Abstract
Kaposi’s sarcoma-associated herpesvirus (KSHV), or human herpesvirus 8, is a newly identified virus with tumorigenic potential. Here, we cloned and expressed the DNA polymerase (Pol-8) of KSHV and its processivity factor (PF-8). Pol-8 bound specifically to PF-8 in vitro. Moreover, the DNA synthesis activity of Pol-8 was shown in vitro to be strongly dependent on PF-8. Addition of PF-8 to Pol-8 allowed efficient synthesis of fully extended DNA products corresponding to the full-length M13 template (7,249 nucleotides), whereas Pol-8 alone could incorporate only several nucleotides. The specificity of PF-8 and Pol-8 for each other was demonstrated by their inability to be functionally replaced by the DNA polymerases and processivity factors of herpes simplex virus 1 and human herpesvirus 6.
Kaposi’s sarcoma-associated herpesvirus (KSHV), or human herpesvirus 8 (HHV-8), is strongly implicated as being critical in the development of Kaposi’s sarcoma (6; reviewed in reference 13). In addition, KSHV has been associated with body cavity-based lymphoma (4), multifocal Castleman’s disease (27), and multiple myeloma (23, 26; reviewed in references 3, 19 and 20). KSHV is a gamma-herpesvirus with homology to herpesvirus saimiri (HVS) and Epstein-Barr virus (EBV). Recent sequencing of the 140-kb KSHV genome (25) revealed at least 81 open reading frames (ORFs), including ORF9 and ORF59, which are predicted to encode viral DNA polymerase and processivity factor, respectively. Processivity factors complex with DNA polymerases, allowing them to synthesize extended stretches of DNA without dissociating from the template. Here, we report the cloning and expression of the DNA polymerase (Pol-8) and processivity factor (PF-8) of KSHV and demonstrate in vitro that they physically associate and, together, are required for processive DNA synthesis.
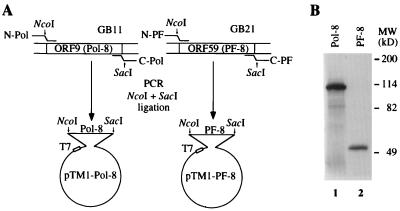
Two genomic DNA clones of KSHV (28), GB11 (containing ORF9) and GB21 (containing ORF59), were used to clone Pol-8 and PF-8, respectively. As depicted in Fig. 1A, the 3,038-bp Pol-8 gene was amplified from GB11 by PCR with two primers, N-Pol (5′-ATTACCATGGATTTTTTCAATCCATTTA-3′), creating a NcoI site flanking the 5′ end, and C-Pol (5′-ATAAGAGCTCTAGGGCGTGGGAAAAG-3′), creat- ing a SacI site flanking the 3′ end of the Pol-8 coding region. The 1,190-bp PF-8 gene was amplified from GB21 by PCR with two primers, N-PF (5′-TATTCCATGGTAATGCCTGTGGATTTTCACT-3′), creating a NcoI site flanking the 5′ end, and C-PF (5′-TATAGAGCTCAAATCAGGGGGTTAAATG- 3′), creating a SacI site flanking the 3′ end of the PF-8 coding region. Both of the genes were cloned into the NcoI and SacI sites of the pTM1 expression vector, and their ORFs were confirmed by DNA sequencing. Proteins were synthesized in vitro with the Promega T7-TNT coupled reticulocyte lysate system. As shown in Fig. 1B, a protein of approximately 114 kDa corresponding to the predicted 1,012-amino-acid ORF of Pol-8 was synthesized from pTM1-Pol-8 (lane 1), and a protein of approximately 50 kDa corresponding to the predicted 396-amino-acid ORF of PF-8 was synthesized from pTM1-PF-8 (lane 2).
FIG. 1.
Cloning and expression of Pol-8 and PF-8. (A) The coding regions of Pol-8 and PF-8 were amplified by PCR from two KSHV genomic DNA clones, GB11 and GB21, respectively, with oligonucleotides containing appended NcoI and SacI sites, and introduced into pTM1 expression vectors under the control of a T7 promoter. (B) Pol-8 and PF-8 were expressed in vitro with the Promega T7-TNT coupled transcription-translation system. [35S]methionine-labeled Pol-8 and PF-8 proteins were fractionated on an SDS–7.5% polyacrylamide gel and examined by autoradiography. The molecular mass (MW) of the protein markers is indicated.
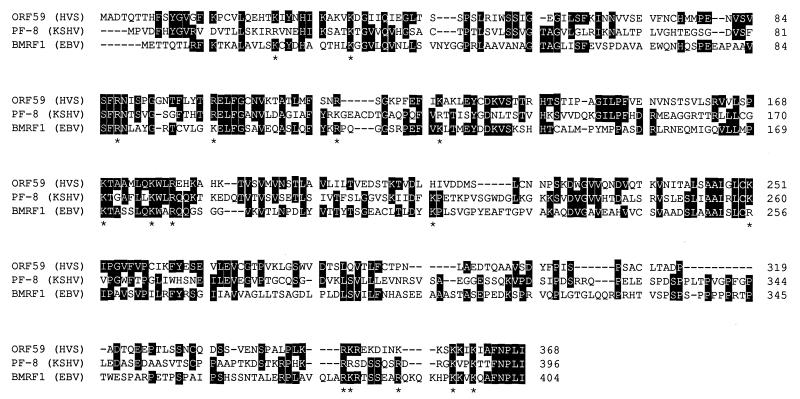
The structure and mechanism of herpesvirus processivity factors remain largely uncharacterized. The amino acid sequence of PF-8 (Fig. 2, middle line) revealed no common structural or functional motifs, except for the presence of a number of potential phosphorylation sites (data not shown). Compared to the amino acid sequences of all known herpesvirus processivity factors, PF-8 displays the greatest homology to ORF59 (31.5% identity) of HVS (2) and to BMRF1 (28.5% identity) of EBV (7, 17, 22, 29). It is noted that KSHV, EBV, and HVS are all gamma-herpesviruses. Compared to the processivity factors of other human herpesviruses, PF-8 exhibits an amino acid sequence identity of 21.8% to UL42 of herpes simplex virus type 1 (HSV-1) (12, 14, 15); 20.4% to ICP36 of human cytomegalovirus (11, 30); 19.3% to p41 of HHV-6 (1, 5); 17.4% to gene 16 product of varicella-zoster virus (8); and 17.3% to U27 of HHV-7 (21). When the amino acid sequences of PF-8, HVS ORF59, and EBV BMRF1 were analyzed with the ClustalW 1.7 multiple sequence alignment program (Fig. 3), there was an identity of 16.7% among all three sequences. No specific homologous domain is apparent; rather, conserved amino acids are distributed almost evenly throughout the entire sequences of these processivity factors. Interestingly, some of these conserved amino acids are basic residues (Fig. 2 [see asterisks]) which may be important for DNA-binding activity.
FIG. 2.
Sequence alignment of PF-8, HVS ORF59, and EBV BMRF1. The amino acid sequences of three herpesvirus processivity factors, PF-8 of KSHV, ORF59 of HVS, and BMRF1 of EBV, were aligned by the ClustalW 1.7 multiple sequence alignment program. Identical residues are shaded, and conserved basic residues are indicated by asterisks (∗).
FIG. 3.
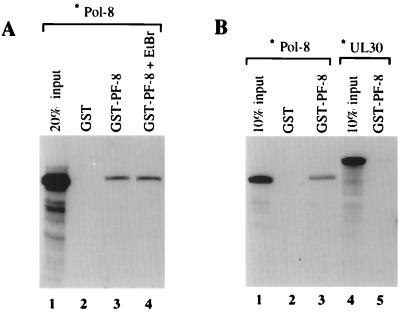
Pol-8 binds to PF-8 in vitro. (A) Pol-8 binds to PF-8 in the presence of ethidium bromide. [35S]methionine-labeled in vitro-translated Pol-8 (∗Pol-8) was incubated with GST or GST–PF-8 in the GST-binding assay in the absence (lanes 2 and 3) or presence (lane 4) of 200 μg of ethidium bromide (EtBr) per ml. Bound proteins were eluted and analyzed by SDS-polyacrylamide gel electrophoresis and autoradiography. (B) The binding of Pol-8 to PF-8 is specific. The binding assay was performed as described for panel A in the presence of ethidium bromide. [35S]methionine-labeled in vitro-translated DNA polymerase of HSV-1 (∗UL30) failed to bind GST–PF-8 (lane 5). Labeled proteins were also loaded directly onto the gels representing 20% (A) or 10% (B) of the input used in the binding assays.
In order to determine if Pol-8 and PF-8 can form a specific complex in vitro, a glutathione S-transferase (GST)-binding assay was employed. The full-length PF-8 gene was subcloned from pTM1-PF-8 into the EcoRI and NotI sites of a GST vector, pGEX4T-2 (Pharmacia). A GST–PF-8 fusion protein was overexpressed in Escherichia coli and purified with glutathionine beads. For the GST-binding assay, 200 ng of either GST or GST–PF-8 fusion protein was incubated with 5 μl of [35S]methionine-labeled in vitro-translated Pol-8 at 30°C for 1 h and then with 50 μl of glutathione beads for 30 min in the presence of 0.1% Nonidet P-40 (NP-40)–100 mM KCl–5 mM MgCl2–50 mM Tris-Cl (pH 7.6). The beads were washed three times with 1% NP-40–0.5 M KCl–5 mM MgCl2–50 mM Tris-HCl (pH 7.6) and then three times with 1% NP-40–0.5% deoxycholate–0.1% sodium dodecyl sulfate (SDS)–100 mM KCl–5 mM MgCl2–50 mM Tris-HCl (pH 7.6). Bound proteins were eluted by being boiled in Laemmli buffer, then fractionated on an SDS–4 to 20% polyacrylamide gel (Bio-Rad), and examined by autoradiography. As shown in Fig. 3A, Pol-8 bound to the GST–PF-8 fusion protein (lane 3) but not GST alone (lane 2). It is important to note that this association between Pol-8 and PF-8 is quite stable since it can withstand the very stringent washing conditions of high salt and detergents as indicated above. In lane 4, ethidium bromide (200 μg/ml) was included in both binding and washing buffers to verify that this interaction was not mediated by the DNA contained in the transcription-translation reaction (18). To demonstrate the specificity of the Pol-8–PF-8 interaction, the DNA polymerase of HSV-1 (UL30) was tested for its ability to complex with GST–PF-8. As shown in Fig. 3B, lane 5, UL30 was unable to bind to GST–PF-8 under the same conditions in which Pol-8 bound to GST–PF-8 (lane 3).
To investigate whether the interaction between Pol-8 and PF-8 is functionally significant, the DNA synthesis activity of Pol-8 in the absence or presence of PF-8 was determined by an in vitro DNA synthesis assay with primed M13 single-stranded DNA (ssDNA) as the template. The primed template was prepared by annealing M13 universal sequencing primer to M13mp18(+) ssDNA (Pharmacia) in the presence of 100 mM NaCl–1 mM EDTA–50 mM Tris-Cl (pH 7.6). Excess primer was removed by filtration through a Centricon-100 spin filter (Amicon). For the DNA synthesis assay (9), 2 μl (unless indicated otherwise) of each in vitro-translated protein was included in a 25-μl reaction mixture containing 100 mM (NH4)2SO4; 20 mM Tris-Cl (pH 7.5); 3 mM MgCl2; 0.1 mM EDTA; 0.5 mM dithiothreitol; 4% glycerol; 40 μg of bovine serum albumin per ml; 60 μM (each) dATP, dGTP, and dTTP; 10 μM [α-32P]dCTP (3,000 Ci/mmol; NEN); and 25 fmol of primed M13 ssDNA template. After incubation for 1 h (unless indicated otherwise) at 37°C, the reactions were terminated by incubating the reaction mixtures at 37°C for 1 h with 50 μl of 1% SDS–10 mM EDTA–10 mM Tris-Cl (pH 8)–200 μg of proteinase K per ml, followed by phenol-chloroform extraction. The reactions were analyzed either for polymerase activity, by measuring incorporation of deoxynucleoside triphosphates (dNTPs) into the DNA products, or for processivity, by measuring the length of newly synthesized DNA strands.
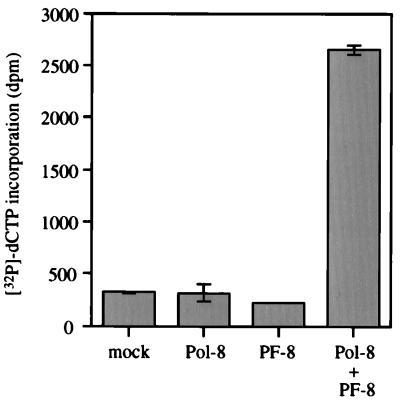
To determine the incorporation of [α-32P]dCTP into synthesized DNA, 5 μl of each reaction mixture was precipitated on ice for 10 min with 50 μl of 5% trichloroacetic acid (TCA)–20 mM sodium pyrophosphate and then captured on a Whatman RF/A glass fiber filter. The filter was washed extensively with 5% TCA–20 mM sodium pyrophosphate, rinsed with 70% ethanol, dried, and counted in a Beckman LS2800 liquid scintillator. The nucleotide incorporation activities of Pol-8 in the absence and in the presence of PF-8 were compared (Fig. 4). While no DNA synthesis activity could be detected with Pol-8 alone or PF-8 alone, significant nucleotide incorporation was achieved upon addition of PF-8 to Pol-8. These results suggest that DNA synthesis activity of Pol-8 strongly depends on the presence of PF-8.
FIG. 4.
DNA synthesis activity of Pol-8 strongly depends on the presence of PF-8. The DNA synthesis activities of mock-translated protein, Pol-8, PF-8, or Pol-8 and PF-8 together were analyzed in the DNA synthesis assay by measuring the incorporation of [α-32P]dCTP (in disintegrations per minute [dpm]) into synthesized DNA products. Mock represents translation from the pTM1 vector lacking a coding insert.
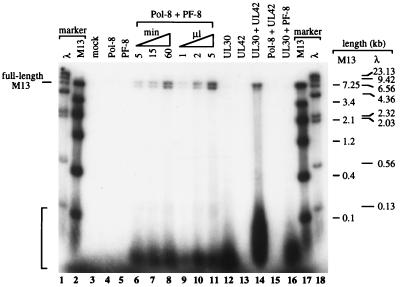
To analyze the processivity of DNA synthesis, the length of the DNA products from the DNA synthesis assay was determined. The DNA products were ethanol precipitated in the presence of 1 M ammonium acetate, resuspended in 50 μl of gel loading buffer (50 mM NaOH, 2.5 mM EDTA, 25% glycerol, 0.025% bromocresol green), and then fractionated on a 1.3% alkaline agarose gel. The gel was dried and examined by autoradiography. As shown in Fig. 5, no products were discernible with Pol-8 alone or PF-8 alone (lanes 4 and 5), whereas addition of PF-8 to Pol-8 resulted in processive DNA synthesis of fully extended M13 product (7,249 nucleotides) (lanes 6 to 11). The minor products of less than 100 nucleotides may arise from obstructed DNA synthesis due to the secondary structure of M13 ssDNA. As expected for an enzymatic synthesis, increasing either the concentrations of Pol-8 and PF-8 (lanes 9 to 11) or the reaction time (lanes 6 to 8) caused an increase in the level of DNA products. These results suggest that the ability of PF-8 to engage Pol-8 in processive DNA synthesis is extremely efficient. To examine the specificity of Pol-8 and PF-8, these KSHV proteins were tested for their ability to function with HSV-1 DNA polymerase (UL30) and its processivity factor (UL42) in the DNA synthesis assay. As expected (14, 15), UL30 alone can synthesize only short DNA products (lane 12), whereas addition of UL42 permits synthesis of full-length M13 product (lane 14). However, UL42 was not able to confer processivity on Pol-8 (lane 15), nor was PF-8 able to confer processivity on UL30 (lane 16). Consistent with this finding was the inability of PF-8 and the processivity factor of HHV-6 (p41) to physically or functionally substitute for one another (data not shown). These results suggest that the functional interactions between herpesvirus DNA polymerases and their processivity factors are specific.
FIG. 5.
PF-8 specifically enables Pol-8 to synthesize fully extended DNA. The DNA synthesis assays were performed for 1 h with 2 μl of each indicated protein, except that in lanes 6 and 7 reaction mixtures were incubated for 5 and 15 min, respectively, and in lanes 9 and 11, reaction mixtures contained either 1 or 5 μl of both Pol-8 and PF-8, respectively. The synthesized DNA products, labeled by [α-32P]dCTP incorporation, were fractionated on a 1.3% alkaline agarose gel which was analyzed by autoradiography. The sizes of 32P-labeled M13 and λ markers are indicated. Mock represents translation from the pTM1 vector lacking a coding insert. The translation efficiencies of all proteins tested in the assay were similar. The bracket at lower left indicates short products.
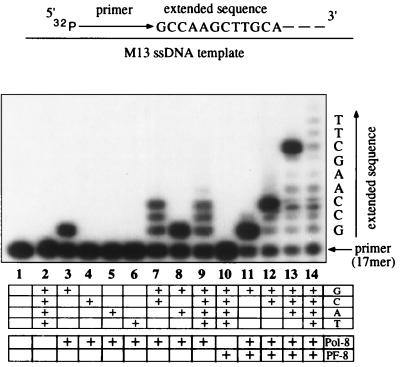
It was important to examine to a higher resolution the DNA synthesis activity of Pol-8 alone since this DNA polymerase might incorporate only several nucleotides, which could explain the failure to detect any DNA products with Pol-8 alone on the agarose gel (Fig. 5, lane 4) or by TCA precipitation (Fig. 4). DNA synthesis assays were performed as described above except that the primer was labeled at the 5′ end with [γ-32P]ATP and annealed to M13 ssDNA template, and various combinations of unlabeled dNTPs were used in the reaction. The synthesized DNA products were fractionated on a 15% urea-polyacrylamide gel and examined by autoradiography. The results are presented in Fig. 6, in which only the bottom of the gel is shown in order to resolve individual nucleotide incorporation extending from the primer. The analysis revealed that Pol-8 alone has significant catalytic activity and displays accuracy of nucleotide incorporation. When each dNTP was individually used in the assay, Pol-8 incorporated only dGTP accurately as the first nucleotide extended from the primer (lanes 3 to 6); when the correct combination of two dNTPs was used, Pol-8 sequentially added the first three expected nucleotides (GCC) (lane 7, compared to lane 8). However, and most importantly, in the presence of all four dNTPs, Pol-8 alone incorporated only the first three nucleotides efficiently (lane 9). While PF-8 alone exhibited no catalytic activity (lane 10), its addition to Pol-8 allowed efficient synthesis of maximum extended products (lanes 11 to 14). It is noted that, in lane 14, full-length DNA product was too large to enter the gel (data not shown). These results demonstrate that, in the absence of PF-8, Pol-8 has significant catalytic activity but very limited processivity.
FIG. 6.
Pol-8 alone has significant catalytic activity but very limited processivity. The DNA synthesis activities of Pol-8, PF-8, and Pol-8 and PF-8 together were compared on a 15% urea-polyacrylamide gel. The DNA synthesis assay employed M13 ssDNA template with an annealed 5′-end 32P-labeled primer and contained different combinations of unlabeled dNTPs. The extended sequence represents the actual M13 sequence immediately following the 3′ end of the primer. Only the bottom of the gel is shown here, since no products were visualized in the top portion of the gel except in lane 14, in which most of the high-molecular-weight products remained in the well.
Pol-8 and PF-8 are the first KSHV replication proteins to be cloned and expressed. PF-8 is required by Pol-8 in order to processively synthesize greatly extended DNA products (>7,000 nucleotides in the in vitro DNA synthesis assay), whereas without PF-8, Pol-8 can incorporate only several nucleotides. This study also shows that PF-8 cannot increase the processivity of the DNA polymerases of HSV-1 or HHV-6, nor can the processivity factors of HSV-1 or HHV-6 increase the processivity of Pol-8. Thus, even though herpesvirus processivity factors do have a degree of sequence homology and might form similar structures, they appear to be specific for their own DNA polymerases. Assuming that the functional interaction between Pol-8 and PF-8 is required for actual viral DNA replication in vivo, as has been suggested for the DNA polymerases and processivity factors of other herpesviruses (10, 16, 24), targeted disruption of this interaction by specifically designed inhibitors could lead to an effective antiviral strategy.
Acknowledgments
We are very grateful to R. Sun (UCLA) for providing the genomic DNA clones of HHV-8 and Y. Yuan (University of Pennsylvania) for his support. We also thank D. Coen (Harvard) for providing HSV-1 reagents and P. Digard and K. Grove for technical advice on the DNA synthesis assay.
The work presented here was partially supported by a University of Pennsylvania Research Foundation Award to R.P.R.
REFERENCES
- 1.Agulnick A D, Thompson J R, Iyengar S, Pearson G, Ablashi D, Ricciardi R P. Identification of a DNA-binding protein of human herpesvirus 6, a putative DNA polymerase stimulatory factor. J Gen Virol. 1993;74:1003–1009. doi: 10.1099/0022-1317-74-6-1003. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht J-C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittmann S, Craxton M A, Coleman H, Fleckenstein B, Honess R W. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brooks L A, Wilson A J, Crooks T. Kaposi’s sarcoma-associated herpesvirus (KSHV)/human herpesvirus 8 (HHV-8)—a new human tumor virus. J Pathol. 1997;182:262–265. doi: 10.1002/(SICI)1096-9896(199707)182:3<262::AID-PATH836>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 4.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 5.Chang C K, Balachandran N. Identification, characterization, and sequence analysis of a cDNA encoding a phosphorprotein of human herpesvirus 6. J Virol. 1991;65:2884–2894. doi: 10.1128/jvi.65.6.2884-2894.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 7.Cho M-S, Milman G, Hayward S D. A second Epstein-Barr virus early antigen gene in BamHI fragment M encodes a 48- to 50-kilodalton nuclear protein. J Virol. 1985;56:860–866. doi: 10.1128/jvi.56.3.860-866.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davison A J, Scott J E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 9.Digard P, Chow C S, Pirrit L, Coen D M. Functional analysis of the herpes simplex virus UL42 protein. J Virol. 1993;67:1159–1168. doi: 10.1128/jvi.67.3.1159-1168.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Digard P, Williams K P, Hensley P, Brooks I S, Dahl C E, Coen D M. Specific inhibition of herpes simplex virus DNA polymerase by helical peptides corresponding to the subunit interface. Proc Natl Acad Sci USA. 1995;92:1456–1460. doi: 10.1073/pnas.92.5.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ertl P F, Powell K L. Physical and functional interaction of human cytomegalovirus DNA polymerase and its accessory protein (ICP36) expressed in insect cells. J Virol. 1992;66:4126–4133. doi: 10.1128/jvi.66.7.4126-4133.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallo M L, Jackwood D H, Murphy M, Marsden H S, Parris D S. Purification of the herpes simplex virus type 1 65-kilodalton DNA-binding protein: properties of the protein and evidence of its association with the virus-encoded DNA polymerase. J Virol. 1988;62:2874–2883. doi: 10.1128/jvi.62.8.2874-2883.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganem D. KSHV and Kaposi’s sarcoma: the end of the beginning? Cell. 1997;91:157–160. doi: 10.1016/s0092-8674(00)80398-0. [DOI] [PubMed] [Google Scholar]
- 14.Gottlieb J, Marcy A I, Coen D M, Challberg M D. The herpes simplex virus type 1 UL42 gene product: a subunit of DNA polymerase that functions to increase processivity. J Virol. 1990;64:5976–5987. doi: 10.1128/jvi.64.12.5976-5987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hernandez T R, Lehman I R. Functional interaction between the herpes simplex-1 DNA polymerase and UL42 protein. J Biol Chem. 1990;265:11227–11232. [PubMed] [Google Scholar]
- 16.Johnson P A, Best M G, Friedmann T, Parris D S. Isolation of a herpes simplex virus type 1 mutant deleted for the essential UL42 gene and characterization of its null phenotype. J Virol. 1991;65:700–710. doi: 10.1128/jvi.65.2.700-710.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiehl A, Dorsky D I. Cooperation of EBV DNA polymerase and EA-D (BMRF1) in vitro and colocalization in nuclei of infected cells. Virology. 1991;184:330–340. doi: 10.1016/0042-6822(91)90849-7. [DOI] [PubMed] [Google Scholar]
- 18.Lai J-S, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy J A. Three new human herpesviruses (HHV6, 7, and 8) Lancet. 1997;349:558–563. doi: 10.1016/S0140-6736(97)80119-5. [DOI] [PubMed] [Google Scholar]
- 20.Neipel F, Albrecht J-C, Fleckenstein B. Cell-homologous genes in the Kaposi’s sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholas J. Determination and analysis of the complete nucleotide sequence of human herpesvirus 7. J Virol. 1996;70:5975–5989. doi: 10.1128/jvi.70.9.5975-5989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearson G R, Vroman B, Chase B, Sculley T, Hummel M, Kieff E. Identification of polypeptide components of the Epstein-Barr virus early antigen complex with monoclonal antibodies. J Virol. 1983;47:193–201. doi: 10.1128/jvi.47.1.193-201.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rettig M B, Ma H J, Vescio R A, Pold M, Schiller G, Belson D, Savage A, Nishikubo C, Wu C, Fraser J, Said J W, Berenson J R. Kaposi’s sarcoma-associated herpesvirus infection of bone marrow dendritic cells from multiple myeloma patients. Science. 1997;276:1851–1854. doi: 10.1126/science.276.5320.1851. [DOI] [PubMed] [Google Scholar]
- 24.Ripalti A, Boccuni M C, Campanini F, Landini M P. Cytomegalovirus-mediated induction of antisense mRNA expression to UL44 inhibits virus replication in an astrocytoma cell line: identification of an essential gene. J Virol. 1995;69:2047–2057. doi: 10.1128/jvi.69.4.2047-2057.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russo J J, Bohenzky R A, Chien M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;91:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Said J W, Rettig M R, Heppner K, Vescio R A, Schiller G, Ma H J, Belson D, Savage A, Shintaku I P, Koeffler H P, Asou H, Pinkus G, Pinkus J, Schrage M, Green E, Berenson J R. Localization of Kaposi’s sarcoma-associated herpesvirus in bone marrow biopsy samples from patients with multiple myeloma. Blood. 1997;90:4278–4282. [PubMed] [Google Scholar]
- 27.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay M F, Clauvel J P, Raphael M, Degos L, Sigaux F. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 28.Sun R, Lin S-F, Gradoville L, Miller G. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1996;93:11883–11888. doi: 10.1073/pnas.93.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsurumi T, Daikoku T, Kurachi R, Nishiyama Y. Functional interaction between Epstein-Barr virus DNA polymerase catalytic subunit and its accessory subunit in vitro. J Virol. 1993;67:7648–7653. doi: 10.1128/jvi.67.12.7648-7653.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiland K L, Oien N L, Homa F, Wathen M W. Functional analysis of human cytomegalovirus polymerase accessory protein. Virus Res. 1994;34:191–206. doi: 10.1016/0168-1702(94)90124-4. [DOI] [PubMed] [Google Scholar]