Abstract
Human immunodeficiency virus type 1 (HIV-1) encodes the transactivator protein Tat, which is essential for viral replication and progression to disease. Here we demonstrate that transcriptional activation by HIV-1 Tat involves p300 or the related cellular transcriptional coactivator CREB binding protein (CBP). Tat transactivation was inhibited by the 12S form of the adenovirus E1A gene product, which inhibits p300 function, and this inhibition was independent of its effect on NF-κB transcription. A biochemical interaction of p300 with Tat was demonstrated in vitro and in vivo by coimmunoprecipitation. The carboxy-terminal region of p300, which binds to E1A, was shown to bind specifically to the highly conserved basic domain of Tat, which also mediates binding to the Tat-responsive region RNA stem-loop structure. The ability of Tat to interact physically and functionally with this coactivator provides a mechanism to assemble a basal transcription complex which may subsequently respond to the effect of Tat on transcriptional elongation and represents a novel interaction between an RNA binding protein and a transcriptional coactivator.
The human immunodeficiency virus type 1 (HIV-1) transactivator of transcription (Tat) stimulates viral gene expression through an RNA element in the viral long terminal repeat (LTR) (14, 17). For optimal transactivation of HIV gene expression, Tat requires specific upstream transcription factors, including Sp1 (13), TATA binding protein (18, 39), Tat-associated kinase (TAK) (11, 43), TFIIH (9, 30), Tip (16), and RNA polymerase II (4, 24, 42). An important effect of Tat is to increase the processivity of RNA polymerase by phosphorylation of its carboxy-terminal domain, which likely occurs in concert with host cellular cofactors (19). Among the factors which associate with Tat, TAK has been shown to interact with Tat in vitro and in vivo through its activation domain (44, 45). The ability of Tat to regulate transcriptional elongation is likely related to its ability to interact with the basal transcription complexes responsible for initiation, and Tat has been shown to influence transcriptional initiation. For example, Tat interacts with the preinitiation complex through its basic domain in a Tat-responsive region (TAR)-independent manner, an association that is likely of functional significance (8), and TFIIH has been shown to regulate its function (3, 9, 30). Though of indirect importance in its effect on HIV gene expression, the assembly of the relevant basal complex is required for Tat activation (9, 13, 15, 18, 39, 42).
Among the factors associated with the basal transcription complex, p300 and the related CREB binding protein (CBP) (2) have emerged as coactivators for a broad group of cellular transcription factors (for reviews, see references 5 and 34). p300, identified initially by its interaction with E1A (6), is highly related to CBP (23); both molecules are large nuclear phosphoproteins that respond to changes in cell growth (5) and integrate into diverse signal transduction pathways (12, 28, 37). To date, the cellular cofactors that bind to Tat have not been implicated in chromatin remodeling. Because inhibitors of histone deacetylation were recently shown to activate integrated HIV-1 LTR (38) and p300 affects histone acetylation (1, 29, 44), we explored the possibility that Tat interacts with p300.
MATERIALS AND METHODS
Plasmids.
The chloramphenicol acetyltransferase (CAT) reporter constructs HIV-CAT and ΔκB HIV-CAT and the Rous sarcoma virus (RSV) expression vector have been previously described (22, 26). The wild-type adenovirus 12S E1A (12S E1A wt) RSV expression plasmid and the mutant Δ2/36 as well as the mutant 2G3N 12S E1A, both used as 12S E1A Δp300 plasmids, were amplified by PCR from the 12S E1A wt expression plasmid previously reported (25) and cloned into pRSVGAL4.1, using the HindIII and BamHI sites. Glutathione S-transferase–(GST)-Tat and the corresponding deletion mutant were generated by PCR from plasmid pcTat/Rev (36) and cloned into pGEX-CD1. Cytomegalovirus (CMV)-Tat was generated by cloning wild-type Tat into the expression vector described earlier (32). GST-Tat (48-57) and GST-Tat (NBD) were generated by cloning oligonucleotides into pGEX-CD1 as follows: for the basic domain of Tat, amino acid residues GRKKRRQRRR; and for the basic domain of the N gene (bacteriophage λ), amino acid residues GQTRRRERRA.
Reporter assays.
Jurkat T-leukemia cells were transfected by using DEAE-dextran, cells were harvested after 24 h, and CAT assays were performed as previously described (22). Equal quantities of extract protein were analyzed in each assay. The transfection efficiency of Jurkat cells with DEAE-dextran is, in contrast to transfection of 293 cells with calcium phosphate, constant and reproducible, with standard deviations of ≤10% (22, 27, 33).
Fusion proteins.
GST fusion proteins were expressed in Escherichia coli BL21 cells and purified as reported previously (32). Purified GST was used as the control.
In vitro binding assays.
GST fusion proteins were incubated at 4°C for 1 h with 10 μl of glutathione-Sepharose beads. Beads were washed three times in buffer containing 75 mM KCl and 0.1% Nonidet P-40. Bound proteins were eluted with an equal volume of sodium dodecyl sulfate (SDS) loading buffer, resolved by SDS-polyacrylamide gel electrophoresis (PAGE), and analyzed by Western blotting or, in the case of in vitro-translated proteins, by exposure to X-ray film (Kodak). Deletion analysis of the carboxy-terminal fragments of p300 was performed by digesting 3′pBS-p300 (32) with BstEII, StuI, or StyI prior to in vitro transcription-translation (radiolabeled T7 TNT reticulocyte transcription-translation system; Promega) to produce p300 fragments truncated at amino acid 1909, 1710, or 1542, respectively.
Immunoprecipitation and Western blotting.
p300 immunoprecipitations were performed as described previously (32) with 3 μg of an anti-p300 antibody (14991A; PharMingen) or control rabbit immunoglobulin G (IgG) in the presence of 75 mM NaCl. The assay was terminated by the addition of SDS gel loading buffer to the samples. Proteins were resolved by SDS-PAGE on either a 7% (for p300) or 15% (for Tat) polyacrylamide gel, and then transferred to nitrocellulose membranes for 14 h at 4°C. After incubation with the indicated antibodies for 2 h at room temperature, blots were incubated with secondary antibodies and visualized by enhanced chemiluminescence as suggested by the manufacturer (Amersham). Western blot analysis was performed with anti-p300 (sc584 and sc585; Santa Cruz Biotechnology) anti-CBP (sc583 and sc369; Santa Cruz), and anti-Tat (13-164-100; ABI) antibodies.
RESULTS
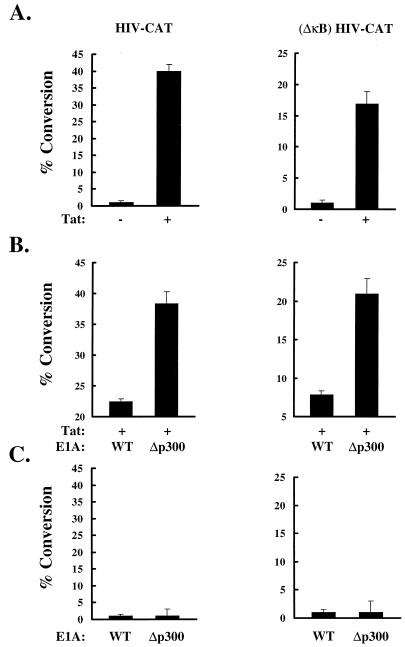
To analyze whether Tat-dependent gene activation was p300 dependent, we examined the ability of 12S E1A to inhibit transcriptional activation of HIV-CAT expression by Tat. Jurkat lymphocytes were cotransfected with HIV-CAT, a 12S E1A expression plasmid, and a Tat expression vector. Activation of the HIV promoter by Tat (Fig. 1A, left) was inhibited by 12S E1A, whereas cotransfection of a 12S E1A mutant, which cannot bind p300 (40), had no effect on Tat activation (Fig. 1B, left). Since the HIV LTR contains two κB sites, and the transcriptional activity of NF-κB can be inhibited by E1A (31), the effect of 12S E1A on Tat-induced gene expression was examined by using an HIV-CAT reporter with mutant κB sites [(ΔκB) HIV-CAT]. 12S E1A wt inhibited κB-independent Tat-induced gene expression, in contrast to the mutant 12S E1A, which did not bind to p300 (Fig. 1B, right). To address the possibility that wild-type or mutant 12S E1A affects Tat expression under these conditions, we repeated the experiment using the same expression plasmid encoding the CAT reporter protein. At the same molar ratio (500-fold mass excess), wild-type or mutant 12S E1A did not alter CAT expression significantly, suggesting that the effect of 12S E1A on Tat transcriptional function is not due to changes in the expression of Tat (Fig. 1, legend). No transcriptional activation of HIV-CAT or (ΔκB) HIV-CAT was observed by 12S E1A wt or mutant 12S E1A itself (Fig. 1C). These data raised the possibility that p300 can potentiate Tat-dependent gene activation.
FIG. 1.
12S E1A wt inhibits Tat induced HIV-1 gene expression. Jurkat T-leukemia cells were transfected with HIV-CAT (3 μg) or (ΔκB) HIV-CAT (3 μg) and RSV-Tat (10 ng) (A) or in combination with RSV-12S E1A wt (5 μg) or 12S E1A Δp300 (5 μg) (B). RSV-12S E1A wt (5 μg) and 12S E1A Δp300 (5 μg) were also transfected alone with HIV-CAT (3 μg) or ΔκB-HIV CAT (3 μg) (C). A control RSV vector was included such that the same amount of expression plasmid was used in each sample. Error bars indicate standard errors of the means of three independent experiments. A 500-fold mass excess of wild-type or mutant 12S E1A did not alter the expression of RSV-CAT significantly (0.77 or 0.72% conversion, respectively).
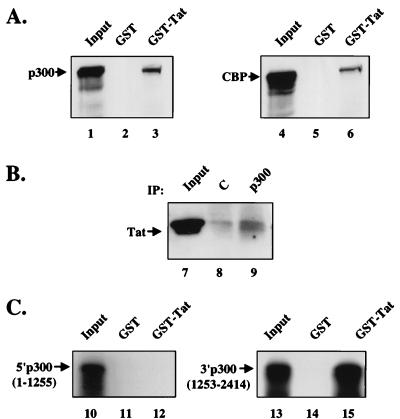
To determine whether p300 and CBP could interact biochemically with Tat, binding assays were performed. Nuclear protein extracts were incubated with a GST-Tat fusion protein (amino acids 1 to 86). p300 and CBP were readily detected by Western blotting of an eluate from a GST-Tat column but not from a GST control (Fig. 2A). This interaction was confirmed in vivo by coimmunoprecipitation studies with polyclonal antibodies directed against p300. Tat was detected at increased levels in anti-p300 immunoprecipitates of nuclear extracts from Tat-transfected 293 cells (Fig. 2B). Background binding of Tat was observed in control immunoprecipitations, likely due to its nonspecific binding to the antiserum. These findings indicated that Tat could interact biochemically with p300 within cells.
FIG. 2.
Tat interacts with p300 and CBP in vitro and in vivo. (A) p300 and CBP from Jurkat nuclear extracts were affinity purified with GST-Tat (lanes 3 and 6) or with GST as a negative control (lane 2 and 5). After three washes, bound proteins were resolved by SDS-PAGE and then subjected to Western blot analysis for p300 (left) or CBP (right). Lanes 1 and 4 represent 20 μg of the input protein. (B) Tat and p300 interact in vivo. Tat was expressed by using a CMV expression plasmid (CMV-Tat) in 293 cells and immunoprecipitated (IP) with a control rabbit IgG (lane 8) and an anti-p300 IgG (lane 9). Bound proteins were resolved by SDS-PAGE and then subjected to Western blot analysis for Tat. Lane 7 represents 25 μg of the input protein. (C) Tat interacts directly with the carboxy-terminal half of p300. In vitro-translated p300 amino-terminal half (left) and carboxy-terminal half (right) were bound to GST-Tat (lanes 12 and 15) or GST (lanes 11 and 14). Lanes 10 and 13 represent 10% of the in vitro-translated input protein.
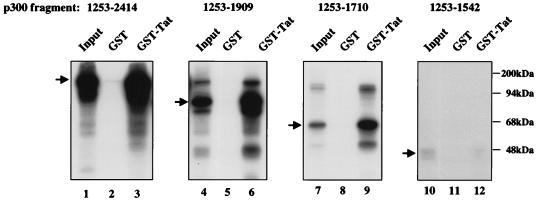
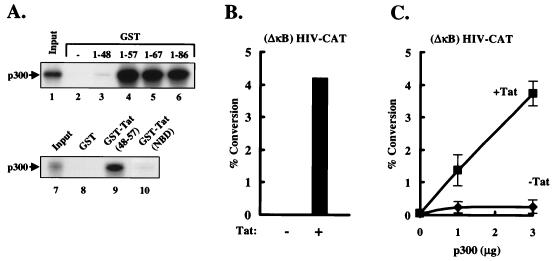
The region of p300 that mediates the Tat interaction was defined by further binding studies with in vitro-translated proteins. Binding of a carboxy-terminal p300 region (amino acids 1542 to 2412) was readily detected (Fig. 2C). In contrast, no detectable binding to GST-Tat was observed with an amino-terminal fragment of p300, indicating that the interaction is due not to nonspecific binding by Tat but to a specific interaction of two distinct domains in p300 and Tat. To confirm the specificity of the p300-Tat interaction, GST-Tat was also used in binding studies with other proteins, including RelA and Stat2. Neither of these proteins bound to GST-Tat under the same buffer conditions (data not shown), confirming the specificity of this interaction. Further localization revealed that the region spanning residues 1542 to 1710 of p300 was sufficient to mediate this interaction (Fig. 3). This carboxy-terminal region of p300 has also been shown to mediate interactions with TFIIB (20), cyclin E/Cdk2 (32), and adenovirus E1A (6). The region of Tat required for this interaction was next defined. A series of deletion mutants was prepared as GST fusion proteins in E. coli and purified with glutathione-Sepharose beads. Only fusion proteins which contained the Tat basic domain (residues 48 to 57) bound p300, including a fusion protein that expressed only this basic domain region (Fig. 4A, top). As a negative control, binding to the basic domain of the antiterminator coded by the N gene of bacteriophage λ was analyzed. This protein has amino acid sequence similarity to the arginine-rich domain of Tat (21) but does not substitute for this domain functionally or in its ability to bind to TAR (35). Binding assays using these two GST fusion proteins revealed that the basic domain of Tat, but not that of the bacteriophage λ N gene, binds to p300 (Fig. 4A, bottom), suggesting that this specific amino acid sequence of Tat mediates binding to p300. Functional experiments using Tat with a mutation in this TAR binding region were not performed because this region is required for binding to TAR and is thus known to be inactive.
FIG. 3.
Tat binds p300 between amino acid residues 1542 and 1710. Tat was immobilized on glutathione beads from bacterial extract which express GST-Tat and incubated with radiolabeled, in vitro-translated, carboxy-terminal deletion mutants of p300 (lanes 3, 6, 9, and 12). After three washes, bound proteins were resolved by SDS-PAGE. GST was used as a negative control (lanes 2, 5, 8, and 11). Lanes 1, 4, 7, and 10 represent 10% of the input protein. In lanes 4, 6, 7, and 9, small amounts of full-length p300 due to incomplete enzymatic digestion of the DNA template were noted.
FIG. 4.
Tat binds p300 through its basic domain. (A, top) The basic domain of Tat is responsible for the p300 interaction. Different GST-Tat deletion mutants were immobilized on glutathione beads from bacterial extract and incubated with radiolabeled, in vitro-translated, carboxy-terminal p300 residues 1253 to 1710 (lanes 3 to 6). GST was used as a negative control (lane 2). Lane 1 represents 10% of the input protein. (A, bottom) The basic domain of Tat specifically binds p300. GST-Tat (48-57) and GST-Tat (NBD) fusion proteins were immobilized on glutathione beads from bacterial extract and incubated with radiolabeled, in vitro-translated, carboxy-terminal p300 residues 1253 to 1710 (lanes 9 and 10). GST was used as a negative control (lane 8). Lane 7 represents 10% of the input protein. (B) Jurkat cells were transfected with (ΔκB) HIV-CAT (3 μg) and Tat (10 ng). A control RSV vector was included such that a total of 10 ng of expression plasmids was used in each sample. Values represent percent conversion over the background level. (C) Transfection of an expression vector encoding p300 stimulates Tat-induced transcriptional activation. Jurkat cells were transfected with (ΔκB) HIV-CAT (3 μg), RSV Tat (10 ng), RSV control (10 ng), and p300 (0, 1, or 3 μg). A control RSV vector was included such that equal moles of RSV vector were used in each sample. pBluescript was used to make up the mass difference. Values (+Tat) represent percent conversion over the level of transcriptional activation by Tat. Error bars indicate standard errors of the means of three independent experiments.
The functional contribution of p300 to Tat activation was investigated further by cotransfection of Jurkat cells with (ΔκB) HIV-CAT and expression vectors for Tat and p300. Expression of p300 alone had little effect on this promoter in the absence of Tat (Fig. 4C); however, expression of p300 increased Tat activation in a dose-dependent manner (Fig. 4C), and this synergistic effect was easily detected with small quantities of Tat expression vector plasmid (10 ng). The interaction of these two proteins therefore correlates with their ability to act in concert functionally.
DISCUSSION
In this study, we report that p300 interacts with the HIV-1 Tat protein and serves as a coactivator of Tat-dependent HIV-1 gene expression. 12S E1A was able to repress Tat-induced HIV gene expression by binding to p300 in an NF-κB-independent manner. The biochemical interaction between p300 and Tat was defined, and functional interaction between Tat and p300 was demonstrated by cotransfection. The region of Tat required for this interaction included the highly conserved basic domain of Tat that mediates binding to the TAR RNA stem-loop structure. These findings suggest that the coactivators p300 and CBP may provide a link between Tat and the basal transcriptional apparatus. To our knowledge, the interaction of Tat with the coactivators p300 and CBP also represents the first demonstration of an RNA binding protein with a transcriptional coactivator.
It has been reported recently that p300 and CBP not only interact with cellular transcription factors but also bind to a histone acetyltransferase (HAT), P/CAF (44), and possesses intrinsic HAT activity (29). The findings described here suggest that these coactivators can serve as adapters for RNA binding proteins and contribute to transcriptional regulation via targeted acetylation of chromatin. It has been suggested that a small amount of Tat protein, possibly packaged in virions, could trigger the remodeling of the chromatin adjacent to the HIV-1 LTR (10), and chromatin remodeling by histone acetylation had previously been implicated in transcriptional activation of the HIV-1 promoter (38). It has also been reported that nucleosomes can negatively influence polymerase elongation, an effect observed too for integrated HIV-1 templates (15).
These observations, together with the findings reported here, suggest that Tat interacts with the preinitiation complex through binding to p300, together with which it can mediate several functions. First, this complex can associate with NF-κB, which further promotes HIV transcription (32). Second, it is likely to induce histone acetylation, which would cause chromatin changes that facilitate transcription and Tat function. Third, this interaction appears to be necessary to assemble the relevant, Tat-responsive RNA polymerase II complex at the promoter. It is important to note that this interaction is relevant to the assembly of a transcription complex which is responsive to Tat, though the p300-CBP complex is unlikely to directly affect the block to transcriptional elongation relieved by Tat. The basic domain of Tat is responsible for binding to p300 as well as TAR, suggesting that dimerized Tat could facilitate the formation of the appropriate preinitiation complex with cellular factors such as P-TEFb, which promotes binding to TAR, and TFIIH, which may alter RNA polymerase activity. Recently, it has been reported that a novel cyclin-related protein, cyclin T, interacts with TAR in combination with Cdk9 and that the resulting complex relieves the block to elongation in a Tat-dependent fashion (41). The finding that Tat can associate with p300 suggests that a HAT activity (from p300 or one of its associated proteins) is recruited to this polymerase complex and further facilitates the profound effects of Tat on transcriptional elongation.
The Tat protein forms dimers (7), and it is thus possible that it interacts simultaneously with p300 and the TAR RNA structure. Alternatively, Tat could bind either TAR or p300 separately. Whether TAR RNA structure would affect the interaction of Tat to p300 in a positive or negative way has yet to be determined. Dimerization of Tat may be responsible for the inability to completely suppress Tat-dependent transcriptional activation by 12S E1A (Fig. 1B), in contrast to the substantial functional synergy between p300 and Tat in stimulating gene expression (Fig. 4C). HIV-1 Tat is therefore likely to exert effects at multiple steps, including its association with p300 in the preinitiation complex, followed by its binding to TAR and recruitment of additional components to the transcriptional elongation complex to enhance HIV gene expression and viral replication.
ACKNOWLEDGMENTS
We thank Nancy Barrett and Donna Gschwend for manuscript preparation, Lisa K. Felzien for critical reading of the manuscript, and other members of the Nabel lab for helpful advice and comments.
M.O.H. is supported by the Commission for the Advancement of Young Scientists and Scholars of the University of Zurich, Switzerland.
REFERENCES
- 1.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 2.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 3.Cujec T P, Okamoto H, Fujinaga K, Meyer J, Chamberlin H, Morgan D O. The HIV transactivator TAT binds to the CDK-activating kinase and activates the phosphorylation of the carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:2645–2657. doi: 10.1101/gad.11.20.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cujec T P, Cho H, Maldonado E, Meyer J, Reinberg D, Peterlin B M. The human immunodeficiency virus transactivator Tat interacts with the RNA polymerase II holoenzyme. Mol Cell Biol. 1997;17:1817–1823. doi: 10.1128/mcb.17.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckner R. p300 and CBP as transcriptional regulators and targets of oncogenic events. Biol Chem. 1996;377:685–688. [PubMed] [Google Scholar]
- 6.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 7.Frankel A D, Bredt D S, Pabo C O. Tat protein from human immunodeficiency virus forms a metal-linked dimer. Science. 1988;240:70–73. doi: 10.1126/science.2832944. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Martinez L F, Ivanov D, Gaynor R B. Association of Tat with purified HIV-1 and HIV-2 transcription preinitiation complexes. J Biol Chem. 1997;272:6951–6958. doi: 10.1074/jbc.272.11.6951. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Martinez L F, Mavankal G, Neveu J M, Lane W S, Ivanov D, Gaynor R B. Purification of a Tat-associated kinase reveals a TFIIH complex that modulates HIV-1 transcription. EMBO J. 1997;16:2836–2850. doi: 10.1093/emboj/16.10.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrich D, Ulich C, Garcia-Martinez L F, Gaynor R B. Tat is required for efficient HIV-1 reverse transcription. EMBO J. 1997;16:1224–1235. doi: 10.1093/emboj/16.6.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrmann C H, Rice A P. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J Virol. 1995;69:1612–1620. doi: 10.1128/jvi.69.3.1612-1620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horvai A E, Xu L, Korzus E, Brard G, Kalafus D, Mullen T-M, Rose D W, Rosenfeld M G, Glass C K. Nuclear integration of JAK/STAT and Ras/AP-1 signaling by CBP and p300. Proc Natl Acad Sci USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeang K T, Chun R, Lin N H, Gatignol A, Glabe C G, Fan H. In vitro and in vivo binding of human immunodeficiency virus type 1 Tat protein and Sp1 transcription factor. J Virol. 1993;67:6224–6233. doi: 10.1128/jvi.67.10.6224-6233.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones K A. Taking a new TAK on Tat transactivation. Genes Dev. 1997;11:2593–2599. doi: 10.1101/gad.11.20.2593. [DOI] [PubMed] [Google Scholar]
- 15.Jones K A, Peterlin B M. Control of RNA initiation and elongation at the HIV-1 promoter. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 16.Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 tat transactivator. Virology. 1996;216:357–366. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- 17.Kao S Y, Calman A F, Luciw P A, Peterlin B M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- 18.Kashanchi F, Piras G, Radonovich M F, Duvall J F, Fattaey A, Chiang C M, Roeder R G, Brady J N. Direct interaction of human TFIID with the HIV-1 transactivator tat. Nature. 1994;367:295–299. doi: 10.1038/367295a0. [DOI] [PubMed] [Google Scholar]
- 19.Kato H, Sumimoto R, Pognonec P, Chen C H, Rosen C A, Roeder R G. HIV-1 Tat acts as a processivity factor in vitro in conjunction with cellular elongation factors. Genes Dev. 1992;6:655–666. doi: 10.1101/gad.6.4.655. [DOI] [PubMed] [Google Scholar]
- 20.Kwok R P S, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G E, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 21.Lazinski D, Grzadzielska E, Das A. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell. 1989;59:207–218. doi: 10.1016/0092-8674(89)90882-9. [DOI] [PubMed] [Google Scholar]
- 22.Leung K, Nabel G J. HTLV-1 transactivator induces interleukin-2 receptor expression through an NF-κB-like factor. Nature. 1988;333:776–778. doi: 10.1038/333776a0. [DOI] [PubMed] [Google Scholar]
- 23.Lundblad J R, Kwok R P S, Laurance M E, Harter M L, Goodman R H. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 24.Mavankal G, Ignatius Ou S H, Oliver H, Sigman D, Gaynor R B. Human immunodeficiency virus type 1 and 2 Tat proteins specifically interact with RNA polymerase II. Proc Natl Acad Sci USA. 1996;93:2089–2094. doi: 10.1073/pnas.93.5.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moran E, Zerler B, Harrison T M, Mathews M B. Identification of separate domains in the adenovirus E1A gene for immortalization activity and the activation of virus early genes. Mol Cell Biol. 1986;6:3470–3480. doi: 10.1128/mcb.6.10.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 27.Nabel G J, Rice S A, Knipe D M, Baltimore D. Alternative mechanisms for activation of human immunodeficiency virus enhancer in T cells. Science. 1988;239:1299–1302. doi: 10.1126/science.2830675. [DOI] [PubMed] [Google Scholar]
- 28.Nakajima T, Fukamizu A, Takahashi J, Gage F H, Fisher T, Blenis J, Montminy M. The signal-dependent coactivator CBP is a nuclear target for pp90. Cell. 1996;86:465–474. doi: 10.1016/s0092-8674(00)80119-1. [DOI] [PubMed] [Google Scholar]
- 29.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 30.Parada C A, Roeder R G. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature. 1996;384:375–378. doi: 10.1038/384375a0. [DOI] [PubMed] [Google Scholar]
- 31.Parker S F, Perkins N D, Gitlin S D, Nabel G J. A cooperative interaction of human T-cell leukemia virus type 1 Tax with the p21 cyclin-dependent kinase inhibitor activates the human immunodeficiency virus type 1 enhancer. J Virol. 1996;70:5731–5734. doi: 10.1128/jvi.70.8.5731-5734.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Regulation of NF-κB by cyclin-dependent kinases associated with the p300 co-activator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 33.Schmid R M, Perkins N D, Duckett C S, Andrews P C, Nabel G J. Cloning of an NF-κB subunit which stimulates HIV transcription in synergy with p65. Nature. 1991;352:733–736. doi: 10.1038/352733a0. [DOI] [PubMed] [Google Scholar]
- 34.Shikama N, Lyon J, La Thangue N B. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- 35.Subramanian T, Govindarajan R, Chinnadurai G. Heterologous basic domain substitutions in the HIV-1 Tat protein reveal an arginine-rich motif required for transactivation. EMBO J. 1991;10:2311–2318. doi: 10.1002/j.1460-2075.1991.tb07768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tiley L S, Madore S J, Malim M H, Cullen B R. The VP16 transcription activation domain is functional when targeted to a promoter-proximal RNA sequence. Genes Dev. 1992;6:2077–2087. doi: 10.1101/gad.6.11.2077. [DOI] [PubMed] [Google Scholar]
- 37.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 38.Van Lint C, Emiliani S, Ott M, Verdin E. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 1996;15:1112–1120. [PMC free article] [PubMed] [Google Scholar]
- 39.Veschambre P, Simard P, Jalinot P. Evidence for functional interaction between the HIV-1 Tat transactivator and the TATA box binding protein in vivo. J Mol Biol. 1995;250:169–180. doi: 10.1006/jmbi.1995.0368. [DOI] [PubMed] [Google Scholar]
- 40.Wang H-G H, Rikitake Y, Carter M C, Yaciuk P, Abraham S E, Zerler B, Moran E. Identification of specific adenovirus E1A N-terminal residues critical to the binding of cellular proteins and to the control of cell growth. J Virol. 1993;67:476–488. doi: 10.1128/jvi.67.1.476-488.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei P, Garber M E, Fang S-M, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 42.Wu-Baer F, Sigman D, Gaynor R B. Specific binding of RNA polymerase II to the human immunodeficiency virus trans-activating region RNA is regulated by cellular cofactors and Tat. Proc Natl Acad Sci USA. 1995;92:7153–7157. doi: 10.1073/pnas.92.16.7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang X, Herrmann C H, Rice A P. The human immunodeficiency virus Tat proteins specifically associate with TAK in vivo and require the carboxyl-terminal domain of RNA polymerase II for function. J Virol. 1996;70:4576–4584. doi: 10.1128/jvi.70.7.4576-4584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang X-J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 45.Zhu Y, Pe’ery T, Peng J, Ramanathan Y, Marshall N, Marshall T. Transcription elongation factor P-TEFb is required for HIV-1 Tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]