Abstract
We have examined the structure and fusion potential of the duck hepatitis B virus (DHBV) envelope proteins by treating subviral particles with deforming agents known to release envelope proteins of viruses from a metastable to a fusion-active state. Exposure of DHBV particles to low pH triggered a major structural change in the large envelope protein (L), resulting in exposure of trypsin sites within its S domain but without affecting the same region in the small surface protein (S) subunits. This conformational change was associated with increased hydrophobicity of the particle surface, most likely arising from surface exposure of the hydrophobic first transmembrane domain (TM1). In the hydrophobic conformation, DHBV particles were able to bind to liposomes and intact cells, while in their absence these particles aggregated, resulting in viral inactivation. These results suggests that some L molecules are in a spring-loaded metastable state which, when released, exposes a previously hidden hydrophobic domain, a transition potentially representing the fusion-active state of the envelope.
Hepadnaviruses are enveloped viruses which primarily infect (replicate in) the liver. Following entry of the virus into the hepatocyte, the incoming small, partially double-stranded DNA genome is repaired to form a covalently closed circular DNA which serves as the replicative template for the synthesis of viral mRNA as well as the pregenomic RNA which is converted to the mature DNA genome by reverse transcription within cytoplasmic nucleocapsids. Virion assembly occurs by interactions of these capsids with the envelope proteins which are synthesized on and span the endoplasmic reticulum. Virions, along with empty subviral particles (SVPs), are assembled and bud into the lumen of a post-endoplasmic reticulum–pre-Golgi compartment, from where they are exported by the cellular vesicular transport pathway (7, 19).
While hepadnavirus genome replication has been well documented, the mechanism by which these viruses enter the host hepatocyte remains far from understood. This is particularly apparent for the prototype human hepatitis B virus (HBV), which lacks a suitable in vitro infection system. However, considerable progress has been possible in the duck HBV (DHBV) infection model, a model instrumental in the description of most of the key steps of the replication cycle and amenable to study of the early events leading to the establishment of productive virus infection in vitro. In this system, a DHBV binding glycoprotein, gp180, a member of the carboxypeptidase D family, has been functionally characterized as the attachment receptor and an important part of a more complex uptake system (13, 14, 26, 27). This primary receptor has been further characterized as a Golgi-resistant protein, which cycles to and is endocytosed from the plasma membrane (3), thereby adding to earlier evidence suggesting that DHBV follows an endocytic route for cellular uptake (12). However, significant deficits in the body of knowledge accumulated for this avian hepadnavirus include the identification of a coreceptor or factor predicted to confer host specificity on the virus, as well as the mechanism leading to virus-cell membrane fusion.
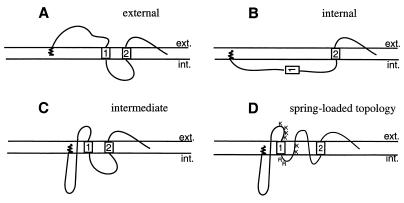
Concomitant with the lack of understanding of these processes is a lack of knowledge of the structural organization of the hepadnavirus envelope itself. With the hallmark of viral entry identified as a major conformational change of a viral envelope protein, typically involving exposure of a previously hidden hydrophobic fusion domain (6, 28), the large surface protein (L) with its external receptor binding pre-S domain appears to be the surface component of the envelope most likely to participate in such a function. This envelope protein, as well as the small (S) surface protein, is encoded by a single open reading frame comprised of the S domain and an upstream pre-S extension; the L protein thus consists of the membrane-anchored S domain with a hydrophilic N-terminal pre-S extension. Unique to hepadnaviruses is the complex, mixed pre-S topology of the L protein molecules, which develops during envelopment and particle maturation as the result of a partial, posttranslational pre-S translocation. This mixed topology provides for functionally diverse L protein molecules, such that external pre-S domains are available for receptor binding (11, 15) (Fig. 1A) while internal pre-S domains serve as a matrix for capsid envelopment (4) as well as exerting various regulatory functions (10, 22, 24) (Fig. 1B). Potentially extending the functional repertoire of L still further through alternate structural forms, a third L topology with a unique membrane-traversing pre-S domain has been identified in mature particles of the DHBV (9) (Fig. 1C). The recent identification of an additional membrane-traversing fold between the first transmembrane (TM1) and second transmembrane (TM2) domains (8) suggests that highly folded L molecules may form a spring-loaded, metastable structure (Fig. 1D) with the potential to participate in viral fusion with the host cell.
FIG. 1.
Models of L protein topology. The first transmembrane domain and the transmembrane anchor domain in S are indicated by boxes 1 and 2, respectively, but the third predicted but uncharacterized C-terminal transmembrane region is not shown. The N-terminal myristate is represented by the spiral. (A) Model of the external topology with an exposed-translocated N-terminal pre-S domain. (B) Model of the internal L topology, present immediately after synthesis, with pre-S and transmembrane domain 1 cytosolically disposed. (C) Model of the intermediate topology, identified in mature particles, with a small part of the C terminus of pre-S exposed to the particle surface while the remainder is proposed to traverse the particle membrane and be located internally. The loop region between TM1 and TM2 is shown as cytoplasmically disposed according to previously assumed models. (D) Proposed spring-loaded topology. The loop region between TM1 and TM2 is shown as membrane embedded with part of the loop exposed to the particle surface as described by Grgacic et al. (8). The trypsin cleavage sites at the C-terminal exposed region of pre-S and within the loop are shown by the letters K and R. ext., external; int., internal.
Several studies (5, 18, 29) have shown that viral proteins involved in entry, such as influenza virus hemagglutinin protein, are in a metastable state which can be altered to a fusion-competent, thermodynamically stable state through deforming agents, such as urea and heat. These agents have been shown to mimic physiological conditions, such as low pH encountered in the late endosome or receptor-coreceptor binding, dramatically affecting envelope conformation of some viruses upon entry. As a means of understanding hepadnavirus entry, we have examined the topological changes in DHBV envelope proteins after exposure of particles to various deforming agents, such as heat, dithiothreitol (DTT), or low pH. By analysis of the pattern of trypsin cleavage products generated after digestion of particles in different conformations, in addition to their infectivity and sedimentation properties, we have identified a major structural change in the L protein in which a hydrophobic domain of L is externalized to the particle surface, enabling particles to bind liposome membranes.
MATERIALS AND METHODS
Western blot analysis.
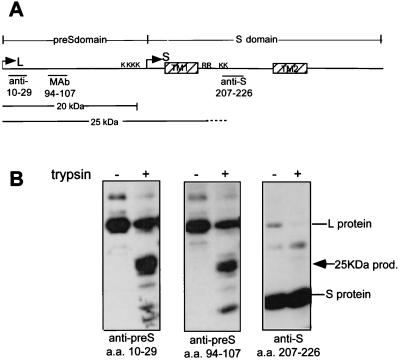
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12.5% acrylamide) and transferred to a nitrocellulose membrane (Schleicher and Schuell) using a Trans-Blot SD semidry transfer cell (Bio-Rad). Membranes were blocked for 1 h with 3% skim milk in TBST (100 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.3% Tween 20). L and L protein products were detected with polyclonal or monoclonal antiserum to pre-S or S domains as indicated by amino acid numbers in the diagram in Fig. 4A. Anti-S domain antiserum detects both L and S proteins due to their shared carboxy-terminal S domains. Membranes were probed with specific antiserum for 1 h in 1% skim milk–TBST, washed with TBST, and probed with horseradish peroxidase-conjugated species-specific antibodies (Amersham) in 1% skim milk–TBST. After a final wash in TBST (three times for 10 min each), protein bands were visualized by enhanced chemiluminescence (ECL; Amersham).
FIG. 4.
The trypsin product generated by low-pH or heat treatment is a 25-kDa extended pre-S product. (A) Untreated and low-pH-treated particles (pH 5, 16 h) were digested with trypsin, and the 25-kDa product was mapped by sequential Western blotting with a series of antisera with epitopes in the pre-S or S domain as indicated. (B) The positions of the epitopes of anti-10–29, anti-94–107, and anti-207–226 are shown in the schematic diagram of the L protein. The identity of the major trypsin cleavage products detected at neutral pH (20 kDa) and low pH (25 kDa) is shown below. MAb, monoclonal antibody.
Treatment of particles with deforming agents and digestion with trypsin.
SVPs were prepared from DHBV-positive serum by a sucrose step gradient as described previously (11). SVPs were stored in small aliquots at −20°C for single use only.
(i) Low-pH treatment.
Ten microliters of SVP preparation (equivalent to approximately 500 ng of L protein and 2 μg of S protein), serum virus, or serum virus pelleted through a 20% sucrose cushion (0.2 ml) was diluted with an equal volume of 50 mM Na acetate–150 mM NaCl adjusted so as to achieve a final pH of either 4 or 5 as required. Particles were incubated at pH 4 for 15 min or pH 5 for 16 h at either 4 or 37°C and at completion neutralized with 1.5 M Tris-HCl (pH 8.8)–150 mM NaCl.
(ii) Heat or DTT treatment.
Particles were incubated for 15 min at 50, 55, or 60°C and then briefly cooled on ice prior to trypsin digestion. Particles for direct trypsin analysis were incubated at 37°C for the prescribed times at a final concentration of 10 mM DTT. Serum virus, used for infection of primary duck hepatocyte (PDH) cultures, was treated with 5 mM DTT.
(iii) Trypsin digestion.
Trypsin efficiently cleaves the pre-S domain of particles at a cluster of lysine residues at amino acid (aa) 154 and 159 to 161 to generate a pre-S product of approximately 20 kDa. Another major trypsin site exists at lysines 204 and 206, between the first two transmembrane domains of the S domain, but is accessible to cleavage only in the presence of detergent (Fig. 1D; see also Fig. 4A). Treated and untreated particles were digested with trypsin (Sigma) (20 μg/ml) for 1 h at 37°C. Trypsin digestion was stopped by addition of aprotinin (final concentration, 2 μg/ml) and boiling in Laemmli buffer.
Infection of PDHs.
PDH cultures were prepared as described previously (20). PDHs seeded onto 12-well multiplates were infected for 4 h with a serum virus pool pretreated with low-pH buffers or DTT as described above. Samples were neutralized with an equal volume of 200 mM Tris-HCl (pH 8.8) prior to infection, and cells were harvested at day 9 postinfection for intracellular viral DNA analysis by dot blot hybridization as described previously (23).
Sucrose gradient separation of virions and SVPs.
Eight milliliters of DHBV-positive serum was pelleted for 3 h at 38,000 rpm in an SW40 rotor (Beckman) through 3 ml of 20% sucrose onto a 2-ml 70% sucrose cushion. Fractions were analyzed by SDS-PAGE and Coomassie blue staining, and 0.2 ml of the S protein-containing fraction was treated with deforming agents as described above. Samples were diluted in phosphate-buffered saline (PBS) to 4 ml and centrifuged for 4 h at 38,000 rpm through a 20 to 70% sucrose step gradient (11). Fractions were collected from the bottom of the gradient and analyzed for envelope proteins and viral DNA by SDS-PAGE and silver staining and DNA dot blot hybridization, respectively.
Liposome membrane binding.
Liposomes were obtained as a preliposome formulation (Sigma) composed of l-α-phosphatidylcholine β-oleoyl-V-palmitoyl (POPC), l-α-phosphatidyl-dl-glycerol,dioleoyl (PGDO), and cholesterol. The preparation was reconstituted in 1 ml of filtered PBS according to product instructions. Fifteen microliters of the preparation was mixed with 10 μl of SVP to which an equal volume of deforming agent was added. The reaction mixture was incubated for 15 min at 37°C and then resuspended with 67% (wt/vol) sucrose in PBS to a final volume of 200 μl and overlaid with a step gradient of 2 ml of 25% sucrose and 2 ml of 10% sucrose. Following centrifugation for 5 h at 45,000 rpm in an SW60 rotor (Beckman), the liposomes floated to the top of the gradient from where 0.25-ml fractions were collected. Fractions were methanol precipitated, and SVPs were detected by SDS-PAGE and Western blotting with an anti-pre-S antiserum.
RESULTS
A trypsin cleavage site is exposed following treatment of DHBV particles with low pH, heat, or DTT.
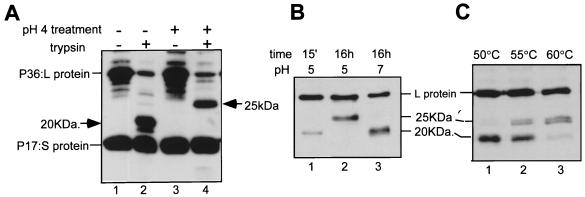
In this study, we have used SVPs (11), which consist of the same ratio and topology of surface proteins as capsid-containing particles and which are produced in a 1,000-fold excess, making them a usable source of particles for study. Gradient-purified SVPs were examined for any conformational or topological changes in the envelope proteins caused by treatment at low pH or heat, under conditions of limited trypsin digestion. In this assay, a proportion of the full-length L (P36) protein remains resistant to protease attack in untreated intact control particles (Fig. 2A, lane 2) even after extended periods of digestion, which, in keeping with previous observations (25; E. V. L. Grgacic, unpublished data), represents the internal topology. Another topological form of L, the intermediate-topology form (Fig. 1C), is susceptible to trypsin cleavage at a cluster of lysine residues at aa 154 and 159 to 161, near the C terminus of the pre-S domain, generating a pre-S product of approximately 20 kDa (Fig. 2A, lane 2) (9, 25). However, a larger pre-S-reactive trypsin product of approximately 25 kDa was observed following treatment at low pH at 37°C (Fig. 2A, lane 4, and 2B, lane 2) or at temperatures above 50°C at neutral pH (Fig. 2C, lanes 2 and 3). Generation of the 25-kDa trypsin product occurred following a 15-min treatment at pH 4 (Fig. 2A, lane 4), while at pH 5 it did not occur after 15 min but required more prolonged periods (16 h, Fig. 2B, compare lanes 1 and 2). At pH 7, however, a 16-h incubation at 37°C did not result in an altered trypsin product (Fig. 2B, lane 3). Treatment at 55°C resulted in a mixture of the 20- and the 25-kDa cleavage products (Fig. 2C, lane 2), while at 60°C the 25-kDa product was the predominant product detected (lane 3). Treatment of particles with 4 M urea, an agent shown to result in changes mimicking those created by low pH and heat in influenza virus (5), resulted in total digestion of the DHBV envelope proteins (data not shown). Furthermore, binding to the primary attachment receptor, gp180, itself or together with low-pH or reducing conditions did not elicit a similar conformational change in L or any change in the trypsin cleavage pattern (data not shown).
FIG. 2.
Trypsin digestion of particles treated with low pH or elevated temperatures results in a 25-kDa trypsin product. (A) Untreated particles or particles pretreated at pH 4 for 15 min and neutralized were digested with trypsin for 1 h at 37°C. The trypsin products were assessed by Western blotting with anti-pre-S and S domain antisera. Lanes: 1, untreated particles; 2, untreated particles digested with trypsin; 3, pH 4-treated particles; 4, pH 4-treated particles digested with trypsin. (B) Kinetics of pH 5 treatment. Lanes: 1, particles treated at pH 5 for 15 min and digested with trypsin; 2, particles treated at pH 5 for 16 h and digested with trypsin; 3, particles incubated at neutral pH for 16 h and digested with trypsin. (C) Heat treatment of particles prior to trypsin digestion. Lanes: 1, 50°C for 15 min at neutral pH; 2, 55°C at neutral pH; 3, 60°C at neutral pH. Trypsin products were detected by Western blotting with an anti-pre-S antiserum.
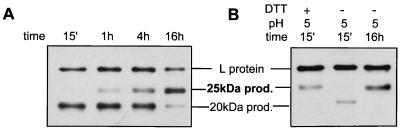
Previously, we have shown that the cysteine-containing region between TM1 and TM2 forms a membrane-embedded loop (8), which may contribute to a metastable L structure and thus be susceptible to reducing agents. We therefore examined the effect that the reducing agent DTT would have on the trypsin cleavage pattern of L. Particles were incubated with 10 mM DTT for various times at 37°C as indicated (Fig. 3A) and then subjected to trypsin digestion as described above. DTT treatment for 15 min did not result in an altered trypsin cleavage pattern (Fig. 3A, lane 1). However, partial conversion to the 25-kDa cleavage product occurred after 4 h, and at 16 h, the 25-kDa product predominated (Fig. 3A, lanes 3 and 4). To assess whether DTT would also enhance the rate of this transition at pH 5, which required some 16 h, particles were incubated at pH 5 with 10 mM DTT at 37°C. DTT triggered a rapid change to the 25-kDa trypsin product when particles were incubated at pH 5 for 15 min (Fig. 3B, lane 1) or with the mildly acidic pHs of 6 and 6.5 (data not shown).
FIG. 3.
The 25-kDa trypsin product is generated following reduction of particles with DTT. (A) Particles were incubated with 10 mM DTT at neutral pH for the times indicated and then digested with trypsin. (B) Increased kinetics of appearance of the 25-kDa trypsin product following combined DTT and pH 5 treatment of particles. Particles were treated at pH 5 with or without DTT for the times indicated and digested with trypsin, and products were detected by Western blotting with an anti-pre-S antiserum.
The 25-kDa pre-S trypsin product is generated by cleavage in the membrane-traversing loop between TM1 and TM2.
The origin of the 25-kDa trypsin product was mapped by sequential Western blotting using antisera recognizing specific epitopes in the L amino acid sequence (Fig. 4A). As shown in Fig. 4B, the 25-kDa product was detected using the antiserum to aa 10 to 29 and aa 94 to 107 in pre-S but did not react with an antiserum recognizing aa 207 to 226 in the S domain. This latter epitope lies immediately downstream of two lysine residues (aa 204 and 206) which are not surface exposed in any known topology of L (Fig. 1) and which require the presence of detergent for cleavage, generating a C-terminal, 12.9-kDa S product (8, 9). Accordingly, the 25-kDa product spans an extended region comprising all of pre-S and extending into the S domain, including the first transmembrane domain, to trypsin sites upstream of TM2. Cleavage may occur at a pair of either lysine or arginine residues within this domain (Fig. 4A), but the size of the pre-S product is more consistent with cleavage at the lysine residues. Following generation of this 25-kDa pre-S product, only a small amount of the expected 12.9-kDa carboxy-terminal fragment is detectable by ECL upon longer exposure (data not shown), suggesting that further cleavage at two potential trypsin sites (K267 and K289) in the external loop downstream of TM2 may be causing the complete degradation of this fragment.
Low pH induces a conformational change which increases the surface hydrophobicity, resulting in viral particle aggregation.
The detection of an altered trypsin cleavage pattern of the envelope proteins is indicative of conformational changes occurring following treatment with deforming agents. To assess whether such changes substantially altered virus particle structure, serum virus, pelleted through a 20% sucrose cushion, was either left untreated or exposed to pH 5 or 10 mM DTT or heated to 60°C before sedimentation over a preformed 20 to 70% sucrose gradient.
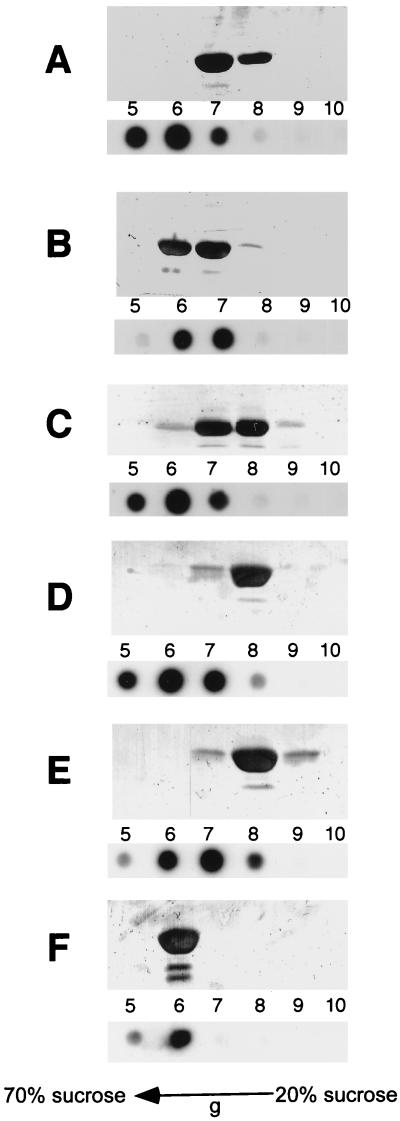
Gradient fractions were examined for virions by dot blot of viral DNA, and SVPs were detected by SDS-PAGE and silver staining for S protein (Fig. 5). Under the sedimentation conditions used (see Materials and Methods), DNA-containing particles and SVPs sediment according to their slightly differing densities, with substantial but incomplete separation of the two entities (Fig. 5A, pH 7.4, fractions 5 to 7 and 7 and 8, respectively). However, after pH 5 treatment for 16 h at 37°C (or pH 4 for 15 min at 37°C), both forms cosedimented, indicative of mixed aggregates of virus particles (Fig. 5B, fractions 6 and 7). The observed aggregation of these viral particles suggests that a major conformational change is occurring, which causes an increase in the hydrophobicity of the particle surface.
FIG. 5.
Viral particles and SVPs aggregate following low-pH or trypsin treatment. Serum virus (0.25 ml), pelleted through a 20% sucrose cushion, was incubated under various conditions prior to sedimentation in a preformed 20 to 70% sucrose gradient. Viral DNA dot blot hybridization (bottom panels) and silver-stained SDS-PAGE analysis of S protein associated with SVPs (top panels) are shown for gradient fractions 5 to 10. (A) pH 7.4 for 16 h; (B) pH 5 for 16 h; (C) 60°C for 15 min; (D) 10 mM DTT, 16 h; (E) 10 mM DTT at pH 5 for 15 min; (F) 10 mM DTT, 16 h, followed by pH 4 for 15 min.
In contrast, virus particles treated with DTT (Fig. 5D), DTT in combination with pH 5 (Fig. 5E), or heat (Fig. 5C) did not aggregate but maintained a sedimentation profile similar to that of the untreated particles. Thus, although heat treatment or DTT facilitated the generation of the 25-kDa trypsin product, either alone or with the help of mildly acidic pH, such treatments failed to generate the hydrophobic conformation, indicating the absence of a major or further conformational rearrangement of L. To examine whether the conformation of the reduced particles still enabled low-pH aggregation to occur, reduced particles (DTT, 16 h at 37°C) were subsequently treated at pH 4 for 15 min and sedimented in a sucrose gradient. Such treatment resulted in virions and SVPs aggregating (Fig. 5F), indicating that reduced particles exhibit an alternative conformational change, primarily affecting the cysteine-containing loop region, while retaining competency for the low-pH conformational change.
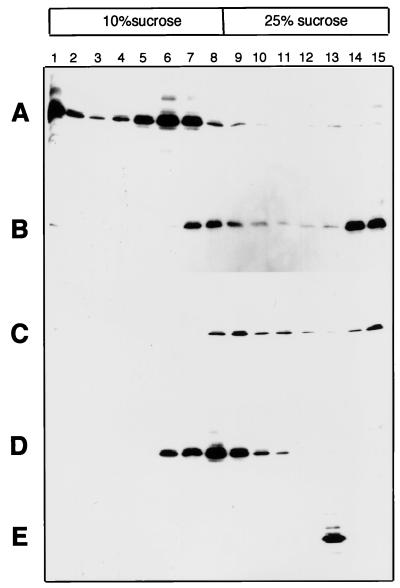
Particles triggered to the hydrophobic L conformation bind to liposome membranes.
The exposure of hydrophobic domains and consequent particle aggregation lead us to speculate that these particles may be primed for membrane association or fusion. To examine whether such a potential existed, particles were incubated with liposomes under various conditions. The mixture was overlaid with a sucrose step gradient in which liposomes float to the top under centrifugation, and particles were detected in gradient fractions by Western blotting. Particles incubated with liposomes at pH 4 were detected in the top half of the gradient in association with the liposome fractions (Fig. 6A). In contrast, particles exposed to DTT at pH 5, like untreated particles, were located in the bottom half of the step gradient (Fig. 6B and C). Particles forced to aggregate through the low-pH conformational change prior to incubation with liposomes partially floated up the gradient, being detected in fractions within the 20 to 10% interface (Fig. 6D). Their partial association with liposomes appears to reflect their general hydrophobic character with a degree of aggregation also evident in a proportion of the untreated particles. In the absence of liposomes, aggregates banded as one fraction near the bottom of the gradient (Fig. 6E).
FIG. 6.
Low-pH-triggered particles bind to liposome membrane vesicles. Liposome membrane vesicles were incubated for 15 min at 37°C with SVPs under the following conditions prior to flotation of the vesicles in a sucrose step gradient as described in Materials and Methods: pH 4 (A), pH 5 with 10 mM DTT (B), pH 7.4 (C), SVP aggregation by prior pH 4 treatment (D), and SVP aggregation by prior pH 4 treatment in the absence of liposome membranes (E).
Formation of the hydrophobic conformation causes viral inactivation.
The structural changes resulting from various deforming treatments were also examined for their effect on viral infectivity (Table 1). Serum virus treated with low-pH conditions at 37°C, either pH 4 for 15 min or pH 5 for 16 h, failed to infect PDHs, as determined by the absence of production of intracellular viral DNA 9 days postinfection. Low-pH treatment at 4°C, a temperature not causing any detectable conformational changes of the envelope, did not affect viral infectivity. In contrast to low pH, DTT treatment, either alone or in combination with a short exposure to pH 5, had no effect on viral infectivity. These data (Table 1) are all in keeping with particle aggregation correlating with a loss in infectivity. Heat treatment of DHBV, previously known to result in a loss of infectivity (1), was not paralleled by particle aggregation. The mechanism by which viruses are inactivated by heat is not understood but may be the result of destabilization of the virus particle.
TABLE 1.
Correlation between structural changes and virus inactivationa
| DTT | pH | Time | Temp (°C) | Virus inactivationb | Virus aggregationc | 25-kDa trypsin productd |
|---|---|---|---|---|---|---|
| − | 4 | 15 min | 37 | + | + | + |
| − | 5 | 15 min | 37 | − | NDf | − |
| − | 5 | 16 h | 4 | − | ND | − |
| − | 5 | 16 h | 37 | + | + | + |
| − | 7 | 16 h | 37 | − | − | − |
| + | 7 | 15 min | 37 | − | ND | − |
| + | 5 | 15 min | 37 | − | − | + |
| + | 7 | 16 h | 37 | − | − | + |
| − | 7 | 15 min | 60 | + | − | +e |
Pelleted serum virus was incubated at the pH and temperature and for times indicated with or without 10 mM DTT and then used to infect PDH cultures.
Viral inactivation was measured by loss of infectivity of PDHs by dot blot hybridization of intracellular viral DNA day 9 postinfection.
Aggregation of treated virus was detected by cosedimentation of DNA-containing particles and SVPs in a sucrose gradient as described for Fig. 5.
Generation of the 25-kDa trypsin product, representative of exposure of trypsin target sites within the cysteine-containing loop.
Data are from the work of Adcock et al. (1).
ND, not done.
The loss of infectivity of viral particles which have aggregated, as observed here, following exposure to low pH is consistent with similar inactivation in several viruses by particle aggregation in the absence of a target membrane (5, 17).
DISCUSSION
To date, our understanding of the hepadnavirus envelope is that it is an unusual densely packed protein shell of predominantly S protein subunits with the lipid content reduced and devoid of unit membrane structure (19). It is not known what effect these apparent constraints on the envelope would have on the mobility of the individual protein subunits during the viral entry process. This study has examined the conformational changes which the envelope proteins may undergo during entry, through the use of deforming agents which have been shown to mimic the release of other viral envelope proteins from their metastable state to a membrane binding, fusion-active state (5, 18). Our data indicate that some L molecules of DHBV may form a similar spring-loaded, metastable structure which is ready to undergo a major conformational change analogous to fusion activation. Low-pH treatment of particles resulted in exposure of a novel trypsin cleavage site normally hidden from the particle surface, generating a product of 25 kDa, comprised of the entire DHBV pre-S domain plus the first transmembrane region within the S domain through cleavage at K204 and K206. This conformational change of the L molecules represents a specific change and not a general denaturation of the DHBV particle, since S subunits remained unaffected and only some L subunits were altered, as evidenced by their trypsin cleavage. Moreover, at 4°C, low pH had no effect on the trypsin cleavage pattern, particle aggregation, or viral infectivity.
Previous studies have indicated that low-pH treatment of HBV particles increases the exposure of pre-S domains to pre-S-specific antibody binding (4) and susceptibility to protease digestion, presumably through forced translocation of pre-S domains from the intermediate folded topology (Fig. 1C) (9). In contrast, we show that a major conformational change is occurring in the S domain, exposing previously hidden trypsin cleavage sites in the S loop region, while precluding access to cleavage sites within pre-S. The generation of the 25-kDa extended pre-S product occurred with low pH or DTT or by heating particles above 55°C. For all these treatments, the resulting conformational change appears to have obscured the trypsin target sites exposed at the C terminus of pre-S in the precursor intermediate topology (Fig. 1C), since there was a loss of the ability to generate the 20-kDa pre-S product. This may be due to translocation of the membrane-traversing pre-S domain to a protease-resistant conformation or may be the result of these trypsin sites being obscured by the release of the adjacent TM1 plus part of the loop region of the S domain. While these effects on pre-S appear to be universal with all the deforming agents, they differed on the S domain, either by rendering the particle surface hydrophobic by extrusion of the TM1 domain (low pH, Fig. 5B), or by changing it to a nonhydrophobic intermediate conformation following reduction (Fig. 5D and E). The latter, less drastic conformational change in L may occur through the partial extrusion of TM1 or through exposure of the hydrophilic face of the amphipathic TM1 helix to the particle surface. This structural difference may reflect the loss or alterations in what appear to be intramolecular disulfide interactions important for the stability of the membrane-traversing loop between TM1 and TM2 (E. V. L. Grgacic, H. Schaller, C. Kuhn, and D. A. Anderson, submitted for publication) which may affect the release of TM1 into an alternative orientation. The ability of reduced viral particles to maintain infectivity and of such particles to adopt a hydrophobic conformation upon subsequent exposure to low pH implies that reduced particles may form a transitional, reversible conformation.
Particles exposed to liposome membranes under low-pH conditions which rapidly trigger the hydrophobic conformation resulted in membrane binding (Fig. 6A), consistent with their aggregation and loss of infectivity in the absence of a target membrane (Fig. 5B and Table 1). A similar low-pH-dependent membrane binding occurred with cell monolayers, detected by particle association with the membrane fraction following NP-40 extraction (data not shown). Since the change to the hydrophobic conformation at pH 5 required some 16 h, pH 4, which effected a rapid change to this conformation, was used. While the conditions used to trigger this membrane association do not represent physiological conditions encountered in the cell, they may mimic the conformational changes needed to generate an increase in surface hydrophobicity, which for many viruses is seen as a precursor of membrane fusion (28). Thus, the same changes which caused viral aggregation also appear to prime the particle for membrane association and possibly membrane fusion. However, in the cell, a cofactor other than low pH is required for conversion to this membrane binding conformation, since previous studies, using various lysomotropic agents, have discounted low pH as an entry requirement (12, 20), despite evidence that DHBV is endocytosed (2, 12). Notwithstanding these data, one cannot rule out the possibility that moderately low pH, which would remain even in the presence of such agents, may affect the kinetics of viral entry.
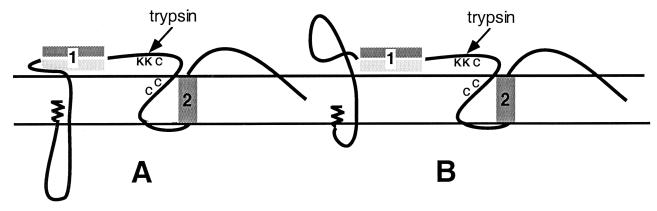
The model we propose for the release of metastable L molecules would involve the translocation and exposure of the hydrophobic face of the first transmembrane domain in addition to trypsin sites downstream in the cysteine-containing loop (Fig. 7). The pre-S domain would either remain largely membrane embedded (Fig. 7A) or adopt a protease-resistant conformation upon translocation (Fig. 7B). Although this change to a hydrophobic conformation facilitates membrane association, it is not clear whether it also exposes a potential fusion domain. Interestingly, it has been shown, for HBV at least that this region contains a fusion domain consensus sequence common to several enveloped viruses, including human immunodeficiency virus type 1 and influenza virus (16). Peptides of this putative fusion sequence have been shown to induce fusion with liposomes in vitro in a pH-dependent manner (21). Moreover, the ability of protease-pretreated HBV to infect nonpermissive HepG2 cells by incubation at pH 5.5 for 12 h (16) suggests that such low-pH conditions may at least partially mimic a fusion-competent conformation, pointing to TM1 as the important structural element in hepadnavirus fusion.
FIG. 7.
Models of the fusion topology. The proposed fusion topologies are shown with the hydrophobic face of TM1 (shaded) and part of the cysteine-containing loop region exposed to the surface. The pre-S domain is shown as either remaining untranslocated (A) or translocated to the particle surface in a protease-resistant conformation (B). The trypsin sites (K204 and K206) utilized to generate the 25-kDa trypsin product following treatment with deforming agents are shown by the letter K. The three cysteine residues in the membrane-embedded loop are shown by the letter C.
ACKNOWLEDGMENTS
This study was supported in part by a fellowship from the Zentrum für Molekulare Biologie, Heidelberg, and the Research Fund of the Macfarlane Burnet Centre for Medical Research.
We thank D. Anderson for critical reading of the manuscript.
REFERENCES
- 1.Adcock W L, MacGregor A, Davies J R, Hattarki M, Anderson D A, Goss N H. Chromatographic removal and heat inactivation of hepatitis B virus during the manufacture of human albumin. Biotechnol Appl Biochem. 1998;28:169–178. [PubMed] [Google Scholar]
- 2.Breiner K M, Schaller H. Cellular receptor traffic is essential for productive duck hepatitis B virus infection. J Virol. 2000;74:2203–2209. doi: 10.1128/jvi.74.5.2203-2209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breiner K M, Urban S, Schaller H. Carboxypeptidase D (gp180), a Golgi-resident protein, functions in attachment and entry of avian hepatitis B viruses. J Virol. 1998;72:8098–8104. doi: 10.1128/jvi.72.10.8098-8104.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruss V, Lu X, Thomssen R, Gerlich W H. Post-translational alterations in transmembrane topology of the hepatitis B virus large envelope protein. EMBO J. 1994;13:2273–2279. doi: 10.1002/j.1460-2075.1994.tb06509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carr C M, Chaudhry C, Kim P S. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc Natl Acad Sci USA. 1997;94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dimitrov D S. How do viruses enter cells? The HIV coreceptors teach us a lesson of complexity. Cell. 1997;91:721–730. doi: 10.1016/s0092-8674(00)80460-2. [DOI] [PubMed] [Google Scholar]
- 7.Ganem D. Hepadnaviridae and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2703–2737. [Google Scholar]
- 8.Grgacic E V L, Kuhn C, Schaller H. Hepadnavirus envelope topology: insertion of a loop region in the membrane and role of S in L protein translocation. J Virol. 2000;74:2455–2458. doi: 10.1128/jvi.74.5.2455-2458.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo J-T, Pugh J C. Topology of the large surface protein of duck hepatitis B virus suggests a mechanism for membrane translocation during particle morphogenesis. J Virol. 1997;71:1107–1114. doi: 10.1128/jvi.71.2.1107-1114.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hildt M, Weber O, Schaller H. Glucagon treatment interferes with an early step of duck hepatitis B virus infection. J Virol. 1998;72:2600–2606. doi: 10.1128/jvi.72.4.2600-2606.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klingmuller U, Schaller H. Hepadnavirus infection requires interaction between the viral pre-S domain and a specific hepatocellular receptor. J Virol. 1993;67:7414–7422. doi: 10.1128/jvi.67.12.7414-7422.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kock J, Borst E-M, Schlicht H-J. Uptake of duck hepatitis B virus into hepatocytes occurs by endocytosis but does not require passage of the virus through an acidic intracellular compartment. J Virol. 1996;70:5827–5831. doi: 10.1128/jvi.70.9.5827-5831.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuroki K, Cheung R, Marion P L, Ganem D. A cell surface protein that binds avian hepatitis B virus particles. J Virol. 1994;68:2091–2096. doi: 10.1128/jvi.68.4.2091-2096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuroki K, Eng F, Ishikawa T, Turck C, Harada F, Ganem D. gp180, a host cell glycoprotein that binds duck hepatitis B virus particles, is encoded by a member of the carboxypeptidase gene family. J Biol Chem. 1995;270:15022–15028. doi: 10.1074/jbc.270.25.15022. [DOI] [PubMed] [Google Scholar]
- 15.Le Seyec J, Chouteau P, Cannie I, Guguen-Guillouzo C, Gripon P. Infection process of the hepatitis B virus depends on the presence of a defined sequence in the pre-S1 domain. J Virol. 1999;73:2052–2057. doi: 10.1128/jvi.73.3.2052-2057.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu X, Block T M, Gerlich W H. Protease-induced infectivity of hepatitis B virus for a human hepatoblastoma cell line. J Virol. 1996;70:2277–2285. doi: 10.1128/jvi.70.4.2277-2285.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marsh M, Pelchen-Mathews A. The endocytic pathway and virus entry. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 18.Meyer W J, Gidwitz S, Ayers V K, Schoepp R J, Johnston R E. Conformational alteration of Sindbis virion glycoproteins induced by heat, reducing agents, or low pH. J Virol. 1998;66:3504–3513. doi: 10.1128/jvi.66.6.3504-3513.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nassal M. Hepatitis B virus morphogenesis. Curr Top Microbiol Immunol. 1996;214:297–337. doi: 10.1007/978-3-642-80145-7_10. [DOI] [PubMed] [Google Scholar]
- 20.Rigg R J, Schaller H. Duck hepatitis B virus infection of hepatocytes is not dependent on low pH. J Virol. 1992;66:2829–2836. doi: 10.1128/jvi.66.5.2829-2836.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodriguez-Crespo I, Nunez I E, Gomez-Gutierrez J, Yelamos B, Albar J P, Peterson D L, Galvilanes F. Phospholipid interactions of the putative fusion peptide of hepatitis B virus surface antigen S protein. J Gen Virol. 1995;76:301–305. doi: 10.1099/0022-1317-76-2-301. [DOI] [PubMed] [Google Scholar]
- 22.Rothman K, Schnoelzer M, Radziwill G, Hildt E, Moelling K, Schaller H. Host cell-virus cross talk: phosphorylation of the duck hepatitis B virus large envelope protein mediates intracellular signaling. J Virol. 1998;72:10138–10147. doi: 10.1128/jvi.72.12.10138-10147.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sprengel R, Kuhn C, Manso C, Will H. Cloned duck hepatitis B virus DNA is infectious in Pekin ducks. J Virol. 1984;52:932–937. doi: 10.1128/jvi.52.3.932-937.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Summers J, Smith P M, Huang M, Yu M. Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J Virol. 1991;65:1310–1317. doi: 10.1128/jvi.65.3.1310-1317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swameye I, Schaller H. Dual topology of the large envelope protein of duck hepatitis B virus: determinants preventing pre-S translocation and glycosylation. J Virol. 1997;71:9434–9441. doi: 10.1128/jvi.71.12.9434-9441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tong S, Li J, Wands J R. Interaction between duck hepatitis B virus and a 170-kilodalton cellular protein is mediated through a neutralizing epitope of the pre-S region and occurs during viral infection. J Virol. 1995;69:7106–7112. doi: 10.1128/jvi.69.11.7106-7112.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urban S, Breiner K M, Fehler F, Klingmueller U, Schaller H. Avian hepatitis B virus infection is initiated by the interaction of a distinct pre-S subdomain with the cellular receptor gp180. J Virol. 1998;72:8089–8097. doi: 10.1128/jvi.72.10.8089-8097.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White J M. Membrane fusion. Science. 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- 29.Wien M W, Curry S, Filman D J, Hogle J M. Structural studies of poliovirus mutants that overcome receptor defects. Nat Struct Biol. 1997;4:666. doi: 10.1038/nsb0897-666. [DOI] [PubMed] [Google Scholar]