Abstract
We have tested a panel of pediatric and adult human immunodeficiency virus type 1 (HIV-1) primary isolates for the ability to employ the following proteins as coreceptors during viral entry: CCR1, CCR2b, CCR3, CCR4, CCR5, CCR8, CXCR4, Bonzo, BOB, GPR1, V28, US28, and APJ. Most non-syncytium-inducing isolates could utilize only CCR5. All syncytium-inducing viruses used CXCR4, some also employed V28, and one (DH123) used CCR8 and APJ as well. A longitudinal series of HIV-1 subtype B isolates from an infected infant and its mother utilized Bonzo efficiently, as well as CCR5. The maternal isolates, which were syncytium inducing, also used CXCR4, CCR8, V28, and APJ.
Human immunodeficiency virus type 1 (HIV-1) enters CD4+ cells by fusion at the plasma membrane after interactions with CD4 and coreceptor molecules (6, 8, 23, 53). The first coreceptor to be identified was the CXC chemokine receptor CXCR4 (30), closely followed by the CC chemokine receptor CCR5 (3, 17, 21, 24, 25). These two proteins are generally considered to be the most important coreceptors used by HIV-1 strains of the T-tropic and M-tropic phenotypes, respectively (6, 8, 23, 53) (see reference 7 for a discussion of how HIV-1 phenotype is related to coreceptor usage). There is strong epidemiological support for this view in the case of CCR5 (19, 38, 46, 63, 80).
At least nine other chemokine receptors, or structurally related molecules, have also been described as supporting HIV-1 env-mediated membrane fusion or viral entry in vitro. These include CCR2b (1, 24, 31), CCR3 (2, 5, 17, 24, 34, 59–61), BOB/GPR15 (20, 29), Bonzo/STRL33/TYMSTR (20, 45, 47), GPR1 (29), CCR8 (37), US28 (57), V28/CX3CR1 (58, 61), and APJ (16, 27). There is good evidence that CCR3 can be used efficiently by a significant fraction of HIV-1 isolates in vitro, provided the expression of this protein in transfected cells is boosted (2, 5, 59, 61).
Several studies have compared coreceptor usage by primary HIV-1 isolates of several genetic subtypes, but the emphasis has usually been on studying CCR1 to CCR5 and CXCR4 (9, 18, 65, 69, 76, 79). In general, the results were similar to those from more limited studies of the individual receptors; other than the occasional use of CCR3, only CCR5 and CXCR4 were efficiently used by the majority of the isolates tested (9, 18, 65, 69, 76, 79). It is therefore not clear how general, and how efficient, the use by HIV-1 of the more recently identified potential coreceptors actually is. Knowing the range of coreceptors that could be used by HIV-1 in vivo is important for the development of antiviral drugs aimed at inhibiting HIV-1 entry. How many different potential coreceptors must be blocked? To address this question, we have compared the usage of CCR1, CCR2b, CCR3, CCR4, CCR5, CCR8, CXCR4, Bonzo, BOB, GPR1, V28, US28, and APJ by collections of primary HIV-1 isolates of both adult and pediatric origins. We tested pediatric isolates because the unusual characteristics of HIV-1 infection in infants could, in principle, be influenced by a cell tropism pattern that is in turn, determined by the use of coreceptors other than CCR5 or CXCR4 (10, 70, 77).
env complementation assay of coreceptor use.
We first used an env complementation assay to measure the entry of a luciferase-containing HIV-1 reporter virus into U87-CD4 cells transiently transfected with a pcDNA3.1 plasmid encoding either CCR1, CCR2b, CCR3, CCR5, CCR8, CXCR4, Bonzo, BOB, or GPR1 (Table 1). U87-CD4 cells transfected with the unmodified pcDNA3.1 plasmid were used as a control. Entry mediated by seven different HIV-1 envelope glycoproteins was assessed 2 days later by the measurement of luciferase activity in triplicate, essentially as described previously (18, 21, 25, 26). The sources of the env genes have been recorded elsewhere (25, 26), except for Gun1 and Gun1V, which were cloned from isolates provided by Paul Clapham and Hiroo Hoshino (49, 72).
TABLE 1.
env complementation assay of HIV-1 entry into U87-CD4 cells transfected with different coreceptors
| Complemented viral envelopea | Input p24 (ng/ml)b | Viral output (cps/ng of p24)c
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| pcDNA3.1 (background) | CCR1 | CCR2 | CCR3 | CCR5 | CCR8 | CXCR4 | BONZO | BOB | GPR1 | ||
| HXB | 340 | 212 | 64 | 85 | 43 | 162 | 32 | 702,159 | 74 | 95 | 974 |
| NDK | 242 | 268 | 702 | 104 | 89 | 290 | 44 | 851,117 | 74 | 164 | 1,726 |
| Gun1 | 288 | 163 | 263 | 353 | 125 | 543,788 | 125 | 589,937 | 125 | 50 | 6,137 |
| Gun1V | 185 | 3,053 | 2,917 | 2,176 | 1,409 | 1543 | 1,594 | 96,032 | 5,643 | 4,514 | 38,393 |
| DH123 | 370 | 185 | 153 | 68 | 166 | 93,892 | 107 | 103,787 | 749 | 49 | 78 |
| JRFL | 370 | 94 | 410 | 137 | 188 | 431,348 | 154 | 292 | 240 | 772 | 240 |
| ADA | 270 | 230 | 703 | 180 | 94 | 434,213 | 391 | 173 | 133 | 94 | 67 |
HIV-1 clone from which the env gene was derived.
HIV-1 p24 antigen content of inoculum. Note that the infectivity-to-particle ratio of env-complemented HIV-1 is low, which accounts for the apparently high p24 antigen input.
HIV-1 replication in cells expressing the coreceptors indicated is expressed as luciferase activity in counts per second per nanogram of input p24 antigen. Values considered indicative of significant HIV-1 entry over the background are in boldface.
Only CCR5 and/or CXCR4 were used efficiently by five of the seven test viruses (boldface values in Table 1). Gun1 and, to a greater extent, Gun1V were able to enter via GPR1 as well as by CXCR4. Note that Gun1V weakly entered U87-CD4 cells transfected with the control plasmid or with several coreceptors, as reported previously (49). This probably reflects limited usage by Gun1V of a coreceptor endogenous to U87-CD4 cells, probably GPR1 (29).
Coreceptor use in stably transfected GHOST cells.
Human osteosarcoma HOS-CD4 cells stably transduced with different potential coreceptors and the green fluorescent protein (GFP) reporter gene under the control of the HIV-2 long terminal repeat (designated GHOST cells) were used to assess the replication of a much larger panel of pediatric and adult primary isolates and clones (Table 2). These cells have been described elsewhere (41, 74). In total, 14 pediatric isolates and 13 viruses of adult origin were tested against 13 different cell lines (GHOST cells expressing CCR1, CCR2b, CCR3, CCR4, CCR5, CCR8, CXCR4, Bonzo, BOB, GPR1, V28, US28, or APJ). The pediatric isolates were obtained from the Pediatric AIDS Foundation’s Ariel Project (11); the sources of most of the adult viruses have been described elsewhere (74, 75). A viral inoculum of 10 ng of p24 per well (24-well plate) was used, with the endpoint being HIV-1 replication, assayed by p24 antigen production on day 6 postinfection.
TABLE 2.
Coreceptor usage of pediatric and adult primary HIV-1 isolates in GHOST-CD4 cells expressing different coreceptors
| Isolatea | Phenotypeb | Usage of coreceptorc:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CCR1 | CCR2 | CCR3 | CCR4 | CCR5 | CCR8 | CXCR4 | BONZO | BOB | GPR1 | V28 | US28 | APJ | ||
| P1 22-211-V4 | NSI | − | − | − | − | + | − | − | − | − | − | − | − | − |
| P2 22-204-V2 | NSI | − | − | − | − | + | − | − | − | − | − | − | − | − |
| P3 19-245-V6 | NSI | − | − | − | − | ++ | − | − | − | − | − | − | − | − |
| P4 03-212-V8 | NSI | − | − | − | − | + | − | − | − | − | − | − | − | − |
| P5 11-213-V4 | NSI | − | − | − | − | +++ | − | − | − | − | − | − | − | − |
| P6 02-217-V3 | NSI | − | − | − | − | +++ | − | − | +++ | − | − | − | − | − |
| P7 22-216-V5 | NSI | − | − | − | − | + | − | − | − | − | − | − | − | − |
| P8 02-236-V6 | NSI | − | − | − | − | ++ | − | − | − | − | − | − | − | − |
| P9 22-236-V6 | NSI | − | − | − | − | +++ | − | − | − | − | − | − | − | − |
| P10 03-237-V6 | NSI | − | − | − | − | ++ | − | − | − | − | − | − | − | − |
| P11 22-202-V3 | NSI | − | − | − | − | + | − | − | − | − | − | − | − | − |
| P13 19-242-V3 | NSI | − | − | − | − | + | − | − | − | − | − | − | − | − |
| P14 22-207-V6 | NSI | − | − | − | − | ++ | − | − | − | − | − | − | − | − |
| P15 22-237-V4 | NSI | − | − | − | − | +++ | − | − | − | − | − | − | − | − |
| 301657 | NSI | − | − | − | − | ++ | − | − | − | − | − | − | − | − |
| 301714 | NSI | − | − | − | − | +++ | − | − | − | − | − | − | − | − |
| 301073 | NSI | − | − | − | − | + | − | − | − | − | − | − | − | − |
| 301056 | NSI | − | − | − | − | +++ | − | − | − | − | − | − | − | − |
| 301660 | NSI | − | − | − | − | ++ | − | − | − | − | − | − | − | − |
| 301727 | NSI | − | − | − | − | + | − | − | − | − | − | − | − | − |
| MWB | NSI | − | − | − | − | ++ | − | − | − | − | − | − | − | − |
| JRFL | NSI | − | − | − | − | ++ | − | − | − | − | − | − | − | − |
| SF162 | NSI | − | − | − | − | +++ | − | − | − | − | − | − | − | − |
| 301593d | SI | − | − | − | − | +++ | − | ++ | − | − | − | + | − | − |
| AD73 | SI | − | − | − | − | ++ | − | ++ | − | − | − | + | − | − |
| DH123d | SI | − | − | − | − | +++ | +++ | +++ | − | − | − | +++ | − | ++ |
| NL4-3d | SI | − | − | − | − | − | − | ++ | − | − | − | − | − | − |
| SIVlib | NSI | − | − | − | − | +++ | − | − | +++ | +++ | + | − | − | − |
HIV-1 isolate or clone used. Viruses P1 through P15 are of pediatric origin. SIVlib is a SIV isolate.
Isolates that do not replicate and induce syncytia in MT-2 cells are designated NSI, and isolates that do induce syncytia are designated SI.
HIV-1 replication in GHOST-CD4 cells expressing the coreceptors indicated is expressed on the following scale: −, p24 antigen undetectable (<0.15 ng/ml); +, p24 antigen ranging from 0.15 to 0.50 ng/ml; ++, p24 antigen ranging from 0.5 to 2 ng/ml; +++, p24 antigen at >2 ng/ml. The GHOST-CD4 cells also express a GFP reporter gene. Activation of this gene was compared with p24 antigen production in CCR5-expressing cells; a p24 production level of + to ++ is equivalent to activation of the GFP reporter in 1 to 10% of the cells, while +++ corresponds to GFP activation in 15 to 40% of the cells.
This isolate replicated to a significant extent in GHOST-CD4 cells with no transfected coreceptor, probably because it used endogenous CXCR4. This “background” level of p24 antigen was subtracted from those derived from coreceptor-transfected cells to yield the net value recorded.
Again, with rare exceptions, only CCR5 and CXCR4 were used efficiently by the HIV-1 isolates. Classified according to the ability to replicate on MT-2 cells (73), with one exception, the non-syncytium-inducing (NSI) viruses all used CCR5 only. In contrast, the syncytium-inducing (SI) isolates had a broader pattern of coreceptor recognition (Table 2). All four SI viruses used CXCR4, and three used CCR5 as well. Two of the SI viruses (301593 and AD-73) also utilized V28 weakly, and the DH123 clone (66) used CCR8, V28, and APJ efficiently, in addition to CCR5 and CXCR4. A simian immunodeficiency virus isolate (SIVlib) was also tested (13); it replicated in cells expressing CCR5, Bonzo, BOB, and (to a lesser extent) GPR1, consistent with earlier studies (4, 13, 20, 27, 29, 48, 61).
Overall, most of the GHOST cell lines supported the entry of at least one HIV-1 or SIV strain, confirming the expression of the transfected coreceptor at a functional level. In addition, the CCR1-, CCR2b-, CCR3-, CCR4-, CCR5-, CXCR4-, Bonzo-, and BOB-expressing GHOST cell lines have each been shown to support the entry of at least three different HIV-2 primary isolates, confirming that the coreceptors are properly expressed on these cells (54). No such positive control exists for the GHOST-US28 cells. In the absence of monoclonal antibodies specific for these and other potential coreceptors, we cannot compare the levels to which they are expressed on the surfaces of GHOST cells and relevant primary cells.
A pediatric HIV-1 isolate uses Bonzo efficiently.
A single pediatric isolate (P6 02-217-V3) replicated efficiently in GHOST cells expressing CCR5 or Bonzo (Table 2). Since this was the only virus in the test panel to utilize Bonzo, we tested six longitudinally obtained isolates from the same male infant (P6) and three from its mother (M6). The mother was resident in Florida when she became HIV-1 infected through heterosexual intercourse, but she has since died of AIDS. Infant P6 was M6’s second child, an older boy having been also vertically infected with HIV-1 (no samples were available from either the elder child or his father at the time this study was performed).
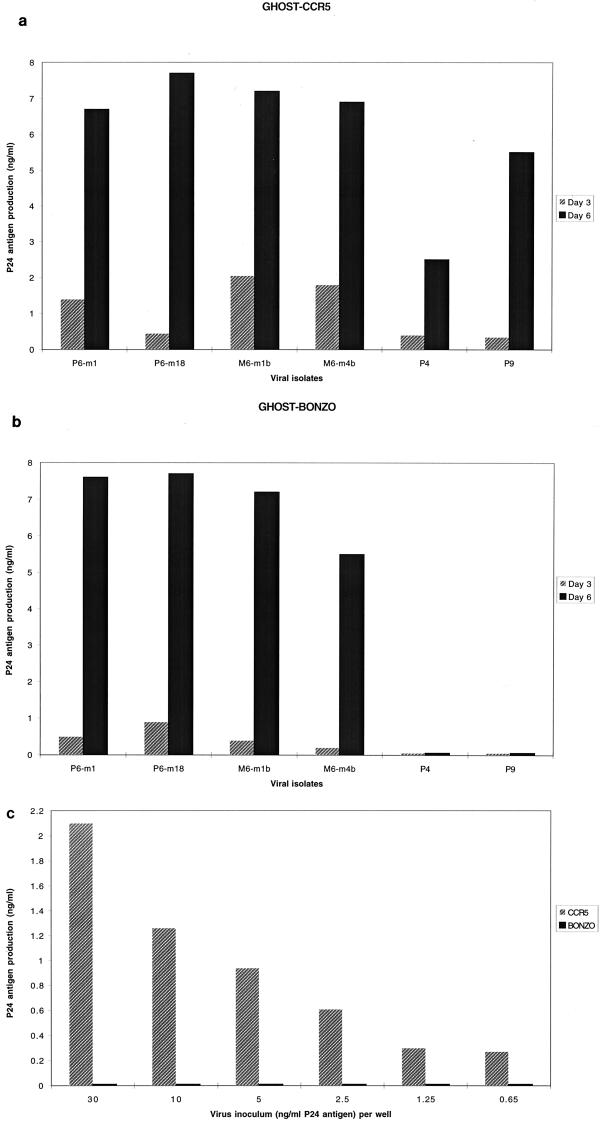
The infant isolates (P6 series; m indicates the month postbirth) were all of the NSI phenotype, and the maternal isolates (M6 series) were all SI (Table 3). The first five infant isolates and all three from the mother used both CCR5 and Bonzo, and the last isolate from the infant used only CCR5. In addition to CCR5, Bonzo, and CXCR4, the maternal isolates utilized CCR8, V28, and APJ (Table 3). With the exception of the last isolate from the infant (P6-m36), the replication of the infant and maternal isolates in Bonzo-expressing GHOST cells was robust, being comparable to the replication of the same isolates in the CCR5-expressing cells (Fig. 1a and b and data not shown). The last-obtained infant isolate (P6-m36) did not, however, replicate in Bonzo-expressing GHOST cells at any inoculum size tested, although its replication in CCR5-expressing cells was strong (Fig. 1c). Presumably, and for reasons unknown, phenotypic evolution in the infected infant has led to the loss of Bonzo usage but the retention of CCR5 usage.
TABLE 3.
Coreceptor usage of sequential primary HIV-1 isolates from a mother-child transmission paira
| Isolateb | Phenotype | Coreceptor usage
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CCR1 | CCR2 | CCR3 | CCR4 | CCR5 | CCR8 | CXCR4 | BONZO | BOB | GPR1 | V28 | US28 | APJ | ||
| P6 02-217-V3-m1 | NSI | − | − | − | − | +++ | − | − | +++ | − | − | − | − | − |
| P6 02-217-V4-m2 | NSI | − | − | − | − | +++ | − | − | +++ | − | − | − | − | − |
| P6 02-217-V6-m6 | NSI | − | − | − | − | +++ | − | − | +++ | − | − | − | − | − |
| P6 02-217-V8-m12 | NSI | − | − | − | − | +++ | − | − | +++ | − | − | − | − | − |
| P6 02-217-V9-m18 | NSI | − | − | − | − | +++ | − | − | +++ | − | − | − | − | − |
| P6 02-217-V14-m36 | NSI | − | − | − | − | +++ | − | − | − | − | − | − | − | − |
| M6-V2-m4b | SI | − | − | − | − | +++ | +++ | +++ | +++ | − | − | +++ | − | ++ |
| M6-V3-m1b | SI | − | − | − | − | +++ | +++ | +++ | +++ | − | − | +++ | − | ++ |
| M6-V6-m6a | SI | − | − | − | − | +++ | + | +++ | ++ | − | − | + | − | + |
The experimental design is described in the footnotes to Table 2.
Designations m1 through m36 for isolates from infant P6 indicate the number(s) of months that elapsed between the infant’s birth and the time of virus isolation. The isolates with the prefix M6 were obtained from the mother. The suffixes 4b and 1b indicate that the isolates were obtained 4 months and 1 month before delivery, respectively; the suffix 6a indicates that the isolate was obtained 6 months after delivery.
FIG. 1.
Use of CCR5 and Bonzo by HIV-1 isolates from a mother and her child. The replication of two isolates (m1 and m18) from infant P6 and of two isolates (m1b and m4b) from his mother (M6) in GHOST cells expressing CCR5 (a) or Bonzo (b) is indicated by the amount of p24 antigen production at 3 days (shaded bars) and 6 days (black bars) postinfection. Infant isolates P4 and P9 are also shown for comparison (see Tables 2 and 3). (c) Replication of different inocula of the last-obtained isolate (m36) from infant P6 in GHOST cells expressing CCR5 (shaded bars) or Bonzo (black bars), as determined by p24 antigen production 4 days postinfection.
Most HIV-1 isolates do not use Bonzo efficiently (Tables 1 and 2). However, coreceptor usage by HIV-2 is broad in comparison to that by HIV-1 (10, 20, 28, 36, 50, 54, 58, 71), and some HIV-2 strains use Bonzo quite well (20, 54). We therefore considered it possible that the P6 and M6 isolates might contain HIV-2 in addition to HIV-1, perhaps as a result of dual infection of the mother. Although culture supernatants from these isolates were weakly positive in commercial HIV-2 antigen assays, so were some known HIV-1 isolates (data not shown). Antigenic similarities between the HIV-1 and HIV-2 core antigens reduce the significance of such findings. Serology studies (Western blots using commercial reagents) on the mother and infant revealed no evidence of HIV-2 infection (data not shown). Furthermore, no HIV-2 sequences could be amplified from peripheral blood mononuclear cells (PBMC) infected with P6 or M6 isolates by using two different sets of nested PCR primer pairs that have each successfully amplified HIV-2 gag and env fragments from primary PBMC DNA in other studies (14, 32).
In contrast, when nested PCR primers designed to amplify the C2-to-C4 env region of all genetic subtypes of the HIV-1 M group were used (43, 44), nucleotide sequences associated with HIV-1 subtype B were obtained. The maternal and infant isolates clustered tightly together in a phylogenetic analysis (100 of 100 bootstrap replicates, using the neighbor-joining program of PHYLIP), consistent with the known vertical transmission. BLAST was used to demonstrate that the sequences obtained from the mother and infant were not a contaminant of pre-existing strains (no sequence was more than 95% similar to anything found in GenBank) (68). The recombination identification program RIP was used to show that, over the region tested, there was no evidence of recombination between the B subtype and any other subtype—the sequence was clearly subtype B throughout (68). More detailed analyses of the genetic sequences and the coreceptor usage properties of clones derived from the HIV-1 isolates from this maternal-infant transmission case are in progress.
Conclusions.
We have found that efficient use of coreceptors other than CCR5 and CXCR4 by HIV-1 is rare in vitro. We observed expanded coreceptor recognition only by SI viruses that can use both CCR5 and CXCR4, including SI viruses from an infected mother who transmitted NSI viruses to her child (Tables 2 and 3). The expanded tropism of SI viruses has been documented previously (9, 16, 18, 27, 65, 69, 76, 79).
One aspect of our conclusions should be tempered. The expression of CCR3 is known to be unusually inefficient in transfected cells, in comparison to other seven-transmembrane-spanning receptors (2, 5, 17, 61). In principle, this could mean that some viruses able to use CCR3 in vivo might not be able to enter CCR3-transfected cells in vitro, leading to false-negative conclusions about CCR3 usage. However, we used CCR3-specific murine monoclonal antibody 7B11 (35) to determine the extent of CCR3 expression on both GHOST-CCR3 cells and mitogen-activated PBMC. CCR3 was detected by this antibody on 25% of the GHOST-CCR3 cells but on only about 1% of activated PBMC (data not shown). The rarity of CCR3-expressing cells among PBMC has been reported previously (62). The fluorescence intensity of CCR3 staining on the GHOST cells was comparable to, or greater than, the intensity on the rare CCR3-positive cells in PBMC (data not shown). Thus, it is unlikely that the GHOST-CCR3 cells would fail to detect any CCR3 usage by viruses that entered activated PBMC via this coreceptor. The possibility remains that CCR3 is an important coreceptor on a minor subset of CD4+ T cells in vivo (62).
None of the isolates we tested used CCR1, CCR2b, CCR4, BOB (GPR15), or US28 for entry into transfected GHOST cells. The failure of any HIV-1 isolate to use CCR4 is notable in the context of a report that macrophage-derived chemokine, a CCR4 ligand (39), inhibits the replication of several NSI and SI HIV-1 isolates in vitro (55). The only isolates able to use Bonzo (also known as STRL33 or TYMSTR) were genetically related SI and NSI strains from a maternal-infant vertical transmission case. The HIV-1 SI isolates all used CXCR4, as expected from multiple previous studies (9, 18, 65, 69, 76, 79). However, the SI isolates also had an expanded tropism for other coreceptors. The M6 series of maternal SI isolates, along with three other SI viruses, used V28 (CX3CR1), albeit to various extents. Only the M6 isolates and the DH123 molecular clone entered CCR8- or APJ-expressing GHOST cells efficiently (Tables 2 and 3), although we were unable to demonstrate CCR8 use by DH123 in transiently transfected U87-CD4 cells (Table 1). This difference may reflect the larger infectious inoculum used with the GHOST cells. Only Gun1V and, to a lesser extent, the related Gun1 virus entered GHOST cells via GPR1. In general, the usage of GPR1, CCR8, V28, and APJ by a subset of SI viruses is in accord with earlier reports on these coreceptors (16, 27, 29, 37, 58, 61).
Positive use of Bonzo, BOB, and GPR1 was, however, shown by SIVlib, which confirms the functional expression of these coreceptors on the GHOST cell lines. Bonzo, BOB, and GPR1 are known to be highly efficient coreceptors for a range of strains from the SIV grouping (4, 20, 29), as is CCR5 (13, 15, 20, 27, 48, 61). Likewise, several HIV-2 primary isolates can enter CCR1-, CCR2b-, and CCR4-expressing GHOST cells, confirming that they also express the transfected coreceptor appropriately (54).
What does a negative result mean in in vitro assays of HIV-1 replication in transfected cells? Previous reports have described efficient use by HIV-1 of several of the “newer” coreceptors (16, 20, 27, 29, 37, 45, 47, 57, 58). Often, but not always, the viruses able to use these coreceptors were “T-tropic” or SI strains. We can confirm that SI viruses can indeed use multiple coreceptors in vitro (Tables 1 and 2). When comparing different experimental systems, a relevant issue is, we believe, one of efficiency of coreceptor usage. In several earlier reports, the assays most commonly employed were those of env-mediated membrane fusion, sometimes in association with measurements of the entry of env-complemented, reporter gene-expressing HIV-1 pseudotypes. These assays can be quite sensitive. However, we have noted circumstances in which a positive result in a membrane fusion assay is not mirrored in a virus entry assay using the same envelope glycoproteins and coreceptors (26). This is probably because virus entry has a more stringent requirement for high-affinity virus-receptor interactions, because of the lower number of envelope glycoprotein complexes on virions than on the surfaces of fusing cells. Overexpression of lower-affinity coreceptors in cell-cell fusion systems can compensate for the reduced efficiency of individual coreceptors, but this is not necessarily predictive of cell-free virus entry (26).
In an env complementation test of virus entry, what is a significant result? The luciferase expression assay can have an extremely wide dynamic range (Table 1). A level of virus entry that is 10-fold over the background in the presence of a particular coreceptor is clearly a demonstration that the virus is capable of interacting with that coreceptor. But if the level of entry is still 100- to 1,000-fold lower than that achieved with CXCR4 or CCR5, as is often the case, that needs to be taken into account when judging whether the interaction is likely to be biologically significant. For example, as shown in Table 1, HIV-1 NDK enters U87-CD4-GPR1 cells 7-fold more efficiently than it enters parental U87-CD4 cells, but entry via GPR1 is 493-fold less efficient than via CXCR4. We believe that the latter value is the more relevant of the two. In the present study, we have mostly used virus replication as an endpoint (Tables 2 and 3). Our use of a large inoculum in these assays reduces the possibility of a false-negative result. In terms of viral output, we are unlikely to detect a weak level of replication that is 100-fold less than what we score as positive (+) (Table 2). However, HIV-1 entry and replication at such a low level are not likely to be important.
The existence of “natural knockouts” for CCR5 (19, 37, 45, 61, 78), combined with the multiplicity of studies that demonstrate ligand-sensitive, efficient use of this coreceptor in vitro (3, 9, 17, 18, 21, 24, 25, 40, 65, 69, 75, 76, 79), has led to the firm conclusion that CCR5 is of paramount importance for HIV-1 transmission and replication in vivo (6, 8, 23, 53). The acquisition of efficient, ligand-sensitive CXCR4 use in vitro by HIV-1 variants isolated at a time when there are alterations in CD4 cell loss and a more rapid disease course in vivo (9, 18, 65, 69) speaks to the physiological importance of this coreceptor. But are other coreceptors important in vivo? Tissue expression patterns are a relevant factor to consider. To be involved in the maintenance of high viral loads, a coreceptor would have to be expressed on activated CD4+ T cells (56). Whether this occurs is far from certain for many of the coreceptors described to date. A significant problem is the unavailability of specific chemokine ligands and/or monoclonal antibodies for many coreceptors. This makes it difficult to judge the importance of individual coreceptors in cells other than transfectants. Blocking HIV-1 entry with the CCR3 ligand eotaxin was a major contribution to demonstrating that CCR3 was a coreceptor in microglia (34), although there is now no consistent agreement as to the relative importance of CCR3 and CCR5 in these cells (33, 67). HIV-1 inhibition studies have been performed with the I-309 ligand for CCR8, but only in CCR8-transfected cells (37). However, wherever it is expressed in vitro (and perhaps also in vivo), the presence of a potential coreceptor on a cell (CD4+ or CD4−) does not prove that it can be used efficiently in that cellular context (22).
Many SI isolates can clearly use multiple coreceptors in vitro. Are these coreceptors used by SI viruses in vivo? Of note is the fact that an SI isolate from an individual homozygous for defective CCR5 alleles was shown to use only CXCR4 when it was tested for the ability to enter GHOST cells expressing CCR1, CCR2b, CCR3, CCR4, CCR5, CXCR4, BOB, Bonzo, or GPR1 (51). Thus, inability to use CCR5 in vivo does not necessarily drive HIV-1 to use a coreceptor other than CXCR4. In addition, any alterations in envelope glycoprotein structure that might be necessary for a HIV-1 strain to switch from using CCR5 or CXCR4 to using a coreceptor such as BOB or Bonzo, whether under drug selection pressure or spontaneously, would have to be compatible with the retention of resistance to humoral immunity. The latter is also a property of the envelope glycoproteins, and a neutralization-sensitive variant may well not persist in the host (52, 74).
Overall, we believe that there is no compelling evidence for the importance of coreceptors other than CCR5 and CXCR4 when considering HIV-1 infection of blood cells of the lymphoid and monocyte lineages in vivo. A proviso is that the expression and use of different coreceptors in other cell types, for example, in the brain or thymus, could contribute to some facets of HIV-1 pathogenesis in adults or in children (6, 8, 23, 53).
We observed that a mother (M6) who was infected with SI viruses able to use CXCR4 as well as CCR5 transmitted only NSI, CCR5-using viruses to her infant (Tables 2 and 3). This pattern of phenotypic selection has been previously documented in maternal-infant transmission of HIV-1 (11, 64), although the converse pattern was noted in a single case of HIV-2 vertical transmission (12). As noted above, both the maternal and infant isolates also used Bonzo, and the maternal SI isolates used several coreceptors. However, with this exception, we noted nothing unusual about the coreceptor usage patterns of pediatric HIV-1 isolates that could distinguish them from adult isolates. Overall, the characteristics of the developing immune system probably have more influence on the nature of pediatric HIV-1 infection than do any unusual properties of the infecting HIV-1 strains (42, 78).
Acknowledgments
We are very grateful to Alexandra Trkola for helpful discussions on virology, to Bette Korber for serious advice on genetics, and to Beatrice Hahn for consultations. We appreciate the contributions of the donors of HIV-1 isolates and clones used in this study, including the participants in the Pediatric AIDS Foundation’s ARIEL Project. We particularly thank Zhi-wei Chen and Preston Marx for gifts of SIVlib and HIV-2 primers, Hiroo Hoshino and Paul Clapham for the Gun1 and Gun1V isolates, WenKai Xiang for provision of cell lines, Paul McHardy and Hugh Jarce for technical support, and Ana Puga for clinical information on the HIV-1 vertical transmission case.
This study was supported by NIH grant AI41420 and by the Pediatric AIDS Foundation. V.N.K. is supported by a postdoctoral fellowship from the Damon Runyon/Walter Winchell Foundation, D.R.L. is an Investigator of the Howard Hughes Medical Institute, and J.P.M. is an Elizabeth Glaser Scientist of the Pediatric AIDS Foundation.
REFERENCES
- 1.Alkhatib G, Ahuja S S, Light D, Mummidi S, Berger E A, Ahuja S K. CC chemokine receptor 5-mediated signaling and HIV-1 co-receptor activity share common structural determinants. Critical residues in the third extracellular loop support HIV-1 fusion. J Biol Chem. 1997;272:19771–19776. doi: 10.1074/jbc.272.32.19771. [DOI] [PubMed] [Google Scholar]
- 2.Alkhatib G, Berger E A, Murphy P M, Pease J E. Determinants of HIV-1 co-receptor function on CC chemokine receptor 3. Importance of both extracellular and transmembrane/cytoplasmic regions. J Biol Chem. 1997;272:20420–20426. doi: 10.1074/jbc.272.33.20420. [DOI] [PubMed] [Google Scholar]
- 3.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 4.Alkhatib G, Liao F, Berger E A, Farber J M, Peden K C W. A new SIV coreceptor, STRL33. Nature. 1997;388:238. doi: 10.1038/40789. [DOI] [PubMed] [Google Scholar]
- 5.Bazan H A, Alkhatib G, Broder C C, Berger E A. Patterns of CCR5, CXCR4, and CCR3 usage by envelope glycoproteins from human immunodeficiency virus type 1 primary isolates. J Virol. 1998;72:4485–4491. doi: 10.1128/jvi.72.5.4485-4491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger, E. A. 1997. HIV entry and tropism: the chemokine receptor connection. AIDS 11(Suppl. A):S3–S16. [PubMed]
- 7.Berger E A, Doms R W, Fenyö E-M, Korber B T M, Littman D R, Moore J P, Sattentau Q J, Schuitemaker H, Sodroski J, Weiss R A. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 8.Bieniasz P D, Cullen B R. Chemokine receptors and human immunodeficiency virus infection. Front Biosci (Online) 1998;3:44–58. doi: 10.2741/a265. [DOI] [PubMed] [Google Scholar]
- 9.Björndal Å, Deng H, Jansson M, Fiore J R, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman D R, Fenyö E M. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bron R, Klasse P J, Wilkinson D, Clapham P R, Pelchen-Matthews A, Power C, Wells T N C, Kim J, Peiper S C, Hoxie J A, Marsh M. Promiscuous use of CC and CXC chemokine receptors in cell-to-cell fusion mediated by a human immunodeficiency virus type 2 envelope protein. J Virol. 1997;71:8405–8415. doi: 10.1128/jvi.71.11.8405-8415.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao Y, Krogstad P, Korber B T, Koup R A, Muldoon M, Macken C, Song J L, Jin Z, Zhao J-Q, Clapp S, Chen I S Y, Ho D D, Ammann A J the Ariel Project Investigators. Maternal HIV-1 viral load and vertical transmission of infection: the Ariel Project for the prevention of HIV transmission from mother to infant. Nat Med. 1997;3:549–552. doi: 10.1038/nm0597-549. [DOI] [PubMed] [Google Scholar]
- 12.Cavaco-Silva P, Taviera N C, Rosado L, Lourenço M H, Moniz-Pereira T, Douglas N W, Daniels R S, Santos-Ferreira M O. Virological and molecular demonstration of human immunodeficiency virus type 2 vertical transmission. J Virol. 1998;72:3418–3422. doi: 10.1128/jvi.72.4.3418-3422.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z-W, Gettie A, Ho D D, Marx P A. Primary SIVsm isolates use the CCR5 co-receptor from sooty mangabeys naturally infected in West Africa: a comparison of coreceptor usage of primary SIVsm, HIV-2 and SIVmac. Virology. 1998;245:113–124. doi: 10.1006/viro.1998.9174. [DOI] [PubMed] [Google Scholar]
- 14.Chen Z-W, Luckay A, Sodora D L, Telfer P, Reed P, Gettie A, Kanu J M, Sadek R F, Yee J, Ho D D, Zhang L, Marx P A. Human immunodeficiency virus type 2 (HIV-2) seroprevalence and characterization of a distinct HIV-2 genetic subtype from the natural range of simian immunodeficiency virus-infected sooty mangabeys. J Virol. 1997;71:3953–3960. doi: 10.1128/jvi.71.5.3953-3960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Z-W, Zhou P, Ho D D, Landau N R, Marx P A. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choe H, Farzan M, Konkel M, Martin K, Sun Y, Marcon L, Cayabyab M, Berman M, Dorf M E, Gerard N, Gerard G, Sodroski J. The orphan seven-transmembrane receptor Apj supports the entry of primary T-cell-line-tropic and dualtropic human immunodeficiency virus type 1. J Virol. 1998;72:6113–6118. doi: 10.1128/jvi.72.7.6113-6118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard G, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 18.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1 infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O’Brien S J Hemophilia Growth and Development Study; Multicenter AIDS Cohort Study; Multicenter Hemophilia Cohort Study; San Francisco City Cohort; ALIVE Study. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion of the CKR5 structural allele. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 20.Deng H, Unutmaz D, Kewalramani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 21.Deng H K, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major coreceptor 5 for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 22.Dittmar M T, McKnight A, Simmons G, Clapham P R, Weiss R A, Simmonds P. HIV-1 tropism and co-receptor use. Nature. 1997;385:495–496. doi: 10.1038/385495a0. [DOI] [PubMed] [Google Scholar]
- 23.Doms R W, Peiper S C. Unwelcome guests with master keys: how HIV uses chemokine receptors for cellular entry. Virology. 1997;235:179–190. doi: 10.1006/viro.1997.8703. [DOI] [PubMed] [Google Scholar]
- 24.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S, Parmentier M, Collman R G, Doms R W. A dual-tropic, primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-2b as fusion cofactors. Cell. 1996;85:1149–1159. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 25.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 26.Dragic T, Trkola A, Lin S W, Nagashima K A, Kajumo F, Zhao L, Olson W C, Wu L, Mackay C R, Allaway G P, Sakmar T P, Moore J P, Maddon P J. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J Virol. 1998;72:279–285. doi: 10.1128/jvi.72.1.279-285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edinger A L, Hoffman T L, Sharron M, Lee B, Yi Y, Choe W, Kolson D L, Mitrovic B, Zhou Y, Faulds D, Collman R G, Hesselgesser J, Horuk R, Doms R W. An orphan seven-transmembrane domain receptor expressed widely in the brain functions as a coreceptor for human immunodeficiency virus type 1 and simian immunodeficiency virus. J Virol. 1998;72:7934–7940. doi: 10.1128/jvi.72.10.7934-7940.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N C, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 29.Farzan M, Choe H, Martin K, Marcon L, Hofmann W, Karlsson G, Sun Y, Barrett P, Marchand N, Sullivan N, Gerard N, Gerard C, Sodroski J. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J Exp Med. 1997;186:405–411. doi: 10.1084/jem.186.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane G protein coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 31.Frade J M R, Llorente M, Mellado M, Alcami J, Gutierrez-Ramos J C, Zaballos A, del Real G, Martinez A C. The amino-terminal domain of the CCR2 chemokine receptor acts as a coreceptor for HIV-1 infection. J Clin Invest. 1997;100:497–502. doi: 10.1172/JCI119558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao F, Yue L, Robertson D L, Hill S C, Hui H, Biggar R J, Neequaye A E, Whelan T M, Ho D D, Shaw G M, Sharp P M, Hahn B. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J Virol. 1994;68:7433–7447. doi: 10.1128/jvi.68.11.7433-7447.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghorpade A, Xia M Q, Hyman B T, Persidsky Y, Nukuna A, Bock P, Che M, Limoges J, Gendelman H E, Mackay C R. Role of the β-chemokine receptors CCR3 and CCR5 in human immunodeficiency virus type 1 infection of monocytes and microglia. J Virol. 1998;72:3351–3361. doi: 10.1128/jvi.72.4.3351-3361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, Mackay C R, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 35.Heath H, Qin S, Rao P, Wu L, LaRosa G, Kassam N, Ponath P D, Mackay C R. Chemokine receptor usage by human eosinophils. The importance of CCR3 demonstrated using an antagonistic monoclonal antibody. J Clin Invest. 1997;99:178–184. doi: 10.1172/JCI119145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hill C M, Deng H, Unutmaz D, Kewalramani V N, Bastiani L, Gorny M K, Zolla-Pazner S, Littman D R. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J Virol. 1997;71:6296–6304. doi: 10.1128/jvi.71.9.6296-6304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horuk R, Hesselgesser J, Zhou Y, Faulds D, Halks-Miller M, Harvey S, Taub D, Samson M, Parmentier M, Rucker J, Doranz B J, Doms R W. The CC chemokine I-309 inhibits CCR8-dependent infection by diverse HIV-1 strains. J Biol Chem. 1998;273:386–391. doi: 10.1074/jbc.273.1.386. [DOI] [PubMed] [Google Scholar]
- 38.Huang Y, Paxton W A, Wolinsky S M, Neumann A U, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbaksh K, Kuntsman K, Erickson D, Landau N R, Phair J, Ho D D, Koup R A. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 39.Imai T, Chantry D, Raport C J, Wood C L, Nishimura M, Godiska R, Voshie O, Gray P W. Macrophage-derived chemokine is a functional ligand for the CC chemokine receptor 4. J Biol Chem. 1998;273:1764–1768. doi: 10.1074/jbc.273.3.1764. [DOI] [PubMed] [Google Scholar]
- 40.Jansson M, Popovic M, Karlsson A, Cocchi F, Rossi P, Albert J, Wigzell H. Sensitivity to inhibition by β-chemokines correlates with biological phenotypes of primary HIV-1 isolates. Proc Natl Acad Sci USA. 1996;93:15382–15387. doi: 10.1073/pnas.93.26.15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.KewalRamani, V. N., B. Volsky, D. S. Kwon, W. Xiang, D. Unutmaz, R. E. Sutton, and D. R. Littman. Unpublished data.
- 42.Kirschner D E, Mehr R, Perelson A S. Role of the thymus in pediatric HIV-1 infection. J Acquired Immune Defic Syndr. 1998;18:95–109. doi: 10.1097/00042560-199806010-00001. [DOI] [PubMed] [Google Scholar]
- 43.Kostrikis L G, Bagdades E, Cao Y, Zhang L, Dimitriou D, Ho D D. Genetic analysis of human immunodeficiency virus type 1 strains from patients in Cyprus: identification of a new subtype designated subtype I. J Virol. 1995;69:6122–6130. doi: 10.1128/jvi.69.10.6122-6130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kostrikis L G, Shin S, Ho D D. Genotyping HIV-1 and HCV strains by a combinatorial DNA melting assay (COMA) Mol Med. 1998;4:443–453. [PMC free article] [PubMed] [Google Scholar]
- 45.Liao F, Alkhatib G, Peden K W C, Sharma G, Berger E A, Farber J M. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–378. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 47.Loetscher M, Amara A, Oberlin E, Brass N, Legler D F, Loetscher P, D’Apuzzo M, Meese E, Rousset D, Virelizier J-L, Baggiolini M, Arenzana-Seisdedos F, Moser B. TYMSTR, a putative chemokine receptor selectively expressed in activated T cells, exhibits HIV-1 coreceptor function. Curr Biol. 1997;7:652–660. doi: 10.1016/s0960-9822(06)00292-2. [DOI] [PubMed] [Google Scholar]
- 48.Marcon L, Choe H, Martin K A, Farzan M, Ponath P D, Wu L, Newman W, Gerard N, Gerard C, Sodroski J. Utilization of C-C chemokine receptor 5 by the envelope glycoproteins of a pathogenic simian immunodeficiency virus, SIVmac239. J Virol. 1997;71:2522–2527. doi: 10.1128/jvi.71.3.2522-2527.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKnight A, Wilkinson D, Simmons G, Talbot S, Picard L, Ahuja M, Marsh M, Hoxie J A, Clapham P R. Inhibition of human immunodeficiency virus fusion by a monoclonal antibody to a coreceptor (CXCR4) is both cell type and virus strain dependent. J Virol. 1997;71:1692–1696. doi: 10.1128/jvi.71.2.1692-1696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKnight A, Dittmar M T, Moniz-Periera J, Ariyoshi K, Reeves J D, Hibbitts S, Whitby D, Aarons E, Proudfoot A E I, Whittle H, Clapham P R. A broad range of chemokine receptors are used by primary isolates of human immunodeficiency virus type 2 as coreceptors with CD4. J Virol. 1998;72:4065–4071. doi: 10.1128/jvi.72.5.4065-4071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michael N L, Nelson J A E, KewalRamani V N, Chang G, O’Brien S J, Mascola J R, Volsky B, Louder M, White II G C, Littman D R, Swanstrom R, O’Brien T R. Exclusive and persistent use of the entry coreceptor CXCR4 by human immunodeficiency virus type 1 from a subject homozygous for CCR5 Δ32. J Virol. 1998;72:6040–6047. doi: 10.1128/jvi.72.7.6040-6047.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moore, J. P., and D. D. Ho. 1995. HIV-1 neutralization: the consequences of viral adaptation to growth on transformed T cells. AIDS 9(Suppl. A):S117–S136. [PubMed]
- 53.Moore J P, Trkola A, Dragic T. Co-receptors for HIV-1 entry. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 54.Owen S M, Ellenberger D, Rayfield M, Wiktor S, Michel P, Grieco M H, Gao F, Hahn B H, Lal R B. Genetically divergent strains of human immunodeficiency virus type 2 use multiple coreceptors for viral entry. J Virol. 1998;72:5425–5432. doi: 10.1128/jvi.72.7.5425-5432.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pal R, Garzino-Demo A, Markham P D, Burns J, Brown M, Gallo R C, DeVico A L. Inhibition of HIV-1 infection by the β-chemokine MDC. Science. 1997;278:695–698. doi: 10.1126/science.278.5338.695. [DOI] [PubMed] [Google Scholar]
- 56.Perelson A, Neumann A U, Markowitz M, Leonard J, Ho D D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 57.Pleskoff O, Treboute C, Brelot A, Heveker N, Seman M, Alizon M. Identification of a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1 entry. Science. 1997;276:1874–1878. doi: 10.1126/science.276.5320.1874. [DOI] [PubMed] [Google Scholar]
- 58.Reeves J D, McKnight A, Potempa S, Simmons G, Gray P W, Power C A, Wells T, Weiss R A, Talbot S J. CD4-independent infection by HIV-2 (ROD/B): use of the 7-transmembrane receptors CXCR-4, CCR-3, and V28 for entry. Virology. 1997;231:130–134. doi: 10.1006/viro.1997.8508. [DOI] [PubMed] [Google Scholar]
- 59.Ross T M, Cullen B R. The ability of HIV type 1 to use CCR-3 as a coreceptor is controlled by envelope V1/V2 sequences acting in conjunction with a CCR-5 tropic V3 loop. Proc Natl Acad Sci USA. 1998;95:7682–7686. doi: 10.1073/pnas.95.13.7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rubbert A, Combadiere C, Ostrowski M, Arthos J, Dybul M, Machado E, Cohn M A, Hoxie J A, Murphy P M, Fauci A S, Weissman D. Dendritic cells express multiple chemokine receptors used as coreceptors for HIV entry. J Immunol. 1998;160:3933–3941. [PubMed] [Google Scholar]
- 61.Rucker J, Edinger A L, Sharron M, Samson M, Lee B, Berson J F, Yi Y, Margulies B, Collman R G, Doranz B J, Parmentier M, Doms R W. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sallusto F, Mackay C R, Lanzavecchia A. Selective expression of the eotaxin receptor CCR3 by human T helper 2 cells. Science. 1997;277:2005–2007. doi: 10.1126/science.277.5334.2005. [DOI] [PubMed] [Google Scholar]
- 63.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C-M, Saragosti S, Lapoumèroulie C, Cogniaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection of Caucasian individuals bearing mutant alleles of the CCR5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 64.Scarlatti G, Hodara V, Rossi P, Muggiasca L, Bucceri A, Albert J, Fenyö E M. Transmission of human immunodeficiency virus type 1 (HIV-1) from mother to child correlates with viral phenotype. Virology. 1993;197:624–629. doi: 10.1006/viro.1993.1637. [DOI] [PubMed] [Google Scholar]
- 65.Scarlatti G, Tresoldi H, Björndal A, Fredriksson R, Colognesi C, Deng H K, Maluati M S, Plebani A, Siccardi A G, Littman D R, Fenyö E M, Lusso P. In vitro evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;11:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 66.Shibata R, Hoggan M D, Broscius C, Englund G, Theodore T S, Buckler-White A, Arthur L O, Israel Z, Schultz A, Lane H C, Martin M A. Isolation and characterization of a syncytium-inducing, macrophage/T-cell line-tropic human immunodeficiency virus type 1 isolate that readily infects chimpanzee cells in vitro and in vivo. J Virol. 1995;69:4453–4462. doi: 10.1128/jvi.69.7.4453-4462.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shieh J T C, Albright A V, Sharron M, Gartner S, Strizki J, Doms R W, Gonzalez-Scarano F. Chemokine receptor utilization by human immunodeficiency virus type 1 isolates that replicate in microglia. J Virol. 1998;72:4243–4249. doi: 10.1128/jvi.72.5.4243-4249.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Siepel A C, Halpern A L, Macken C, Korber B. A computer program designed to rapidly screen for HIV-1 intersubtype recombinant sequences. AIDS Res Hum Retroviruses. 1995;11:1413–1416. doi: 10.1089/aid.1995.11.1413. [DOI] [PubMed] [Google Scholar]
- 69.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sleasman J W, Aleixo L F, Morton A, Skoda-Smith S, Goodenow M M. CD4+ memory T cells are the predominant population of HIV-1-infected lymphocytes in neonates and children. AIDS. 1996;10:1477–1484. doi: 10.1097/00002030-199611000-00004. [DOI] [PubMed] [Google Scholar]
- 71.Sol N, Ferchel F, Braun J, Pleskoff O, Tréboute C, Ansart I, Alizon M. Usage of the coreceptors CCR-5, CCR-3, and CXCR-4 by primary and cell line-adapted human immunodeficiency virus type 2. J Virol. 1997;71:8237–8244. doi: 10.1128/jvi.71.11.8237-8244.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takeuchi Y, Akutsu M, Murayama K, Shimizu N, Hoshino H. Host range mutant of human immunodeficiency virus type 1: modification of cell tropism by a single point mutation at the neutralization epitope in the env gene. J Virol. 1991;65:1710–1718. doi: 10.1128/jvi.65.4.1710-1718.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tersmette M, de Goede R E Y, Al B J M, Winkel I N, Gruters R A, Cuypers H T, Huisman H G, Miedema F. Differential syncytium-inducing capacity of human immunodeficiency virus isolates: frequent detection of syncytium-inducing isolates in patients with acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. J Virol. 1988;62:2026–2032. doi: 10.1128/jvi.62.6.2026-2032.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trkola A, Ketas T, KewalRamani V N, Endorf F, Binley J M, Katinger H, Robinson J, Littman D R, Moore J P. Neutralization sensitivity of human immunodeficiency virus type 1 primary isolates to antibodies and CD4-based reagents is independent of coreceptor usage. J Virol. 1998;72:1876–1885. doi: 10.1128/jvi.72.3.1876-1885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trkola A, Paxton W A, Monard S P, Hoxie J A, Siani M A, Thompson D A, Wu L, Mackay C R, Horuk R, Moore J P. Genetic subtype-independent inhibition of human immunodeficiency virus type 1 replication by CC and CXC chemokines. J Virol. 1998;72:396–404. doi: 10.1128/jvi.72.1.396-404.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tscherning C, Alaeus A, Fredriksson R, Björndal A, Deng H, Littman D R, Fenyö E M, Albert J. Differences in chemokine coreceptor usage between genetic subtypes of HIV-1. Virology. 1998;241:181–188. doi: 10.1006/viro.1997.8980. [DOI] [PubMed] [Google Scholar]
- 77.Ullum H, Lepri A C, Victor J, Skinhoj P, Phillips A N, Pedersen B K. Increased losses of CD4+CD45RA+ cells in later stages of HIV infection is related to increased risk of death: evidence from a cohort of 347 HIV-infected individuals. AIDS. 1997;11:1479–1485. doi: 10.1097/00002030-199712000-00012. [DOI] [PubMed] [Google Scholar]
- 78.Yang L P, Riley J L, Carroll R G, June C H, Hoxie J, Patterson B K, Ohshima Y, Hodes R J, Delepesse G. Productive infection of neonatal CD8(+) T lymphocytes by HIV-1. J Exp Med. 1998;187:1139–1144. doi: 10.1084/jem.187.7.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang L, Carruthers C D, He T, Huang Y, Cao Y, Wang G, Hahn B, Ho D D. HIV type 1 subtypes, coreceptor usage, and CCR5 polymorphism. AIDS Res Hum Retroviruses. 1997;13:1357–1366. doi: 10.1089/aid.1997.13.1357. [DOI] [PubMed] [Google Scholar]
- 80.Zimmerman P A, Buckler-White A, Alkhatib G, Spalding T, Kubofcik J, Combadiere C, Weissman D, Cohen O, Rubbert A, Lam G, Vaccarezza M, Kennedy P E, Kumaraswami V, Giorgi J V, Detels R, Hunter J, Chopek M, Berger E A, Fauci A S, Nutman T B, Murphy P M. Inherited resistance to HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: studies in populations with contrasting clinical phenotypes, defined racial background, and quantified risk. Mol Med. 1997;3:23–26. [PMC free article] [PubMed] [Google Scholar]