar, and 100 mg of adenine and 50 mg of tryptophan per liter) containing 0.5 M KCl.
TABLE 1.
S. cerevisiae strains
| Strain | Genotype | Source or reference |
|---|---|---|
| W303-1A | MATaade 2-1 his3-11,15 trp1-1 ura3-1 leu2-3,112 can1-100 | P. P. Slominski |
| W303-1B | MATα ade2-1 his3-11,15 trp1-1 ura3-1 leu2-3,112 can1-100 | P. P. Slominski |
| WB2d | MATα gas1::LEU2; as W303-1B | 37 |
| WAH | MATagas1::HIS3; as W303-1A | This study |
| W303-chs3Δ | MATα cal1 (chs3)::LEU2; as W303-1B | A. Duran |
| W303-chs1Δ | MATachs1::URA3; as W303-1A | This study |
| LF3 | MATa/α CHS3/chs3::LEU2 GAS1/gas1::HIS3; ade2-1/ade2-1 his3-11,15/his3-11,15 trp1-1/trp1-1 ura3-1/ura3-1 leu2,3-112/leu2,3-112 can1-100/can1-100 | This study |
| LF4 | MATa/α CHS1/chs1::URA3 GAS1/gas1::LEU2; ade2-1/ade2-1 his3-11,15/his3-11,15 trp1-1/trp1-1 ura3-1/ura3-1 leu2,3-112/leu2,3-112 can1-100/can1-100 | This study |
| Y604 | MATaura3-52 lys2-801ade2-101 trp1-901 his3-Δ200 | 31 |
| Y1306 | MATaCHS3::3XHA; ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 | 31 |
| Y1306ΔG | MATaCHS3::3XHA gas1::HIS3; as Y1306 | This study |
| LF5 | MATa/α chs1::URA3 chs3::LEU2 gas1::HIS3; ade2-1/ade2-1 his3-11,15/his3-11,15 trp1-1/trp1-1 ura3-1/ura3-1 leu2,3-112 leu2,3-112 can1-100/can1-100 | This study |
The following Escherichia coli strains were used: JM101 [Δ (lac-proAB) thi strA supE endA sbcB hsdR (F′ traD36 proAB laqIq lacZΔM15)], DH5α [F′ endA1 hsdR17(rk−mk−) supE44 thi-1 recA1 gyrANaIr) relA1 Δ(lacZYA-orgF)U169 deoR(φ80 dlacΔ(lacZ)M15)], and TOP10 [F′ mcrA Δ(mrr hsdRMS mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 deoR araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG] (Invitrogen, Carlsbad, Calif.).
DNA manipulations.
Recombinant DNA manipulations were performed using standard techniques (30). Transformation of yeast cells was carried out using the lithium acetate procedure (14) or the Saccharomyces cerevisiae EasyComp Transformation Kit (Invitrogen).
Plasmid and strain construction.
The S. cerevisiae haploid strain WB2d was generated from the wild-type strain W303-1B by gene replacement (37). In order to obtain the gas1::HIS3 mutation we used PCR to synthesize a linear fragment which lacked almost the whole of the GAS1 open reading frame (ORF). The upstream primer (5′-TGC GGA CGA TGT TCC AGC GAT TGA AGT TGT TGG TAA TAA GGT CCT GTT CCC TAG CAT GTA-) was designed to join 40 bp corresponding to the region from nucleotides 63 to 102 of the GAS1 ORF with the 5′ end from nucleotides 66 to 83 of HIS3. The downstream primer (5′-AGA CTT GGA AGA AGA CCC CGA AGC GTT AGA AGA GGC AGT ACT TGC CAC CTA TCA CCA CCA-) was designed to join 40 bp corresponding to the region from nucleotides 1470 to 1509 of the GAS1 ORF with the 3′ end from nucleotides 1446 to 1426 of HIS3. These primers were used to amplify a ∼1.2-kbp fragment from YEp6. This PCR product was transformed into W303-1A and Y1306. His+ transformants were selected, and correct substitution was tested by PCR analysis. Immunoblot analysis further confirmed the absence of the GAS1 gene product.
To construct W303-chs1Δ and WAH-chs1Δ, the BamHI/BglII fragment (∼2.4 kbp) of pHV149 (32), which carries the chs1::URA3 allele, was used to transform strains W303-1A and WAH. Correct substitution at the CHS1 locus was verified by PCR analysis and the absence of CSI activity.
CHS1 and CHS2 radiolabeled RNA probes were obtained using the TOPO TA Cloning Kit Dual Promoter (Invitrogen). Plasmids pCHS1 and pCHS2 were constructed by inserting the following PCR fragments, covering the ORF regions, into the pCRII-TOPO vector: a 1.3-kbp fragment from pMS1 (3) for CHS1 (from nucleotides 531 to 1821 from ATG), and a 1.5-kbp fragment from pUC19-CHS2 (M. H. Valdivieso, unpublished data) for CHS2 (nucleotides 787 to 2253 from ATG). ACT1 and CHS3 probes were obtained from pACT and pCAL1 plasmids. pACT was obtained by cloning the 1.5-kbp HindIII-BamHI fragment of the ACT1 gene into the HindIII-and BamHI-digested pGEM-3Zf(+). Plasmid pCAL1 was constructed by inserting the BglII-HindIII fragment of ca. 0.9 kbp of the CHS3 gene into the pGEM-Blue plasmid cut with BamHI and HindIII.
RNA extraction and Northern analysis.
Total RNA was prepared according to the method of selective precipitation with LiCl (12). Northern analysis was performed as previously described, using nonradioactive or 32P-radiolabeled single-stranded RNA probes generated by in vitro transcription (25). After hybridization at 50°C, blots were twice washed at 50°C in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 15 min, once in 1× SSC–0.1% sodium dodecyl sulfate (SDS) for 30 min, and twice in 0.1× SSC–0.1% SDS for 30 min. Two final washes were carried out at 68°C in 0.2× SSC–0.1% SDS for 15 min and 0.2× SSC for 2 min. Densitometric quantification of mRNA was performed by using a computer program. mRNA loading was normalized using the hybridization signal of ACT1.
Preparation of total extracts and membrane fractionation.
For total extracts, 2 × 108 cells were collected by filtration, washed, and resuspended in ice-cold deionized water supplemented with a Protease Inhibitor Cocktail (Boehringer Mannheim), one capsule in 25 ml, and 1 mM phenylmethylsulfonyl fluoride. After a 2-min centrifugation at 4°C, the pellets were frozen in dry ice-acetone and stored at −80°C. After thawing, 400 μl of SB-minus buffer (0.0625 M Tris-HCl pH 6.8; 5% SDS), supplemented with the protease inhibitors, was added to each pellet. After the addition of glass beads, cells were broken by shaking on a vortex for four cycles of 1 min alternated with 1 min in ice. After 5 min of centrifugation, the clarified lysate was withdrawn, quickly frozen, and stored at −80°C until use. For the determination of protein concentration, 15-μl aliquots of lysates in duplicate and the DC Protein Assay (Bio-Rad) were used. For SDS-polyacrylamide gel electrophoresis (PAGE) analysis, appropriate amounts of a concentrated solution were added to the lysate in order to bring the samples to a final concentration of 10% glycerol, 5% β-mercaptoethanol, and 0.002% bromophenol blue (BFB). Before the loading, samples were denatured at 100°C for 2 min.
Membranes were prepared as described by Orlean (22), except that the protease inhibitors were present during the whole procedure. Then, 2 × 109 cells were collected by centrifugation, washed once with cold distilled water and then again with TM buffer (50 mM Tris-HCl, pH 7.5; 2.5 mM MgCl2), and finally resuspended in 1.5 ml of TM buffer. After mechanical breakage, a pooled cell-wall-free extract was obtained as described above and then centrifuged at 60,000 × g for 45 min at 4°C, yielding supernatant (S) and pellet (P) fractions. The P fraction was resuspended in 200 to 400 μl of SB-minus buffer containing protease inhibitors, whereas the S fraction was concentrated with Centricon-10. After we determined the protein concentrations, aliquots of the P fraction were supplemented with the appropriate amounts of glycerol, β-mercaptoethanol, and BFB, whereas an equal volume of double-strength SDS sample buffer (0.0625 M Tris-HCl, pH 6.8; 2.3% SDS; 5% β-mercaptoethanol; 10% glycerol) was added to aliquots of the S fraction. Samples were denatured at 100°C for 2 min.
Electrophoresis and immunoblotting.
Proteins were resolved by SDS-PAGE on 7 or 8% polyacrylamide slab gels. Immunodecoration was carried out as previously described (26). Mouse anti-hemagglutinin (HA) monoclonal HA.11 antibodies (Babco) were used at a 1:1,000 dilution in TBS (0.01 M Tris–0.9% NaCl, pH 7.4) containing 5% bovine serum albumin (BSA) and 0.5% Tween 20 and anti-Gas1p rabbit polyclonal antibodies at a 1:3,000 dilution in TBS, 5% BSA, and 0.1% Tween 20. Horseradish peroxidase-conjugated anti-mouse antibodies (Amersham) or anti-rabbit antibodies (Zymed) were diluted to 1:5,000 and 1:10,000, respectively, in TBS–5% BSA–0.2% Tween 20. Binding was visualized with the ECL Western Blotting Detection Reagent (Amersham) according to the manufacturer's instructions.
Measurement of chitin levels.
Pellets corresponding to 5 × 109 cells were collected, resuspended in 4.5 ml of H2O, and divided into three equal aliquots (one was used for the determination of dry weight, and the other two were centrifuged), and the pellets stored at −20°C until use. After three extractions with 3% NaOH at 75°C, the alkali-insoluble pellet was neutralized and treated for 16 h with 4 mg of Zymolyase 100T per ml at 37°C. The chitin present in the indigestible material of the alkali-insoluble fraction was measured as described previously (24). The micrograms of glucosamine were normalized to the milligrams of dry weight.
Measurement of CS and chitinase activities.
For CS activity measurements, cell extracts were obtained according to the protocol described previously (2). CS assays were performed in Tris (pH 7.5) in the presence of Mg2+, Co2+, or Co2+ plus Ni2+ in order to discriminate among the three different activities, as described earlier (7). For CSI activity, MES buffer at pH 6.3 was also used (7). The determination of CS activity in cells permeabilized with digitonin was accomplished as described previously (13), except that CSIII activity was measured in the presence of 50 mM Tris (pH 7.5), Co2+, and Ni2+. Chitinase activity assays were performed as described previously (17).
Microscopic techniques.
Chitin was visualized by fluorescence microscopy after being stained with 2 mg of Calcofluor White (CF) per ml (24).
RESULTS
CSI activity and CHS1 mRNA increases are not responsible for the hyperaccumulation of chitin in gas1Δ cells.
CS activities were measured in the wild-type (W303-1B) and gas1Δ (WB2d) strains in the presence or absence of trypsin. The results (Table 2) reveal that in the absence of trypsin, the CSI activity doubles in the gas1Δ mutant with respect to the isogenic strain and that trypsin treatment elicits a six- to sevenfold increase in CSI activity in both strains, a finding in agreement with the zymogenic properties of this enzyme. On the contrary, the CSII and CSIII activities were similar in both strains.
TABLE 2.
CS activities in the gas1 null mutant and its isogenic strain
| Strain | CS activity (mU/mg of protein)a
|
|||||
|---|---|---|---|---|---|---|
| CSI
|
CSII
|
CSIII
|
||||
| − trypsin | + trypsin | − trypsin | + trypsin | − trypsin | + trypsin | |
| W303-1B | 38.5 ± 7.9 | 266.5 ± 26.5 | 2.7 ± 0.1 | 6.3 ± 0.3 | 8.7 ± 2 | 14.2 ± 1.7 |
| WB2d | 75.2 ± 2.7 | 428.0 ± 37 | 2.9 ± 0.1 | 6.1 ± 0.5 | 7.7 ± 0.8 | 14.8 ± 1.8 |
Assays were performed on crude membrane preparations in the presence of 50 mM Tris (pH 7.5) with (+) or without (−) trypsin.
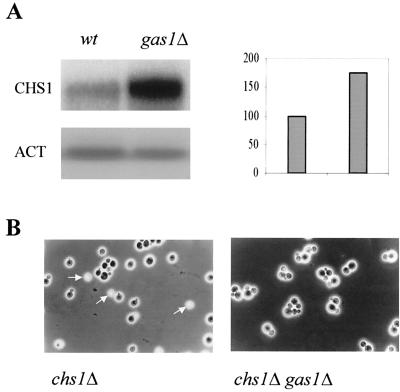
We then analyzed the expression of the CHS1, -2, and -3 genes in the gas1Δ mutant. The level of CHS1 mRNA undergoes an increase of about 70% in gas1Δ cells (Fig. 1A), whereas the expression of the CHS3 and CHS2 genes was unchanged (data not shown). The increase in the CHS1 mRNA level is in good agreement with the increase of CSI activity in the gas1Δ mutant.
FIG. 1.
(A) Northern blot analysis of CHS1 expression in gas1Δ cells. Total RNA was extracted from the gas1Δ mutant (WB2d) and its isogenic strain (W303-1B) when cells in YNB−glucose at 30°C reached a density of about 8 × 106 to 107 cells/ml. Ca. 8 μg of RNA was loaded into each lane. Hybridization was performed with 32P-labeled CHS1 or ACT1 antisense RNA probes. The autoradiograms were quantitated by densitometry. (B) Suppression of the lysis bud phenotype on the chs1Δ gas1Δ mutant. Cells were examined by phase-contrast microscopy. The arrows indicate the refractile cells.
In order to determine whether the induction of CSI activity is involved in the increase in cell wall chitin levels in the gas1Δ mutant, we analyzed the tetrads obtained after sporulation of the chs1Δ gas1Δ heterozygous diploid LF4 (see Table 1). The spores obtained after dissection of 19 asci germinated normally, and all of them were viable. Two tetratype tetrads were analyzed in greater depth. No additional detrimental effect on the growth rate or morphological modification of gas1Δ cells were brought about by the chs1Δ mutation. Staining with CF revealed that the surface fluorescence of gas1Δ cells was very intense and was not changed by introduction of the chs1 null mutation (data not shown).
The chitin present in the zymolyase-undigestible material of the alkali-insoluble fraction was measured. The amounts of chitin were 3.2 ± 0.5, 2.8 ± 0.15, 35 ± 5.8, and 28 ± 4.7 μg of glucosamine/mg (dry weight) of cells for the wild-type, chs1Δ, gas1Δ, and gas1Δ chs1Δ spores, respectively. In the gas1Δ chs1Δ mutant the chitin level was about 20% lower than in gas1Δ, but it was still 10-fold higher than in chs1Δ.
This result and the genetic analysis indicate that despite the increase in the in vitro CSI activity, Chs1p is not the major enzyme responsible for in vivo chitin synthesis in the gas1 null mutant.
The gas1 null mutation suppresses the lysed-bud phenotype of chs1 null mutants.
When chs1Δ cells are grown in unbuffered minimal medium, numerous small refractile buds can be observed by phase-contrast microscopy (3, 4). The refractile cells have been shown to be lysed cells because of the lack of the Chs1p repair function, which does not counterbalance the acid-induced chitinase activity after cell separation (4, 5). Surprisingly, in the chs1Δ gas1Δ double mutant this phenotype could not be observed (Fig. 1B).
We wondered whether the suppression in the lysed-bud phenotype of the chs1 gas1 null mutant might be a consequence of a decrease in chitinase activity induced by the gas1Δ mutation. Thus, we measured chitinase activity in the spores of a tetrad. Unexpectedly, the secreted chitinase activity was stimulated in the presence of the gas1Δ mutation, with activities of 0.12 ± 0.01 nmol/107 cells/min for the wild-type spores and 0.18 ± 0.01, 0.4 ± 0.15, and 0.32 ± 0.12 nmol/107 cells/min for the chs1Δ, gas1Δ, and chs1Δ gas1Δ spores, respectively. Thus, suppression of the lysis-bud phenotype cannot be due to a decrease in chitinase activity. Moreover, this result strongly points to the notion that the increase in chitin accumulation in gas1Δ cells must be due to an increase of the synthesis of this polymer and not to a reduction in its degradation.
Chs3p is responsible for the increase in cell wall chitin levels in the gas1 null mutant.
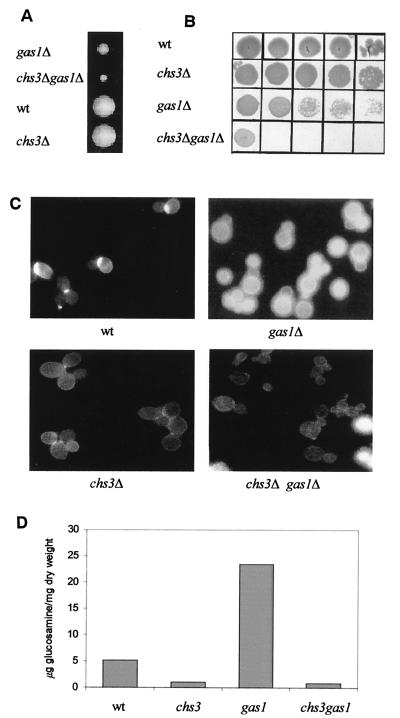
We checked whether deletion of the CHS3 gene might have any effect on the chitin synthesis process in the gas1Δ mutant. A modification of the CHS3 locus by plasmid targeting had provided the first indication that the phenotype of gas1Δ cells is severely affected in the double mutant (24). In order to avoid any residual activity of Chs3p, we used a construct in which a large portion of the CHS3 gene had been replaced by the LEU2 gene. A heterozygous chs3Δ gas1Δ diploid (LF3) was sporulated, dissection of 20 asci was carried out, and 76 spores were examined. Growth was scored at different times during incubation of the spores at 24°C. Most of the spores gave rise to visible colonies after 48 to 72 h, whereas microcolonies appeared after a further 72 h of incubation (Fig. 2A). The scoring of the phenotype revealed that all of the microcolonies belonged to the His+ Leu+ class and were therefore chs3Δ gas1Δ double mutants. Inoculation of chs3Δ gas1Δ spores in liquid YNB-glucose medium had detrimental consequences. The double-mutant cells appeared to be greatly damaged, exhibited an aberrant morphology with many irregularly shaped cells, and were unable to grow. After a prolonged incubation of 4 to 5 days, the double-mutant spores started to grow weakly and in stationary phase rapidly lost viability compared to the wild-type or chs3Δ spores (Fig. 2B). After CF staining of chs3Δ gas1Δ cells only a faint fluorescence was detectable where the primary septum is produced (Fig. 2C). This finding is consistent with the specific loss of Chs3p function. Moreover, many cells were dead and became permeable to the dye (data not shown). The double-mutant cells progressively adapted to growth in liquid medium, and the growth rate defect was gradually suppressed, although not completely. This adaptation did not occur through restoration of the cell wall chitin and is probably due to the selection of second-site suppressors.
FIG. 2.
Semilethal effects of the double inactivation of CHS3 and GAS1 genes. (A) Representative tetratype tetrads from LF-3 (chs3Δ gas1Δ heterozygous diploid) 8 days after dissection. (B) Cell viability assay. Stationary-phase cells from the first inoculum of spores in YNB−glucose at 30°C were concentrated to a value of 8 A450. Then, 5 μl of this suspension and four subsequent 10-fold serial dilutions were spotted onto YEPDAT plates. (C) CF staining of four representative spores. Magnifications: upper panels and lower left panel, ×3,200; lower right panel, ×2,000. (D) Effects of CHS3 deletion on chitin levels.
Inactivation of CHS3 was found to dramatically reduce the level of chitin in the double mutant compared to the single gas1 null mutant (Fig. 2D). This result indicates that Chs3p is indeed responsible for the increase in chitin accumulation induced by inactivation of the GAS1 gene. The CSI, CSII, and CSIII activities of the spores of two tetrads were analyzed; no changes in in vitro CSIII or CSII activities were detected in the gas1Δ mutant spores compared to the GAS1 ones. As previously shown (Table 2), the CSI activity increased in the spores carrying the gas1 null mutation. Moreover, a further twofold increase in CSI activity was found in the chs3 gas1 double mutant compared to the gas1 single mutant (results not shown).
Analysis of Chs3p level in gas1Δ cells.
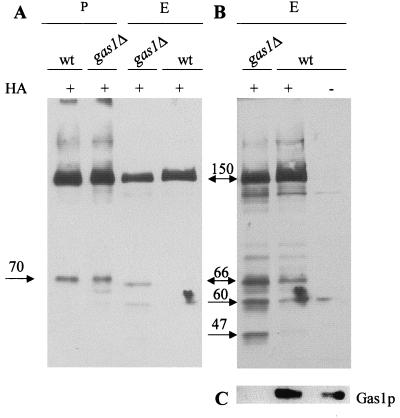
We studied whether Chs3p might be differentially expressed in gas1Δ cells with respect to controls. We introduced the gas1::HIS3 mutation into the Y1306 strain, which expresses Chs3p fused at the C-terminal with three HA epitopes (31). By Western blot analysis using anti-HA monoclonal antibodies, we detected the HA-tagged Chs3 protein of 150 kDa in total extracts. Its level slightly decreased in gas1Δ cells compared to those of the isogenic strain (Fig. 3A). Upon overexposure of the blots additional smeared fragments of about 66 and 60 kDa were detected, and one of about 47 kDa was present only in the mutant (Fig. 3B). The extent of recovery of these fragments was not always the same across a series of experiments, but the pattern was qualitatively reproducible. These results suggest that Ha-tagged Chs3p is more susceptible to proteolytic degradation in the gas1Δ mutant than in the wild type.
FIG. 3.
Western blot analysis of Chs3p in gas1Δ cells. (A) Total extracts (E; 80 μg) or membrane fractions (P; 40 μg) from Y604, Y1306, or Y1306ΔG (gas1Δ) were subjected to SDS-PAGE. Equal loadings were verified by Ponceau-S staining of the blotted proteins. Blots were immunodecorated with anti-HA monoclonal antibody HA.11. (B) Prolonged exposure of the total extracts shown in panel A. (C) Immunoblot with anti-Gas1p polyclonal antibodies of total extracts shown in panel B. The numbers indicate the molecular masses (in kilodaltons) of the relevant polypeptides.
Membrane and supernatant fractions were obtained with the same protocol used to determine the CSIII activity. In the membrane fraction (P), the 150-kDa polypeptide was the prominent band and its level did not show any appreciable differences between wild-type and gas1Δ strains (Fig. 3A). No enrichment of the 47- or 60- to 66-kDa polypeptides was observed, suggesting that they are probably unstable. Analysis of the supernatant fractions did not reveal any relevant species recognized by the antibodies used (data not shown).
CSIII activity increases in gas1 mutant cells permeabilized with digitonin.
In order to measure CSIII activity under more physiological conditions, whole cells permeabilized with digitonin were used (13). To avoid the high CSI activity level detected under these conditions, we constructed a set of strains that were deleted of the CHS1 gene and were either wild type or mutant for the CHS3 and/or GAS1 genes. A heterozygous diploid (LF5) was constructed by crossing chs1Δ cells with the suppressed LF3-13D spore (chs3Δ gas1Δ) and then sporulated. Using this method we were able to detect an increase in CSIII activity in gas1Δ cells (30.4 ± 5.2 mU/107 cells in the chs1Δ gas1Δ mutant spore compared to 9.6 ± 2.3 mU/107 cells in the chs1Δ spore). No activity was detected in the chs1Δ chs3Δ or the chs1Δ chs3Δ gas1Δ control strains.
DISCUSSION
The aim of this study was to establish the role of the different CSs present in S. cerevisiae in the increase in chitin accumulation in the gas1 mutant. Of the three CS activities, only CSI activity was affected by the presence of the gas1Δ mutation. The basal activity (measured without adding trypsin) doubled in the gas1Δ mutant. In contrast to this, the phenotype of the chs1Δ gas1Δ double mutant was indistinguishable from the single gas1Δ mutant in growth properties and morphology. Furthermore, the chitin level was only slightly decreased. We can therefore exclude the possibility that the increase in CSI would be implicated in the chitin response of the mutant. The induction of CSI could be explained in terms of the need to counterbalance the increase in chitinase activity that was unexpectedly found in the mutant. CSI is involved in the repair of damage to the cell wall caused by excessive chitinase activity an acidic pH. The gas1 null mutant could require a higher chitinase activity at the time of cytokinesis due to the higher chitin content in the cell wall. The increase in CSI could subsequently compensate for this increase but would not be essential since the chs1 gas1 double mutant does not show any worsening of the phenotype of gas1Δ cells. In addition, it is relevant to note that in α-factor-treated cells, a well-known condition in which an increase in chitin occurs, CHS1 mRNA was also found to be induced (1), and the expression of myc-Chs1p is approximately threefold higher than in untreated cells (38). Nevertheless, no direct participation of Chs1p in shmoo formation or mating was found, suggesting secondary roles for these changes in the conjugation process (32).
In addition to the lack of any compromise of the growth rate in the chs1Δ gas1Δ mutant, we observed a clear suppression of the small-bud-lysis phenotype typical of chs1Δ cells. Since this phenotypic trait has been ascribed to the loss of the repair function of Chs1p at the birth scar, we interpret this result as being a consequence of the presence of an increased chitin deposition also found in the daughter cells, which could reduce the detrimental effects of the lack of CSI activity after cell division.
Biochemical analysis revealed that CSII and CSIII activities do not change in the mutant. However, we analyzed in detail the phenotypes of yeast cells carrying deletions in the GAS1 and CHS3 genes. The double-mutant cells were severely affected in germination, and the double mutation led to detrimental effects in liquid medium. The progressive adaptation of the cells suggested that suppressors were selected, as was also described for other double null mutants in cell-wall-related genes (for example, kre6 skn1 [29]). Analysis of the chitin content of double-mutant cells clearly demonstrated that the bulk of chitin induced by the presence of the gas1Δ mutation is produced by CSIII. Since chitinase activity is increased, it can be ruled out that inhibition of degradation of this polysaccharide would contribute to the increase in its accumulation.
We attempted to determine at which level chitin synthesis is regulated in the gas1 mutant. The CHS3 mRNA level did not change in the gas1Δ mutant, excluding the idea that a transcriptional regulation would be involved. The protein level in total membrane did not change significantly between the mutant and the control. The presence of Chs3p-derived polypeptides of lower mobility in total protein extracts from gas1 cells indicates an increased turnover of the Chs3p full-length protein. For the time being, it is not possible to say whether this effect is associated with a possible activation of the protein, with increased mobilization of the protein through the endocytotic pathway, or whether it might simply be an indirect consequence of pleiotropic effects of the mutation. Preliminary experiments have indicated that the same proteolytic fragments are detectable when the HA-tagged Chs3p is overproduced by the GAL1-GAL10 promoter, indicating that they probably represent endogenous products of proteolysis, which for some reason are slightly stimulated in the mutant.
It is well known that also under other conditions of increase in chitin levels, such as treatment with CF or sporulation, no increase in the Chs3p level is found. By contrast, with α-factor treatment of the Chs3p level is sixfold higher, although this does not change the in vitro activity (9, 11). Thus, there is no correlation between the levels of protein and chitin synthesis, probably because other factors, such as posttranslational modifications, mobilization of the enzyme, or interaction with proteins limit the enzymatic activity. It is relevant to note here that a recent study has proposed that the stress-related chitin synthesis probably has a unique targeting and activation mechanism (23). Interestingly, the deposition of chitin by Chs3p in a fks1Δ mutant was found to be independent from Chs6p (23). Moreover, a putative cell wall sensor protein, Mid2, which functions upstream of the cell integrity pathway, appears to specifically modulate the accumulation of chitin in response to cell wall stress (16).
We measured CSIII activity in cells permeabilized with digitonin. This method was described to measure CSI activity (13); however, the facts that no activity was detected in the chs3Δ strains and that we obtained reproducible results confirm that it can also be used to detect CSIII activity. Although it is possible that the twofold increase in CSIII activity, detected under these conditions, could be due to different effects of digitonin in the control and the gas1 strains because of their difference in cell wall structure, the results obtained are in agreement with genetic data demonstrating a requirement for CSIII activity in chitin accumulation in the gas1 mutant. The ability to detect such an increase is lost in membrane fractions. This suggests that in the case of the gas1 null mutant a posttranslational modification, an interaction with a regulatory molecule, or a specific ion requirement is not preserved in the membrane preparations used for testing CSIII activity according to the method currently available. In order to understand this posttranslational regulation, experiments to determine the role of other genes involved in the regulation of cell wall biosynthesis in the increase in chitin levels in the gas1Δ mutant are in progress.
ACKNOWLEDGMENTS
We thank B. Santos, M. Snyder, and C. Roncero for strains and plasmids; S. Piatti for the help in tetrad analysis; A. Turchini for technical assistance; and A. Grippo for preparing the figures.
This work has been partially financed by grants MURST-Università di Milan Cofin 1999 and MURST 60% 1999 (L.P.), by Azioni Integrate Italia-Spagna (L.P. and A.D.), and by grant BIO98-0814-C02-02 from the Comision Interministerial Cientifica y Técnica, Madrid, Spain (M.H.V. and A.D.). L.F. was a recipient of a fellowship from Prassis-Sigma Tau Italy.
REFERENCES
- 1.Appeltauer U, Achstetter T. Hormone-induced expression of the CHS1 gene from Saccharomyces cerevisiae. Eur J Biochem. 1989;181:243–247. doi: 10.1111/j.1432-1033.1989.tb14718.x. [DOI] [PubMed] [Google Scholar]
- 2.Arellano M, Cartagena-Lirola H, Nasser Hajibagheri M A, Duran A, Valdivieso M H. Proper ascospore maturation requires the chs1+ chitin synthase gene in Schizosaccharomyces pombe. Mol Microbiol. 2000;35:79–89. doi: 10.1046/j.1365-2958.2000.01678.x. [DOI] [PubMed] [Google Scholar]
- 3.Bulawa C E, Slater M, Cabib E, Au-Young J, Sburlati A, Lee Adair W, Jr, Robbins P W. The S. cerevisiae structural gene for chitin synthase is not required for chitin synthesis in vivo. Cell. 1986;46:213–225. doi: 10.1016/0092-8674(86)90738-5. [DOI] [PubMed] [Google Scholar]
- 4.Cabib E, Sburlati A, Bowers B, Silverman S J. Chitin synthase 1, an auxiliary enzyme for chitin synthesis in S. cerevisiae. J Cell Biol. 1989;108:1665–1672. doi: 10.1083/jcb.108.5.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabib E, Silverman S J, Shaw J A. Chitinase and chitin synthase 1: counterbalancing activities in cell separation of S. cerevisiae. J Gen Microbiol. 1992;138:97–102. doi: 10.1099/00221287-138-1-97. [DOI] [PubMed] [Google Scholar]
- 6.Cabib E, Shaw J A, Mol P C, Bowers B, Choi W-J. Brambl and Marzluf (ed.) 1996. Chitin biosynthesis and morphogenetic processes; pp. 243–267. The mycota III. Biochemistry and molecular biology. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 7.Choi W J, Cabib E. The use of divalent cations and pH for the determination of specific yeast chitin synthases. Anal Biochem. 1994;219:368–372. doi: 10.1006/abio.1994.1278. [DOI] [PubMed] [Google Scholar]
- 8.Choi W J, Sburlati A, Cabib E. Chitin synthase 3 from yeast has zymogenic properties that depend on both the CAL1 and CAL3 genes. Proc Natl Acad Sci USA. 1994;91:4727–4730. doi: 10.1073/pnas.91.11.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi W J, Santos B, Duran A, Cabib E. Are yeast chitin synthases regulated at the transcriptional or the posttranscriptional level? Mol Cell Biol. 1994;14:7685–7694. doi: 10.1128/mcb.14.12.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chuang J S, Schekman R W. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J Cell Biol. 1996;135:597–610. doi: 10.1083/jcb.135.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cos T, Ford R A, Trilla J A, Duran A, Cabib E, Roncero C. Molecular analysis of Chs3p participation in chitin synthase III activity. Eur J Biochem. 1998;256:419–426. doi: 10.1046/j.1432-1327.1998.2560419.x. [DOI] [PubMed] [Google Scholar]
- 12.Federoff H J, Cohen J D, Eccleshall T L, Needleman R B, Buchferer B A, Giacalone J, Marmur J. Isolation of maltase structural gene from Saccharomyces carlsbergensis. J Bacteriol. 1982;149:1064–1070. doi: 10.1128/jb.149.3.1064-1070.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez M P, Correa J V, Cabib E. Activation of chitin synthase in permeabilized cells of Saccharomyces cerevisiae. J Bacteriol. 1982;152:1255–1264. doi: 10.1128/jb.152.3.1255-1264.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hille J K, Jan A, Donald G, Griffiths E. DMSO-enhanced whole yeast transformation. Nucleic Acids res. 1991;19:5791. doi: 10.1093/nar/19.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapteyn J C, Ram A F J, Groos E M, Kollar R, Montijn R C, Van Der Ende H, Llobell A, Cabib E, Klis F M. Altered extent of cross-linking of β1,6-glucosylated mannoproteins to chitin in Saccharomyces cerevisiae mutants with reduced cell wall β1,3-glucan content. J Bacteriol. 1997;179:6279–6284. doi: 10.1128/jb.179.20.6279-6284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ketela T, Green R, Bussey H. Saccharomyces cerevisiae Mid2p is a potential cell wall stress sensor and upstream activator of the PKC1-MPK1 cell integrity pathway. J Bacteriol. 1999;181:3330–3340. doi: 10.1128/jb.181.11.3330-3340.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuranda M J, Robbins P W. Cloning and heterologous expression of glycosidase genes from S. cerevisiae. Proc Natl Acad Sci USA. 1987;84:2585–2589. doi: 10.1073/pnas.84.9.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuranda M J, Robbins P W. Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J Biol Chem. 1991;266:19758–19767. [PubMed] [Google Scholar]
- 19.Madden K, Snyder M. Cell polarity and morphogenesis in budding yeast. Annu Rev Microbiol. 1998;52:687–744. doi: 10.1146/annurev.micro.52.1.687. [DOI] [PubMed] [Google Scholar]
- 20.Mouyna I, Fontaine T, Vai M, Monod M, Fonzi W A, Diaquin M, Popolo L, Hartland R P, Latgé J-P. GPI-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J Biol Chem. 2000;275:14882–14889. doi: 10.1074/jbc.275.20.14882. [DOI] [PubMed] [Google Scholar]
- 21.Orlean P. Biogenesis of yeast wall and surface components. In: Pringle J R, Broach J R, Jones E W, editors. Molecular and cellular biology of the yeast Saccharomyces cerevisiae. III. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1997. pp. 229–362. [Google Scholar]
- 22.Orlean P. Two chitin synthases in Saccharomyces cerevisiae. J Biol Chem. 1987;262:5732–5739. [PubMed] [Google Scholar]
- 23.Osmond B C, Specht C A, Robbins P W. Chitin synthase III: synthetic lethal mutant and “stress related” chitin synthesis that bypasses the CSD3/CHS6 localization pathway. Proc Natl Acad Sci USA. 1999;96:11206–11210. doi: 10.1073/pnas.96.20.11206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Popolo L, Girardelli D, Bonfante P, Vai M. Increase in chitin as an essential response to defects in assembly of cell wall polymers in the ggp1Δ mutant of Saccharomyces cerevisiae. J Bacteriol. 1997;179:463–469. doi: 10.1128/jb.179.2.463-469.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popolo L, Cavadini P, Vai M, Alberghina L. Transcript accumulation of the GGP1 gene, encoding a yeast GPI-anchored glycoprotein, is inhibited during arrest in the G1 phase and during sporulation. Curr Genet. 1983;24:382–387. doi: 10.1007/BF00351845. [DOI] [PubMed] [Google Scholar]
- 26.Popolo L, Grandori R, Vai M, Lacanà E, Alberghina L. Immunochemical characterization of gp115, a yeast glycoprotein modulated by the cell cycle. Eur J Cell Biol. 1988;47:173–180. [PubMed] [Google Scholar]
- 27.Popolo L, Vai M. The Gas1 glycoprotein, a putative wall polymer cross-linker. Biochim Biophys Acta. 1999;1426:385–400. doi: 10.1016/s0304-4165(98)00138-x. [DOI] [PubMed] [Google Scholar]
- 28.Ram A F J, Kapteyn J C, Montijn R C, Caro L H P, Douwes J E, Baginsky W, Mazur P, Van Den Ende H, Klis F M. Loss of the plasma membrane-bound protein Gas1p in Saccharomyces cerevisiae results in the release of β1,3-glucan into the medium and induces a compensation mechanism to ensure cell wall integrity. J Bacteriol. 1998;180:1418–1424. doi: 10.1128/jb.180.6.1418-1424.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roemer T, Delaney S, Bussey H. SKN1 and KRE6 define a pair of functional homologues encoding putative membrane proteins involved in β-glucan synthesis. Mol Cell Biol. 1993;13:4039–4048. doi: 10.1128/mcb.13.7.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Santos B, Snyder M. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J Cell Biol. 1997;136:95–110. doi: 10.1083/jcb.136.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santos B, Duran A, Valdivieso M H. CHS5, a gene involved in chitin synthesis and mating in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:2485–2496. doi: 10.1128/mcb.17.5.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw J A, Mol P C, Bowers B, Silverman S J, Valdivieso M H, Duran A, Cabib E. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1991;114:111–123. doi: 10.1083/jcb.114.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silverman S J, Sburlati A, Slater M L, Cabib E. Chitin synthase 2 is essential for septum formation and cell division in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1988;85:4735–4739. doi: 10.1073/pnas.85.13.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trilla J A, Cos T, Duran A, Roncero C. Characterization of CHS4 (CAL2), a gene of Saccharomyces cerevisiae involved in chitin biosynthesis and allelic to SKT5 and CSD4. Yeast. 1997;13:795–807. doi: 10.1002/(SICI)1097-0061(199707)13:9<795::AID-YEA139>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 36.Trilla J A, Duran A, Roncero C. Chs7p, a new protein involved in the control of protein export from the endoplasmic reticulum that is specifically engaged in the regulation of chitin synthesis in Saccharomyces cerevisiae. J Cell Biol. 1999;145:1153–1163. doi: 10.1083/jcb.145.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vai M, Gatti E, Lacanà E, Popolo L, Alberghina L. Isolation and deduced amino acid sequence of gene encoding gp115, a yeast glycophospholipid-anchored protein containing a serine-rich region. J Biol Chem. 1991;266:12242–12248. [PubMed] [Google Scholar]
- 38.Ziman M, Chuang J S, Schekman R W. Chs1p and Chs3p, two proteins involved in chitin synthesis, populate a compartment of the Saccharomyces cerevisiae endocytic pathway. Mol Biol Cell. 1996;7:1909–1919. doi: 10.1091/mbc.7.12.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ziman M, Chuang J S, Tsung M, Hamamoto S, Schekman R. Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. Mol Cell Biol. 1998;9:1565–1576. doi: 10.1091/mbc.9.6.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]