Abstract
Recombinant Sendai viruses were prepared which cannot express their Cprime, C, or Cprime plus C proteins due to mutation of their respective start codons ([Cprime-minus], [C-minus] and [double mutant], respectively). The [Cprime-minus] and [C-minus] stocks were similar to that of wild-type (wt) virus in virus titer and plaque formation, whereas the double-mutant stock had a much-reduced PFU or 50% egg infective dose/particle ratio and produced very small plaques. Relative to the wt virus infection, the [Cprime-minus] and [C-minus] infections of BHK cells resulted in significantly greater accumulation of viral RNAs, consistent with the known inhibitory effects of the Cprime and C proteins. The double-mutant infection, in contrast, was delayed in its accumulation of viral RNAs; however, once accumulation started, overaccumulation quickly occurred, as in the single-mutant infections. Our results suggest that the Cprime and C proteins both provide a common positive function early in infection, so that only the double mutant undergoes delayed RNA accumulation and exhibits the highly debilitated phenotype. Later in infection, the same proteins appear to act as inhibitors of RNA accumulation. In infections of mice, [Cprime-minus] was found to be as virulent as wt virus whereas [C-minus] was highly attenuated. These results suggest that the Cprime and C proteins cannot be functionally equivalent, since C can replace Cprime for virulence in mice whereas Cprime cannot replace C.
The paramyxomyxovirus C proteins were first found in Sendai virus (SeV)-infected cells and, since they were apparently absent in virions, were termed nonstructural proteins (reviewed in reference 22). Subsequent work with specific antisera showed that the C proteins could be detected in virions, associated with nucleocapsids. They are, however, greatly underrepresented in virions relative to their intracellular amounts (22, 27, 38). Furthermore, unlike the other virion proteins, which are translated from separate transcriptional units, the C proteins are translated from the P (phosphoprotein) gene mRNA, from an alternate open reading frame (ORF) which overlaps the N-terminal end of the P protein ORF (14) (Fig. 1).
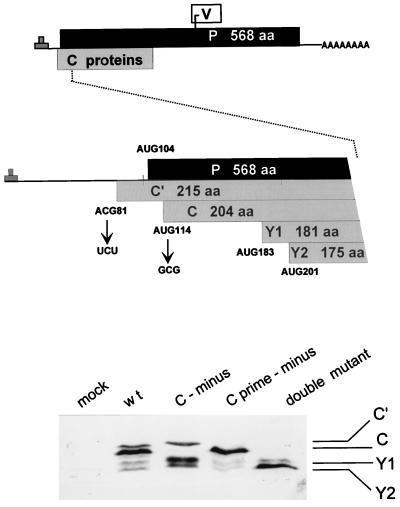
FIG. 1.
ORF organization and expression of the SeV P gene. The three ORFs expressed as proteins (P, C, and V) are shown as horizontal boxes, drawn roughly to scale (top). A blow-up of the 5′ end of the mRNA is shown in the middle, and the five ribosomal start sites in this region are indicated. The mutations used to eliminate expression from the Cprime and C protein start codons are shown. The effects of these mutations on C protein expression in BHK cells infected with a 1:5 dilution of the wt (SeVZ), [C-minus], [Cprime-minus], and double-mutant virus stocks (see the text) were examined by immunoblotting with a polyclonal rabbit anti-C antiserum (bottom). aa, amino acids.
The expression of the paramyxovirus C proteins can also be complex. SeV is a member of the newly renamed Respirovirus genus (formerly the Paramyxovirus genus) of the subfamily Paramyxovirinae. This subfamily also includes the Morbillivirus (e.g., measles virus) and the Rubulavirus (e.g., mumps virus and simian virus 5) genera. The more distantly related pneumoviruses, such as respiratory syncytial virus, are now classified in a separate subfamily (Pneumovirinae) of the Paramyxoviridae (24). The members of the Paramyxovirinae have been recently reclassified based on their P gene organizations, including the presence of a C ORF. In SeV-infected cells, the C proteins are relatively abundant and are the major products of this P gene on a molar basis. There are four independently initiated SeV C proteins (Fig. 1) expressed via a non-AUG start site (C′ or Cprime), and there are both scanning-dependent (C) and scanning-independent (Y1/Y2) ribosomal initiation (reviewed in reference 7). However, morbilliviruses such as measles virus express only a single C protein, and in significantly smaller amounts than that of P (1, 3). Furthermore rubulaviruses do not contain a C ORF at all (32). The optional nature of this ORF is also apparent in other mononegaviruses. For the pneumoviruses, an overlapping ORF is absent in the P gene of respiratory syncytial virus, but the pneumonia virus of mice P gene does contain a C-like ORF, which expresses two small basic proteins (2). Similarly, for the more distantly related Rhabdoviridae, vesiculoviruses like vesicular stomatitis virus express two C proteins whereas lyssaviruses like rabies virus do not contain a C ORF of any size (30). We note that the term “C protein” refers only to the origin of its coding sequence with reference to that of SeV; it is not as yet clear whether the different proteins carry out similar functions.
The SeV C proteins (Cprime, C, Y1, and Y2) are relatively small basic proteins (175 to 215 residues), whose intracellular localization shows little or no specificity (27, 38). Only very small amounts of the C proteins are found bound to nucleocapsids intracellularly or in reconstitution experiments with transfected cell extracts (unpublished data); this presumably accounts for their vast underrepresentation in virions. Nevertheless, consistent with their presence on nucleocapsids, the C proteins appear to play a role in viral RNA synthesis. When mRNA synthesis is studied with transfected cell extracts containing the P and L proteins, elimination of C protein coexpression (by placing a stop codon just downstream of AUG201/Y2 [P/Cstop]) was found to strongly increase mRNA synthesis in vitro. Moreover, its reexpression from a separate plasmid eliminated this increase (8). The ability of C to negatively affect viral RNA synthesis depended on its coexpression with P and L, and C may therefore act during formation of the P3-L polymerase complex. Similarly, when SeV minigenome replication is reconstituted in transfected cells by using P genes which do (P/Cwt) and do not (P/Cstop) express C proteins, C protein coexpression was also found to be strongly inhibitory, in a promoter-specific manner. The genomic promoter (which also contains the start site for N mRNA synthesis) was particularly sensitive to inhibition by C protein, in contrast to genome synthesis from the antigenomic promoter (6). The C proteins also have the remarkable property of preferentially inhibiting the replication of minigenomes containing mutant promoters (26) or minigenomes which were not of hexamer length (31). In this respect, the C proteins appeared to function as viral fidelity (or stringency) factors. In infectious-virus recoveries from DNA, C protein coexpression (from the P gene support plasmid needed to initiate viral RNA synthesis) must also be repressed for a successful outcome (6, 12). It is possibly this fidelity function of C which needs to be repressed. The temporary lack of stringency of the viral polymerase may be beneficial here, since our full-length clone expresses an antigenome transcript with three extra G residues at its 5′ end and is therefore not of hexamer length.
More recently, the SeV C proteins have been found to play a positive role in infections of mice (a natural host). These conclusions were based on studies of SeVMVC, an avirulent cell culture-derived mutant of the highly virulent SeVM, which differs from SeVM by only two substitutions; F170S in the protein encoded by the C gene (silent in the P ORF) and E2050A in the protein encoded by the L gene (18, 37). To examine which amino acid substitutions were responsible for the avirulence, rSeVZ carrying the C gene of either the virulent M strain (rSeVZ-CM) or the avirulent MVC strain (rSeVZ-CMVC) within the virulent Z strain background were prepared. Only the chimeric rSeVZ carrying the CMVC gene was found to be avirulent, suggesting that the normal function of the C proteins was required for virulence in mice, i.e., for multiple cycles of virus replication (13). These experiments offered the first evidence that the SeV C proteins provide a function required for virus replication, at least in mice.
This paper reports the preparation and characterization of rSeV strains which specifically do not express the Cprime, C, or Cprime plus C proteins, by mutating their ribosomal start codons. Our results suggest that (i) the functions of the C proteins are complex, in that they display both positive and negative effects on viral RNA synthesis during the course of the infection, and (ii) the functions of the individual C proteins must be partly different.
MATERIALS AND METHODS
Cell cultures and viruses.
LLC-MK2 and BHK cell lines were grown in minimal essential medium supplemented with 5% fetal calf serum. Wild-type (wt) and rSeV stocks were prepared in the allantoic cavity of 9-day-old embryonated chicken eggs and harvested after 3 days at 33°C. Titer determination and plaque isolation of rSeV were carried out for 3 to 9 days at 33°C in LLC-MK2 cells covered with a 0.3% agarose overlay containing 1.2 μg of acetyl-trypsin per ml. The 50% egg infective dose (EID50) of each viral stock was determined by injecting serial dilutions into the allantoic cavity of hen eggs as above. The presence of virus in the allantoic fluids was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% polyacrylamide) and staining with Coomassie brilliant blue on material pelleted at 10,000 × g for 30 min through a TNE (10 mM Tris-Cl, pH 7.4, 100 mM NaCl, 1 mM EDTA)–25% glycerol cushion in an Eppendorf centrifuge. To determine the 50% tissue culture infective dose (TCID50), 4-cm petri dishes of BHK cells were infected in duplicate with serial dilutions of the virus stocks, and the cytopathic effect was monitored visually until 40 h postinfection (p.i.). At this time, cytoplasmic extracts were prepared and the levels of N protein contained in three different amounts of the extracts were determined by Western blotting with monoclonal anti-N877 antibody (25).
Generation of mutant SeV (DNA) genomes.
pGem-P/C carrying TCT81/C′ and GCG114/C mutations in the initiation codons of the P/C mRNA have been described previously (8). pGem-P/C combining these mutations was constructed by fusion PCR on pGem P/C-TCT81/C′ DNA. The product was digested with SacI (in the upstream polylinker) and EagI (position 1005 on the P/C mRNA) and then subcloned into pGem P/C-TCT/C′. Sendai viruses containing the various mutations in the C protein start sites were recovered from DNA by taking advantage of the recombinogenic nature of the vaccinia virus-based recovery system, as previously described (12, 13). Briefly, pFL3 (full-length DNA clone) was modified to contain a deletion encompassing most of the N and P genes (pFL3-Δ1), such that a rSeV could not be generated simply by the recombination of pFL3-Δ1 DNA with that of the individual pGEM-N and pGEM-P support plasmids. A shuttle vector (pN/PXho/M) containing the deleted region plus flanking sequences into which the mutated P/C genes were cloned was then constructed as follows. PCR was performed with a positive-sense primer carrying an XhoI site at position 69 (of the untranslated region of the P/C mRNA) and a negative-sense primer complementary to position 1178. The products were digested with XhoI and EagI and inserted into the same sites of pN/PXhoI/M. The various mutated P/C genes were then recovered in viruses via recombination of the various pN/PXho/M vectors with pFL3-Δ1. In all cases, the relevant regions were verified by reverse transcription-PCR and DNA sequencing.
Generation of rSeV with an alternate expression of C proteins.
The recovery system was essentially as described previously (12, 13). Briefly, one 9-cm dish of HeLa cells (ca. 80% confluent) was infected with 3 PFU of vaccinia virus TF7-3 per cell (11). The cultures were transfected 1 h later with 1.5 μg of pGEM-L, 2.5 μg of pGEM-N, 4.5 μg of pGEM-HAP/Coff (which do not express C proteins), 15 μg of pFL3-Δ1, and 5 μg of one of the pN/PXho/M shuttle vectors carrying the desired mutation. At 24 h later, 1-β-d-arabinofuranosylcytosine (100 μg/ml) was added to inhibit vaccinia virus replication, and 24 h after that, the cells were scraped into their medium and injected into two or three 10-day-old embryonated chicken eggs. At 3 days later, the allantoic fluids were harvested and reinjected undiluted into eggs. For further passages, the stocks were generally diluted 1/1,000 before injection. The presence of viruses in the allantoic fluids was determined as above.
Primer extension analysis of viral RNAs.
Total RNA was extracted from virus-infected cells with Trizol (Gibco-BRL) and analyzed by primer extension using with Moloney murine leukemia virus reverse transcriptase (Gibco-BRL) and 200,000 cpm of each of the following 32P-5′- primers, which had been purified on sequencing gels: NP126 (5′-126CGGCCATCGTGAACTTTGGC107-3′ [numbers refer to map coordinates of the entire genome]) for estimation of antigenomes and N mRNAs and L15,270 (5′-15,270GAAGCTCCGCGGTACC15,285-3′) for genomes.
Immunoblotting.
C protein levels were determined by Western blotting of cytoplasmic extracts of BHK cells infected with the various viral stocks, using a rabbit anti-C polyclonal antiserum with the CSPD light detection system (Boehringer). Immunoblotting was also carried out on viruses released from cell cultures, purified by being pelleted through a TNE–25% glycerol cushion from the medium of infected BHK cells, with monoclonal anti-N877 antibody (25).
Determination of mouse pathogenicity.
Specific-pathogen-free 7-week-old (female) C57BL/6 mice (Iffo Credo) were infected intranasally with 106 PFU of the various viruses. The mice were observed for symptoms and weighed daily. Groups of three mice were sacrificed on days 2, 5, 7, and 8 or 9, and the number of PFU present in 10% lung homogenates was determined on LLC-MK2 cells. The lung homogenates were also injected into 9-day-old embryonated hen eggs and passaged twice. BHK cells were infected with this allantoic fluid, and viral protein expression was monitored by immunoblotting for the N and C proteins. Total RNA was also extracted from the lung homogenates with Trizol (Gibco-BRL). cDNA was prepared with Moloney murine leukemia virus reverse transcriptase (Gibco) and amplified with the Expand high-fidelity PCR system (Boehringer) with the primers mentioned above. The PCR product DNA was sequenced.
Specific-pathogen-free, 3-week-old male mice [ICR/CRJ(CD-1) strain; Clea, Inc.] were infected intranasally with 25 μl of serially diluted virus under ether anesthesia. The mice were observed for symptoms and weighed daily. The 50% lethal dose (LD50) was calculated by the method of Reed and Muench (29). The cell infectious unit (CIU) was essentially equivalent to PFU.
RESULTS
Failure to recover a totally C-minus Sendai virus.
Measles virus (MeV) expresses a single C protein from its P gene, and Radecke and Billeter (28) have recovered a rMeV which cannot express its C protein due to the introduction of a stop codon into this ORF. This rMeV-Cminus strain replicated normally in cell culture, indicating that the MeV C protein does not play an essential role in the cellular replication of this paramyxovirus, at least in cell culture. A similar situation has also been found for the more distantly related rhabdovirus vesicular stomatitis virus (21). We have also attempted to recover a rSeV which cannot express any of its four C proteins, by placing a stop codon in the C ORF just downstream of the Y2 start codon (ATG201/Y2) to terminate all four C proteins (8). A full-length SeV genome expression plasmid containing the Cstop mutation was constructed by exchanging the SalI fragment of pFL3 (which includes the N-terminal region of the P/C gene) with one containing the Cstop mutation (Fig. 2). As a positive control, the SalI fragment was always exchanged with itself at the same time. Virus recoveries from pFL3 and pFL3-Cstop were carried out in parallel (by using the pGEM-P/Cstop support plasmid, so that the Cstop mutation of pFL3-Cstop could not be lost by recombination), and the resulting viruses were amplified in hen eggs. In four separate attempts, sufficient virus was present in the pFL3 recoveries that 60 μl of the second-passage allantoic fluid showed a strong pattern of SeV structural proteins when analyzed by SDS-PAGE and Coomassie blue staining. This corresponded to reference stocks containing ca. 109 to 1010 EID50/ml (Fig. 3). However, we were unable to detect virus from the pFL3-Cstop transfections by this test, even when 200 μl of sixth-passage allantoic fluid was examined.
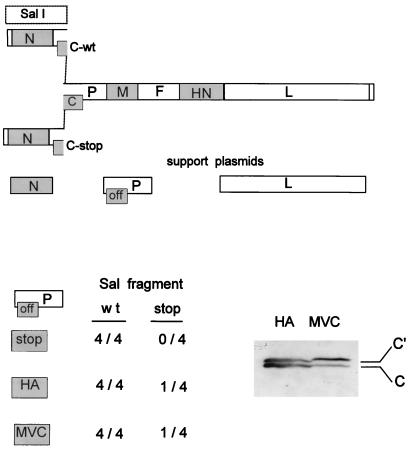
FIG. 2.
Inability to recovery a SeV strain which cannot express any of its C proteins. The reconstruction of full-length SeV DNA in which the C ORF remains open (C-wt) or is closed just after AUG201/Y2 (C-stop) by exchanging SalI fragments is shown schematically at the top. The three pGEM support plasmids expressing N, P, and L, needed to recover infectious virus from the full-length DNA, are shown just below; “off” in the box representing the C ORF indicates that C either is not expressed or is not completely functional (see the text). A summary of the results of the virus recoveries obtained with both SalI fragments and three different pGEM-P support plasmids (stop, HA, and MVC [see the text]) is shown at the bottom. When virus was recovered from full-length DNA containing the C-stop mutation, this virus was used to infect BHK cells, and the presence or absence of C expression was examined by immunoblotting (bottom right). Y1 or Y2 protein expression was not detected in this infection.
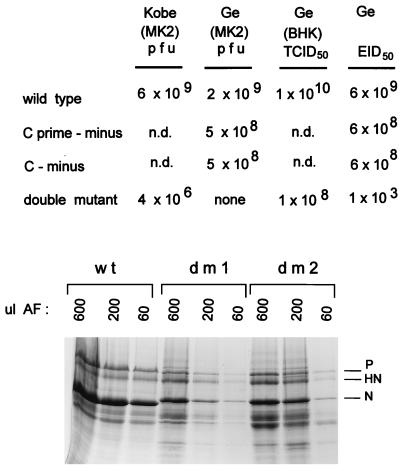
FIG. 3.
Titer determination of the various SeV stocks. Egg allantoic fluid stocks of the various mutant SeV strains and wt virus (SeVZ) were subjected to titer determination by (i) plaque formation on MK2 cells with stocks prepared in Kobe and Geneva (Ge), (ii) TCID50 determination on BHK cells, and (iii) EID50 determination in hen eggs (top). All values listed are per milliliter. Relative amounts of virus proteins present in 600, 200, and 60 μl of allantoic fluid (AF) were determined by SDS-PAGE and staining with Coomassie brilliant blue (bottom). dm1 and dm2 are independently recovered double-mutant viruses (dm2 was subjected to titer determination [above]). The N proteins of the mutant viruses migrate slightly slower than that of SeVZ, as expected, because these viruses were prepared in the rSeVZ-NH background to mark them as recombinants.
As mentioned above, expression of (wt) C proteins from the pGEM-P/C support plasmid adversely affects the recovery of rSeV from DNA. However, we and others have successfully used other pGEM-P support plasmids in rSeV recoveries (6, 19), namely, (i) pGEM-HAP/Coff, in which the influenza virus HA tag was fused between the Cprime and P protein start codons in one continuous ORF, which eliminates almost all C protein expression; and (ii) pGEM-P/CMVC, containing the gene of the avirulent cell culture-derived mutant virus. The CMVC proteins have mostly lost their ability to inhibit viral RNA synthesis, and their electrophoretic mobility can also be distinguished from those coded for by the silenced C ORF of pGEM-HAP/Coff (6). We were particularly interested in the results with pGEM-HAP/Coff, which performs better than the other P/C gene support plasmids in genome amplification and virus recoveries (data not shown, and see below). Virus recoveries from pFL3 and pFL3-Cstop were again carried out in parallel with either the pGEM-HAP/Coff or pGEM-P/CMVC support plasmid. rSeV was routinely recovered from pFL3 as before, and in these cases the virus could also be recovered at low frequency (one in four attempts) from the pFL3-Cstop transfections. However, in both of these cases it is possible that the Cstop mutation of pFL3-Cstop was lost by recombination with the pGEM-P/C support plasmids, even though the conditions for recombination were not favorable in either case and recombination would be expected to occur only at low frequency. Virus recovered from these pFL3-Cstop transfections was therefore used to infect BHK cells, and the presence or absence of C proteins intracellularly was monitored by immunoblotting. As shown in Fig. 2, the Cprime and C proteins were clearly expressed by the recovered viruses. Moreover, their electrophoretic mobility in each case was identical to that of the particular pGEM-P/C used; i.e., these viruses had presumably arisen by DNA recombination between pFL3-P/Cstop and the P/C gene support plasmid. Therefore, we have been unable to prepare a totally C-minus virus that can be amplified in eggs, even after multiple passages. While this paper was under review, we learned that Kurotani et al. (21a) were eventually able to prepare a virus slightly different from Cstop [named 4C(−)], which was detectable only at extremely low PFU.
rSeV expressing two or three C proteins.
To eliminate the expression of individual C proteins, we mutated their start codons individually or in combination, as detailed in Fig. 1. The mutations were first constructed in the pGEM-P/C support plasmids, so that these genes could be controlled for their ability to express the various C proteins. Mutation of ACG81/C′ to TCT and ATG114/C to GCG eliminated expression of the Cprime and C proteins respectively, as expected (data not shown [cf. Fig. 1]). Mutation of ATG114/C to GCG introduces an Asp4Gly substitution in the P protein, but the only suitable P-silent mutation (ATG114/C to ACG) is unreliable in this case. The Asp4Gly substitution had no noticeable effect on the ability of P to support minigenome RNA synthesis in transfected cells. Our attempts to eliminate Y1 and Y2 expression by simultaneously mutating ATG183/Y1 and ATG201/Y2 to ACG (both silent in the P ORF) were unsuccessful. These mutations had only a minor effect in reducing Y1 and Y2 expression. Further, their simultaneous mutation to GCG more strongly reduced but did not eliminate Y1 and Y2 expression (data not shown). The Y proteins appear to be initiated by an unusual mechanism, in which the codon-anticodon interactions at the ribosomal start site are considerably less important than at standard start sites (23). Hence, we have so far been unable to prepare rSeV strains which express Cprime and C but not Y1 and Y2.
Recombinant SeV-[Cprime-minus] was routinely recovered from the transfections with either pGEM-HAP/Coff or pGEM-P/Cstop as the support plasmid, and this virus grew to the same levels as that recovered from the wt control (by the second passage in eggs). Immunoblots of BHK cells infected with this virus showed that Cprime protein expression was specifically ablated whereas C, Y1, and Y2 protein levels were normal (Fig. 1). Recombinant SeV-[C-minus] was also routinely recovered from the transfections but only with pGEM-HAP/Coff as the support plasmid. Immunoblots of BHK cells infected with [C-minus] showed that C protein expression was specifically ablated whereas Cprime protein accumulation was normal. However, Y1, and Y2 protein levels had risen strongly, so that their combined level easily replaced that of the missing C protein (Fig. 1). [Cprime/C-minus], the start codon double mutant, in contrast, was recovered from the transfections in only one-third of the attempts and again only with pGEM-HAP/Coff as the support plasmid, and it reached titers suitable for high-multiplicity cell culture infection by passage 4 (see below). Immunoblots of extracts of cells infected with [Cprime/C-minus] showed that Cprime and C protein expression was specifically ablated and that Y2 protein levels alone had risen strongly. It is possible that some of the increase in Y1 and Y2 levels when C or Cprime plus C expression is ablated is simply due to increased initiation at ATG183/Y1 and ATG201/Y2 when ATG114/C is no longer functional. The increasing difficulty in recovering rSeV strains which do not express the Cprime, C, and Cprime plus C proteins is in line with our inability to recover a totally C-minus virus.
Titers of the rSeV stocks.
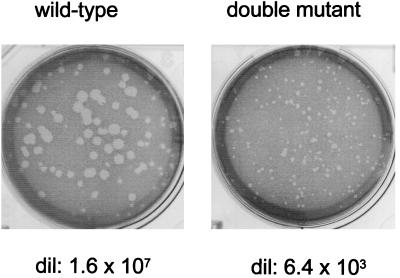
Allantoic fluid stocks suitable for high-multiplicity infections of cell cultures (5 × 108 to 20 × 108 PFU/ml) were routinely prepared in recoveries of [Cprime-minus] and [C-minus], as in the parallel rSeVZ or rSeVZ-NH wt controls [Ge (MK2), Fig. 3]. These virus stocks also contained roughly similar amounts of virus proteins (data not shown; see Materials and Methods). [Cprime/C-minus] (the double mutant) was recovered twice (dm1 and dm2), and although fourth- passage stocks of these viruses contained considerable amounts of viral proteins (only ca. 10-fold less than the wt stock [Fig. 3]), we were unable to determine the titers of the double-mutant stocks in Geneva. In contrast to the three other rSeV strains, which formed clear plaques 1 to 2 mm in diameter by day 4, the double mutant did not form visible plaques by day 5, when our serum-free monolayers began to disintegrate. A fresh stock of dm2, however, did form very small plaques by day 7 in Kobe (Fig. 4), albeit with a titer of only 4 × 106 PFU/ml, i.e., 3 log units lower than that for the wt control (6 × 109 PFU/ml) (Fig. 3).
FIG. 4.
Plaque formation of recombinant wt and double-mutant virus. Serial dilutions of wt (rSeVZ-NH) and dm2Kobe stocks were allowed to form plaques for 9 days at 33°C on LLC-MK2 cells in the presence of trysin and the absence of serum. An example of the plaques formed by each virus and its dilution is shown.
The PFU titer (on MK2 cells) of the dm2 stock, however, was inconsistent with the finding that 107 BHK cells infected with 1 ml of a 1/5 dilution of dm2Ge accumulated roughly as much C proteins as did those infected with 10 PFU of wt virus (or either of the single mutants) per cell (Fig. 1). We therefore estimated the titer of the dm2Ge stock by infecting BHK cells with serial dilutions and determining the levels of N protein intracellularly at 40 h pi (see Materials and Methods), i.e., its TCID50. The dm2Ge stock was estimated to contain ca. 108 (BHK) TCID50/ml, and our wt stock was estimated to contain 1010 (BHK) TCID50/ml. The relative infectivities of these stocks were further estimated by performing limiting dilutions in eggs (EID50 determinations; see Materials and Methods). The wt stock contained 6 × 109 EID50/ml, whereas the dm2Ge stock contained only 1 × 103 EID50/ml. Thus, in contrast to the wt and single-mutant virus stocks, whose PFU, TCID50, and EID50 titers were relatively constant (2 × 109 to 10 × 109 and 5 × 108 to 6 × 108 IU/ml, respectively), the titer of the dm stock depended heavily on the method by which the numbers of infectious units were determined (Fig. 3). Relative to the wt virus stocks, the dm stocks contained ca. 10-fold less protein, 100-fold less (BHK) TCID50, 1,000-fold less PFU, and 1,000,000-fold less EID50.
Infection of cells in culture.
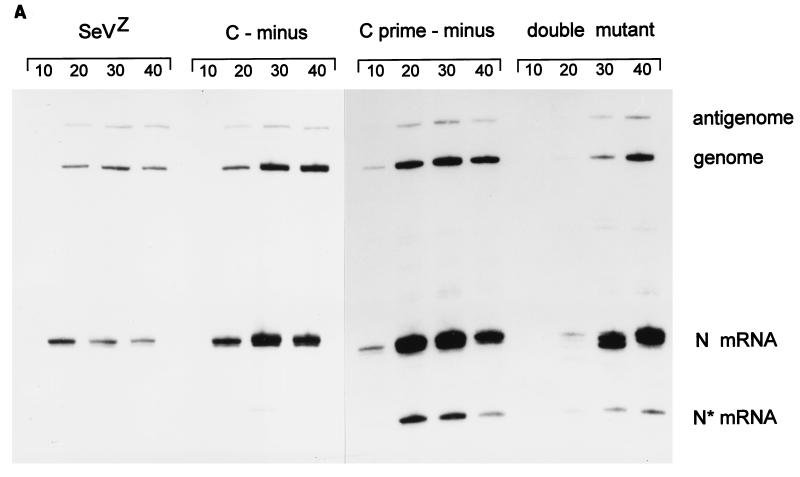
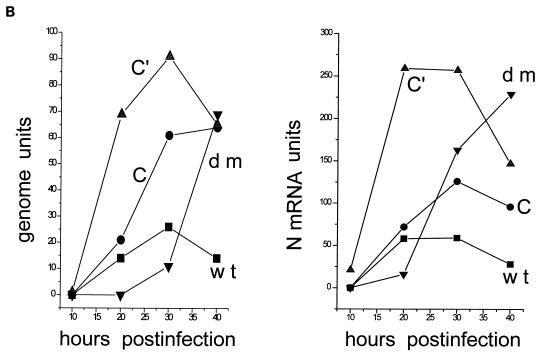
Parallel 4-cm-diameter BHK cultures (ca. 107 cells) were infected with 2 (MK2) PFU of wt, [Cprime-minus], or [C-minus] per cell and 2 (BHK) TCID50 of dm2Ge per cell [2 PFU of wt virus per cell is equivalent to 10 to 20 (BHK) TCID50/cell (Fig. 3)]. Infected cultures were harvested at various times after infection, and the relative amounts of viral genomes, antigenomes, and N mRNAs present in equal samples of the total RNA (one-quarter of each dish) were estimated by duplex primer extension (Fig. 5A and B). Relative to the wt infection, the [Cprime-minus] and [C-minus] infections led to the accumulation of significantly more viral RNA, and this was most apparent in the [Cprime-minus] infections, where the accumulation occurred earlier and to higher levels. The double-mutant infection, in contrast, led to the accumulation of relatively little viral RNA by 20 h p.i. From 20 to 30 h p.i., however, a large increase in the amounts of the three viral RNAs occurred in this infection. In particular, N mRNAs had now accumulated to higher levels than in the wt and [C-minus] infections (but remained below those of the [Cprime-minus] infection) and the genome and antigenome levels approached those of the wt infection by 30 h p.i. N mRNA levels continued to increase until 40 h p.i. in the double-mutant infection and reached levels equivalent to the peak of the [Cprime-minus] infection, whereas the N mRNA levels decreased from 30 to 40 h p.i. infection in the three other infections. Similarly, genomes continued to accumulate from 30 to 40 h p.i. in the double-mutant infection, such that their abundance was equal to those in the [Cprime-minus] and [C-minus] infections at this time, which were all fourfold higher than in the wt infection at 40 h p.i. Independent isolates of all three mutant viruses (e.g., dm1 and dm2) behaved similarly in cell culture infections (data not shown).
FIG. 5.
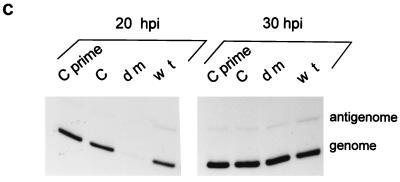
Accumulation of viral RNAs in the mutant and wt virus-infected BHK cells. Parallel 4-cm-diameter BHK cultures (ca. 107 cells) were infected with 2 (MK2) PFU or 20 (BHK) TCID50 of wt, [Cprime-minus], or [C-minus] per cell and 2 (BHK) TCID50 of dm2Ge per cell. Infected cultures were harvested at various times after infection, as indicated above the lanes. The relative amounts of viral genomes, antigenomes, and N mRNAs present in equal samples of the total RNA (from one-quarter of each dish) were estimated by duplex primer extension with NP126, whose 5′ end is 126 and 70 nucleotides from that of the antigenome and N mRNA, respectively, and L15,270, whose 5′ end is 114 nucleotides from that of the genome. (A) The extended primers were separated on an 8% sequencing gel. N* mRNA is presumably a degradation product of the N mRNA. (B) The intensities of the various bands were determined in a PhosphorImager, and their values are plotted in terms of genomes and N and N* mRNA. (C) Accumulation of genomes and antigenomes in cultures infected individually with 2 (BHK) TCID50 of each virus per cell (or 0.2 PFU of wt, [Cprime-minus] or [C-minus] per cell), as above. hpi, hours p.i.
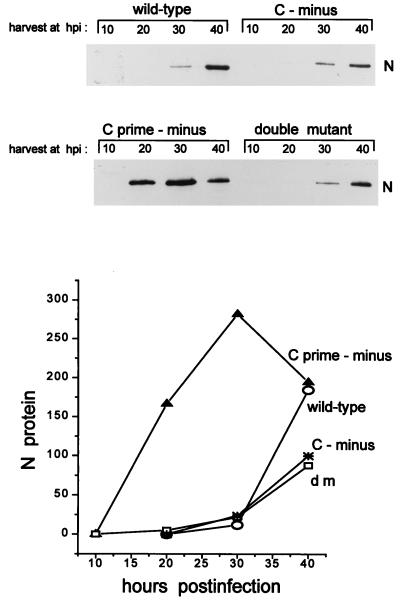
The supernatants of the various infections were also harvested, and the relative numbers of virus particles in these fluids were estimated by immunoblotting for N protein (see Materials and Methods). As shown in Fig. 6, the [C-minus] and dm2Ge infections liberated slightly less virion N protein into the medium than did the wt infection, but with similar kinetics, even though both these mutant infections led to fourfold more viral genomes intracellularly than did the wt infection (Fig. 5A and B). The [Cprime-minus] infection, in contrast, liberated virion N protein into the medium at earlier times and in larger amounts than did the wt infection, consistent with the amounts of Cprime-minus genomes intracellularly. The [C-minus] and double-mutant infections may thus be defective in virus maturation; if this is so, this defect is presumably due to the absence of the C protein. These results also suggest that the inhibition of viral RNA synthesis at the beginning of the wt and [C-minus] infections is due primarily to the presence of Cprime.
FIG. 6.
Accumulation of viral N protein in the culture supernatants. The supernatants of the infected cultures in Fig. 5A and B were harvested at the times indicated. Virus was pelleted through a glycerol cushion (see Materials and Methods), dissolved in sample buffer, separated by SDS-PAGE (10% polyacrylamide), and immunoblotted with an anti-N monoclonal antibody. The intensities of the various bands were determined in a densitometer, and their values are plotted below the gels. hpi, hours p.i.
The wt, [Cprime-minus], and [C-minus] infections were all carried out at 2 to 20 IU/cell, and the vast majority of these cells were thus infected. The dm2-infected cultures must then have been infected with at least 1 IU/cell [consistent with the (BHK) TCID50 titer of the stock], since these infections lead to accumulation of the same levels of genomes as those found in the other mutant infections at 40 h p.i. The dm2 infections in Fig. 5A and B were nevertheless initiated at a 10-fold-lower (BHK) TCID50/cell than the other infections, and some of the 10-h delay in RNA accumulation in the experiment in Fig. 5B may be due to the lower MOI. To determine whether the simultaneous absence of the Cprime and C proteins also played a role in this delay, cultures infected with 2 TCID50 of each of the four viruses per cell were also compared as above. As shown in Fig. 5C, the dm infection again led to a significantly smaller number of genomes than did the three other infections at 20 h p.i., but the difference had been made up by 30 h p.i. The simultaneous absence of both the Cprime and C proteins during infection thus appears to result in a delay in the accumulation of the viral RNAs.
Infection of mice.
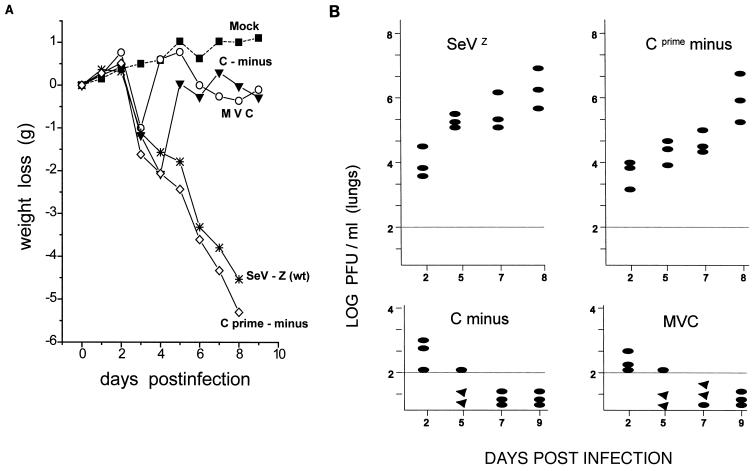
Groups of 7-week-old, specific-pathogen-free C57BL/6 mice were inoculated intranasally with 106 PFU of [Cprime-minus] or [C-minus], as well as SeVZ as a virulent wt virus control. As an avirulent virus control, the same mice were also infected with SeVMVC, which is avirulent for 3-week-old (outbred) ICR mice (18, 37). All the mice were weighed daily as a general indicator of their health (Fig. 7A), since loss of body weight is a useful indicator of SeV pathogenicity in these animals (20). Groups of three mice were sacrificed at various times p.i., and the virus titers (PFU on MK2 cells) in their lungs homogenates were determined.
FIG. 7.
Infections of C57BL/6 mice. Groups of 7-week-old C57BL/6 mice (weighing ca. 20 g) were infected intranasally with 106 PFU of either SeVZ (wt), SeVMVC, or the recombinant [C-minus] or [Cprime-minus]. Phosphate-buffered saline was used for the mock-infected mice. (A) The body weights of the mice were measured daily and are given as weight loss relative to the weight on day 0. Three mice from each of the infected groups were sacrificed at the times indicated, and the PFU present in 10% lung homogenates were determined on LLC-MK2 cells. (B) The titers for each of the three mice are shown individually. Triangles (below the limit of detection by this method [102 PFU/lobe]) indicate that injecting these homogenates into eggs and passaging twice does lead to detectable virus; ovals indicate that virus could not be detected by this procedure.
As shown in Fig. 7A, [Cprime-minus] was highly virulent like the SeVZ-wt control, since these virus-infected mice all steadily lost weight starting on day 3 and either died or were killed in extremis by day 8. Mice infected with [C-minus] or SeVMVC also began to lose weight starting on day 3. However, in contrast to the [Cprime-minus]- and wt-infected animals, all these mice stopped losing weight by day 4 or 5 and then quickly regained weight, so that their body weight was similar to that of the mock-infected control animals by day 5 or 6. None of these mice died, and they all appeared to be in good health at day 8. Consistent with these results, infectious virus accumulated during the course of the [Cprime-minus] and wt infections, reaching 106 PFU/lobe (but was somewhat delayed in the [Cprime-minus] infection). On the other hand, detectable virus (>102 PFU/lobe) was present only on day 2 of the [C-minus] or SeVMVC infections and was absent by day 5 (Fig. 7B). As a more sensitive test for the presence of virus, eggs were infected with the lung homogenates and passaged twice. Two of three mice were still virus positive on day 5 of the [C-minus] or SeVMVC infections (Fig. 7B), but all three were virus negative by day 9. The virus present in the lungs was amplified and used to infect BHK cells. In all cases, the pattern of C protein accumulation was found to be identical to that of the rSeV that had initiated the mouse infection (data not shown [cf. Fig. 1]). The relevant region of the viral genomes was also amplified by RT/PCR and sequenced, and in all cases the mutations introduced to alter the start codons were found to be stable (not shown). The LD50s of these viruses for 3-week-old (outbred) ICR mice were determined (Table 1). Relative to rSeVZ-NH (our wt control), the LD50 of [Cprime-minus] was very similar whereas that of [C-minus] had increased by ca. 2 log units, consistent with the results (weight loss and lung virus titer) obtained in inbred C57BL/6 mice. None of the mice died after infection with the highest titer of the dm2Kobe stock.
TABLE 1.
Mouse pathogenicity of rSeV-C protein variants
| Inoculum (CIU/mouse) | No. of mice which survived/total no. of mice given indicated virus
|
||
|---|---|---|---|
| rSeVZ - NH | C-minus | Cprime-minus | |
| 1.25 × 107 | NDa | 0/2 | ND |
| 1.25 × 106 | ND | 0/4 | ND |
| 1.25 × 105 | ND | 3/4 | ND |
| 1.25 × 104 | 0/4 | 4/4 | 0/4 |
| 1.25 × 103 | 4/4 | ND | 3/4 |
| 1.25 × 102 | 4/4 | ND | 4/4 |
| 1.25 × 101 | 2/2 | ND | ND |
| LD50 (CIU/mouse) | 6.9 × 103 | 5.0 × 105 | 5.0 × 103 |
ND, not determined.
Thus, rSeV strains which cannot specifically express Cprime but whose expression of C, Y1, and Y2 is normal, grows to high titers in mouse lungs like wt virus and remains highly virulent for these animals. On the other hand, rSeV strains which specifically cannot express C but which express Cprime normally and whose levels of Y1 and Y2 are increased is quickly cleared from the mouse lungs and is no longer highly virulent for these animals. The C protein (i.e., that initiated at ATG114) is thus specifically required for virulence in mice.
DISCUSSION
It has been suspected for some time that the paramyxovirus C proteins play an important role in natural infections. In contrast to the V proteins (which are coded for in part by the other ORF that overlaps the P ORF [Fig. 1]) (32, 34), C proteins are expressed in all viruses of the Respirovirus and Morbillivirus genera. Furthermore, in long-term persistent MeV infections of the human brain (subacute sclerosing parencephalitis), where expression of the M protein and the cytoplasmic tail of the F protein are often lost by point mutations which close these ORFs, the C protein ORF has always remained intact (4). The paramyxovirus C proteins have nevertheless remained a riddle, since it has never been clear why some viruses express four C proteins and some express only one. More recently, characterization of the C gene of an avirulent mutant virus has provided evidence that these proteins are required for multiple cycles of replication in mice but dispensable for growth in hen eggs or cell culture (13). This latter property, however, is unlikely to represent the pressure maintaining the integrity of this overlapping ORF, especially in reference strains that have been passaged in eggs for countless generations. This paper provides more direct evidence that the SeV C proteins play an important role in virus replication at the cellular level, as well as hinting at why this virus expresses a nested set of four C proteins. We have been unable to prepare a totally C-minus virus which grows to detectable levels in eggs, even though we have recovered rSeV strains which cannot express their V and/or W proteins by the same approach (9, 10). We have, however, been able to prepare rSeV strains that cannot express their Cprime, C, or Cprime plus C proteins, by mutating their ribosomal start codons. The characterization of their infections of cell culture (and eggs) has added insight into the possible function(s) of these proteins.
In one respect, our results agree with previous findings that coexpression of either Cprime or C individually (along with the P and L proteins) leads to an inhibition of RNA synthesis in transfected cell extracts whereas coexpression of Y1 and Y2 was not inhibitory (8). These results suggested that one function of Cprime or C is to negatively affect viral mRNA synthesis. In this view, viral transcription is less inhibited in the [C-minus] infection than in the wt infection due to the absence of C, and the overaccumulation of Y1/Y2 cannot compensate for this absence. Similarly, transcription has apparently been even less inhibited in the [Cprime-minus] infections, presumably because Cprime is the more inhibitory of the two. However, in contrast to [Cprime-minus] infections, where the viral RNAs overaccumulate precociously, the simultaneous absence of both Cprime and C in the double-mutant infections leads to a delay in viral RNA accumulation. This suggests that Cprime and C also provide a positive function without which the onset of viral RNA accumulation is delayed. We presume that either the Cprime or C proteins (but not the Y proteins) can supply this required function in natural infections. Viruses unable to express either protein individually would have this function provided by the remaining Cprime or C protein and would not undergo a delay in viral RNA accumulation.
Although viral RNA synthesis is delayed in the double-mutant infection, once started, it quickly leads to the overaccumulation of the viral RNAs as seen in the [Cprime-minus] infection, consistent with the absence of the inhibitory Cprime (and C) protein. The remaining Y1 and Y2 proteins apparently cannot compensate for the absence of Cprime and C in this delay, but this may be misleading. It is presumably the presence of Y1 and Y2 expression in the Cprime and C-minus infection that accounts for the relative viability of the double-mutant virus with respect to rSeV-Cstop. Y1 and Y2 may to some extent also provide the required function, but less efficiently, accounting for the delay in the onset of viral RNA accumulation. Furthermore, we note that in contrast to transfected-cell systems, where (ectopic) C protein expression inhibits minigenome replication in a promoter-specific manner, we have seen no evidence for this in natural virus infections. Although mutation in the C gene (13, 18) or elimination of Cprime and/or C expression (Fig. 5) leads to significantly higher levels of mRNA, the ratios of antigenomes to genomes remain unaltered. The inhibitory effects of C may thus act primarily on mRNA transcription in natural virus infections, as opposed to acting on antigenome synthesis. We also note that the simultaneous ablation of all C protein expression (Cstop) does not adversely affect genome amplification in transfected cells (6, 31), as it does during natural virus infections. We presume that the positive function of C required early in natural virus infection is somehow bypassed in the transfected-cell system, where SeV protein expression is no longer under the control of the viral genome.
The view that Cprime and C provide a positive function for virus replication is supported by the specific infectivities of these virus stocks prepared in eggs. Viruses in which Cprime or C have been individually ablated grow only slightly less well in eggs than wt virus does (i.e., the titers of their stocks are only 4- and 10-fold lower by PFU and EID50, respectively [Fig. 3]). The double-mutant virus stock, however, contains 3 log units less (MK2) PFU than does the wt virus and 6 log units less EID50 (even though the amount of virus protein in these allantoic fluids is reduced by only 1 log unit). As before, either Cprime or C (but not the Y proteins) can presumably supply this required function (for growth in eggs), and so only the double mutant displays the highly debilitated phenotype. One explanation for the exaggerated effect on EID50 is that amplification of the infectious unit is more exponential here and may require more cycles of replication to be detected. Infections that liberate fewer virus particles (Fig. 6) and with lower specific infectivities (Fig. 3) might be expected to be associated with a particularly low EID50. Nevertheless, the double-mutant stocks would contain almost as many virions as the wt virus stocks would when eggs are inoculated with high doses, since fewer cycles of replication are needed to reach these levels.
In spite of the similar abilities of [Cprime-minus] and [C-minus] to be amplified in eggs, [Cprime-minus] accumulated to high levels in the lungs of these infected mice and remained highly virulent for this animal host (like SeVZ) whereas infectious virus did not accumulate in the lungs of [C-minus]-infected mice, and this virus infection was strongly attenuated. These results further suggest that the Cprime and C proteins cannot be functionally equivalent in all respects, since C can apparently replace Cprime for virulence in mice whereas Cprime (or Y1 and Y2) cannot replace C for this property. This latter result is somewhat surprising, since Cprime contains all the sequences of C plus an 11-amino-acid N-terminal extension. This difference in virulence may be due to less [C-minus] than [Cprime-minus] being liberated during the initial infection of the mouse respiratory epithelium, similar to what is seen in BHK cells (Fig. 6), allowing the host antiviral defense mechanisms enough time to operate. Some of these antiviral defense mechanisms (e.g., natural killer cells) would presumably be absent from hen eggs.
The view of the SeV C proteins that emerges from this study is that their expression is complex because the different C proteins carry out nonoverlapping functions to some extent. For example, Cprime and C (but not Y1 and Y2) act as inhibitors of viral RNA synthesis, and C (but not Cprime, Y1, or Y2) is specifically required for virulence (multiple cycles of replication) in an animal host. Furthermore, some (but not all) of these proteins are also apparently required for an essential function(s) in virus replication at the cellular level, which remains to be elucidated. This functional complexity, due to a nested set of C proteins, is made possible by the translational gymnastics used by this mRNA to direct ribosomal initiation at the various start codons (7). Cprime is initiated at a non-ATG codon (ACG81), where the bases at positions +5 and +6, in addition to those at positions −3 and +4, play an important role (5, 15). P, V, and W (all ATG104) and C (ATG114) are initiated by leaky scanning ribosomes, since mutating ACG81 to ATG ablates initiation from ATG104/P and ATG114/C. Initiation from ATG183/Y1 and ATG201/Y2, in contrast, does not appear to be due to a leaky scanning mechanism, since the translation of Y1 and Y2 in these constructs is either unaffected or enhanced (7). Rather, initiation from ATG183/Y1 and ATG201/Y2 appears to be due to a shunting mechanism, in which ribosomes are translocated from a region upstream of ACG81 directly to an acceptor site, which includes ATG183/Y1 and ATG201/Y2 (23). The tripartite leader of adenovirus late mRNAs mediates a similar shunting mechanism, and in this case shunting is promoted by heat shock (39). Translation of the four SeV C proteins does not respond to heat shock (data not shown). However, the relative amounts of the various P gene products sometimes differ widely in cell culture infections (e.g., Y1 and Y2 are sometimes undetectable [Fig. 2] and the ratio of Cprime to C varies severalfold). Translation of the four SeV C proteins might respond to other stress-related modifications of translational capacity, such as those induced by interferon (35). Different mechanisms of translational initiation (non-ATG codons, leaky scanning, and ribosomal shunting) may have different requirements for initiation factors. If so, the relative amounts of the various P gene products may be determined in part by the stress-related modifications of the host cell translational apparatus. These products would then be well placed to act as sensors of the host antiviral defense mechanisms, particularly those of the innate immune system, and to thwart this defense accordingly.
Our results suggest that Cprime and C provide a positive function for viral RNA accumulation early in intracellular virus replication but that they act as inhibitors at later times. Although these two properties of the C proteins act in opposite directions, it is possible that they are, in fact, the same property. Whereas the relative ratios of P, L, and the N:RNA template are constant throughout the virus life cycle, the C proteins are very abundant at the later stages of infection but are largely excluded from virions. Intracellular viral RNA synthesis thus begins with relatively little C in proportion to P, L, and N:RNA, and this ratio increases as the infection proceeds. The opposite effects of C early and late in infection could then be due to the different ratios of C to the other viral components involved in RNA synthesis. Exactly how C acts in modulating RNA synthesis is far from clear. The C proteins (as glutathione S-transferase fusion proteins) bind the L protein (17) but apparently act through P, since their inhibitory effects (on viral RNA synthesis in transfected cells) are relieved by overexpressing P and not by overexpressing L (8, 31). We have suggested that high concentrations of C act as viral fidelity factors by somehow causing the viral polymerase to be more selective in its choice of promoters and that this results in a less active enzyme (31). It is possible that the low concentrations of C found intracellularly at the start of the replicative cycle interact with the P-L polymerase in a similar manner but that these limited interactions have a positive (as opposed to a negative) effect on polymerase activity. If so, the trace amounts of these proteins found in virions could play an important role in the initiation of viral RNA accumulation in infected cells.
It is also possible that the C proteins exert their positive effects indirectly, by helping to form active polymerase complexes intracellularly prior to virion release. The vif protein (viral infectivity factor) is a late-gene product of most lentiviruses and acts during virus assembly to allow the formation of particles competent for the early steps of infection (reviewed in reference 33). The effect of vif is probably indirect, since (like the SeV C proteins) there is very little vif protein in virions. Human immunodeficiency virus type 1 strains with mutations in the vif gene efficiently produce virus particles, but these fail to carry out proviral DNA synthesis due to improperly packed nucleoprotein cores (16, 36). Cprime/C-minus SeV prepared in eggs have higher particle/PFU ratios than do wt or single-mutant virions (Fig. 3). The delay in viral RNA accumulation seen at the start of the double-mutant infections may then be due not only to the paucity of C proteins at this time but also to improperly assembled viral cores [P + L + N:RNA] contained in these virions.
REFERENCES
- 1.Alkhatib G, Massie B, Briedis D J. Expression of bicistronic measles virus P/C mRNA by using hybrid adenoviruses: levels of C protein synthesized in vivo are unaffected by the presence or absence of the upstream P initiator colon. J Virol. 1988;62:4059–4069. doi: 10.1128/jvi.62.11.4059-4069.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barr J, Chambers P, Harriott P, Pringle C R, Easton A J. Sequence of the phosphoprotein gene of pneumonia virus of mice: expression of multiple proteins from two overlapping reading frames. J Virol. 1994;68:5330–5334. doi: 10.1128/jvi.68.8.5330-5334.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellini W J, Englund G, Rozenblatt S, Arnheiter H, Richardson C D. Measles virus P gene codes for two proteins. J Virol. 1985;53:908–919. doi: 10.1128/jvi.53.3.908-919.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billeter M A, Cattaneo R, Spielhofer P, Kaelin K, Huber M, Schmid A, Baczko K, ter Meulen V. Generation and properties of measles virus mutations typically associated with subacute sclerosing panencephalitis. Ann N Y Acad Sci. 1994;724:367–377. doi: 10.1111/j.1749-6632.1994.tb38934.x. [DOI] [PubMed] [Google Scholar]
- 5.Boeck R, Kolakofsky D. Positions +5 and +6 can be major determinants of the efficiency of non-AUG initiation codons for protein synthesis. EMBO J. 1994;13:3608–3617. doi: 10.1002/j.1460-2075.1994.tb06668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cadd T, Garcin D, Tapparel C, Itoh M, Homma M, Roux L, Curran J, Kolakofsky D. The Sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter-specific fashion. J Virol. 1996;70:5067–5074. doi: 10.1128/jvi.70.8.5067-5074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curran J, Latorre P, Kolakofsky D. Translational gymnastics on the Sendai virus P/C mRNA. Semin Virol. 1998;8:351–357. [Google Scholar]
- 8.Curran J, Marq J-B, Kolakofsky D. The Sendai virus nonstructural C proteins specifically inhibit viral mRNA synthesis. Virology. 1992;189:647–656. doi: 10.1016/0042-6822(92)90588-g. [DOI] [PubMed] [Google Scholar]
- 9.Delenda C, Hausmann S, Garcin D, Kolakofsky D. Normal cellular replication of Sendai virus without trans-frame nonstructural V protein. Virology. 1997;228:55–62. doi: 10.1006/viro.1996.8354. [DOI] [PubMed] [Google Scholar]
- 10.Delenda C, Taylor G, Hausmann S, Garcin D, Kolakofsky D. Sendai viruses with altered P, V, and W protein expression. Virology. 1998;242:327–337. doi: 10.1006/viro.1998.9027. [DOI] [PubMed] [Google Scholar]
- 11.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcin D, Pelet T, Calain P, Roux L, Curran J, Kolakofsky D. A highly recombinogenic system for the recovery of infectious Sendai paramyxovirus from cDNA: generation of a novel copy-back nondefective interfering virus. EMBO J. 1995;14:6087–6094. doi: 10.1002/j.1460-2075.1995.tb00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcin D, Itoh M, Kolakofsky D. A point mutation in the Sendai virus accessory C proteins attenuates virulence for mice, but not virus growth in cell culture. Virology. 1997;238:424–431. doi: 10.1006/viro.1997.8836. [DOI] [PubMed] [Google Scholar]
- 14.Giorgi C, Blumberg B M, Kolakofsky D. Sendai virus contains overlapping genes expressed from a single mRNA. Cell. 1983;35:829–836. doi: 10.1016/0092-8674(83)90115-0. [DOI] [PubMed] [Google Scholar]
- 15.Grunert S, Jackson R J. The immediate downstream codon strongly influences the efficiency of utilization of eukaryotic translation initiation codons. EMBO J. 1994;13:3618–3630. doi: 10.1002/j.1460-2075.1994.tb06669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Höglund S, Ohagen A, Lawrence K, Gabudzda D. Role of vif during packing of the core of HIV-1. Virology. 1994;201:349–355. doi: 10.1006/viro.1994.1300. [DOI] [PubMed] [Google Scholar]
- 17.Horikami S M, Hector R E, Smallwood S, Moyer S A. The Sendai virus C protein binds the L polymerase protein to inhibit viral RNA synthesis. Virology. 1997;235:261–270. doi: 10.1006/viro.1997.8702. [DOI] [PubMed] [Google Scholar]
- 18.Itoh M, Isegawa Y, Hotta H, Homma M. Isolation of an avirulent mutant of Sendai virus with two amino acid mutations from a highly virulent field strain through adaptation to LLC-MK2 cells. J Gen Virol. 1997;78:3207–3215. doi: 10.1099/0022-1317-78-12-3207. [DOI] [PubMed] [Google Scholar]
- 19.Kato A, Kiyotani K, Sakai Y, Yoshida T, Nagai Y. The paramyxovirus Sendai virus V protein encodes a luxury function required for viral pathogenesis. EMBO J. 1997;16:678–587. doi: 10.1093/emboj/16.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiyotani K, Takao S, Sakaguchi T, Yoshida T. Immediate protection of mice from lethal Sendai virus infection by a temperature sensitive mutant (HVJpi) possessing homologous interfering capacity. Virology. 1990;177:65–74. doi: 10.1016/0042-6822(90)90460-9. [DOI] [PubMed] [Google Scholar]
- 21.Kretzschmar E, Peluso R, Schnell M J, Whitt M A, Rose J K. Normal replication of vesicular stomatitis virus without C proteins. Virology. 1996;216:309–316. doi: 10.1006/viro.1996.0066. [DOI] [PubMed] [Google Scholar]
- 21a.Kurotani A, Kiyotani K, Kato A, Shioda T, Saki Y, Mizumoto K, Yoshida T, Nagai Y. The paramyxovirus, Sendai virus, C proteins are categorically nonessential gene products but silencing their expression severely impairs viral replication and pathogenesis. Genes Cells. 1998;3:111–124. doi: 10.1046/j.1365-2443.1998.00170.x. [DOI] [PubMed] [Google Scholar]
- 22.Lamb R A, Paterson R G. The nonstructural proteins of paramyxoviruses. In: Kingsbury D W, editor. The paramyxoviruses. New York, N.Y: Plenum Press; 1991. pp. 181–210. [Google Scholar]
- 23.Latorre, P., D. Kolakofsky, and J. Curran. The Sendai virus Y proteins are initiated by a ribosomal shunt. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 24.Murphy F A, Fauquet C M, Bishop D H L. Sixth report of the international committee on taxonomy of viruses. Arch Virol Suppl. 1994;10:268–274. [Google Scholar]
- 25.Orvell C, Grandien M. The effects of monoclonal antibodies on biological activities of structural proteins of Sendai virus. J Immunol. 1982;129:2779–2787. [PubMed] [Google Scholar]
- 26.Pelet T, Delenda C, Gubbay O, Garcin D, Kolakofsky D. Partial characterization of a Sendai virus replication promoter and the rule of six. Virology. 1996;224:405–414. doi: 10.1006/viro.1996.0547. [DOI] [PubMed] [Google Scholar]
- 27.Portner A, Gupta K C, Seyer J M, Beachey E H, Kingsbury D W. /87. Localization and characterization of Sendai virus nonstructural C and C′ proteins by antibodies against synthetic peptides. Virus Res. 1986;6:109–121. doi: 10.1016/0168-1702(86)90043-2. [DOI] [PubMed] [Google Scholar]
- 28.Radecke F, Billeter M A. The nonstructural C protein is not essential for multiplication of Edmunston B strain measles virus in cultured cells. Virology. 1996;217:418–421. doi: 10.1006/viro.1996.0134. [DOI] [PubMed] [Google Scholar]
- 29.Reed L J, Muench H. A simple method of estimating fifty per cent endpoints. Am J Hyg. 1938;23:493–497. [Google Scholar]
- 30.Spiropoulou C F, Nichol S T. A small highly basic protein is encoded in overlapping frame within the P gene of vesicular stomatitis virus. J Virol. 1993;67:3103–3110. doi: 10.1128/jvi.67.6.3103-3110.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tapparel C, Hausmann S, Pelet T, Curran J, Kolakofsky D, Roux L. Inhibition of Sendai virus genome replication due to promoter-increased selectivity: a possible role for the accessory C proteins. J Virol. 1997;71:9588–9599. doi: 10.1128/jvi.71.12.9588-9599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas S M, Lamb R A, Paterson R G. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell. 1988;54:891–902. doi: 10.1016/S0092-8674(88)91285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trono D. HIV accessory proteins: leading roles for the supporting cast. Cell. 1995;82:189–192. doi: 10.1016/0092-8674(95)90306-2. [DOI] [PubMed] [Google Scholar]
- 34.Vidal S, Curran J, Kolakofsky D. Editing of the Sendai virus P/C mRNA by G insertion occurs during mRNA synthesis via a virus-encoded activity. J Virol. 1990;64:239–246. doi: 10.1128/jvi.64.1.239-246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vilcek J, Sen G C. Interferons and other cytokines. In: Fields B N, Knipe D N, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 375–399. [Google Scholar]
- 36.von Schwedler U, Song J, Aiken C, Trono D. vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J Virol. 1993;67:4945–4955. doi: 10.1128/jvi.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X-L, Itoh M, Hotta H, Homma M. A protease activation mutant, MVCES1, as a safe and potent live vaccine derived from currently prevailing Sendai virus. Virology. 1994;68:3369–3373. doi: 10.1128/jvi.68.5.3369-3373.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamada H, Hayata S, Omata-Yamada T, Taira H, Mizumoto K, Iwasaki K. Association of the Sendai virus C protein with nucleocapsids. Arch Virol. 1990;113:245–253. doi: 10.1007/BF01316677. [DOI] [PubMed] [Google Scholar]
- 39.Yueh A, Schneider R J. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 1996;10:1557–1567. doi: 10.1101/gad.10.12.1557. [DOI] [PubMed] [Google Scholar]