Abstract
The small nuclear structures known as ND10 or PML nuclear bodies have been implicated in a variety of cellular processes including response to stress and interferons, oncogenesis, and viral infection, but little is known about their biochemical properties. Recently, a ubiquitin-specific protease enzyme (named HAUSP) and a ubiquitin-homology family protein (PIC1) have been found associated with ND10. HAUSP binds strongly to Vmw110, a herpesvirus regulatory protein which has the ability to disrupt ND10, while PIC1 was identified as a protein which interacts with PML, the prototype ND10 protein. We have investigated the role of ubiquitin-related pathways in the mechanism of ND10 disruption by Vmw110 and the effect of virus infection on PML stability. The results show that the disruption of ND10 during virus infection correlates with the loss of several PML isoforms and this process is dependent on active proteasomes. The PML isoforms that are most sensitive to virus infection correspond closely to those which have recently been identified as being covalently conjugated to PIC1. In addition, a large number of PIC1-protein conjugates can be detected following transfection of a PIC1 expression plasmid, and many of these are also eliminated in a Vmw110-dependent manner during virus infection. These observations provide a biochemical mechanism to explain the observed effects of Vmw110 on ND10 and suggest a simple yet powerful mechanism by which Vmw110 might function during virus infection.
ND10, also known as PML nuclear bodies or PODs, are small punctate nuclear structures that contain a subset of at least 6 cellular proteins (2, 3, 12, 17, 32, 55). Although the functions of ND10 are unknown, their appearance is modulated by a number of diverse stimuli, including stress, interferon treatment, and infection by several DNA viruses (11, 30, 38, 40). Of particular note is the finding that the cancerous blast cells in patients with promyelocytic leukemia have a chromosomal translocation which results in the production of a fusion protein containing the N-terminal part of PML linked to the retinoic acid receptor alpha (RARα) (9, 18, 25, 28, 45). The consequence of the expression of this aberrant protein is the alteration of the normal distribution of the ND10 proteins from about 5 to 20 discrete structures per nucleus to a much larger number of smaller foci distributed in both the nucleus and cytoplasm. Treatment of such cells with retinoic acid results in the proteasome-dependent destruction of the PML-RARα fusion protein, normal differentiation of the tumor cells, and restoration of the normal ND10 distribution (12, 31, 56, 59). Therefore, the distribution of ND10 and the constituent proteins correlates in some way with the physiology of the cell. Accordingly, there has been much recent interest in the biological role of ND10, the properties of the constituent proteins, and the interactions between them.
Infection with several DNA viruses also causes a dramatic disturbance of ND10. The parental genomes of herpes simplex virus type 1 (HSV-1), cytomegalovirus, and adenovirus preferentially migrate to the periphery of ND10 (21, 22, 41). Following expression of viral proteins, the ND10 proteins are dispersed or relocated into novel structures (1, 7, 11, 16, 22, 30, 39). In the case of HSV-1, the dispersal of the ND10 proteins early in infection is entirely dependent on the expression of the viral immediate-early (IE) protein Vmw110 (a positive regulator of gene expression), which rapidly localizes to ND10 and of itself is able to disrupt them (16, 39). Mutational analysis has demonstrated that sequences in the C-terminal 150 residues of Vmw110 are required for the localization of Vmw110 at ND10, and a characteristic zinc binding domain (termed a RING finger) in the N-terminal part of the protein is required for ND10 disruption (39).
HSV-1 viruses with lesions in Vmw110 are viable, but they exhibit a multiplicity-, cell-type-, and cell cycle-dependent defect in the onset of productive infection (5, 46, 54, 58). The mutant viral genomes which fail to initiate the viral transcription program attain a quiescent state from which they can be reactivated by provision of exogenous Vmw110 (19, 61). Therefore, it has been suggested that Vmw110 influences the balance between the lytic and latent states of the virus, such that in its absence the latter is favored. These observations in cultured cells have been supported by studies in mouse models which have shown that Vmw110-deficient viruses reactivate poorly from latency (6, 34). Mutations in Vmw110 which affect its ability to migrate to and disrupt ND10 are also detrimental to its roles in augmenting viral gene expression and stimulating the reactivation of latent viral genomes (19, 39). Accordingly, it is an attractive hypothesis that the interactions between Vmw110 and ND10 play an important role in influencing the virus-host relationship.
Most of the evidence linking ND10 and viral regulatory proteins has been acquired by immunofluorescence microscopy, and there is little information on direct protein-protein interactions or the biochemical fate of ND10 proteins following virus infection. Recently it has been shown that Vmw110 binds strongly and specifically to a ubiquitin-specific protease, named HAUSP, which is also a component of a subset of ND10 (17, 42, 43). This finding raised the question of whether a ubiquitin-related mechanism is involved in the disruption of ND10 by HSV-1 infection. Intriguingly, yeast two-hybrid analysis has shown that a ubiquitin-homology family member (PIC1, also known as SUMO-1, Sentrin, and UBL-1 [reviewed in references 23 and 48]) interacts with PML and is again a component of a subset of ND10 (3). The primary PIC1 translation product appears to be, like that of ubiquitin, a precursor with a C-terminal extension (in this case only four residues) which by analogy would be expected to be cleaved to allow conjugation to other proteins. Indeed, it has recently been shown that PIC1 can be covalently linked to the nuclear import factor RanGAP1 (36, 37, 47) and a large number of nuclear proteins (26; see also below). Three very recent studies have clearly shown that PML is also a target for PIC1 conjugation and that the conjugates form an array of high-molecular-weight isoforms (27, 44, 53).
These observations led us to explore whether there was any connection between the ability of Vmw110 to disrupt ND10 and the proteasome-mediated protein degradation pathway. The relevant questions are whether the stability of PML (or PML-PIC1 conjugates) is affected by expression of Vmw110 during virus infection and, if so, whether PIC1 conjugates might be a substrate for HAUSP. We found that Vmw110 induces the disappearance of the high-molecular-weight isoforms of PML, and the loss of these bands is dependent on active proteasome-dependent degradation pathways. Recent publications have demonstrated that it is highly likely that these bands comprise PML-PIC1 conjugates. In addition, we found that the amounts of many other high-molecular-weight PIC1 conjugates are also significantly reduced during virus infection. Inhibition of proteasome-mediated proteolysis eliminated the destruction of ND10 during HSV-1 infection. These observations link the observed effect of Vmw110 on ND10 to a biochemical mechanism and suggest that Vmw110 could achieve its biological functions at least in part by altering the stability of selected cellular proteins.
MATERIALS AND METHODS
Plasmids and bacteria.
The pCI expression plasmids used in this study were based on the pCIneo vector (Promega), in which the human cytomegalovirus enhancer-promoter region lies upstream of an intron sequence, a multicloning region, and the simian virus 40 late poly(A) signal. A PIC1 coding region with an N-terminal c-Myc epitope tag sequence (3) was inserted into the cloning region to give plasmid pCIPIC1. Plasmid pPIC1GST was constructed by using oligonucleotides to link the C-terminal double-glycine codons of the N-terminally Myc-tagged PIC1 coding region to the initiating methionine codon of glutathione S-transferase (GST) in a T7 expression vector based on pET8c, thus allowing the expression of a fusion protein with PIC1 linked to GST in a way analogous to the ubiquitin-GST (Ub-GST) fusion protein expressed by plasmid pRB307 (a kind gift from Rohan Baker). Plasmids pACYC-HAUSP and pACYC-UBP2 express HAUSP and UBP2, respectively, from pACYC replicons with tac promoters (17). Plasmids were grown in Escherichia coli DH5, and large-scale preparations were made by using the boiling method and CsCl purification.
Viruses and cells.
HSV-1 strain 17 syn+ was the wild-type strain used in these studies. Vmw110 mutant viruses dl1403, FXE, D12, D13, D14, and E52X have been described previously (13, 43, 54). Additional mutant viruses E58X, A8X, and A78 have lesions as described in the text and Table 1; their detailed characterization will be described elsewhere. All viruses were grown and titrated in baby hamster kidney (BHK) cells propagated in Glasgow modified Eagle’s medium (MEM) containing penicillin (100 U/ml), and streptomycin (100 μg/ml) and supplemented with 10% newborn calf serum and 10% tryptose phosphate broth. HEp-2 cells were grown in Dulbecco’s MEM supplemented with 10% fetal calf serum and antibiotics as described above. HFL cells were grown in Dulbecco’s MEM supplemented with 5% fetal calf serum, 5% newborn calf serum, 1% nonessential amino acids, and antibiotics as described above.
TABLE 1.
Properties of mutant Vmw110 proteins expressed by the viruses used in the study and their effects on ND10 in HEp-2 cells
| Virus | Vmw110 amino acids | Punctate distribution?a | ND10 disruption?b | HAUSP binding?c | Comments |
|---|---|---|---|---|---|
| 17+ | 1–775 | Yes | Yes | Yes | Wild type |
| dl1403 | 1–105 | No | No | No | Taken as null mutant |
| FXE | Δ106–149 | Increased | No | Yes | RING finger deletion |
| E52X | 1–594 | No | No | No | |
| E58X | 1–633 | No | No | No | |
| A8X | 1–646 | No | No | Minimal | |
| D12 | Δ594–633 | Yes | Yes | Minimal | |
| A78 | Δ592–646 | Reduced | Reduced | Minimal | |
| D13 | Δ634–679 | No | No | Yes | |
| D14 | Δ680–719 | No | No | Yes |
Most Vmw110 proteins exhibit a degree of diffuse staining, which is reduced by the FXE mutation. The D12 and A78 mutations result in proteins which can still colocalize with PML at ND10 at early times of infection, in all cell types tested.
This is a difficult assay as it is variable from cell to cell, with time of infection and in different cell types. Most wild-type-infected cells have lost ND10 by 4 h. At very late times of infection, gross structural changes to the nucleus invalidate this assay, but after extended expression in transfected cells, the dl1403 and FXE alleles have no disruptive effect on ND10 and the FXE protein stably colocalizes with PML. With the exceptions of D12 and A78 (discussed in the text), the other mutations can cause some disturbance to ND10 in HEp-2 cells, but this occurs very much more slowly than with the wild type.
As detected by coimmunoprecipitation. Trace amounts of HAUSP have sometimes been detected in experiments using A8X, D12, and A78, but these are very substantially reduced compared to the wild type.
Electroporation.
HEp-2 cells were trypsinized, resuspended in complete medium, pelleted, and washed twice with serum-free medium before being resuspended in serum-free medium at a concentration of 6 × 106 cells per ml. Plasmid DNA (20 μg) and 0.8 ml of cells were added to a 4-mm electroporation cuvette, incubated on ice for 10 min then mixed again, and pulsed in a Hybaid electroporator at a setting of 400 V. The cells were incubated on ice for a further 10 min before being diluted into fresh complete medium and seeded into Linbro wells at a nominal density of 2 × 105 cells per well for Western blotting samples and half of that for immunofluorescence. The cells were used for experimentation the following day.
Infections and cell extracts.
HEp-2 cells, seeded in Linbro wells the previous day, were infected with HSV-1 by adsorption in 0.1 ml of medium at multiplicities as indicated in the figure legends. After 1 h, an additional 0.5 ml of medium was added and infection was allowed to proceed at 37°C. When lactacystin lactone (Boston Biochem Inc.) or MG132 (Calbiochem) was used, the cells were preincubated with medium containing the drug for 30 min before infection; then all subsequent incubations were performed in the presence of drug. Samples for use in immunofluorescence were treated as described below. Samples for Western blotting were washed in phosphate-buffered saline (PBS) and then resuspended directly into sodium dodecyl sulfate (SDS)-gel loading buffer. When proteasome inhibitors had been used, the loading buffer was supplemented with 5 mM N-ethylmaleimide.
Western blotting.
SDS-polyacrylamide gels (7.5% acrylamide) were prepared and run in the Bio-Rad MiniProtean II apparatus, and then proteins were electrophoretically transferred to nitrocellulose membranes according to the manufacturer’s recommendations. After blocking in PBS containing 0.1% Tween 20 (PBST) and 5% dried milk overnight at 4°C, the membranes were incubated with primary antibody in PBST–5% dried milk at room temperature for 4 h and then washed in PBST at least six times before incubation with horseradish peroxidase-conjugated secondary antibody in PBST–2% dried milk at room temperature for 1 h. After extensive washing, the filters were soaked in Amersham ECL (enhanced chemiluminescence) reagent and exposed to film. Antibodies were stripped from the membranes following the Amersham ECL protocol, and the membranes were reprobed as necessary.
Antibodies.
Anti c-Myc monoclonal antibody (MAb) 9E10 was purchased from Santa Cruz Biotechnology Inc. Anti-PML MAb 5E10 (55) was a generous gift from R. van Driel. The anti-UL42 and anti-Vmw110 MAbs Z1F11 and 11060 and the anti-PML rabbit serum r8 have been described previously (3, 15, 51). Horseradish peroxidase-conjugated sheep anti-mouse and goat anti-rabbit antibodies were purchased from Sigma Immunochemicals.
Immunofluorescence.
HEp-2 cells were fixed with formaldehyde (5% [vol/vol] in PBS containing 2% sucrose) and permeabilized with 0.5% Nonidet P-40 in PBS with 10% sucrose. The primary antibodies were diluted in PBS containing 1% newborn calf serum. Anti-Vmw110 MAb 11060 and anti-PML rabbit serum r8 were used at dilutions of 1/2,000 and 1/1,000 respectively. After incubation for 1 h, the coverslips were washed several times in PBS–1% calf serum and then incubated with goat anti-mouse fluorescein isothiocyanate-labeled and goat anti-rabbit tetramethylrodamine isothiocyanate-labeled secondary antibodies (Sigma) at dilutions of 1/100. After staining, the coverslips were mounted and examined by using appropriate narrow band filters in a Nikon Microphot-SA microscope adapted with a Digital Pixel CCD digital camera. Captured images were prepared for printing by using Photoshop.
Cleavage of ubiquitin and PIC1 fusion proteins in bacteria.
E. coli Novablue (DE3) strains carrying the substrate plasmids pPIC1GST and pRB307 were established, and derivatives which carried in addition either pACYC-HAUSP or pACYC-UBP2 were made. The former two plasmids express ampicillin resistance on pBR322 replicons and are therefore compatible with the pACYC184 replicons expressing chloramphenicol resistance of the latter two plasmids. The bacteria were grown to mid-log phase, and expression of substrate and enzyme activities were induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG). The bacteria were harvested 2 h later. With the PIC1-GST substrate, whole-cell extracts were analyzed on a 12.5% polyacrylamide gel and the Myc-tagged proteins were detected by Western blotting using MAb 9E10. With the tag at the N-terminal end, this detects the PIC1-GST fusion protein and any liberated N-terminal fragments but not the C-terminal GST polypeptide. To analyze cleavage of the Ub-GST fusion protein, soluble protein extracts were prepared and the Ub-GST substrate and GST product were purified on glutathione-agarose beads as described previously (42). The purified proteins were analyzed on 10% polyacrylamide gels and detected by staining with Coomassie blue.
Detection of a PIC1-RanGAP1 hydrolase activity in Xenopus egg extracts and immunodepletion of a HAUSP-related Xenopus protein.
A GST-PIC1 fusion protein expression construct was produced by using a pGEX-2TK vector (Pharmacia) and a PIC1 cDNA (obtained from the IMAGE consortium [35]) that had been mutated to truncate the last four amino acids from its C terminus (49). Full-length PIC1 must be processed to remove the C-terminal four amino acids in order to produce a form of the protein that can be conjugated to target substrates (26). The truncated form of GST-PIC1 is readily conjugated to substrate protein in reticulocyte lysates and Xenopus egg extracts (49). Bacterial expression and purification of GST-PIC1 were carried out as described previously (49). Membrane-free soluble Xenopus protein extracts were fractionated from interphase egg extracts as described previously (8, 52). Samples of the extracts (50 μl) were depleted of HAUSP by using 5 μl of anti-HAUSP rabbit serum r201 and 20 μl of protein A-Sepharose beads; after incubation at 4°C for 1 h, the beads were removed by centrifugation. Mock depletion was carried out by using the same protocol, replacing r201 with the corresponding preimmune serum. The extracts were stored at −80°C. To detect RanGAP1-PIC1 isopeptidase activity using GST-PIC1, a Xenopus RanGAP1 cDNA (48) was cloned into a pT7 vector to give an open reading frame initiated at the original methionine start codon. Radiolabeled RanGAP1 protein was produced by using a TnT-coupled transcription-translation kit (Promega) in the presence of GST-PIC1 (50 ng/μl) and l-[35S]methionine for 1.5 h at 30°C. The products of the reaction were purified on glutathione-Sepharose beads and washed extensively with PBS with 0.1% Triton X-100. Isopeptidase activity was assayed by incubation of 5 μl of the glutathione-Sepharose beads carrying GST-PIC1-RanGAP1 conjugates with 2 μl of untreated extract, mock-depleted extract, or HAUSP-depleted extract for 10 min at 30°C. The products were immediately analyzed on a SDS–4 to 20% polyacrylamide gel followed by autoradiography.
RESULTS
HSV-1 infection causes the rapid loss of high-molecular-weight isoforms of PML.
Infection by HSV-1 results in the rapid disruption of ND10 and the apparent dispersal of the constituent proteins (16, 38). While it has been clear that PML-derived immunofluorescence in infected cells is substantially reduced, the fate of the PML protein was uncertain. Because two reports have shown that both the PML-RARα fusion protein (59) and PML itself (60) are subject to degradation following treatment of cells with all-trans retinoic acid and arsenic oxide, respectively, we set out to determine if HSV-1 infection also caused the actual loss of PML. The primary transcript of the PML gene is subject to extensive alternative splicing, and the encoded proteins are subsequently modified posttranslationally to produce a plethora of isoforms with gel mobilities varying from about 65 to over 180 kDa (45, 55). These can be detected by Western blotting using MAb 5E10, which recognizes an epitope in a region of PML present in most isoforms (31, 55).
HEp-2 cells were infected with HSV-1 at increasing multiplicities of infection and harvested 4 h later. Western blotting showed that many multiple high-molecular-weight PML isoforms were lost or substantially reduced following virus infection and that the effect could be seen at a moderate virus dose of 5 PFU per cell (Fig. 1A). The reduction in the band intensities was substantial even as early as 2 h postinfection (data not shown), and it is pertinent that this timing correlates well with the observed virus-induced disruption of ND10 (38). It is noteworthy that these results are not an artifact arising from the use of MAb 5E10 since a similar pattern of high-molecular-weight bands is recognized by anti-PML rabbit serum r8 and these bands are also eliminated during virus infection (data not shown). There appears to be some isoform- or modification-dependent specificity to this process since major PML isoforms at about 70 kDa, and particularly the major bands at about 130 kDa, were not as greatly affected. However, appropriate exposure of the blots indicated that these bands also decreased in intensity in response to virus infection (see also Fig. 5).
FIG. 1.
HSV-1 infection stimulates the loss of several high-molecular-weight PML isoforms. (A) HEp-2 cells were infected with HSV-1 at multiplicities of 5, 10, 20, and 50 PFU per cell as indicated and harvested 4 h later. The samples were analyzed by Western blotting using anti-PML MAb 5E10 in comparison with an uninfected control (M). Positions of molecular weight markers are indicated on the right in kilodaltons, and PML isoforms most sensitive to elimination are indicated by arrows. (B) The same filter was reprobed with MAb Z1F11 to compare the efficiency of virus infection by detection of the early viral protein UL42.
FIG. 5.
Correlation of ND10 disruption and PML isoform loss in different cell lines. BHK cells (A) or HFL cells (B) were infected with the indicated viruses at multiplicities of 20 PFU per cell, and total cell proteins were harvested 4 h later. The proteins were separated by SDS-polyacrylamide gel electrophoresis and PML was detected by Western blotting using MAb 5E10. Positions of molecular weight markers are indicated in kilodaltons, and the PML isoforms in HFL cells equivalent to those in HEp-2 cells indicated in Fig. 1 and 2 are marked by arrows in panel B. A similar but not identical pattern of isoform bands is present in BHK cells (A). In parallel, BHK cells (C, D, G, and H) or HFL cells (E, F, I, and J) were infected with wild-type virus (C to F) or mutant D12 (G to J) and stained for Vmw110 and PML. The paired panels (for example, C and D) show Vmw110 staining on the left and PML staining on the right. The bar indicates 5 μm.
An important question concerns the nature of these endogenous PML bands. The complexity of the PML isoforms detected on Western blots, the low amount of total PML in the cell, and the difficulty of isolating it in soluble form have hampered attempts to study the nature and biochemical properties of endogenous PML. However, three recent studies used various methods to induce increased expression of PML (including induction of endogenous PML with interferon, induction from stably integrated or transiently transfected genes, or use of a cell line engineered to express PML at high levels), with or without simultaneous increased expression of PIC1. All three describe the formation of a number of high-molecular-weight PML-PIC1 conjugate bands with gel mobilities that appear extremely similar to the pattern of endogenous high-molecular-weight PML bands shown in Fig. 1 (27, 44, 53). Importantly, a similar pattern of endogenous PML bands was found in untreated cells after prolonged exposure of the Western blots (53), conditions which closely parallel those used in our experiments. Accordingly, comparison with these other studies leads to the reasonable conclusion that the high-molecular-weight isoforms of PML that are lost during virus infection represent PML-PIC1 conjugates. A more direct proof of this conclusion would require immunoprecipitation of the endogenous PML-PIC1 conjugates, but the poor solubility of the protein, its low amounts in the cell, and the fact that many PIC1 conjugates appear to be extremely labile in vitro have so far made this experiment impossible.
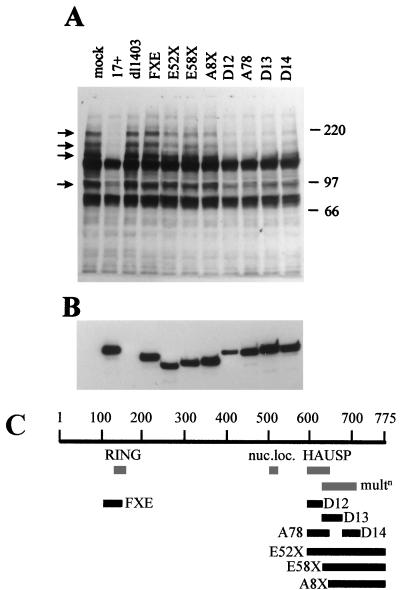
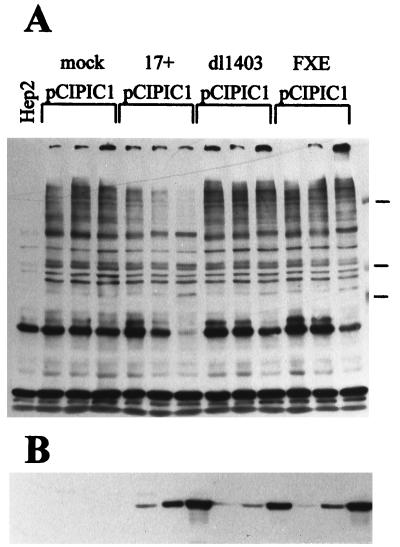
Expression of Vmw110 and its ability to interact normally with ND10 are essential for efficient loss of the PML isoforms.
Since the HSV-1 IE protein Vmw110 initially colocalizes with PML at ND10 and then causes their disruption, it seemed possible that Vmw110 is involved in the loss of the PML isoforms. HEp-2 cells were infected with wild-type and several Vmw110 mutant viruses, whose genotypes are explained in Table 1 and depicted in Fig. 2C. The cells were harvested 4 h after infection and analyzed by Western blotting. The results (Fig. 2A) clearly show that (i) Vmw110 is absolutely essential for the loss of the high-molecular-weight isoforms of PML (because dl1403 had no effect), (ii) the RING finger domain of Vmw110 is also essential (virus FXE had no effect), (iii) the complete C-terminal region of Vmw110 containing the HAUSP binding and ND10 localization sequences is also very important (virus E52X had little effect), (iv) the ability of Vmw110 to migrate to ND10 is required for normally efficient loss of the PML isoforms (viruses E58X, A8X, D13, and D14 were all less active than wild type), but (v) Vmw110 mutants which bind HAUSP poorly yet migrate to ND10 are still capable of inducing the loss of the PML isoforms, although somewhat less efficiently than wild-type Vmw110 (compare viruses D12 and A78 with wild type). Probing of the same filter with MAb 11060 indicated that the mutant viruses expressed similar levels of Vmw110 (Fig. 2B). An important factor of this experiment is that the differences between the mutant viruses is partly kinetic, in that virus E52X (for example) induces increased loss of the PML isoforms as infection progresses. However, viruses dl1403 and FXE were completely defective in this assay, even late in infection (data not shown). These results generally correlate well with the ability of the mutant forms of Vmw110 to disrupt ND10 early in virus infection (39, 43; see also below). The possible role of HAUSP in this process is discussed further below.
FIG. 2.
Vmw110 is absolutely required for the virally induced loss of high-molecular-weight PML isoforms. (A) HEp-2 cells were infected with viruses as indicated (as described in the text) at a multiplicity of 20 PFU per cell and harvested 4 h later. The samples were analyzed by western blotting using anti-PML MAb 5E10 in comparison with an uninfected control (mock). Positions of molecular weight markers are indicated on the right in kilodaltons, and PML isoforms most sensitive to elimination are indicated by arrows. (B) The same filter was reprobed with MAb 11060 to compare the efficiency of the virus infections by detection of Vmw110. (C) A schematic representation of the viruses used in this experiment. The 775-codon open reading frame of Vmw110 is depicted, with the locations of the RING finger, nuclear localization signal (nuc.loc.), HAUSP binding region, and sequences implicated in multimerization (multn) indicated below. The solid bars indicate the extents of the deleted sequences in the mutant viruses, as labeled.
Involvement of the proteasome in the loss of the PML isoforms.
Since the loss of the high-molecular-weight PML isoforms occurred rapidly, it seemed likely that this was a proteasome-dependent process. The ubiquitin-mediated protein degradation pathway is being increasingly recognized as a common mechanism of regulating the amount of a protein within the cell, and its activation can lead to very rapid elimination of the target protein (reviewed in references 20 and 57). The pathway involves initial conjugation of a ubiquitin monomer to the target protein, followed by the polymerization of the ubiquitin chain to produce a tag which targets the substrate to the proteasome for rapid degradation. Inhibition of the proteolytic activity of the proteasome results in the accumulation of multiubiquitinated polypeptides.
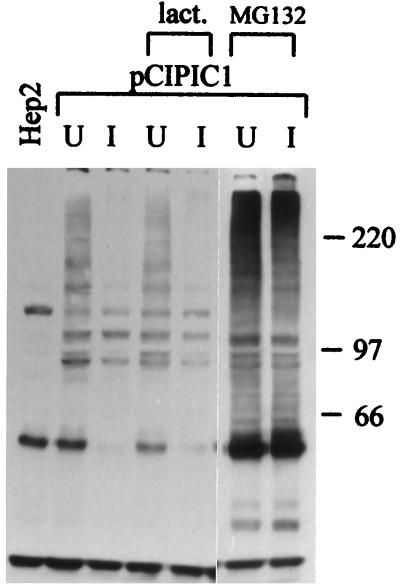
Addition of the proteasome inhibitors lactacystin lactone or MG132 to the culture medium eliminated the virus-induced loss of the PML isoforms (Fig. 3). Although there was no evidence of accumulation of a ladder of ubiquitinated forms of PML in the drug-treated samples in response to viral infection, it is possible that their appearance is masked by the size and complexity of the various isoforms. Probing of the same blot with MAb Z1F11 indicated that at the multiplicity of virus infection used in this experiment, the inhibitors had no effect on the efficiency of expression of UL42, a viral protein of the early kinetic class (data not shown). This finding demonstrates that the effect of MG132 was not due to interference with viral gene expression.
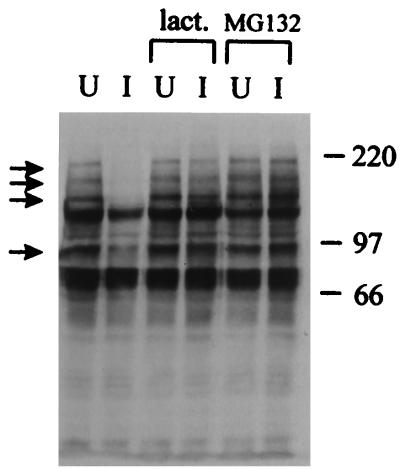
FIG. 3.
Inhibitors of proteasome activity eliminate the virally induced loss of the high-molecular-weight PML isoforms. HEp-2 cells were infected (I) or left uninfected (U) with HSV-1 strain 17+ at a multiplicity of 20 PFU per cell in the absence of drug or the presence of lactacystin lactone (lact.; 5 μM) or MG132 (5 μM) as indicated. The cells were harvested 4 h later, and the samples were analyzed by Western blotting using anti-PML MAb 5E10. Positions of molecular weight markers are indicated on the right in kilodaltons, and PML isoforms most sensitive to elimination are indicated by arrows.
Proteasome inhibitors stabilize ND10 in virus-infected cells.
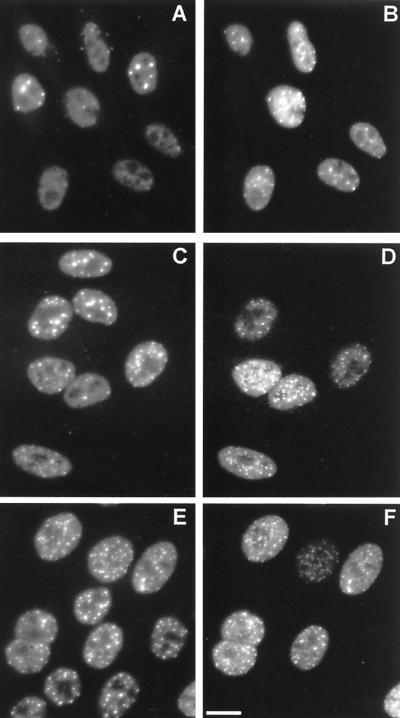
The correlation between the virally induced destruction of ND10 and the Vmw110-induced, proteasome-mediated loss of the high-molecular-weight isoforms of PML suggests that proteasome activity may be essential for the effect of the virus on ND10. HEp-2 cells were infected with HSV-1 in the presence and absence of MG132 and lactacystin lactone and stained 4 h later for PML and Vmw110. Comparison of the infected and uninfected cells in each situation clearly showed that both proteasome inhibitors greatly reduced the ability of the virus to disrupt ND10, thus providing a direct mechanistic link between the loss of the PML isoforms and the ND10 disruption. In the absence of drug, the uninfected cells (upper right and middle-leftmost cells in Fig. 4A) showed the normal punctate accumulations of PML in ND10, while most of the infected cells had no trace of ND10. In the presence of lactacystin lactone, the infected cells clearly retained ND10 (Fig. 4C), and this effect was even more striking in the presence of MG132 (Fig. 4E). Interestingly, MG132 by itself clearly affected ND10, causing an increase in their number and prominence (compare the uninfected cells in Fig. 4A and E). Virus infection in the presence of MG132 led to extensive colocalization of PML with Vmw110 (Fig. 4E and F), a situation reminiscent of RING finger mutant forms of Vmw110 which migrate to ND10 but do not disrupt them. Taken with the results of Fig. 3, these observations suggest that the loss of the PML isoforms correlates with and probably precedes ND10 disruption.
FIG. 4.
Proteasome inhibitors inhibit the disruption of ND10 during HSV-1
Virus-induced disruption of ND10 correlates with the loss of high-molecular-weight PML isoforms in different cell types.
Many of the results presented above suggest an attractive model which would connect the loss of the PML isoforms and the proteasome-dependent disruption of ND10 by Vmw110 with its ability to bind to the ubiquitin-specific protease HAUSP. However, Fig. 2 shows that the role of HAUSP in this effect is not clear, since the HAUSP binding-deficient mutants D12 and A78 retain the ability to stimulate the loss of the PML bands, at least in HEp-2 cells. In a previous publication we showed that the HAUSP binding-negative mutant D12 was defective in the disruption of ND10 in BHK cells (43), and at first sight this result is not consistent with either the loss of the PML isoforms or the time course of ND10 disruption in HEp-2 cells infected with virus D12 (Fig. 2 and data not shown). Therefore we have investigated the correlation between loss of the PML isoforms and ND10 disruption using the HAUSP binding-negative mutant D12 in different cell types. In these experiments, we compared the results for primary human fibroblasts (HFL cells) with those for the BHK cells that had been used previously. The wild-type virus efficiently disrupted ND10 in both cell types, and this correlated with the loss of the PML isoforms (Fig. 5A to F). Figures 5C and D show a group of six cells, three of which have been infected. The PML staining indicates clear loss of ND10 from the infected cells but not from their uninfected neighbors. Similarly, compare the three infected HFL cells (the middle one of which expresses only very low amounts of Vmw110) with the uninfected cell at the top of Fig. 5D. In contrast, virus D12 had a markedly reduced ability both to disrupt ND10 and induce the loss of the isoforms in BHK cells but not HFL cells (Fig. 5A, B, and G to J). Figure 5A shows that virus D12 induced only a marginal loss of the PML isoforms in BHK cells, and this correlated with the retention of ND10 in most cells 4 h after infection (for example, the lowest cells in Fig. 5G and H). In contrast, all D12-infected HFL cells, even those with expressing low amounts of the D12 protein (compare the lowest two cells in Fig. 5I and J), had lost ND10. The Western blot analysis performed in parallel showed that D12 was as efficient as the wild type in stimulating the loss of the PML bands from HFL cells (Fig. 5B). A feature of the D12 phenotype is an increase in the rate at which the protein accumulates in the cytoplasm (Fig. 5G). However, the failure to disrupt ND10 and stimulate PML loss is not due this property since at earlier times of infection the localization of D12 mutant protein in the nucleus and at ND10 is similar to that of the wild-type protein in HEp-2, BHK, and HFL cells (reference 43 and data not shown). These results further underline the correlation between ND10 disruption and PML isoform loss, and they suggest that the ability of Vmw110 to bind to HAUSP contributes to these effects in a cell-type (or species)-dependent manner. It should be noted that BHK cells express a protein of the same size and antigenicity as HAUSP which also binds to Vmw110 (reference 42 and results not shown).
The reduced ability of virus D12 to disrupt ND10 and induce the degradation of the PML isoforms in BHK but not HFL cells correlates with its plaque-forming abilities in the two cell types; unlike most Vmw110 mutant viruses, it forms plaques more efficiently in HFL than BHK cells (data not shown).
HSV-1 infection results in the loss of several high-molecular-weight PIC1 conjugates.
Given that the high-molecular-weight PML isoforms which disappear during HSV-1 infection have gel mobilities extremely similar to those of defined PML-PIC1 conjugate proteins, and that PIC1 can also be conjugated to a number of other proteins (26), we decided to investigate the fate of other PIC1 conjugates during virus infection. Plasmid pCIPIC1 (which expresses an epitope-tagged version of PIC1; see Materials and Methods) was electroporated into HEp-2 cells which were subsequently infected with wild-type virus, the Vmw110 deletion mutant dl1403, and the RING finger mutant FXE. Samples were taken at 4, 8, and 22 h after infection and analyzed by Western blotting.
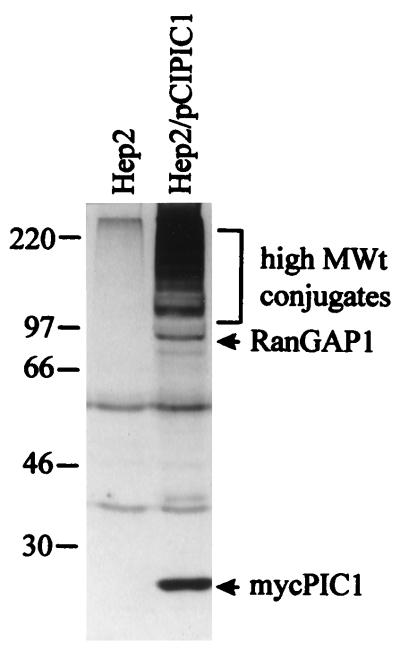
Uninfected electroporated cells expressed a PIC1 protein of about 18 kDa (the size expected considering the gel mobility of the endogenous monomeric PIC1 plus the epitope tag) and a number of higher-molecular-weight bands culminating in a complex pattern of a multitude of high-molecular-weight polypeptides (Fig. 6). The prominent band at about 90 kDa probably corresponds to a form of RanGAP1 which is covalently conjugated to PIC1 (36, 37), while the higher-molecular-weight bands have been identified as other PIC1 conjugates (26).
FIG. 6.
High-level expression of an epitope-tagged form of PIC1 and formation of PIC1-conjugated proteins. HEp-2 cells were electroporated with plasmid pCIPIC1 and harvested 40 h later. The samples were analyzed by Western blotting using MAb 9E10 in comparison with control untreated cells. Positions of molecular weight markers, the monomeric Myc-tagged PIC1, a band likely to represent PIC1 conjugated to RanGAP1, and a multitude of other high-molecular-weight PIC1 conjugate proteins are indicated.
In response to HSV-1 infection, many of the high-molecular-weight PIC1 conjugate bands disappeared (Fig. 7). This effect was slower than that seen with the high-molecular-weight isoforms of PML, becoming almost complete by 22 h after infection; however, it should be noted that these cells express artificially high levels of PIC1, and so endogenous PIC1 conjugates in untreated cells may be eliminated much faster. It is evident that there is a degree of specificity to this process since there was no reduction in a number of the PIC1 bands, including that likely to correspond to RanGAP1. This result is consistent with the observation that the PIC1 conjugation status of endogenous RanGAP1 (as determined by Western blotting with a RanGAP1-specific antibody) is not affected during virus infection (data not shown). The results with the Vmw110 mutant viruses showed that this effect was again totally dependent on Vmw110 and the integrity of its RING finger (Fig. 7). The same set of virus mutants as used for Fig. 2 was analyzed in a similar experiment, and all were less active than the wild type in reducing the PIC1 conjugate bands (data not shown).
FIG. 7.
HSV-1 infection leads to the loss of several high-molecular-weight PIC1-conjugated proteins. HEp-2 cells were electroporated with plasmid pCIPIC1 and the following day either left uninfected (mock) or infected at a multiplicity of 20 PFU per cell with wild-type virus (17+), Vmw110 deletion mutant dl1403, or Vmw110 RING finger mutant FXE. Samples were taken at 4, 8, and 22 h after infection, loaded from left to right in each set of three lanes, and analyzed by Western blotting using the anti-Myc tag MAb 9E10 (A) and anti-UL42 MAb Z1F11 (B) to control for infection. The leftmost lane contains a sample from untreated HEp-2 cells. Positions of the 220-, 97-, and 66-kDa molecular weight markers are shown to the right in panel A.
The loss of the PIC1 conjugates was inhibited by MG132 (Fig. 8), an observation which suggests that a similar mechanism may operate for both this effect and the Vmw110-induced loss of the PML isoform bands. A striking feature of this experiment was that MG132 resulted in greatly increased levels of the PIC1 conjugate bands. This result implies that these polypeptides are subject to turnover, directly or indirectly, by a proteasome-dependent pathway. This point is discussed further below. Lactacystin lactone did not inhibit the degradation of the PIC1 conjugates, but the drug would be expected to be hydrolyzed to an inactive form early in the relatively long time course of this experiment (10).
FIG. 8.
Proteasome inhibitor MG132 inhibits the virus-induced loss of high-molecular-weight PIC1 conjugates. HEp-2 cells were electroporated with plasmid pCIPIC1 and the following day either left uninfected (U) or infected at a multiplicity of 50 PFU per cell with wild-type virus (I) in the presence of 10 μM lactacystin lactone (lact.) or 5 μM MG132 as marked. Samples were taken 22 h after infection and analyzed by Western blotting using MAb 9E10. The positions of the molecular weight markers are shown in kilodaltons on the right.
Does HAUSP cleave PIC1 conjugates?
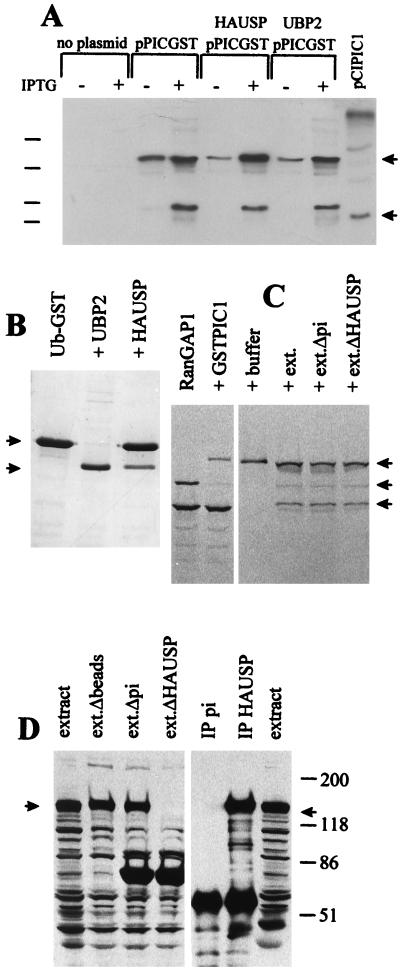
Because of the observed colocalization of PIC1 and HAUSP in ND10, and the direct implication of the ubiquitin pathway in the stability of PIC1 conjugates and ND10 themselves, it is a reasonable and intriguing question whether PIC1 conjugates are substrates for HAUSP. Indeed, precisely this question has been posed by other workers in two recent publications (23, 44). We have taken four approaches to this question. First, we asked whether HAUSP could bind to immobilized PIC1 in vitro, but the results were negative (data not shown). Second, there was no evidence that HAUSP could cleave the C-terminal four residues from the primary PIC1 translation product in an assay where clear cleavage of the analogous ubiquitin precursor by HAUSP was evident (data not shown). Third, we constructed a PIC1-GST model substrate expression plasmid and compared the ability of HAUSP to cleave it in parallel with an equivalent Ub-GST model substrate. The results clearly showed that while HAUSP cleaved Ub-GST, it could not cleave PIC1-GST (Fig. 9A and B). It is possible that cleavage of PIC1 conjugates by HAUSP is context dependent, requiring a natural PIC1 conjugate or an isopeptide linkage. We were able to test these possibilities in one instance since there is an activity in Xenopus egg extracts which efficiently cleaves PIC1 from RanGAP1, and also a protein which is strongly related to HAUSP in terms of size, immunoprecipitation by anti-HAUSP r201 serum (Fig. 9D), and interaction with the C-terminal region of Vmw110 (data not shown). Depletion of this protein from egg extracts by immunoprecipitation had no effect on the PIC1 cleavage activity (Fig. 9C and D), and there was no increased activity in the HAUSP immunoprecipitate (data not shown). However, these results do not exclude the possibility that any hypothetical PIC1 cleavage activity of HAUSP is both context dependent and highly substrate specific.
FIG. 9.
Evidence that HAUSP does not cleave PIC1 conjugates. (A) Bacterial strains were established and enzyme activity was induced as described in Materials and Methods section. The far right-hand lane contains proteins expressed by pCIPIC1 in HEp-2 cells (Fig. 6). The arrows indicate the substrate band (upper arrow) and the Myc-tagged PIC1 protein with the precursor C terminus (i.e., 4 residues longer than the predicted specific cleavage product) (lower arrow). Induction of HAUSP and UBP2 activities has no effect on the amount of the PIC1-GST fusion protein substrate, and there is no evidence for a specific cleavage product. A dominant breakdown product appears as readily in the control as in the HAUSP- and UBP2-expressing cells. Positions of the 46-, 30-, 21-, and 14-kDa molecular weight markers are indicated on the left. (B) Cleavage of Ub-GST by HAUSP and UBP2. An experiment analogous to that in panel A was conducted except that the substrate was Ub-GST expressed by plasmid pRB307. Only the IPTG-induced lanes are shown. The arrows on the left indicate the Ub-GST substrate and the GST product. Cleavage is complete in bacteria carrying UBP2. The partial cleavage in bacteria carrying HAUSP is probably due to the toxicity of this protein in bacteria, leading to instability of the expression plasmid. (C) Conjugation of GSP-PIC1 to RanGAP1 and assay of isopeptidase activity on the purified product. The left-most lane shows 35S-labeled RanGAP1 synthesized in reticulocyte lysates, which appears as a 65-kDa unmodified protein and an 88-kDa endogenous PIC1-conjugated form (49). In the presence of excess GST-PIC1, RanGAP1 is preferentially conjugated with GST-PIC1 to produce a 120-kDa protein. The right-hand four lanes show the 120-kDa substrate (purified on glutathione beads) incubated with buffer, untreated egg extract (ext.), mock-depleted egg extract, or HAUSP-depleted egg extract, as described in Materials and Methods. The arrows on the right indicate the 120-kDa GST-PIC1-RanGAP1 substrate, the 88-kDa RanGAP1-endogenous PIC1 conjugate (which is formed by reconjugation of released RanGAP1 with endogenous PIC1), and the 65-kDa free form of RanGAP1. (D) Depletion of Xenopus HAUSP from egg extracts. The left-hand four lanes show proteins detected by Western blotting using anti-HAUSP r201 serum in untreated egg extracts, in extracts treated with Sepharose beads, and in extracts treated with beads charged with preimmune (pi) r201 antibodies and immune r201 antibodies, respectively. The extracts analyzed in the latter two lanes were used for the incubations in the rightmost two lanes in panel C. On the right, the proteins in the immunoprecipitate (IP) pellets obtained with preimmune and immune r201 sera are shown. The arrows indicate the position of the approximately 130-kDa Xenopus HAUSP homolog.
DISCUSSION
This paper demonstrates that expression of Vmw110 during HSV-1 infection leads to the proteasome-dependent loss of several modified PML isoforms, a process which correlates with the disruption of ND10. These observations provide a biochemical explanation of the observed effects of Vmw110 on ND10, and coupled with the Vmw110-dependent elimination of several high-molecular-weight PIC1 conjugate proteins, these data strongly suggest a novel yet simple way in which Vmw110 could modify the intracellular environment.
Before discussing the implications of these results in detail, two important questions must be addressed. The first is the nature of the high-molecular-weight isoforms of PML which are the subject of this study, and the second is their fate during virus infection. Because endogenous PML is present in tiny quantities and a large proportion appears to be insoluble, many laboratories have been unable to characterize endogenous PML biochemically despite extensive efforts. However, as discussed earlier, the sizes and pattern of gel mobilities of the PML isoform bands in question appear extremely similar to those for PML-PIC1 conjugate proteins that have been defined in a variety of expression systems (27, 44, 53), and it is a reasonable conclusion that the endogenous high-molecular-weight PML isoform bands are also PIC1 conjugates. If so, the disappearance of these bands during virus infection could represent simple deconjugation of the PIC1 by an activity analogous to a ubiquitin-specific protease, or the conjugate proteins could be degraded. In theory, such degradation could occur either directly because of the PIC1 modification or indirectly after ubiquitination of the proteins (before or after PIC1 deconjugation). If simple deconjugation occurs, the levels of the unconjugated protein and monomeric PIC1 would be expected to increase. The low levels of the endogenous PML-PIC1 conjugates make it impossible to test this possibility in normal cells. However, in the experiment using cells expressing high levels of tagged PIC1 (Fig. 7), there was no increase in monomeric PIC1 despite significant reductions in the conjugate proteins (data not shown); thus, in this case there appears to be a real reduction of at least the PIC1 moiety. In the case of PML, particularly in BHK and HFL cells (Fig. 5), there is also an observable reduction in all PML-related bands in response to viral infection. These results, taken with the observation of the inhibition of the loss of the bands by proteasome inhibitors, strongly suggest that protein degradation was occurring. However, an important proviso is that these data do not suggest that PIC1 modification acts analogously to ubiquitination in targeting proteins to the proteasome since it is clear that many PIC1 conjugates are stable. Rather, expression of Vmw110 induces an activity which results in preferential loss of many PIC1-conjugated proteins. This point is discussed further below.
Muller et al. (44) have recently suggested that PIC1 conjugation of PML directs the protein to ND10. This is an attractive model which is pertinent to the experiments described here. Indeed, it is possible that Vmw110 simply induces the deconjugation of PML and that this is sufficient to disrupt ND10. However, there are a number of other observations that must be borne in mind. First, PIC1 conjugation cannot be a simple ND10 localization signal since PIC1-conjugated RanGAP1 is located at the nuclear membrane. Second, the published evidence (and our unpublished data) are not consistent with a model in which the majority of endogenous PML is conjugated to PIC1. Third, overexpressed PIC1 becomes conjugated to many nuclear proteins (Fig. 6) yet is largely located diffusely in the nucleoplasm, with only a proportion localizing at ND10 (3, 4). Fourth, overexpression of PML leads to rapid accumulation of PML in giant ND10 (4), but only a small proportion appears to be PIC1 conjugated (our unpublished data). Accordingly, while the evidence indicates that PML-PIC1 conjugates are located at ND10 (44), PIC1 conjugation appear to be neither necessary nor sufficient for such localization. However, it is easy to envisage a situation in which an important event is the initial localization of PML-PIC1 conjugates at ND10, followed by the accumulation of PML and other proteins through other protein-protein interactions.
As a working hypothesis, we propose that the disruption of ND10 by Vmw110 proceeds as follows. (i) Sequences in the C-terminal 150 residues of Vmw110 promote its efficient migration to ND10 (because mutants D13, D14 and A8X, for example, give a diffuse staining pattern). (ii) The Vmw110 RING finger region itself, or a function dependent on the RING finger, induces a process which leads to the ubiquitination and proteasome-dependent degradation of several PML isoforms and PIC1 conjugates (because the RING finger mutant FXE is completely deficient in these activities). It is tempting to speculate that the PIC1-conjugated forms of PML are preferentially eliminated and that this is sufficient to disrupt ND10. (iii) HAUSP might normally protect these substrates from ubiquitination, but either inhibition of its activity or its sequestration by binding to Vmw110 diminishes this protection. The activity induced by the RING finger region must be dominant because mutants D12 and A78X have greatly reduced abilities to bind to HAUSP but are still able to promote the degradation of the PML isoforms and disrupt ND10, albeit at somewhat reduced efficiencies.
This scenario is consistent with all available data on the activities of Vmw110 and its various mutants, in particular the essential nature of the RING finger and the observation that the ability to bind to HAUSP contributes to but is not absolutely essential for Vmw110 function (reference 43 and unpublished data).
This study arose from the observations that Vmw110 binds to HAUSP, that both proteins could colocalize with PML at ND10, and that the ubiquitin-like protein PIC1 also binds to PML and colocalizes at ND10. We wished to explore the possible connections between these observations, the most obvious being whether PIC1 can be covalently conjugated to PML and whether such conjugates could be substrates for HAUSP. We have confirmed very recent observations (27, 44, 53) that PIC1 can be conjugated to PML (data not shown), but we have been unable to find any evidence for cleavage of PIC1 conjugates by HAUSP (Fig. 9). Despite the lack of evidence for a direct biochemical connection between PIC1 and HAUSP at ND10, the observations that Vmw110 induces the proteasome-dependent loss of several PML isoforms and PIC1 conjugates suggest an alternative but consistent explanation. It is possible that proteins which are subject to PIC1 conjugation can alternatively be ubiquitinated; in the latter case they would be targeted for degradation, but in the former they may be stabilized by protection from ubiquitination. An attractive model is that PIC1 or ubiquitin could be conjugated at the same lysine residues, resulting in mutually exclusive conjugation processes. This model would be more flexible if both PIC1 conjugation to and cleavage from its substrates were relatively rapid processes; indeed both activities can be observed at high levels in vitro (36, 37).
Vmw110 has been shown to activate gene expression by a mechanism that does not require specific promoter sequences, to stimulate reactivation of quiescent viral genomes in cultured cells, and to be required for efficient reactivation of latent virus in mouse ganglia (reviewed in reference 14). It is possible that Vmw110 exerts these effects directly at the level of transcription; indeed, a recent study has ruled out any posttranscriptional mechanism (25). However, it is also feasible that Vmw110 acts pretranscriptionally by modifying cellular factors which in turn have a direct or indirect effect on transcription. The observations that Vmw110 disrupts ND10 and binds to a ubiquitin-specific protease suggest that an indirect mechanism is more likely than a direct interaction of Vmw110 with the transcription initiation complex. The results of this study suggest a simple yet powerful mechanism by which Vmw110 could exert its effects. We have shown that Vmw110 induces the elimination of several PIC1 conjugates and isoforms of PML. Given the evidence that many cellular proteins can be conjugated to PIC1 (reference 26 and this study), Vmw110 could alter the stability of many of the endogenous PIC1-conjugated proteins (or other proteins), leading to far-reaching changes in the intracellular environment. It has already been observed that Vmw110 stimulates the loss of DNA-dependent protein kinase from the cell (33) and stabilizes cyclin D3 (30), and it seems likely that further examples will be forthcoming. This model could also underlie cell survival observations obtained with a family of recombinant viruses that are mutated in multiple IE genes (and therefore do not enter the lytic cycle). The results showed that a virus deficient in Vmw110, Vmw175, and Vmw63 was significantly less detrimental to cell survival than a related virus which was Vmw175 and Vmw63 deficient but retained expression of Vmw110 (50). The underlying prediction from our observations is that changes in the stability of specific cellular proteins leads to stimulation of gene expression and the increased likelihood of the onset of virus infection. While the details of how this might occur remain to be defined, the results presented here provide strong evidence that this mechanism operates during the Vmw110-induced disruption of ND10. Future studies may identify other candidate substrate proteins which are physiologically relevant to virus-host interactions.
ACKNOWLEDGMENTS
We thank Duncan McGeoch, Chris Preston, and Patrick Lomonte for constructive criticisms. The following people kindly made available several key reagents: Roel van Driel (anti-PML MAb 5E10); Rohan Baker (pRB307 and a UBP2 expression construct).
This work was supported by the Medical Research Council, the Imperial Cancer Research Fund, and the National Institute of Child Health and Human Development.
REFERENCES
- 1.Ahn J-H, Hayward G S. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol. 1997;71:4599–4613. doi: 10.1128/jvi.71.6.4599-4613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ascoli C A, Maul G G. Identification of a novel nuclear domain. J Cell Biol. 1991;112:785–795. doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boddy M N, Howe K, Etkin L D, Solomon E, Freemont P S. PIC1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 4.Boddy M N, Duprez E, Borden K L B, Freemont P S. Surface residue mutations of the PML RING finger domain alter the formation of nuclear-matrix-associated PML bodies. J Cell Sci. 1997;110:2197–2205. doi: 10.1242/jcs.110.18.2197. [DOI] [PubMed] [Google Scholar]
- 5.Cai W, Schaffer P A. A cellular function can enhance gene expression and plating efficiency of a mutant defective in the gene for ICP0, a transactivating protein of herpes simplex virus type 1. J Virol. 1991;65:4078–4090. doi: 10.1128/jvi.65.8.4078-4090.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai W, Astor T D, Liptak L M, Cho C, Coen D, Schaffer P A. The herpes simplex virus type 1 regulatory protein ICP0 enhances replication during acute infection and reactivation from latency. J Virol. 1993;67:7501–7512. doi: 10.1128/jvi.67.12.7501-7512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carvalho T, Seeler J-S, Ohman K, Jordan P, Petterson U, Akusjarvi G, Carmo-Fonseca M, Dejean A. Targeting of adenovirus E1A and E4-orf3 proteins to nuclear matrix-associated PML bodies. J Cell Biol. 1995;131:45–56. doi: 10.1083/jcb.131.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dasso M, Nishitani H, Kornbluth S, Nishimoto T, Newport J W. RCC1, a regulator of mitosis, is essential for DNA replication. Mol Cell Biol. 1992;12:3337–3345. doi: 10.1128/mcb.12.8.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De The H, Lavau C, Marcho A, Chomienne C, Degos L, DeJean A. The PML-RARα fusion mRNA generated by the t15;17. translocation in promyelocytic leukaemia encodes a functionally altered RAR. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 10.Dick L R, Cruikshank A A, Destreee A T, Grenier L, McCormack T A, Melandri F D, Nunes S L, Palombella V T, Parent L A, Plamondon L, Stein R L. Mechanistic studies on the inactivation of the proteasome by lactacystin in cultured cells. J Biol Chem. 1997;272:182–188. doi: 10.1074/jbc.272.1.182. [DOI] [PubMed] [Google Scholar]
- 11.Doucas V, Ishov A, Romo A, Juguilon H, Weitzman M D, Evans R M, Maul G G. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 12.Dyck J A, Maul G G, Jr Miller W H, Chen J D, Kakizuka A, Evans R M. A novel macromolecular structure is a target of the promyelocytic-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 13.Everett R D. Construction and characterisation of herpes simplex virus type 1 mutants with defined lesions in immediate-early gene 1. J Gen Virol. 1989;70:1185–1202. doi: 10.1099/0022-1317-70-5-1185. [DOI] [PubMed] [Google Scholar]
- 14.Everett R D, Preston C M, Stow N D. Functional and genetic analysis of the role of Vmw110 in herpes simplex virus replication. In: Wagner E K, editor. The control of herpes simplex virus gene expression. Boca Raton, Fla: CRC Press Inc.; 1991. pp. 50–76. [Google Scholar]
- 15.Everett R D, Cross A, Orr A. A truncated form of herpes simplex virus type 1 immediate-early protein Vmw110 is expressed in a cell-type dependent manner. Virology. 1993;197:751–756. doi: 10.1006/viro.1993.1651. [DOI] [PubMed] [Google Scholar]
- 16.Everett R D, Maul G G. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everett R D, Meredith M R, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goddard A D, Borrow J, Freemont P S, Solomon E. Characterization of a zinc finger gene disrupted by the t15;17. in acute promyelocytic leukemia. Science. 1991;254:1371–1374. doi: 10.1126/science.1720570. [DOI] [PubMed] [Google Scholar]
- 19.Harris R A, Everett R D, Zhu X, Silverstein S, Preston C M. The HSV immediate early protein Vmw110 reactivates latent HSV type 2 in an in vitro latency system. J Virol. 1989;63:3513–3515. doi: 10.1128/jvi.63.8.3513-3515.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 21.Ishov A M, Maul G G. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishov A M, Stenberg R M, Maul G G. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J Cell Biol. 1997;138:5–16. doi: 10.1083/jcb.138.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson P R, Hochstrasser M. SUMO-1: ubiquitin gains weight. Trends Cell Biol. 1997;7:408–413. doi: 10.1016/S0962-8924(97)01132-X. [DOI] [PubMed] [Google Scholar]
- 24.Jordan R, Schaffer P A. Activation of gene expression by herpes simplex virus type 1 ICP0 occurs at the level of mRNA synthesis. J Virol. 1997;71:6850–6862. doi: 10.1128/jvi.71.9.6850-6862.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kakizuka A, Miller W H, Umesono K, Warrell R P, Frankel S R, Murty V V V S, Dmitrovsky E, Evans R M. Chromosomal translocation t15;17. in acute promyelocytic leukaemia fuses RARα with a novel putative transcription factor PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 26.Kamitani T, Nguyen H P, Yeh E T H. Preferential modification of nuclear proteins by a novel ubiquitin-like molecule. J Biol Chem. 1997;272:14001–14004. doi: 10.1074/jbc.272.22.14001. [DOI] [PubMed] [Google Scholar]
- 27.Kamitani T, Nguyen H P, Kito K, Fukuda-Kamitani T, Yeh E T H. Covalent modification of PML by the Sentrin family of ubiquitin-like proteins. J Biol Chem. 1998;273:3117–3120. doi: 10.1074/jbc.273.6.3117. [DOI] [PubMed] [Google Scholar]
- 28.Kastner P, Perez A, Lutz Y, Rochette-Egly C, Gaub M-P, Durand B, Lanotte M, Berger R, Chambon P. Structure, localization and transcriptional properties of two classes of retinoic acid receptor alpha fusion proteins in acute promyelocytic leukaemia APL.: structural similarities with a new family of oncoproteins. EMBO J. 1992;11:629–642. doi: 10.1002/j.1460-2075.1992.tb05095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawaguchi Y, van Sant C, Roizman B. Herpes simplex virus regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelly C, van Driel R, Wilkinson G W G. Disruption of PML nuclear bodies during human cytomegalovirus infection. J Gen Virol. 1995;76:2485–2491. doi: 10.1099/0022-1317-76-11-2887. [DOI] [PubMed] [Google Scholar]
- 31.Koken M H M, Puvion-Dutilleul F, Guillemin M, Viron A, Linares-Cruz G, Stuurman N, de Jong L, Szostecki C, Calvo F, Chomienne C, Degos L, Puvion E, de The H. The t15;17. translocation alters a nuclear body in a retinoic acid reversible fashion. EMBO J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korioth F, Gieffers C, Maul G G, Frey J. Molecular characterisation of NDP52, a novel protein of nuclear domain 10 which is redistributed upon infection and interferon treatment. J Cell Biol. 1995;130:1–14. doi: 10.1083/jcb.130.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lees-Miller S P, Long M C, Kilvert M A, Lam V, Rice S A, Spencer C A. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J Virol. 1996;70:7471–7477. doi: 10.1128/jvi.70.11.7471-7477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leib D A, Coen D M, Bogard C L, Hicks K A, Yager D R, Knipe D M, Tyler K L, Schaffer P A. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J Virol. 1989;63:759–768. doi: 10.1128/jvi.63.2.759-768.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lennon G G, Auffray C, Polymeropoulos M, Soares M B. The I.M.A.G.E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 36.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 37.Matunis M J, Couvavas E, Bloebel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1997;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maul G G, Guldner H H, Spivack J G. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate-early gene 1 product ICP0. J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 39.Maul G G, Everett R D. The nuclear location of PML, a cellular member of the C3HC4 zinc binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 40.Maul G G, Yu E, Ishov A M, Epstein A L. Nuclear domain 10 (ND10) associated proteins are present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J Cell Biochem. 1995;59:499–514. doi: 10.1002/jcb.240590410. [DOI] [PubMed] [Google Scholar]
- 41.Maul G G, Ishov A, Everett R D. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type 1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- 42.Meredith M R, Orr A, Everett R D. Herpes simplex virus type 1 immediate-early protein Vmw110 binds strongly and specifically to a 135kD cellular protein. Virology. 1994;200:457–469. doi: 10.1006/viro.1994.1209. [DOI] [PubMed] [Google Scholar]
- 43.Meredith M R, Orr A, Elliott M, Everett R D. Separation of the sequence requirements for HSV-1 Vmw110 multimerisation and interaction with a 135kD cellular protein. Virology. 1995;209:174–187. doi: 10.1006/viro.1995.1241. [DOI] [PubMed] [Google Scholar]
- 44.Muller S, Matunis M J, DeJean A. Conjugation of the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pandolfi P P, Alcalay M, Fagioli M, Zangrilla D, Mencarelli A, Diverio D, Biondi A, Lo Coco F, Rambaldi A, Grignani F, Rochette-Egly C, Gaub M-P, Chambon P, Pellici P G. Genomic variability and alternative splicing generate multiple PML/RARα transcripts that encode aberrant PML proteins and PML/RARα isoforms in acute promyelocytic leukaemia. EMBO J. 1992;11:1397–1407. doi: 10.1002/j.1460-2075.1992.tb05185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sacks W R, Schaffer P A. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J Virol. 1987;61:829–839. doi: 10.1128/jvi.61.3.829-839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saitoh H, Pu P, Cavenagh M, Dasso M. RanBP2 associates with Ubc9 and a modified form of RanGAP1. Proc Natl Acad Sci USA. 1997;94:3736–3741. doi: 10.1073/pnas.94.8.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saitoh H, Pu R T, Dasso M. SUMO-1: wrestling with a new ubiquitin-related modifier. Trends Biochem Sci. 1997;22:374–376. doi: 10.1016/s0968-0004(97)01102-x. [DOI] [PubMed] [Google Scholar]
- 49.Saitoh H, Pu R T, Sparrow D, Shiomi T, Mohun T, Nishimoto T, Dasso M. Ubc9p and the conjugation of SUMO-1 to RanGAP1 and RanBP2. Curr Biol. 1998;8:121–124. doi: 10.1016/s0960-9822(98)70044-2. [DOI] [PubMed] [Google Scholar]
- 50.Samaniego L A, Wu N, DeLuca N A. The herpes simplex virus immediate-early protein ICP0 affects transcription from the viral genome and infected cell survival in the absence of ICP4 and ICP27. J Virol. 1997;71:4614–4625. doi: 10.1128/jvi.71.6.4614-4625.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schenk P, Ludwig H. The 65kd DNA binding protein appears early in HSV-1 infection. Arch Virol. 1988;102:119–123. doi: 10.1007/BF01315568. [DOI] [PubMed] [Google Scholar]
- 52.Smythe C, Newport J W. Systems for the study of nuclear assembly, DNA replication, and nuclear breakdown in Xenopus laevis egg extracts. Methods Cell Biol. 1991;35:449–468. doi: 10.1016/s0091-679x(08)60583-x. [DOI] [PubMed] [Google Scholar]
- 53.Sternsdorf T, Jensen K, Will H. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J Cell Biol. 1997;139:1621–1634. doi: 10.1083/jcb.139.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stow N D, Stow E C. Isolation and characterisation of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate-early polypeptide Vmw110. J Gen Virol. 1986;67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 55.Stuurman N, DeGraaf A, Josso A, Humbel B, DeYong L, van Driel R. A monoclonal antibody recognising nuclear matrix associated nuclear bodies. J Cell Sci. 1992;101:773–784. doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- 56.Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, DeJean A. Retinoic acid regulates aberrant nuclear localization of PML-RARα in acute promyelocytic leukaemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 57.Wilkinson K D. Roles of ubiquitination in proteolysis and cellular regulation. Ann Rev Nutr. 1995;15:161–189. doi: 10.1146/annurev.nu.15.070195.001113. [DOI] [PubMed] [Google Scholar]
- 58.Yao F, Schaffer P A. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J Virol. 1995;69:6249–6258. doi: 10.1128/jvi.69.10.6249-6258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoshida H, Kitamura K, Tanaka K, Omura S, Miyazaki T, Hachiya T, Ohno R, Naoe T. Accelerated degradation of PML-retinoic receptor αPML-RARA. oncoprotein by all-trans-retinoic acid in acute promyelocytic leukaemia: possible role of the proteasome pathway. Cancer Res. 1996;56:2945–2948. [PubMed] [Google Scholar]
- 60.Zhu J, Koken M H M, Quignon F, Chelbi-Alix M K, Degos L, Wang Z Y, Chen Z, de The H. Arsenic-induced PML targeting onto nuclear bodies: implications for the treatment of acute promyelocytic leukaemia. Proc Natl Acad Sci USA. 1997;94:3978–3983. doi: 10.1073/pnas.94.8.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu X, Chen J, Young C S H, Silverstein S. Reactivation of latent herpes simplex virus by adenovirus recombinants encoding mutant IE-0 gene products. J Virol. 1990;64:4489–4498. doi: 10.1128/jvi.64.9.4489-4498.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]