Abstract
We tested chemokine receptor subset usage by diverse, well-characterized primary viruses isolated from peripheral blood by monitoring viral replication with CCR1, CCR2b, CCR3, CCR5, and CXCR4 U87MG.CD4 transformed cell lines and STRL33/BONZO/TYMSTR and GPR15/BOB HOS.CD4 transformed cell lines. Primary viruses were isolated from 79 men with confirmed human immunodeficiency virus type 1 (HIV-1) infection from the Chicago component of the Multicenter AIDS Cohort Study at interval time points. Thirty-five additional well-characterized primary viruses representing HIV-1 group M subtypes A, B, C, D, and E and group O and three primary simian immunodeficiency virus (SIV) isolates were also used for these studies. The restricted use of the CCR5 chemokine receptor for viral entry was associated with infection by a virus having a non-syncytium-inducing phenotype and correlated with a reduced rate of disease progression and a prolonged disease-free interval. Conversely, broadening chemokine receptor usage from CCR5 to both CCR5 and CXCR4 was associated with infection by a virus having a syncytium-inducing phenotype and correlated with a faster rate of CD4 T-cell decline and progression of disease. We also observed a greater tendency for infection with a virus having a syncytium-inducing phenotype in men heterozygous for the defective CCR5 Δ32 allele (25%) than in those men homozygous for the wild-type CCR5 allele (6%) (P = 0.03). The propensity for infection with a virus having a syncytium-inducing phenotype provides a partial explanation for the rapid disease progression among some men heterozygous for the defective CCR5 Δ32 allele. Furthermore, we did not identify any primary viruses that used CCR3 as an entry cofactor, despite this CC chemokine receptor being expressed on the cell surface at a level commensurate with or higher than that observed for primary peripheral blood mononuclear cells. Whereas isolates of primary viruses of SIV also used STRL33/BONZO/TYMSTR and GPR15/BOB, no primary isolates of HIV-1 used these particular chemokine receptor-like orphan molecules as entry cofactors, suggesting a limited contribution of these other chemokine receptors to viral evolution. Thus, despite the number of chemokine receptors implicated in viral entry, CCR5 and CXCR4 are likely to be the physiologically relevant chemokine receptors used as entry cofactors in vivo by diverse strains of primary viruses isolated from blood.
Primate immunodeficiency viruses enter target cells by fusion at the cell surface membrane. Viral entry requires a specific interaction among the viral envelope glycoprotein, the CD4 cell surface molecule, and members of the seven-span transmembrane chemokine receptor family as entry cofactors (1, 9, 12, 15, 16, 20–23, 29, 37, 42). Coreceptor activity has been observed with the CC chemokine receptors CCR2b, CCR3, CCR5, CCR8, and US28; the CXC chemokine receptor CXCR4; and the chemokine receptor-like orphan molecules STRL33 or BONZO or TYMSTR, GPR15 or BOB, and V28 (1, 9, 10, 16–18, 20–23, 36, 42). Nonetheless, among the different chemokine receptors used as entry cofactors, CXCR4 and CCR5 are the major ones utilized by both primary and laboratory strains of human immunodeficiency virus type 1 (HIV-1) (1, 5, 10, 14, 16, 21, 45, 46, 48, 50).
Viruses that differ in their phenotypic properties use different subsets of chemokine receptors as entry cofactors. Our knowledge of chemokine receptor usage stems mostly from HIV-1 isolates that have been extensively propagated in vitro and from recombinant envelope proteins derived from these in vitro viral cultures (1, 10, 16, 21, 22). It is only recently that the chemokine receptor usage by the primary HIV-1 isolates has been extensively studied, albeit with a limited number of viruses (5, 14, 45, 46, 48). It was found that viral isolates obtained during the asymptomatic stages generally used only CCR5 as a coreceptor and were inhibited by RANTES, MIP-1α, and MIP-1β but not SDF-1 (46). In contrast, viral isolates obtained during the later stages of infection have expanded their coreceptor usage primarily from CCR5 to CXCR4, although a small number of isolates has also acquired the ability to use CCR3 (5, 46). A majority of the isolates, however, still retain their CCR5 usage after acquiring new coreceptor usage (5, 44–46, 48). This finding supports the notion that the adaptation of HIV-1 to use CXCR4 or other chemokine receptors during phenotypic evolution may not necessarily be at the expense of CCR5 usage.
Here we tested the chemokine receptor usage of 137 diverse, well-characterized primary virus isolates by monitoring viral replication in U87MG.CD4 transformed cell lines expressing CCR1, CCR2b, CCR3, CCR5, and CXCR4 and HOS.CD4 transformed cell lines expressing STRL33/BONZO/TYMSTR and GPR15/BOB. These 137 primary viruses were isolated from 79 men with incident HIV-1 infection enrolled in the Chicago component of the Multicenter AIDS Cohort Study (MACS) on the basis of a demonstrated time of primary infection and the availability of viably frozen peripheral blood mononuclear cells (PBMC) obtained within 3 months after infection. Fifty-eight of these 79 men also had PBMC samples available from a subsequent time point. All the men had clinical examinations, CD4 T-cell counts, and plasma viral burden measurements performed during approximately 6-month-interval study visits, and all had defined CCR5 wild-type and defective CCR5 Δ32 alleles (28). We isolated virus from approximately 5 × 106 cryopreserved PBMC from the patients by cocultivation with 5 × 106 phytohemagglutinin-stimulated normal donor PBMC, as previously described (6, 27). Culture supernatants were monitored for p24 production on days 4, 7, and 14 by using a commercial enzyme immunoassay (Abbott Laboratories, Abbott Park, Ill.). A culture was considered positive if the p24 value was above a cutoff of 30 pg/ml. We titrated viruses in PBMC to determine the 50% tissue culture infectious dose (TCID50). We also obtained nucleotide sequences of the C2-V3 region of env from these 79 men to assess the genetic identity of the isolated virus. Phylogenetic analyses of these viral sequences revealed distinct lineages for each subject without clustering, indicating the lack of epidemiological linkage and the absence of contamination by prototypic laboratory-adapted virus strains (data not shown).
We determined the phenotypes of the 137 isolated primary viruses by their abilities to induce syncytia in the MT-2 transformed T-cell line, as previously described (13). Among the 79 isolated viruses obtained near the time of primary infection, 71 (89.9%) had a non-syncytium-inducing (NSI) phenotype while only 8 (10.1%) had a syncytium-inducing (SI) phenotype, suggesting that viruses with an NSI phenotype predominate at this time. The proportion of viruses with an SI phenotype is somewhat higher than that in the earlier findings of others (30, 31, 35, 38). This increase in the proportion of viruses with an SI phenotype may reflect the increase in viral pathogenicity and transmissibility over the course of the epidemic. The predominance of a viral population with an NSI phenotype during primary infection supports the hypothesis of selective viral transmission of amplification (11, 35, 51, 52). While this hypothesis does not exclude the transmission of viruses with an SI phenotype, transmission of such viruses is likely to be much less efficient. We find support for this conclusion in the observation that individuals homozygous for the defective CCR5 Δ32 allele can be infected by viruses with an SI phenotype; presumably, viral entry here is mediated through CXCR4 cofactor usage (39, 41, 49). Furthermore, a hybrid construct between simian and human immunodeficiency viruses with exclusive CXCR4 usage in vitro penetrates the genitourinary mucosal barrier and establishes persistent infection in the rhesus macaque model (9a). Thus, the selection pressure exerted during the course of transmission is a likely consequence of both the availability and the level of the CCR5 and/or CXCR4 chemokine receptor expressed on the target cells as well as the phenotypic composition of the infecting virus.
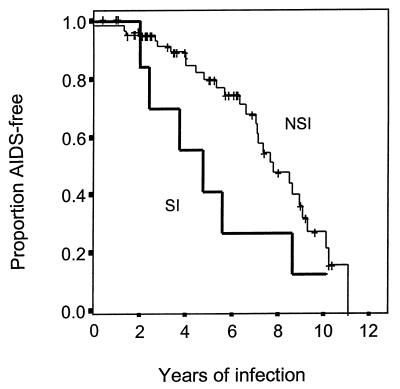
We performed univariant analysis of virologic parameters associated with disease progression by breaking down the cohort according to the NSI and SI viral phenotypes within 3 months of primary infection. AIDS-free survival was determined by Kaplan-Meier analysis, and the level of significance was set at a P value of <0.05. We observed faster progression to AIDS for the eight men who were initially infected with a virus having an SI phenotype than for the others, who were not (Fig. 1). We observed a switch in the phenotype of the isolated virus from NSI to SI for 5 of the 71 men initially infected with an NSI variant. A faster rate of CD4 T-cell loss over time was also observed for these 5 men than for the other 66 men, whose isolated primary viruses maintained an NSI phenotype (data not shown). We also observed a greater tendency for infection with a virus with an SI phenotype in those men heterozygous for the defective CCR5 Δ32 allele (25%) than in those men homozygous for the wild-type CCR5 allele (6%) (P = 0.03). Because the number of viruses with an SI phenotype is relatively small, this finding should not be overinterpreted. Nonetheless, by using a population-based study of men with incident infection we found that rapid disease progression is associated with viruses with the SI phenotype, regardless of whether the men were homozygous for the wild-type CCR5 allele or heterozygous for the defective CCR5 Δ32 allele. This finding may provide some explanations for the bimodal distribution in disease progression among men heterozygous for the defective CCR5 Δ32 allele observed in our previous study (28). We previously found that a lower proportion of these men were AIDS free at 4 to 6 years after primary infection, but a higher proportion were AIDS free at year 10 relative to the men who were homozygous for the wild-type CCR5 allele (28). Here we confirmed that the lower proportion of men heterozygous for the CCR5 Δ32 allele who were AIDS-free at 4 to 6 years after primary infection was the result of infection with a virus having an SI phenotype.
FIG. 1.
Kaplan-Meier plot of time to development of AIDS for all 79 men whose initial blood samples were obtained within 3 months of primary HIV-1 infection. These men are divided into two groups according to the phenotype of the virus. The thick line represents men infected with viruses having SI phenotypes, whereas the thin line represents men infected with viruses having NSI phenotypes. The comparison of these two lines indicates a significant difference in disease progression between the two groups (P < 0.05).
We tested the 137 isolated primary viruses and 35 additional primary viruses representing HIV-1 group M subtypes A, B, C, D, and E and group O and three primary simian immunodeficiency virus (SIV) isolates for different subsets of chemokine receptors used as entry cofactors. A number of these last viral isolates were obtained from the World Health Organization Network for HIV-1 Isolation and Characterization and from the National Institute of Allergy and Infectious Diseases network for characterizing antigenic variation (24, 25, 32, 33, 40). Twenty-eight of the 35 viruses had an NSI phenotype, and 7 had an SI phenotype. SIVmac251 and SIVmac239 were provided by R. C. Desrosiers of the New England Regional Primate Research Center (Southborough, Mass.) and expanded in PBMC obtained from uninfected rhesus macaques. We propagated SIVsmLib1, an isolate originally derived from a pet West African sooty mangabey (7), in CEMx174 cells. The viral titer was quantified by limiting-dilution analysis in CEMx174 cells with a commercially available SIV p27 immunoassay (Cellular Products, Buffalo, N.Y.).
We tested chemokine receptor subset usage by these diverse, well-characterized primary viruses by monitoring viral replication in CCR1, CCR2b, CCR3, CCR5, and CXCR4 U87MG.CD4 transformed cell lines, and STRL33/BONZO/TYMSTR and GPR15/BOB HOS.CD4 transformed cell lines (provided by D. Littman, Skirball Institute for Molecular Medicine, New York, N.Y.). We assessed chemokine receptor usage by inoculating 100 TCID50 of each isolated primary HIV-1 onto 104 U87MG.CD4 cells expressing CCR1, CCR2b, CCR3, CCR5, or CXCR4, as well as HOS.CD4 cells expressing STRL33/BONZO/TYMSTR or GPR15/BOB. After 4 h, unbound virus was removed by three exchanges of culture medium (Dulbecco’s minimal essential medium supplemented with 10% fetal calf serum, glutamine, antibiotics, and 1 μg of puromycin [Sigma Chemicals, St. Louis, Mo.] per ml). The culture supernatant was monitored for HIV-1 p24 antigen production on days 3 and 7 after infection by using a commercial enzyme immunoassay (Abbott Laboratories). A culture was considered positive if the p24 antigen concentration was above 30 pg/ml 7 days after inoculation.
Whereas all isolates of primary HIV-1 used CCR5 and/or CXCR4 for viral entry, we found no viruses that used CCR1, CCR2b, CCR3, STRL33/BONZO/TYMSTR, or GPR15/BOB (Table 1). The restricted use of CCR5 and CXCR4 was observed regardless of whether the virus represented the genetically divergent viral strains within the M group or the even more divergent set of viruses within the O group. In contrast, the isolate of SIVsmLib1 used both STRL33/BONZO/TYMSTR and GPR15/BOB, while SIVmac239 and SIVmac251 preferentially used GPR15/BOB, as entry cofactors in addition to CCR5. Conservation of CCR5 binding across the genetically divergent viral strains within the HIV-1 M and O groups suggests that conserved amino acid residues within the CCR5 binding site and/or the viral envelope structure contribute to the multipoint binding site for gp120 on the chemokine receptor. Amino acid sequence alignments of CCR5 derived from people of diverse genetic backgrounds show that this chemokine receptor is highly conserved (2, 50). Similarly, despite the high frequency of amino acid polymorphism for CCR5 alleles derived from nonhuman primates, including apes, Old World monkeys, New World monkeys, and selected prosimians, we observed amino acid conservation within specific regions, especially within the chemokine receptor ectodomain (4, 9, 34, 43, 50). All of these characterized nonhuman primate CCR5 alleles permit primate immunodeficiency virus fusion and entry (34, 35a, 43) and thus likely permit the cross-species transmission of primate immunodeficiency virus (8, 47).
TABLE 1.
Viral phenotype and coreceptor usagea
| Virus | No. of viruses | Viral phenotype (no.) | Co-receptor usagec
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| CCR1 | CCR2b | CCR3 | CCR5 | CXCR4 | BOB | BONZO | |||
| HIV-1 (MACS) | 137 | NSI (123) | 0 | 0 | 0 | 123 | 0 | 0 | 0 |
| SI (14) | 0 | 0 | 0 | 14 | 14 | 0 | 0 | ||
| HIV-1 (groups M and O) | 35b | NSI (28) | 0 | 0 | 0 | 28 | 0 | 0 | 0 |
| SI (7) | 0 | 0 | 0 | 7 | 7 | 0 | 0 | ||
| SIVsmLib1 | − | − | − | + | − | + | + | ||
| SIVmac239 | − | − | − | + | − | + | − | ||
| SIVmac251 | − | − | − | + | − | + | − | ||
Correlation between phenotype of isolated primary viruses and CC and CXC chemokine receptors or chemokine receptor-like orphan molecules used for virus entry.
Distribution was as follows. Group M subtypes: A, 2; B, 6; C, 19; D, 3; E, 3. Group O, 2.
Number using each coreceptor. For SIV: −, not used; +, used. BOB, GPR15/BOB; BONZO, STRL33/BONZO/TYMSTR.
We also found that all isolates of viruses with an NSI phenotype used CCR5 as an entry cofactor and all isolates of viruses with an SI phenotype used CCR5 and CXCR4. No isolates of primary viruses used CXCR4 exclusively. Viruses that showed an NSI-to-SI phenotypic switch between a time 3 months after primary infection and a subsequent time point were associated with a broadening of chemokine receptor usage, evolving from the exclusive use of CCR5 to use of both CCR5 and CXCR4 as entry cofactors (Table 1). Broadening coreceptor usage from CCR5 to both CCR5 and CXCR4 was associated with a faster rate of disease progression, which is consistent with other studies. Broadening of virus tropism during the progression of disease was not an invariable finding, however. The relatively limited numbers of primary isolates of HIV-1 tested here, especially of the SI phenotype, preclude any possible correlation between CCR3 usage and the particular viral phenotype. Thus, for HIV-1 the viral phenotypic properties correlate perfectly with the specific subset of chemokine receptor used for viral entry (Table 1).
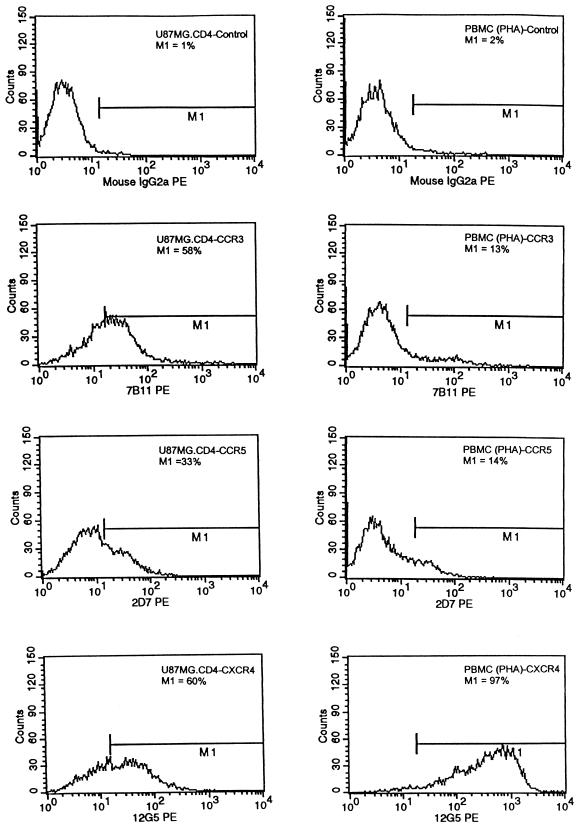
We were unable to identify any primary viruses that used CCR3 as an entry cofactor. Our observations are in contrast to those of others, who found that several Env proteins, including ADA, JR-FL, HXB2, and 89.6, used CCR3 for virus entry (3, 5, 46). The discrepancy likely represents differences in the transformed cell lines tested and the cell surface expression levels of CCR3. Given these discrepant results, we analyzed the levels of cell surface expression of CCR3, CCR5, and CXCR4 by flow cytometry analysis of stained cells with the 7B11, 2D7, and 12G5 CC- and CXC-chemokine-specific phycoerythrin (PE)-conjugated monoclonal antibodies (National Institute of Allergy and Infectious Diseases AIDS Research Reagents Repository Program), respectively (Fig. 2). We found by measuring relative fluorescence intensity that CCR3, CCR5, and CXCR4 all were expressed at the cell surface at comparable levels (58, 33, and 60%, respectively) commensurate with or higher than what we observed for cells derived from men who are homozygous for the wild-type CCR5 allele (Fig. 2). Thus, it is likely that observed differences in chemokine receptor usage for virus entry by isolates of primary viruses or laboratory-adapted strains represent heterogeneity of expression levels and a potential threshold level of chemokine cell surface expression.
FIG. 2.
Flow cytometric analysis for CCR3, CCR5, and CXCR4 in U87MG.CD4 transformed cell lines (left) and phytohemagglutinin-stimulated PBMC (right). The cells were labeled with an anti-CCR3 (7B11), anti-CCR5 (2D7), or anti-CXCR4 (12G5) PE-conjugated antibody or a PE-conjugated isotypic control antibody (mouse immunoglobulin G 2a [IgG2a]). Each histogram is representative of at least two separate experiments.
Our results demonstrate that the expression of CD4 in combination with either CCR5 or CXCR4 is necessary and sufficient for primary HIV-1 entry. Only primary isolates of SIV used the STRL33/BONZO/TYMSTR and GPR15/BOB chemokine receptor-like orphan molecules as entry cofactors. Despite the diversity of viruses isolated at primary infection and at a later time in the course of disease, the use of other members of the chemokine receptor family of seven-span transmembrane G protein-coupled receptors does not appear to influence viral pathogenesis. It should be noted that the viruses used in our work were isolated from peripheral blood; those compartmentalized in privileged anatomic and immunologic sites may use other chemokine receptors for viral entry. The ability of those sequestered viruses to use alternative chemokine receptors may underlie the ability of viruses with specific tropism to infect specific target cells, as demonstrated by the use of CCR3 by neurotropic isolates (26). While broadening of chemokine receptor usage may enable the pathogen to exploit a new ecological niche within the host, data from HIV-1-infected individuals with advanced disease do not support this contention (19, 44, 45, 48). Isolates of primary viruses obtained from HIV-1-infected individuals with advanced disease predominantly use CCR5 and/or CXCR4, although a small number of isolates can also use CCR3 (5, 14, 46). Here, no primary viruses that we isolated used CCR3 for virus entry when expressed at physiologically comparable levels. Thus, it is unlikely that HIV-1 genetic variants, which evolve during the development of disease to use alternative chemokine receptors, are responsible for the ultimate devastation of the immune system. Rather, broadening of chemokine receptor usage is more likely attributable to target cell availability and the threshold level of chemokine cell surface expression, especially in regard to the stoichiometry of the CD4 and chemokine cell surface molecules required for a proficient cell fusion complex. Thus, these results and the results of others strongly suggest that CCR5 and CXCR4 are the physiologically most relevant chemokine receptors used for virus entry by diverse strains of primary viruses isolated from peripheral blood.
Acknowledgments
We thank Robert Doms for helpful discussions and Ariel Ploss for secretarial assistance.
This work was supported by the National Institutes of Health under grants HD-31756, AI-35168, AI45218, AI40387, AI45218, and AI35039; the Aaron Diamond Foundation; and a gift from an anonymous foundation.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: A RANTES, MIP1-alpha, MIP1-beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Ansari-Lari M A, Liu X M, Metzker M L, Rut A R, Gibbs R A. The extent of genetic variation in the CCR5 gene. Nat Genet. 1997;16:221–222. doi: 10.1038/ng0797-221. [DOI] [PubMed] [Google Scholar]
- 3.Bazan H A, Alkhatib G, Broder C C, Berger E A. Patterns of CCR5, CXCR4, and CCR3 usage by envelope glycoproteins from human immunodeficiency virus type 1 primary isolates. J Virol. 1998;72:4485–4491. doi: 10.1128/jvi.72.5.4485-4491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger, E. A. 1997. HIV entry and tropism: the chemokine receptor connection. AIDS 11(Suppl. A):S3–S16. [PubMed]
- 5.Bjorndal A, Deng H, Jansson M, Fiore J R, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman D R, Fenyo E M. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao Y, Qin L, Zhang L, Safrit J, Ho D D. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. N Engl J Med. 1995;332:201–208. doi: 10.1056/NEJM199501263320401. [DOI] [PubMed] [Google Scholar]
- 7.Chen Z, Telfer P, Reed P, Zhang L, Getti A, Ho D D, Marx P A. Isolation and characterization of the first simian immunodeficiency virus from a feral sooty mangabey (Cercocebus atys) in West Africa. J Med Primatol. 1995;24:108–115. doi: 10.1111/j.1600-0684.1995.tb00155.x. [DOI] [PubMed] [Google Scholar]
- 8.Chen Z, Telfer P, Gettie A, Reed P, Zhang L, Ho D D, Marx P A. Genetic characterization of new West African simian immunodeficiency virus SIVsm: geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J Virol. 1996;70:3617–3627. doi: 10.1128/jvi.70.6.3617-3627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Z, Zhou P, Ho D D, Landau N R, Marx P A. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Cheng-Mayer, C. Personal communication.
- 10.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The b-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 11.Cichutek K, Norley S, Linde R, Kreuz W, Gahr M, Lower J, von Wangenheim V, Kurth R. Lack of HIV-1 V3 region sequence diversity in two haemophiliac patients infected with a mutative biologic clone of HIV-1. AIDS. 1991;5:1185–1187. doi: 10.1097/00002030-199110000-00005. [DOI] [PubMed] [Google Scholar]
- 12.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1a, and MIP-1b as the major HIV suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 13.Connor R, Ho D D. Human immunodeficiency virus type 1 variants with increased replicative capacity develop during the asymptomatic stage before disease progression. J Virol. 1994;68:4400–4408. doi: 10.1128/jvi.68.7.4400-4408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalgleish A G, Beverley P C, Clapham P R, Crawford D H, Greaves M F, Weiss R A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 16.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz S, Burkhart M, DiMarzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 17.Deng H K, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 18.Dimitrov D S. How do viruses enter cells? The HIV coreceptors teach us a lesson of complexity. Cell. 1997;91:721–730. doi: 10.1016/s0092-8674(00)80460-2. [DOI] [PubMed] [Google Scholar]
- 19.Dittmar M T, Simmons G, Donaldson Y, Simmonds P, Clapham P R, Schulz T F, Weiss R A. Biological characterization of human immunodeficiency virus type 1 clones derived from different organs of an AIDS patient by long-range PCR. J Virol. 1997;71:5140–5147. doi: 10.1128/jvi.71.7.5140-5147.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual tropic primary HIV-1 isolate that uses fusion and the b-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 21.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 22.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of seven-transmembrane domain, G-protein coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 23.Frade J M R, Llorente M, Mellado M, Alcami J, Gutierrez-Ramos J C, Zaballos A, Real G, Martinez A C. The amino-terminal domain of the CCR2 chemokine receptor acts as coreceptor for HIV-1 infection. J Clin Investig. 1997;100:497–502. doi: 10.1172/JCI119558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao F, Morrison S G, Robertson D L, Thornton C L, Craig S, Karlsson G, Sodroski J, Morgado M, Galvao-Castro B, von Briesen H, Beddows S, Weber J, Sharp P M, Shaw G M, Hahn B H the WHO and NIAID Networks for HIV Isolation and Characterization. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. J Virol. 1996;70:1651–1667. doi: 10.1128/jvi.70.3.1651-1667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao F, Yue L, Craig S, Thornton C L, Robertson D L, McCutchan F E, Bradac J A, Sharp P M, Hahn B H the WHO Network for HIV Isolation and Characterization. Genetic variation of HIV type 1 in four World Health Organization-sponsored vaccine evaluation sites: generation of functional envelope (glycoprotein 160) clones representative of sequence subtypes A, B, C, and E. AIDS Res Hum Retroviruses. 1994;10:1359–1368. doi: 10.1089/aid.1994.10.1359. [DOI] [PubMed] [Google Scholar]
- 26.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, Mackay C R, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 27.Ho D D, Moudgil T, Alam M. Quantitation of human immunodeficiency virus type 1 in the blood of infected persons. N Engl J Med. 1989;321:1621–1625. doi: 10.1056/NEJM198912143212401. [DOI] [PubMed] [Google Scholar]
- 28.Huang Y, Paxton W A, Wolinsky S M, Neumann A U, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau N R, Phair J, Ho D D, Koup R A. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 29.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman J C, Montagnier L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 30.Koot M, van ’t Wout A B, Kootstra N A, de Goede R E, Tersmette M, Schuitemaker H. Relation between changes in cellular load, evolution of viral phenotype, and the clonal composition of virus populations in the course of human immunodeficiency virus type 1 infection. J Infect Dis. 1996;173:349–354. doi: 10.1093/infdis/173.2.349. [DOI] [PubMed] [Google Scholar]
- 31.Koot M, Vos A H, Keet R P, de Goede R E, Dercksen M W, Terpstra F G, Coutinho R A, Miedema F, Tersmette M. HIV-1 biological phenotype in long-term infected individuals evaluated with an MT-2 cocultivation assay. AIDS. 1992;6:49–54. doi: 10.1097/00002030-199201000-00006. [DOI] [PubMed] [Google Scholar]
- 32.Korber B T M, Osmanov S, Esparza J, Myers G the World Health Organization Network for HIV Isolation and Characterization. The World Health Organization global programme on AIDS proposal for standardization of HIV sequence nomenclature. AIDS Res Hum Retroviruses. 1994;10:1355–1358. doi: 10.1089/aid.1994.10.1355. [DOI] [PubMed] [Google Scholar]
- 33.Kostrikis L G, Cao Y, Ngai H, Moore J P, Ho D D. Quantitative analysis of serum neutralization of human immunodeficiency virus type 1 from subtypes A, B, C, D, E, F, and I: lack of direct correlation between neutralization serotypes and genetic subtypes and evidence for prevalent serum-dependent infectivity enhancement. J Virol. 1996;70:445–458. doi: 10.1128/jvi.70.1.445-458.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuhmann S E, Platt E J, Kozak S L, Kabat D. Polymorphisms in the CCR5 genes of African green monkeys and mice implicate specific amino acids in infections by simian and human immunodeficiency viruses. J Virol. 1997;71:8642–8656. doi: 10.1128/jvi.71.11.8642-8656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuiken C L, Lukashov V V, Baan E, Dekker J, Leunissen J A M, Goudsmit J. Evidence for limited within-person evolution of the V3 domain of the HIV-1 envelope in the Amsterdam population. AIDS. 1996;10:31–37. doi: 10.1097/00002030-199601000-00005. [DOI] [PubMed] [Google Scholar]
- 35a.Kunstman, K. J. Unpublished data.
- 36.Liao F, Alkhatib G, Peden K W, Sharma G, Berger E A, Farber J M. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maddon P J, Dalgleish A G, McDougal J S, Clapham P R, Weiss R A, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–385. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 38.Michael N L, Chang G, Louie L G, Mascola J R, Dondero D, Birx D L, Sheppard H W. The role of viral phenotype and CCR-5 gene defects in HIV-1 transmission and disease progression. Nat Med. 1997;3:338–340. doi: 10.1038/nm0397-338. [DOI] [PubMed] [Google Scholar]
- 39.Michael N L, Nelson J A E, KewalRamani V N, Chang G, O’Brien S J, Mascola J R, Volsky B, Louder M, White II G C, Littman D R, Swanstrom R, O’Brien T R. Exclusive and persistent use of the entry coreceptor CXCR4 by human immunodeficiency virus type 1 from a subject homozygous for CCR5 Δ32. J Virol. 1998;72:6040–6047. doi: 10.1128/jvi.72.7.6040-6047.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore J P, McCutchan F E, Poon S W, Mascola J, Liu J, Cao Y, Ho D D. Exploration of antigenic variation in gp120 from clades A through F of human immunodeficiency virus type 1 by using monoclonal antibodies. J Virol. 1994;68:8350–8364. doi: 10.1128/jvi.68.12.8350-8364.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Brien T, Winkler C, Dean M, Nelson J A, Carrington M, Michael N L, White G C., II HIV-1 infection in a man homozygous for CCR5 delta 32. Lancet. 1997;349:1219. doi: 10.1016/s0140-6736(97)24017-1. [DOI] [PubMed] [Google Scholar]
- 42.Pleskoff O, Treboute C, Brelot A, Heveker N, Seman M, Alizon M. Identification of a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1 entry. Science. 1997;276:1874–1878. doi: 10.1126/science.276.5320.1874. [DOI] [PubMed] [Google Scholar]
- 43.Pretet J L, Zerbib A C, Girard M, Guillet J G, Butor C. Chimpanzee CXCR4 and CCR5 act as coreceptors for HIV type 1. AIDS Res Hum Retroviruses. 1997;13:1583–1587. doi: 10.1089/aid.1997.13.1583. [DOI] [PubMed] [Google Scholar]
- 44.Rana S, Besson G, Cook D G, Rucker J, Smyth R J, Yi Y, Turner J D, Guo H-H, Du J-G, Peiper S C, Lavi E, Samson M, Libert F, Liesnard C, Vassart G, Doms R W, Parmentier M, Collman R G. Role of CCR5 in infection of primary macrophages and lymphocytes by macrophage-tropic strains of human immunodeficiency virus: resistance to patient-derived and prototype isolates resulting from the Δccr5 mutation. J Virol. 1997;71:3219–3227. doi: 10.1128/jvi.71.4.3219-3227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rucker J, Edinger A L, Sharron M, Samson M, Lee B, Berson J F, Yi Y, Margulies B, Collman R G, Doranz B J, Parmentier M, Doms R W. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scarlatti G, Tresoldi E, Bjorndal A, Fredriksson R, Colognesi C, Deng H K, Malnati M S, Plebani A, Siccardi A G, Littman D R, Fenyo E M, Lusso P. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 47.Sharp, P. M., D. L. Robertson, F. Gao, and B. H. Hahn. 1994. Origins and diversity of human immunodeficiency viruses. AIDS 8(Suppl.):S27–S42.
- 48.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either LESTR or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Theodorou I, Meyer L, Magierowska M, Katlama C, Rouzioux C the Seroco Study Group. HIV-1 infection in an individual homozygous for CCR5 delta 32. Lancet. 1997;349:1219–1220. [PubMed] [Google Scholar]
- 50.Zhang L, Carruthers C D, He T, Huang Y, Cao Y, Wang G, Hahn B, Ho D D. HIV type 1 subtypes, coreceptor usage, and CCR5 polymorphism. AIDS Res Hum Retroviruses. 1997;13:1357–1366. doi: 10.1089/aid.1997.13.1357. [DOI] [PubMed] [Google Scholar]
- 51.Zhang L Q, MacKenzie P, Cleland A, Holmes E C, Brown A J, Simmonds P. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J Virol. 1993;67:3345–3356. doi: 10.1128/jvi.67.6.3345-3356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]