Abstract
Previously, we reported that human cytomegalovirus (HCMV) infection of fibroblasts markedly affects p53 and other regulatory proteins and inhibits transit through the cell cycle (F. M. Jault, J.-M. Jault, F. Ruchti, E. A. Fortunato, C. Clark, J. Corbeil, D. D. Richman, and D. H. Spector, J. Virol. 69:6697–6704, 1995). Although the p53 steady-state levels are elevated throughout the infection, evidence suggests that the ability of p53 to transactivate some of its downstream targets is compromised. To elucidate the mechanisms governing the accumulation of p53, we examined the synthesis, stability, and localization of the protein in HCMV-infected fibroblasts. Synthesis of p53 was not increased in the infected cells during the first 24 h postinfection. In fact, pulse-chase experiments revealed that synthesis of p53 in infected fibroblasts was lower than in mock-infected cells. However, after an initial decay, the p53 was stabilized. In addition, beginning at approximately 30 h postinfection, p53 was localized to discrete foci within the nuclei of infected cells. The morphology of these foci suggested that they were replication centers. We confirmed that these are sites of DNA replication by demonstrating both incorporation of bromodeoxyuridine and localization of UL44 (the viral polymerase processivity factor) into these centers. The single-stranded DNA binding protein RPA was also sequestered. In contrast, Rb and HCMV IE1 72 remained distributed throughout the infected cell nuclei, indicating specific targeting of certain proteins. Taken together, our results provide two alternative mechanisms to account for the increased steady-state levels of p53 observed in HCMV-infected fibroblasts.
Beginning with the interaction of viral particles with the plasma membrane, human cytomegalovirus (HCMV) disrupts the normal cellular functioning of the host cell. Initial entry or contact of HCMV with the cell leads to a second messenger-type response similar to what occurs during growth regulation via hormones and/or growth factors (for a review, see reference 2). With the stimulation of expression of several proteins involved in preparing the cell for DNA replication, including the proto-oncogene products Fos, Jun, and Myc (4), dihydrofolate reductase (DHFR) (45), thymidine kinase (14), DNA polymerase α (18), ornithine decarboxylase (21), and proliferating-cell nuclear antigen (PCNA) (10), the productive HCMV infection leads to a fully activated state. In addition, early after infection, HCMV induces elevated levels of active cyclins E and B, p53, and hyperphosphorylated Rb but delays active cyclin A expression until late in the infection (23). Several studies have concluded that these HCMV-induced effects inhibit cell cycle transit (6, 10, 23, 30).
In addition to this block in transit, HCMV infection also results in significant genotoxic effects to the infected cell. Major damage to the cellular DNA, including breakage, deletions, and premature condensation, is observed (1). Moreover, recent studies have emphasized the mutagenic potential of the virus (3, 41). One possible mechanism for inducing these genotoxic effects is the interaction of the virus with the tumor suppressor protein p53, which is involved in the regulation of the G1 and G2 cell cycle checkpoints (reviewed in reference 15). Loss of p53 function is associated with a broad spectrum of human cancers (19), and although mice lacking the p53 gene are viable, they are susceptible to multiple types of tumors at a young age (11).
Elevation of p53 protein levels is observed after several potentially damaging events, including UV or gamma ray irradiation, exposure to extreme heat, hypoxia, or starvation (reviewed in reference 28). These elevated levels of p53 can lead either to cell cycle arrest, presumably to allow repair of damaged DNA, or to apoptosis. p53 controls these two outcomes by sequence specific binding to and transactivation of specific target genes, including those coding for p21 (also known as WAF1 or CIP1) (13), GADD45 (27), cyclin G (36), IGF-BP3 (7), bax (32), and mdm2 (25). In addition to p53’s regulation of the DNA damage response through transcriptional activation of particular target genes, there is increasing evidence that it participates in this response by binding to damaged regions with its C-terminal domain and recruiting the repair machinery to the site via protein-protein interactions. To this end, p53 directly interacts with several proteins involved in nucleotide excision repair and/or homologous recombination, including two of the RNA polymerase II coactivator TFIIH components, XPB and XPD (47), CSB (47), RAD51 (44), and the single-stranded DNA binding protein RPA (12, 17, 31, 33, 44).
Several lines of evidence point to interactions between HCMV and p53. As mentioned above, elevated steady-state levels of p53 are observed in HCMV-infected fibroblasts by 24 h postinfection (hpi) (23, 34, 43). Like several other viruses, including simian virus 40 (SV40) (24), adenovirus (8, 52), hepatitis B virus (46), and the high-risk-type human papillomaviruses (HPVs) (40, 49), HCMV encodes two proteins that can interact with p53: the mtrII oncoprotein (UL111a) (in vitro only) (35) and the immediate early protein IE2 86 (UL122) (5, 43). Transient assays demonstrating downregulation of p53-responsive promoters by coexpression of either mtrII or IE2 86 (35, 43) have led to speculation that interactions between these two viral proteins and p53 block the latter’s ability to transactivate one of its main targets, p21 (6, 23). In addition, there are reports that HCMV immediate early proteins IE1 72 and IE2 86 can block the induction of apoptosis (presumably p53 mediated) by tumor necrosis factor α or by the adenovirus E1A protein (53). Whether these proteins act alone or in conjunction with other viral proteins, as is proposed by Bonin and McDougall (5), remains to be determined. Nevertheless, these data suggest that one mechanism by which HCMV may fully activate the infected cell without triggering its death and may also precipitate the genotoxic effects observed is by interfering with the ability of p53 to regulate the repair of damaged cellular DNA through transcriptional activation and protein-protein interactions.
To better understand the effects of the HCMV-p53 interaction, we have undertaken a study to determine the mechanism of p53 accumulation within infected fibroblasts. We have investigated p53’s stability and subcellular localization and show in this work that the elevated steady-state levels of p53 observed in infected fibroblasts are due not to increased levels of synthesis but rather to a stabilization of the protein. Moreover, there is a distinct sequestering of p53 into viral replication centers within the nucleus which may also aid in p53 stabilization. p53 is localized to these viral replication centers as early as 30 hpi and remains there throughout the duration of the infection. The additional localization of RPA but not Rb into these centers suggests a specific redistribution of certain nuclear proteins. Possible mechanisms by which HCMV alters the stability and localization of p53, as well as the role of these events in the viral life cycle, are discussed.
MATERIALS AND METHODS
Cells and virus.
Primary human foreskin fibroblasts were obtained from the University of California, San Diego, Medical Center and propagated in minimal essential medium with Earle’s salts (MEM). The cells were propagated in incubators maintained at 37°C and 5% CO2, and the medium was supplemented with 10% heat-inactivated fetal bovine serum, l-glutamine (2 mM), penicillin (200 U/ml), streptomycin (200 μg/ml), amphotericin B (1.5 μg/ml), and gentamicin sulfate (50 μg/ml). The Towne strain of HCMV was obtained from the American Type Culture Collection (VR 977) and used at a multiplicity of infection of 5.
Cell cycle synchronization and infection conditions.
All experiments using fibroblasts were performed under G0 synchronization conditions. Cells were seeded into flasks and allowed to become confluent. After 2 to 3 days at confluence, cells were reseeded onto 10-cm-diameter dishes at 106 cells/dish. Approximately 1 h after plating, the medium was removed, and the infection inoculum (either virus or an equivalent amount of mock-conditioned medium, diluted 1:1 in fresh culture medium) was added to the cells. At 6 hpi, the inoculum was removed, fresh culture medium was added, and the cells were incubated for various times. All experiments were performed at least twice, and data presented in the present work are representative examples of those trials.
Western time course experiments.
Cells were synchronized, plated, and infected as described above. At the desired time points, cells were trypsinized, washed, and counted. Cells were then resuspended in Laemmli reducing sample buffer (LRSB) (2% sodium dodecyl sulfate [SDS], 10% glycerol, 100 mM dithiothreitol, 60 mM Tris pH 6.8, bromophenol blue dye, aprotinin and leupeptin [2 μg/ml each], and phenylmethylsulfonyl fluoride [1 mM]) at a concentration of 104 cells/μl of buffer. Cells were then sonicated, boiled for 5 min, and spun at 16,000 × g for 10 min at 4°C to pellet any debris. Equal amounts of lysate (usually 25 to 30 μl) were then loaded onto an SDS-polyacrylamide gel electrophoresis minigel. Proteins were then transferred to Immobilon P (Millipore) and detected with mouse anti-p53 primary antibody (Ab) (DO-1) (Santa Cruz Biotechnology), followed by a goat anti-mouse secondary Ab conjugated with horseradish peroxidase (Amersham). Proteins were visualized with enhanced chemiluminescence reagents (Pierce), used according to the manufacturer’s instructions.
Metabolic labeling and pulse-chase immunoprecipitations.
Cells were synchronized, plated, and infected as described above. Prior to the radioactive pulse, normal culture medium was removed and cells were washed twice with phosphate-buffered saline (PBS) and refed with MEM lacking methionine but supplemented with 10% dialyzed fetal bovine serum. After 30 min, this starvation medium was removed and replaced with 3 ml of the same medium supplemented with 300 μCi of Tran35S label (ICN) per plate. For metabolic labeling, cells were incubated in this labeling medium for a period of 2 h. The cells were then harvested as described below. For pulse-chase experiments, cells were incubated in labeling medium for 30 min and then one set of plates was harvested (pulse) and the other plates were washed twice with warm PBS, refed with normal culture medium supplemented with 1 mM cold methionine, and incubated for various lengths of time (chase). To harvest the cells, plates were washed twice in cold PBS and the cells were then scraped in PBS and transferred to 15-ml conical tubes. Cells were pelleted at 4°C, resuspended in 1 ml of cold PBS, and counted. Equal numbers of cells (approximately 5 × 105) were then pelleted and resuspended in 100 μl of cold PBS. Three hundred microliters of RIPA buffer (50 mM Tris [pH 7.4], 120 mM NaCl, 0.1% deoxycholic acid, 2% Nonidet P-40, 0.2% SDS, aprotinin and leupeptin [2 μg/ml each], and dithiothreitol and phenylmethylsulfonyl fluoride [1 mM each]) was added, and the cells were sonicated for 2.5 min. After vortexing for an additional 10 min, the lysate was cleared with a 16,000 × g spin and transferred to a fresh tube containing an additional 600 μl of RIPA buffer. After gentle mixing, an aliquot was removed for analysis of total p53 concentration prior to incubation with Ab. The remaining lysate was mixed with 20 μl of agarose-coupled DO-1 Ab (Santa Cruz Biotechnology), an amount which we had determined would precipitate all of the p53 in the sample, and rocked for 2 h at 4°C. After incubation, 40 μl of a 50% protein A-Sepharose slurry was added, the beads were pelleted, and another aliquot was removed for determination of p53 concentration after incubation with Ab. The beads were washed three times in RIPA buffer, transferred to a fresh tube, and then washed once more. Proteins were eluted from the beads in LRSB and after boiling were electrophoresed through an SDS–10% polyacrylamide gel. Gels were stained and then subjected to fluorography, dried, and autoradiographed. Quantitation of bands was performed with a PhosphorImager (Molecular Dynamics).
Immunofluorescence.
Synchronized cells were seeded onto glass coverslips and infected as described above. At the desired time points, coverslips were washed twice in PBS and the cells were then fixed in a 3% paraformaldehyde solution in PBS. After 10 min, this was replaced with a 1% Triton X-100 solution in PBS. After a 5-min incubation, the cells were rinsed three times in PBS and then stored at 4°C until all samples were taken. Coverslips were then incubated for 10 min in blocking solution (1% normal goat serum in PBS–0.2% gelatin), washed in PBS, and incubated with one of several mouse primary Abs diluted in PBS–0.2% gelatin for 10 min. After extensive washing in PBS, the cells were then incubated with a fluorescein isothiocyanate-conjugated goat anti-mouse secondary Ab (Jackson Labs) (diluted in PBS–0.2% gelatin). Following more washes, the cells were finally incubated in Hoechst dye (Calbiochem) for 2 min to illuminate the DNA, washed and mounted onto slides using glycerol-paraphenylene diamine (an antiphotobleaching agent). For analysis of replication, cells were incubated for 30 min prior to harvesting at 48 hpi in bromodeoxyuridine (BrdU) labeling mix diluted in normal media. Coverslips were then harvested and fixed as described above. Prior to Ab detection, these coverslips were incubated in 4 M HCl diluted in PBS for 10 min to expose the BrdU residues. All slides were analyzed on a Nikon EFD-3 flourescence microscope equipped with Neofluor objectives and an MTT CCD2 camera, and images were captured by using NIH Image 1.59. Collected images were arranged and labeled by using Adobe Photoshop 3.0 and Adobe Illustrator 6.0.2. Monoclonal Abs used for fluorescence included commercially available anti-p53 (DO-1; Santa Cruz Biotechnology), anti-Rb (G3-245; Pharmingen), antitrinitrophenol (Pharmingen), and anti-BrdU (Boehringer Mannheim). Antibodies to HCMV UL44 and IE1 72 were kind gifts from L. Pereira and W. Britt, respectively. Anti-RPA was a kind gift from J. Wang. Note that attempts to colocalize proteins by double labeling using other commercially available Abs (raised in either rabbits or goats) proved unsuccessful due to their lack of avidity and/or their binding by the virally encoded immunoglobulin G Fc receptor-like protein.
RIPA buffer extractions.
Cells were harvested, counted, and pelleted in aliquots of 5 × 105 cells/tube at 48 hpi. The pellets were then resuspended in 10 μl of PBS, to which was added 50 μl of RIPA buffer. Parallel sets of tubes were processed; one with and the other without a 2.5-min sonication before 10 min of vortexing. Thirty microliters of 3× LRSB was added to the cleared lysates, and the pelleted debris was resuspended in an equivalent amount of 1× LRSB. Samples were boiled, and one half of each tube was loaded and electrophoresed through an SDS–10% polyacrylamide gel and processed as described above for Western time course experiments.
RESULTS
Increased steady-state levels of p53 in infected fibroblasts are due to decreased degradation and not increased synthesis.
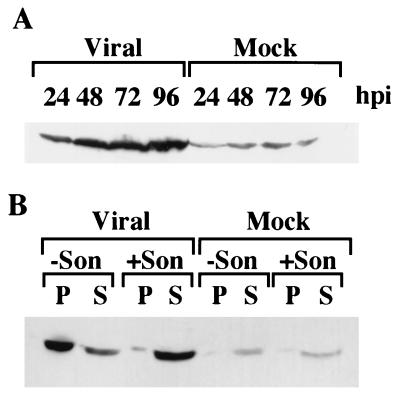
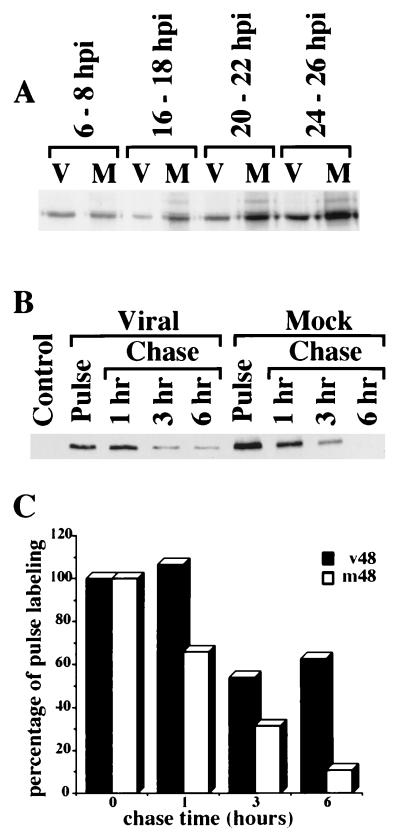
As we had previously reported (23), infection of fibroblasts with HCMV Towne strain induced elevated steady-state levels of p53 in infected cells relative to mock infected cells 24 to 36 hpi (Fig. 1A). These elevated steady-state levels could be due to an increase in the rate of synthesis and/or a decrease in the rate of degradation. In order to distinguish between these alternatives, we metabolically labeled cells every 2 h for the first 24 hpi and then again at 48 hpi. In our initial pilot experiments, we found that resuspension in RIPA buffer alone was not sufficient to extract all of the p53 from the infected cell nuclei (Fig. 1B). We therefore incorporated an additional sonication step in our lysis procedure to ensure complete extraction. Even under these extraction conditions, no burst of protein synthesis was found early in the infection that could account for the increased steady-state levels. A representative sample of these labelings is shown in Fig. 2A. In fact, when cells were pulse-labeled for 30 min at 48 hpi, we found that the synthesis levels of p53 in the infected cells were actually somewhat lower than in their mock-infected counterparts (approximately twofold) (Fig. 2B). Interestingly, analysis of the newly synthesized p53 during the chase period revealed a dramatic difference between the infected and uninfected cells; after an initial decrease to approximately half the incorporated label by 3 h, the labeled p53 in the infected cells became stabilized, with no further decrease observed for up to 6 h of chase. In contrast, the p53 in the mock-infected cells steadily decayed over the span of the experiment, going through approximately three half-lives in 6 h (half-life, ∼2 h). Preliminary experiments performed at 24 hpi revealed the same pattern of lower synthesis but extended stabilization as observed at 48 hpi (data not shown). Therefore, a stabilization rather than an increase in synthesis appeared to be the cause of the elevated steady-state levels of p53 observed in infected fibroblasts.
FIG. 1.
The p53 protein is stabilized in HCMV-infected fibroblasts but is not easily extracted from their nuclei. (A) Confluence-synchronized fibroblasts were collected at the indicated times by trypsinization, counted, and lysed in LRSB as described in Materials and Methods. Each lane of the SDS–15% polyacrylamide gel was loaded with 3 × 105 cell equivalents. Proteins were transferred to Immobilon and detected with the monoclonal Ab DO-1 directed against p53. (B) At 48 hpi, cells were harvested and aliquots of 5 × 105 cells were lysed in RIPA buffer with or without an additional sonication step (+Son and −Son, respectively) to help improve extraction as indicated. Lysates were cleared, and the pelleted debris was resuspended in an equivalent amount of LRSB. One half of each aliquot, either pellet (P) or supernatant (S) was then electrophoresed through an SDS–10% polyacrylamide gel, transferred to Immobilon P, and probed with DO-1 Ab.
FIG. 2.
Stabilization of p53 is due to decreased degradation, not increased synthesis. (A) Cells were metabolically labeled for 2-h periods at the indicated times postinfection as described in Materials and Methods. Approximately 5 × 105 cells were sonicated in RIPA buffer as described in the legend to Fig. 1B to ensure complete extraction of p53, and cleared lysates were incubated with agarose-coupled DO-1 Ab for 2 h at 4°C with gentle rocking. The beads were then washed several times in RIPA buffer and resuspended in LRSB, and the eluted proteins were run on an 10% SDS–polyacrylamide gel, fluorographed, dried, and exposed to film. M, mock-infected cells; V, virally infected cells. (B) Cells were pulse-labeled at 48 hpi, chased for the indicated times as described in Materials and Methods, and then processed as described for panel A. An agarose-coupled Ab to SV40 large-T antigen was used as a control for specificity. (C) Quantitation of data from panel B by using PhosphorImager software. For each set of data, the pulse was taken as 100% incorporation of label. m48, mock-infected cells pulse-labeled at 48 hpi; v48, virally infected cells pulse-labeled at 48 hpi.
p53 is sequestered into discrete foci within the nuclei of infected cells.
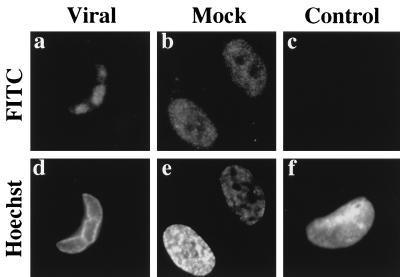
The increased difficulty of extraction of p53 from the infected cell nuclei in the experiments described above led us to suspect that the subcellular localization of p53 might be altered in the infected fibroblasts. To investigate this possibility, cells were seeded onto glass coverslips and then infected or mock infected as described in Materials and Methods. At selected times postinfection, coverslips were removed, washed gently in PBS, fixed in paraformaldehyde, and then permeabilized with Triton X-100. We found this method of fixation coupled with the use of the DO-1 Ab to p53 to be the most reliable and reproducible combination for visualizing the staining pattern of p53 in both the mock-infected and infected cells. Surprisingly, we found that the p53 in infected cells was localized to discrete foci within the nuclei of virally infected cells beginning at approximately 30 hpi (Fig. 3a). These foci increased in size as the infection progressed and by 72 hpi almost completely covered the nucleus (data not shown). This pattern of staining was not apparent in mock-infected cells (Fig. 3b), where p53 was distributed uniformly throughout the entire nucleus. There was also no detectable staining observed when virally infected cells were incubated with a control Ab directed against trinitrophenol (Fig. 3c). These results support our hypothesis that a relocalization of p53 in the infected fibroblasts accounts for the increased difficulty of extraction and potentially for some of the stabilization of p53 observed in these cells.
FIG. 3.
p53 is sequestered into discrete foci in the nucleus of infected fibroblasts. Cells were confluence synchronized, seeded onto glass coverslips, infected, and at 30 hpi were processed as described in Materials and Methods. Viral (a) and mock (b) cells were stained with DO-1 Ab. Viral cells were also stained with a control Ab to trinitrophenol to show specificity of staining (c). Hoechst dye was used to illuminate the DNA of the imaged cells (d through f). Magnification ≈×410. FITC, fluorescein isothiocyanate.
p53 localizes to viral replication centers along with the HCMV protein UL44.
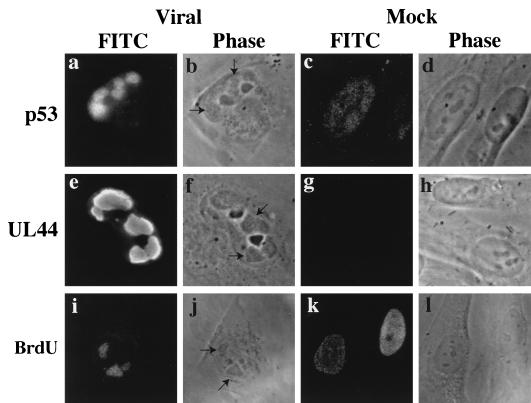
The staining pattern observed for p53 in the infected fibroblasts was reminiscent of that seen for proteins associated with viral replication. Closer examination of these foci in the virally infected cells by phase microscopy at 48 hpi revealed the distinct morphology of the viral replication centers produced by both HCMV and another herpesvirus, herpes simplex virus (HSV) (9) (arrows in Fig. 4b, f, and j). It was also apparent that these structures were not present within the mock-infected cell nuclei (Fig. 4d, h, and l).
FIG. 4.
p53 localizes to viral replication centers in infected fibroblasts as delineated by staining of the HCMV DNA polymerase processivity factor UL44 and BrdU incorporation. At 48 hpi, coverslips were fixed and permeabilized as described in Materials and Methods. (a and c) Staining with DO-1 (anti p53); (e and g) staining with CH16.1 (anti UL44); (i and k) staining with anti BrdU. Arrows in the phase-contrast micrographs point to the electron-dense structures that are viral replication centers. BrdU images are taken from a separate experiment in which, 30 min prior to harvesting at 48 hpi, cells were incubated in media containing BrdU. Magnification ≈ ×370. FITC, fluorescein isothiocyanate.
To confirm that these structures were DNA replication centers, we stained for the HCMV protein UL44, an early protein that associates with the virally encoded DNA polymerase (48), is required for oriLyt dependent viral DNA replication (20, 37, 38), and localizes to these intranuclear inclusions during infection (22). Figure 4 shows that the UL44 protein also localized to these same structures within the virally infected nuclei (Fig. 4e and f [arrows]) and was not present in the mock-infected cells (Fig. 4g). These results confirmed earlier studies which colocalized UL44 and UL112-113, another class of viral proteins required for viral DNA replication (20, 37, 38), to these foci during HCMV infection (22, 51a). Moreover, by pulse-labeling the cells with BrdU, we were able to demonstrate that these foci were the primary sites of active DNA replication within the infected cells (Fig. 4i). We concluded from these results that the p53-containing foci in HCMV-infected cell nuclei were centers of viral replication.
Localization to replication centers is observed with some cellular and viral proteins but not others.
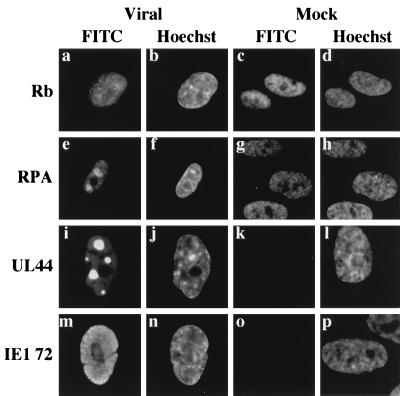
Previously, Wilcock and Lane (50) showed that HSV sequestered p53 as well as certain other host cell proteins, including Rb, PCNA, and RPA into viral replication centers. To determine whether HCMV also sequestered other cellular proteins into these centers and whether this might be a generalized phenomenon for cellular proteins normally present in the nucleus, we used indirect immunofluorescence staining to examine the viral and mock-infected fibroblasts at 30 hpi (Fig. 5); results obtained at 48 hpi were identical (data not shown). We focused on two cellular proteins: Rb, because we had previously observed modulation of both the steady-state levels and phosphorylation state of this protein upon infection, and RPA, because it is a protein involved in both cellular DNA replication and repair and is known to bind to p53. Figure 5 demonstrates that RPA but not Rb (Fig. 5e and a, respectively) localized to the replication centers. The viral immediate early protein IE1 72 (Fig. 5m) also did not localize to these foci, providing further evidence that there was specificity in the process. As noted above, the UL44 protein was clearly sequestered in these centers (Fig. 5i), and this localization could be observed as early as 24 hpi (data not shown). No foci were apparent in any of the mock-infected cells (Fig. 5c, g, k, and o). Furthermore, we observed the identical localization for each of these proteins in another permissive cell line, U373 (data not shown), providing evidence that this was not a cell type-specific phenomenon. Taken together, these results indicate that p53 and RPA are specifically sequestered into the viral replication centers of infected cells beginning at 30 hpi. This sequestering may in part contribute to the stabilization of p53 within infected fibroblasts.
FIG. 5.
Localization to replication centers in infected fibroblasts is observed with some cellular and viral proteins, but not others. Coverslips were processed for immunofluorescence at 30 hpi. Cells were stained with PMG-345 (anti-Rb) (a and c), monoclonal Ab to RPA (e and g), CH16.1 (anti-UL44) (i and k), and monoclonal Ab to IE1 72 (m and o). Hoechst dye was used to illuminate the DNA in all imaged cells. Magnification ≈ ×290. FITC, fluorescein isothiocyanate.
DISCUSSION
Earlier studies have shown that upon infection of permissive fibroblasts with HCMV, there is a marked increase in the steady-state levels of p53, which appears to be wild type in genotype but unable to transactivate at least some of its normal target genes (6, 23, 34, 43). The present study confirms and extends these observations to address the reason for the marked rise in p53 steady-state levels within the infected fibroblast. We have shown that the elevated levels of p53 in infected fibroblasts are due to decreased degradation of the protein rather than increased synthesis. In addition, we find that p53 in HCMV-infected cells is specifically sequestered into viral replication centers within the nucleus beginning at 30 hpi. This sequestering is not a generalized phenomenon and is not cell type specific but is particular for certain cellular and viral proteins.
Many viruses have evolved mechanisms to sequester and/or functionally inactivate p53. SV40 large-T antigen binds to the core region of p53 and blocks its sequence-specific DNA binding (24). The adenovirus 55K E1B protein counters p53-initiated apoptosis (8), presumably by binding to the N terminus of p53 (26) and blocking its transactivation domain (16). Hepatitis B virus X protein can interact in vitro with the C terminus of p53, inhibiting both sequence-specific DNA binding and p53’s ability to complex with XPB (46, 47). Finally, HPV counters the function of p53 by inhibiting its accumulation in infected cells. The E6 proteins of the high risk-type HPV bind in vitro to the core region of p53 (49) and induce a rapid ubiquitin-mediated degradation of the protein with the help of an associated cellular protein, E6 AP (40).
A key question raised by the studies presented here is this: how does HCMV stabilize p53 within the infected cell? The most probable explanation is that one or more proteins either induced or encoded by HCMV bind to p53 and counter the normal ubiquitin-mediated degradation of the protein. In support of this hypothesis, we found that when we infected fibroblasts stably expressing the HPV16 E6 protein, which normally causes a rapid degradation of p53, we saw a marked increase in the steady-state levels of p53 along with sequestering into the replication centers (data not shown).
Two HCMV-encoded proteins (mtrII and IE2 86) may contribute to the enhanced stabilization and apparent inability of p53 to transactivate its normal target genes within infected cells. NIH 3T3 cells stably expressing the nuclear protein mtrII alone have greatly increased p53 steady-state levels and decreased p53 degradation (35). mtrII binds to the N terminus of p53 in vitro and downregulates transactivation by p53 when transiently coexpressed (35). The immediate early protein IE2 86 also appears to block p53-mediated transactivation of a chloramphenicol acetyltransferase reporter construct, at least when transiently cotransfected into smooth-muscle cells (43). IE2 86 forms a complex with p53 in vitro and in vivo (5, 43), and we have evidence that late in infection (after 48 hpi), a small proportion of IE2 86 localizes to the replication centers (data not shown). Therefore, binding by IE2 86 could be a means of transporting p53 into these centers and thus potentially contribute to its stabilization. However, observations by Bonin and McDougall (5) showing that p53 is not stabilized and appears to respond appropriately to DNA damage in a fibroblast line stably expressing IE2 86 indicate that this protein may not be able to affect p53 when expressed alone.
Our observations with HCMV infection of both fibroblasts and U373 cells show sequestering of p53 and RPA into viral replication centers, and PCNA has been localized to these sites by others (10). This is similar to what is observed during productive HSV-1 replication (50). However, unlike HCMV, HSV also sequesters Rb into these centers. Perhaps the key to this difference lies with the ability of HCMV to effectively counter the regulatory functioning of Rb by hyperphosphorylating it very early during infection (23), thus negating the need to sequester it from the cell. However, this does not address the reason why p53 and the other proteins are being sequestered by HCMV. Two alternatives come to mind: either they play an active role in viral replication and/or repair or they are being kept from performing their normal cellular functions.
The first alternative is quite appealing in the case of PCNA and RPA. Both are active participants in cellular DNA replication and could therefore be used in this capacity by the virus as they are for both bovine papillomavirus (29) and SV40 (39, 51) viral replication. RPA and PCNA also participate in the repair of damaged DNA via nucleotide excision repair and homologous recombination (17, 33, 42). p53 may also act in the repair process as an assembly factor, binding first to damaged DNA through its C terminus and then directly to several components of the repair machinery, including RPA, XPB, and XPD (15, 47). The virus could use this assembled complex to aid in the fidelity of its replication. The repair aspect also lends an alternative mechanism for the translocation of p53 into the replication centers. Miller and colleagues (31) hypothesize that an existing complex between RPA and p53 is translocated to sites of DNA damage and then disassociated in the presence of single-stranded DNA. HCMV may actively recruit RPA to help in damage control of viral replication or in excision repair of damage to the viral DNA and, by doing so, passively transport p53 into the centers.
The second alternative comes into play when considering the overall effect achieved by infection with HCMV. It is well established that HCMV infection stimulates gene expression of several cellular proteins that aid in transit into S phase and are required for DNA replication in normal cells: proteins like those encoded by the protooncogenes fos, jun, and myc (2, 4), dihydrofolate reductase (involved in nucleotide biosynthesis) (45), thymidine kinase (14), DNA polymerase α (18), ornithine decarboxylase (involved in pyrimidine biosynthesis) (21), PCNA (involved in DNA replication and repair) (10), and cyclins E and B (23). This activation might well trigger a p53-mediated response of either arrest at the G1 damage checkpoint or apoptosis, both of which would defeat the purpose of achieving concerted viral replication and packaging. We hypothesize that viral protein binding and/or sequestering of wild-type p53 in the fibroblasts could be one mechanism that HCMV has evolved to counter the known p53 damage response, while keeping p53 available for protein-protein interactions and nonspecific DNA binding potentially beneficial for viral replication and repair.
The experiments described in this work begin to provide insight into the mechanisms employed by HCMV to increase steady-state levels of p53 in infected fibroblasts. Other studies are currently in progress to elucidate the functional nature of the interactions between p53 and HCMV proteins and to resolve the question of whether p53 and the other sequestered cellular proteins play a role in HCMV replication and repair or are simply sequestered by the virus to block normal cellular functioning.
ACKNOWLEDGMENTS
This work was supported by NIH grant CA34729 and a grant from the University of California Cancer Research Coordinating Committee. E.A.F. was supported by NIH Training Grant T32AI07036.
We are indebted to Jeff Stack for countless helpful discussions and insightful critique of the manuscript. We also thank John O’Dowd and all the members of the Spector laboratory for critical reading of the manuscript.
REFERENCES
- 1.AbuBakar S, Au W W, Legator M S, Albrecht T. Induction of chromosome aberrations and mitotic arrest by cytomegalovirus in human cells. Environ Mol Mutagen. 1988;12:409–420. doi: 10.1002/em.2860120409. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht T, Fons M P, Bologh I, AbuBakar S, Deng C Z, Millinoff D. Metabolic and cellular effects of human cytomegalovirus infection. Transplant Proc. 1991;23:48–55. [PubMed] [Google Scholar]
- 3.Albrecht T, Fons M P, Deng C Z, Boldogh I. Increased frequency of specific locus mutation following human cytomegalovirus infection. Virology. 1997;230:48–61. doi: 10.1006/viro.1997.8467. [DOI] [PubMed] [Google Scholar]
- 4.Boldogh I, AbuBakar S, Deng C Z, Albrecht T. Transcriptional activation of cellular oncogenes fos, jun, and myc by human cytomegalovirus. J Virol. 1991;65:1568–1571. doi: 10.1128/jvi.65.3.1568-1571.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonin L R, McDougall J K. Human cytomegalovirus IE2 86-kilodalton protein binds p53 but does not abrogate G1 checkpoint function. J Virol. 1997;71:5861–5870. doi: 10.1128/jvi.71.8.5861-5870.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bresnahan W A, Boldogh I, Thompson E A, Albrecht T. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology. 1996;224:150–160. doi: 10.1006/viro.1996.0516. [DOI] [PubMed] [Google Scholar]
- 7.Buckbinder L, Talbott R, Velasco-Miguel S, Takenaka I, Faha B, Seizinger B R, Kley N. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature. 1995;377:646–649. doi: 10.1038/377646a0. [DOI] [PubMed] [Google Scholar]
- 8.Debbas M, White E. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- 9.de Bruyn Kops A, Knipe D M. Preexisting nuclear architecture defines the intranuclear location of herpesvirus DNA replication structures. J Virol. 1994;68:3512–3526. doi: 10.1128/jvi.68.6.3512-3526.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dittmer D, Mocarski E S. Human cytomegalovirus infection inhibits G1/S transition. J Virol. 1997;71:1629–1634. doi: 10.1128/jvi.71.2.1629-1634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C, Jr, Butel J S, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 12.Dutta A, Ruppert J M, Aster J C, Winchester E. Inhibition of DNA replication factor RPA by p53. Nature. 1993;365:79–82. doi: 10.1038/365079a0. . (Comments.) [DOI] [PubMed] [Google Scholar]
- 13.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 14.Estes J E, Huang E-S. Stimulation of cellular thymidine kinases by human cytomegalovirus. J Virol. 1977;24:13–21. doi: 10.1128/jvi.24.1.13-21.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottlieb T M, Oren M. p53 in growth control and neoplasia. Biochim Biophys Acta. 1996;1287:77–102. doi: 10.1016/0304-419x(95)00019-c. [DOI] [PubMed] [Google Scholar]
- 16.Grand R J, Owen D, Rookes S M, Gallimore P H. Control of p53 expression by adenovirus 12 early region 1A and early region 1B 54K proteins. Virology. 1996;218:23–34. doi: 10.1006/viro.1996.0162. [DOI] [PubMed] [Google Scholar]
- 17.He Z, Henricksen L A, Wold M S, Ingles C J. RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature. 1995;374:566–569. doi: 10.1038/374566a0. [DOI] [PubMed] [Google Scholar]
- 18.Hirai K, Watanabe Y. Induction of α-type DNA polymerases in human cytomegalovirus-infected WI-38 cells. Biochim Biophys Acta. 1976;447:328–339. doi: 10.1016/0005-2787(76)90056-3. [DOI] [PubMed] [Google Scholar]
- 19.Hollstein M, Sidransky D, Vogelstein B, Harris C C. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 20.Iskenderian A C, Huang L, Reilly A, Stenberg R M, Anders D G. Four of eleven loci required for transient complementation of human cytomegalovirus DNA replication cooperate to activate expression of replication genes. J Virol. 1996;70:383–392. doi: 10.1128/jvi.70.1.383-392.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isom H C. Stimulation of ornithine decarboxylase by human cytomegalovirus. J Gen Virol. 1979;42:265–278. doi: 10.1099/0022-1317-42-2-265. [DOI] [PubMed] [Google Scholar]
- 22.Iwayama S, Yamamoto T, Furuya T, Kobayashi R, Ikuta K, Hirai K. Intracellular localization and DNA-binding activity of a class of viral early phosphoproteins in human fibroblasts infected with human cytomegalovirus (Towne strain) J Gen Virol. 1994;75:3309–3318. doi: 10.1099/0022-1317-75-12-3309. [DOI] [PubMed] [Google Scholar]
- 23.Jault F M, Jault J-M, Ruchti F, Fortunato E A, Clark C, Corbeil J, Richman D D, Spector D H. Cytomegalovirus infection induces high levels of cyclins, phosphorylated RB, and p53, leading to cell cycle arrest. J Virol. 1995;69:6697–6704. doi: 10.1128/jvi.69.11.6697-6704.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenkins J R, Chumakov P, Addison C, Sturzbecher H W, Wade-Evans A. Two distinct regions of the murine p53 primary amino acid sequence are implicated in stable complex formation with simian virus 40 T antigen. J Virol. 1988;62:3903–3906. doi: 10.1128/jvi.62.10.3903-3906.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Juven T, Barak Y, Zauberman A, George D L, Oren M. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene. 1993;8:3411–3416. [PubMed] [Google Scholar]
- 26.Kao C C, Yew P R, Berk A J. Domains required for in vitro association between the cellular p53 and the adenovirus 2 E1B 55K proteins. Virology. 1990;179:806–814. doi: 10.1016/0042-6822(90)90148-k. [DOI] [PubMed] [Google Scholar]
- 27.Kastan M B, Zhan Q, el-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 28.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 29.Li R, Botchan M R. The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell. 1993;73:1207–1221. doi: 10.1016/0092-8674(93)90649-b. [DOI] [PubMed] [Google Scholar]
- 30.Lu M, Shenk T. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J Virol. 1996;70:8850–8857. doi: 10.1128/jvi.70.12.8850-8857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller S D, Moses K, Jayaraman L, Prives C. Complex formation between p53 and replication protein A inhibits the sequence-specific DNA binding of p53 and is regulated by single-stranded DNA. Mol Cell Biol. 1997;17:2194–2201. doi: 10.1128/mcb.17.4.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 33.Moore S P, Erdile L, Kelly T, Fishel R. The human homologous pairing protein HPP-1 is specifically stimulated by the cognate single-stranded binding protein hRP-A. Proc Natl Acad Sci USA. 1991;88:9067–9071. doi: 10.1073/pnas.88.20.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muganda P, Mendoza O, Hernandez J, Qian Q. Human cytomegalovirus elevates levels of the cellular protein p53 in infected fibroblasts. J Virol. 1994;68:8028–8034. doi: 10.1128/jvi.68.12.8028-8034.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muralidhar S, Doniger J, Medelson E, Araujo J C, Kashanchi F, Azumi N, Brady J N, Rosenthal L J. Human cytomegalovirus mtrII oncoprotein binds to p53 and down-regulates p53-activated transcription. J Virol. 1996;70:8691–8700. doi: 10.1128/jvi.70.12.8691-8700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okamoto K, Beach D. Cyclin G is a transcriptional target of the p53 tumor suppressor protein. EMBO J. 1994;13:4816–4822. doi: 10.1002/j.1460-2075.1994.tb06807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pari G S, Anders D G. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J Virol. 1993;67:6979–6988. doi: 10.1128/jvi.67.12.6979-6988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pari G S, Kacica M A, Anders D G. Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J Virol. 1993;67:2575–2582. doi: 10.1128/jvi.67.5.2575-2582.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prelich G, Kostura M, Marshak D R, Mathews M B, Stillman B. The cell-cycle regulated proliferating cell nuclear antigen is required for SV40 DNA replication in vitro. Nature. 1987;326:471–475. doi: 10.1038/326471a0. [DOI] [PubMed] [Google Scholar]
- 40.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 41.Shen Y, Zhu H, Shenk T. Human cytomegalovirus IE1 and IE2 proteins are mutagenic and mediate “hit-and-run” oncogenic transformation in cooperation with the adenovirus E1A proteins. Proc Natl Acad Sci USA. 1997;94:3341–3345. doi: 10.1073/pnas.94.7.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shivji K K, Kenny M K, Wood R D. Proliferating cell nuclear antigen is required for DNA excision repair. Cell. 1992;69:367–374. doi: 10.1016/0092-8674(92)90416-a. [DOI] [PubMed] [Google Scholar]
- 43.Speir E, Modali R, Huang E-S, Leon M B, Sahwl F, Finkel T, Epstein S E. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science. 1994;265:391–394. doi: 10.1126/science.8023160. [DOI] [PubMed] [Google Scholar]
- 44.Sturzbecher H W, Donzelmann B, Henning W, Knippschild U, Buchhop S. p53 is linked directly to homologous recombination processes via RAD51/RecA protein interaction. EMBO J. 1996;15:1992–2002. [PMC free article] [PubMed] [Google Scholar]
- 45.Wade M, Kowalik T F, Mudryj M, Huang E S, Azizkhan J C. E2F mediates dihydrofolate reductase promoter activation and multiprotein complex formation in human cytomegalovirus infection. Mol Cell Biol. 1992;12:4364–4374. doi: 10.1128/mcb.12.10.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X W, Forrester K, Yeh H, Feitelson M A, Gu J R, Harris C C. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc Natl Acad Sci USA. 1994;91:2230–2234. doi: 10.1073/pnas.91.6.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X W, Yeh H, Schaeffer L, Roy R, Moncollin V, Egly J M, Wang Z, Freidberg E C, Evans M K, Taffe B G, et al. p53 modulation of TFIIH-associated nucleotide excision repair activity. Nat Genet. 1995;10:188–195. doi: 10.1038/ng0695-188. [DOI] [PubMed] [Google Scholar]
- 48.Weiland K L, Oien N L, Homa F, Wathen M W. Functional analysis of human cytomegalovirus polymerase accessory protein. Virus Res. 1994;34:191–206. doi: 10.1016/0168-1702(94)90124-4. [DOI] [PubMed] [Google Scholar]
- 49.Werness B A, Levine A J, Howley P M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 50.Wilcock D, Lane D P. Localization of p53, retinoblastoma and host replication proteins at sites of viral replication in herpes-infected cells. Nature. 1991;349:429–431. doi: 10.1038/349429a0. [DOI] [PubMed] [Google Scholar]
- 51.Wold M S, Weinberg D H, Virshup D M, Li J J, Kelly T J. Identification of cellular proteins required for simian virus 40 DNA replication. J Biol Chem. 1989;264:2801–2809. [PubMed] [Google Scholar]
- 51a.Wright D. Ph.D. thesis. La Jolla, Calif: University of California, San Diego; 1989. [Google Scholar]
- 52.Zantema A, Fransen J A, Davis-Olivier A, Ramaekers F C, Vooijs G P, DeLeys B, Van der Eb A J. Localization of the E1B proteins of adenovirus 5 in transformed cells, as revealed by interaction with monoclonal antibodies. Virology. 1985;142:44–58. doi: 10.1016/0042-6822(85)90421-0. [DOI] [PubMed] [Google Scholar]
- 53.Zhu H, Shen Y, Shenk T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J Virol. 1995;69:7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]