Abstract
Papillomaviruses establish a long-term latency in vivo by maintaining their genomes as nuclear plasmids in proliferating cells. Bovine papillomavirus type 1 encodes two proteins required for viral DNA replication: the helicase E1 and the positive regulator E2. The homodimeric E2 is known to cooperatively bind to DNA with E1 to form a preinitiation complex at the origin of DNA replication. The virus also codes for two short forms of E2 that can repress viral functions when overexpressed, and at least one copy of the repressor is required for stable plasmid maintenance in transformed cells. Employing a tetracycline-regulated system to control E1 and E2 production from integrated loci, we show that the short form of E2 negatively regulates DNA replication. We also found that the short form could repress replication in a cell-free replication system and that the repression requires the DNA binding domain of the protein. In contrast, heterodimers of the short and long forms were activators and, by footprint analysis, were shown to be as potent as homodimeric E2 in loading E1 to its cognate site. DNA binding studies show that when E1 levels are low and are dependent upon E2 for occupancy of the origin site, the repressor can block E1-DNA interactions. We conclude that DNA replication modulation results from competition between the different forms of E2 for DNA binding. Given that heterodimers are active and that the repressor form of E2 shows little cooperativity with E1 for DNA binding, this protein is a weak repressor.
The papillomaviruses have an unusual replication cycle in that they infect the dividing basal cells of epithelial tissue but these cells do not produce progeny virus. Rather, the viral DNA maintains itself as a nuclear plasmid in the dividing cells, and true vegetative replication occurs only as these cells move up the stratified layers to those regions where the differentiated cells, surrounding the infected cell, have departed from the cell cycle (15, 40). The viruses have devised a multifaceted and probably diverse strategy for maintaining the infected cells’ ability to replicate DNA despite a differentiation program that should normally end such a capacity. This is accomplished by at least three viral gene products, each of which deregulates crucial cell cycle and/or apoptotic control programs (15). Evidence from human papillomavirus (HPV) supports the notion that the primary infected cell does maintain at least some of its differentiation potential (5) during this passage to virus production.
Beyond the fact that vegetative replication is tied to the emergence of the infected cells into the upper differentiated layers, little is known about how the plasmid mode of replication switches into one that produces viruses. Bovine papillomavirus type 1 (BPV-1) is a particularly robust family member, and it also infects the fibroblasts as well as basal cells present in the lower levels of the tissue. Although plasmids are maintained in these transformed fibroblasts, which often constitute a major fraction of the fibropapilloma, virus particles only assemble in the epithelial cells. A striking illustration of the tissue and cellular specificity of virion production is provided by transgenic mouse strains maintaining integrated tandem copies of BPV-1. In such mice, viral gene expression becomes sporadically activated in the skin of the young ones and viral DNA excision yields cells with multicopy viral DNA circles; however, no virions are detected (19). The viral oncogenic transformation of immortalized murine cell lines has thus provided a valuable system for studying the virus-cell interaction required for plasmid replication (9).
Because of their small size the papillomaviruses, like the other small DNA tumor viruses, must usurp host cell enzymes for much of their DNA replication. Only two viral factors, the E1 and E2 gene products, are required directly as positive factors for plasmid replication in established cell systems (39). The E1 protein is a DNA binding protein that can assemble by oligomerization into a helicase. E2 is the major regulator of the transcriptional program and as such, for the well-studied bovine model, activates transcription in an autocatalytic manner from four viral promoters. E2 also plays a direct role in replication as it helps target E1 to its cognate binding site by cooperative binding with the E1 monomer. Considerable in vitro and in vivo data support the model that heteromeric protein-protein and protein-DNA interactions are critical for creation of this specific E1-E2-DNA ternary preinitiation complex (reviewed in reference 39). Using an in vitro replication system Yang et al. (43) showed that E2 can activate DNA replication at a limiting concentration of E1 and that this activation was not dependent upon RNA polymerase II activity. Interestingly, as shown most clearly by Ustav et al. (41), only one half of the normally palindromic E2 binding site at the origin site is required in cis for viral replication in vivo, but in engineered constructs intact consensus E2 binding sites are required if the E2 sites are to work from remote plasmid situations to lead E1 onto its cognate sites. Somewhat paradoxically, work from our lab showed that E2 could activate E1’s replication activity in vitro even in recombinant origin DNA constructs that did not have virus-encoded E2 binding sites (42). As we show here, this stimulation is largely due to the vector plasmid backbone, whereas in another vector (that does not contain consensus E2 sites) E2 activation in vitro is indeed sensitive to the viral E2 binding sites in cis.
The in vitro replication system utilizing unfractionated cellular extracts has provided the most direct evidence that key cellular proteins are required for viral DNA replication. Thus, DNA polymerase α-primase interacts with E1, and neutralizing antibody to that protein inactivates in vitro replication (3, 34). Similarly, the cellular single-stranded DNA binding protein, called RPA, is critical in such extracts and can be readily depleted (24). It is widely assumed that papillomaviruses utilize, for the most part, the same set of cellular replication factors utilized by simian virus 40. Muller and colleagues (33) have tested this hypothesis and found that replication of BPV-1 DNA with either their monopolymerase (alpha) or dipolymerase (alpha and delta) system required the other well-characterized replication proteins. However, it may be emphasized that the latter experiments show how the papillomavirus DNA might replicate their DNA, and studies by Melendy et al. (31) indicate that there may be interesting divergences with simian virus 40 in the actual factors required in vivo.
BPV-1 and HPV type 11 (HPV-11) have been shown to encode short forms of the E2 protein, and it seems highly likely that these proteins play key roles in the protracted viral latency described above (6, 7, 20, 21). For BPV-1 one of the two mRNAs for the repressor form is transcribed from a promoter internal to the open reading frame (ORF) (E2C, which is also called E2TR by some researchers), and the other is created by splicing to an acceptor located within the E2 ORF (E8/E2). These proteins maintain the DNA binding and dimerization domains of the enhancer protein, but without the activation domain they serve as natural transcriptional repressors. Overexpression of one of these short forms of E2 results in loss of viral transformation and plasmid maintenance (7, 21). For BPV-1 the two promoters for the short forms are themselves activated by E2, and thus the autocatalytic activation of the BPV-1 transcription program is likely held in check by stimulation of transcription of the repressor (27, 42). The importance of the repressors in the regulation of plasmid copy number in stable plasmid replication achieved by the virus is underscored by a key observation. When synthesis of the E2C form of the repressor is blocked by mutation of the start methionine within the E2 ORF, the resulting viral genome establishes in the transformed cells at higher than normal copy number; in contrast, when both repressor forms are eliminated, stable transformation and plasmid maintenance by the mutant is greatly reduced and transformation may rely upon either integration or plasmid rearrangements (22, 23, 36). These data establish that at least one form of the repressor is required for stable plasmid replication in the murine cell system.
The mechanism(s) by which the repressor form of E2 might regulate DNA replication provides an opportunity to probe more directly the model for how E2 itself might activate viral replication. In this study we show that the E2C protein does repress replication both in vitro and in vivo, whereas heterodimers composed of E2 and E2C stimulate replication in vitro. The data are most consistent with a simple competition model wherein E2 and E2C compete for binding to the viral DNA. As E2C shows no cooperative DNA binding interactions with E1 when assayed by DNase I footprint analysis, we conclude that repression is the consequence of the inadequate loading of E1.
MATERIALS AND METHODS
Plasmids.
pKSO has been previously described (43). The FLAG-E2 expression vector (pAcC13-FE2) was cloned from a previously described SP6-E2 expression vector (18) as follows. SP6-E2 was cut with BglII and SphI, and a double-stranded oligonucleotide (top strand, GATCTACCATGGACTACAAGGACGACGATGACAAGGAGACAGCATG; bottom strand, CTGTCTCCTTGTCATCGTCGTCCTTGTAGTCCATGGTA) coding for the FLAG epitope (Kodak, IBI) and the N-terminal four amino acids of E2 (minus the start methionine) was ligated into those sites (creating SP6-FE2). This FE2 fusion was removed from SP6 with BamHI and BglII and ligated to the pAcC13 vector at the BamHI and BglII sites. The GE2C expression vector was cloned into the pAcC13 vector by standard PCR methods; the Glu epitope has been previously described (12). The 339M mutation (35) was cloned from the pCG-E2-339M vector into the GE2C expression vector by using the KpnI and filled-in SacI sites. The WK33 mutation (10, 18) was cloned from the YEpE2B-WK33 expression vector (gift of E. Androphy) into the SP6-FE2 plasmid by using the SphI and KpnI sites; the FE2-WK33 reading frame was transferred to the pAcC13 vector as described above. After generation of the respective transfer vectors the infectious recombinant baculoviruses were selected by cotransfection and plaque selection in Sf9 cells (Baculogold; Pharmingen). pCLO and pCLOT were constructed by inserting the HindIII to BamHI fragment from pKSO or pKSOT, respectively, into these sites in the pACYC177 vector, which lacks E2 binding sites.
Cell culture.
To facilitate the transient-replication assays described below, we generated a stable cell line expressing BPV-1 E1 and E2 genes placed under control of tetracycline-regulated promoters. The principle of this gene expression system as well as the plasmids utilized was described previously (11). CHO cells were transfected with linearized plasmid pUHD15-1neo containing the tTA (tetracycline-controlled transactivator) gene under control of a cytomegalovirus (CMV) promoter and a neomycin selection marker. Stable transformants were selected with G418 (500 μg/ml). Individual clones were isolated and transiently supertransfected in the presence and absence of tetracycline with pUHC13-3, containing a tTA-responsive luciferase reporter gene. The CHO tTA+ clone showing the highest dynamic range of luciferase regulation was used for integration of the E1 and E2 genes. Both genes were cloned into the multiple cloning site of the tTA-responsive expression vector pUHD10-3. The resulting expression vectors, pUHD10-3/E1 and pUHD10-3/E2, were cotransfected with a hygromycin selection marker into CHO tTA+ cells with selection at a hygromycin concentration of 300 μg/ml. The functionality of the stable clones raised was analyzed by their capacity to promote transient replication of pKSO (see above) in a tetracycline-dependent fashion. The cell line chosen for all subsequent experiments was named CHO tTA-E1/E2 #6. The general tissue culture techniques employed, the transient transfections, and the analysis of replicated DNA have been previously described (23). Sf9 cells were maintained in suspension in Grace’s medium with 10% fetal calf serum, 0.1% pluronic acid, 0.33% yeastolate, and 0.33% lactalbumin. Sf9 cells were infected at a density of 5 × 105 cells/ml; the multiplicity of infection was between 3 and 5 for both single infections and coinfections. Infections were allowed to progress for 48 h before harvesting. Cells were pelleted and washed once with phosphate-buffered saline before lysis.
Protein expression and purification.
The expression and purification of GE1 have been previously described (43). GE2C purification was similar to that of GE1, except that the Sf9 cells were lysed whole (omitting the nuclear isolation-extraction step). FE2 purification was essentially the same as that of GE2C, except that α-FLAG M2 monoclonal antibody-linked resin (Kodak, IBI) was used instead of α-Glu antibody. Batch elution from the α-FLAG column was accomplished with 200 μg of FLAG peptide/ml for 1 h at 4°C. Before elution, all columns were washed with at least 75 column volumes of buffer C (25 mM Tris [pH 8], 1 M LiCl, 1 mM EDTA, 10% glycerol) and at least 30 column volumes of buffer D (25 mM Tris [pH 8], 200 mM NaCl, 1 mM EDTA, 10% glycerol). For the purification of FE2-GE2C heterodimers, extracts from coinfected cells were first passed over the α-FLAG resin column; the column was washed and batch eluted as described above. The eluate was then incubated with α-Glu resin at 4°C with rotation. After the described washes, 20 mM triethylamine elution fractions were collected into tubes containing neutralizing volumes of 1 M HEPES, pH 7.5. Proteins were dialyzed against replication buffer (20 mM KPO4 [pH 7.5], 1 mM EDTA, 1 mM dithiothreitol, 150 mM potassium glutamate, 10% glycerol), frozen in liquid N2, and stored at −70°C.
DNase I protection assay.
Assays were performed as previously described with no carrier DNA (43). Protein-DNA mixtures were incubated at 37°C for 15 min; reactions were performed in a total volume of 100 μl at room temperature. The BamHI to EcoRI DNA fragment used for protection studies was derived from pKSO. Top and bottom strands were end labeled separately with 32P in standard polynucleotide kinase reactions. The order of addition of the three proteins did not affect the appearance of the footprints.
In vitro DNA replication.
Assays were performed essentially as previously described (43). A typical reaction contained 50 ng of pKSO, pCLO, or pCLOT template and 10 μl of FM3A extract (5 to 10 mg/ml). The FM3A extract was prepared as previously described (43). Reactions included an ATP regenerating system, deoxynucleoside triphosphates, and [α-32P]dCTP in a total volume of 25 μl. The amounts of GE1, FE2, and GE2C are indicated in the figure legends; the order of addition of the DNA, GE1, FE2, and GE2C did not affect the outcomes of the experiments. The GE1 protein was diluted to working concentrations with FM3A extract. Data shown are representative of reactions that were repeated several times, with different GE1, FE2, and GE2C preparations.
RESULTS
Chiang et al. (6) have reported that truncated forms of HPV-11 E2 protein can negatively regulate transient amplification of HPV-11 DNA. In those experiments four separate recombinant plasmids had to be introduced into the cells (the positive factors provided by the E1 and E2 expression cassettes, the truncated E2 vector, and the HPV-11 origin plasmid) to monitor amplification at various levels of the repressor form. To extend these observations to BPV and to simplify the transfection protocols, we have created a useful cell line that contains integrated copies of the BPV-1 E1 and E2 genes whose expression is in turn regulated by the bacterial tet operator sequences and the trans-acting tTA gene product in response to the tetracyline concentration in the medium (11).
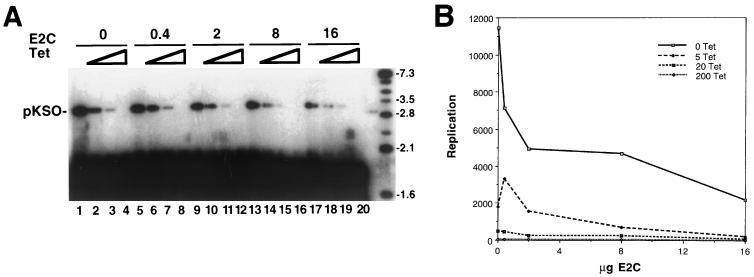
Stable CHO cell lines functionally expressing the tTA gene (coding for a fusion between the tet repressor and the VP16 transcriptional activation domain) were cotransfected with BPV-1 E1 and E2 genes placed under control of a tetracycline-responsive promoter. Positive colonies were selected and screened for their abilities to transiently amplify BPV-1 plasmids in a tetracycline-regulated manner. The advantages of this approach to the study of the E2 repressor are that (i) the complexity of the transient-transfection protocol is reduced, as E1 and E2 are synthesized from stably integrated copies of their genes in a clonal cell line; (ii) the efficiency of cotransfecting the two plasmids required, the origin-containing reporter and the expression vector for E2C, can be close to 100%; and (iii) the expression of E1 and E2 genes can not only be switched on or off but can also be adjusted to intermediate levels by changing the concentration of tetracycline in the culture medium. Using this system, we could turn replication on or off as illustrated in the following in vivo replication experiment. Figure 1 (lanes 1 to 4) shows that the BPV-1 recombinant plasmid pKSO (43) will amplify in such cells when tetracycline is removed from the medium (facilitating E1 and E2 expression). Replication progressively decreased with increasing concentrations of tetracycline, and no DNA accumulation was detected when the tetracycline concentration in the medium was 200 ng/ml. Thus, the activity levels of the BPV-1-encoded replication factors are regulated by the drug.
FIG. 1.
Replication of BPV is regulated by E1 and E2 levels and is negatively modulated by E2C in vivo. The pKSO reporter was transfected into CHO cells that have E1 and E2 genes regulated by tetracycline. The amplification of the reporter under all conditions was monitored by DNA blot analysis at 3 days posttransfection after DNAs were linearized and treated with DpnI to degrade unreplicated reporter. (A) Autoradiogram of the blot hybridized with labeled pKSO probe. Triangles indicate increasing concentration of tetracycline in the medium, i.e., 0, 20, 50, and 200 ng/ml for lanes 1, 2, 3, and 4, respectively. This order was followed for each set of experiments with an increased E2C concentration. The amounts (in micrograms) of the pCMVE2C vector used are shown at the top. The levels of E1 and E2 in this expression system are inversely related to the tetracycline concentration. Repression under equivalent tetracycline induction can be followed, for example, by comparing the signals in lanes 1, 5, 9, 13, and 17. Markers in rightmost lane are molecular size standards. Numbers indicate sizes in kilobases. (B) Replication measured in arbitrary units was plotted by using PhosphorImager readouts from the linear DNA positions shown in panel A. The data show how the signals compared under identical tetracyline levels (0 to 200 ng of tetracycline [Tet]/ml) as a function of input E2C vector. Similar data were obtained in another independent experiment (data not shown).
The repressor construct encoding BPV-1 E2C at different concentrations was cotransfected with the pKSO reporter, and we monitored the level of amplification concomitant with different levels of tetracycline. (In lanes 1 to 20 the total levels of the CMV promoter were kept constant by addition of the empty CMV expression cassette to compensate for various levels of the CMV-E2C plasmid in the transfection mix.) At high levels of the repressor vector a 5- to 10-fold repression of replication could be measured (Fig. 1B). In this experiment the best repression was obtained at intermediate levels of tetracycline or when the E1 and E2 proteins were not fully induced (compare lanes 2 and 18). The high level of expression of the E2C protein obtained with the CMV vector did not affect cell growth, and thus, the effects observed could not be attributed to E2C-mediated toxicity or cell cycle arrest; we also note that the E2 expression cassette does not produce detectable levels of the E2C protein in transient assays or from its integrated position (data not shown). We conclude from these experiments that the E2C protein can negatively modulate BPV-1 transient replication but that it is not a very potent replication repressor.
Heterodimers of E2-E2C are activators of in vitro DNA replication.
One reasonable mechanism by which the E2C protein might repress BPV-1 DNA replication is through formation of inactive heterodimers with the full-length E2. While dimers of E2 are stable against dissociation even at picomolar concentrations, it has been previously shown that heterodimers of short and long fragments of E2 can be created by cotranslation of the respective mRNA in vitro. Moreover, while mixing of the short and long forms does not create heterodimers, denaturation in 5 M urea followed by subsequent dialysis does result in heterodimeric species (30).
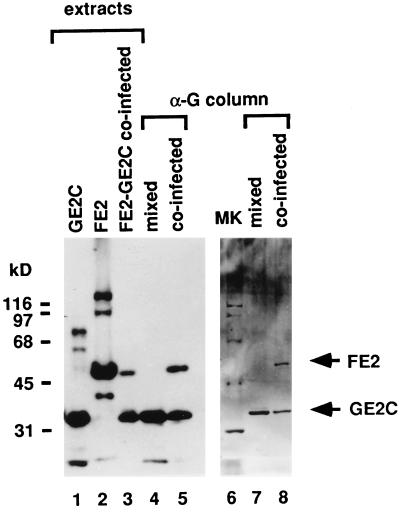
To explore this possibility we sought to purify E2-E2C heterodimers and to assay their potential for replication activation side by side with full-length E2 or E2C homodimers. To achieve this goal two different sorts of recombinant baculoviruses were created. The first virus (FE2) expresses a full-length E2 protein fused to the FLAG epitope (14), and the other (GE2C) encodes an E2C protein fused to the Glu epitope (12). Figure 2, lanes 1 to 3, shows the proteins in the crude extracts of infected Sf9 cells as detected in a Western blot with E2 monoclonal antibody. The cells were infected with either GE2C or FE2 separately or were coinfected with both viruses. The extracts from the coinfected cells were initially passed over an α-FLAG monoclonal antibody column, washed extensively, and batch eluted with FLAG peptide. The eluate containing the peptide, FE2 homodimers, and FE2-GE2C heterodimers was then incubated with the α-Glu resin; the resin was transferred to a column and washed extensively, and the remaining affinity-bound FE2-GE2C heterodimers were eluted and collected. Western blot and silver-stained analyses of this heterodimer preparation are shown in Figure 2, lanes 5 and 8, respectively. As a control, extracts from Sf9 cells infected singly with either GE2C or FE2 were mixed in equal proportion and then passed through the α-G agarose resin. After washes identical to those for the coinfected material, the eluted proteins were visualized by Western blotting (Fig. 2, lane 4) and by silver staining the material on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (Fig. 2, lane 7). In these circumstances the FLAG-tagged E2 could not be detected.
FIG. 2.
Purification of FE2-GE2C heterodimers. Full-length FE2 is not retained on the α-Glu (α-G) monoclonal antibody column. Lanes 1 to 5 show an autoradiographic film of an SDS–12% PAGE enhanced chemiluminescence Western blot probed with the E2-specific B202 monoclonal antibody. Lanes 6 to 8 show a silver-stained SDS–12% PAGE gel containing purified preparations. Whole-cell extracts of singly infected Sf9 cells expressing GE2C (lane 1) and FE2 (lane 2) and of coinfected cells expressing both FE2 and GE2C (lane 3) are shown. Some unreduced protein appears in the extract preparation as well as degradation products of E2, but in all cases the major band in the extracts is either E2 or E2C. The purified material is homogeneous. Proteins retained by the α-Glu column from a mixture of GE2C and FE2 extracts (lanes 4 and 7) and from an FE2-GE2C coinfected extract (lanes 5 and 8) are also shown. Arrows indicate the mobilities of FE2 and GE2C. Lane 6, molecular mass standards (in kilodaltons shown at left).
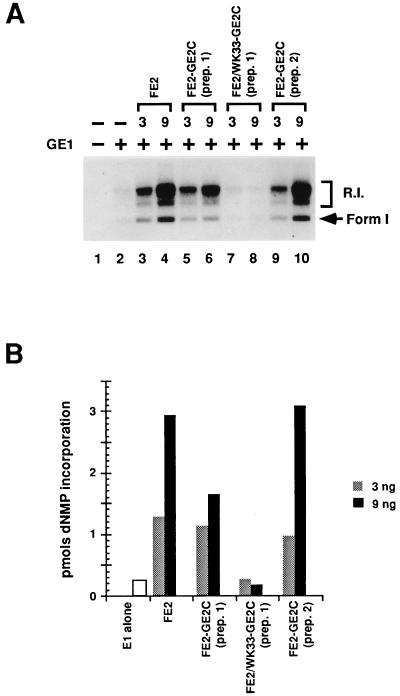
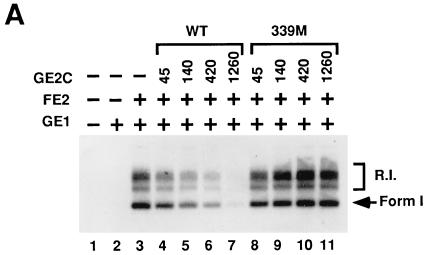
FE2-GE2C heterodimers and FE2 homodimers were tested in parallel in the BPV-1 in vitro DNA replication assay. As shown in Fig. 3, the heterodimers were as potent as the homodimers of E2 in activating E1-dependent DNA replication. Two independent heterodimer preparations were compared to the homodimers, and synthesis levels were quantitated (Fig. 3B). The heterodimer preparations showed equivalent activation compared to untagged E2, and homodimers of GE2C showed no activation of in vitro DNA replication (see below and data not shown). Furthermore, as the titrations of the heterodimers and the homodimers were essentially identical (Fig. 3 and data not shown), these experiments are not consistent with the counterargument that trace quantities of homodimers serve as activators. As a control, heterodimers of GE2C with a FLAG-tagged full-length E2 mutant, WK33, were prepared. The WK33 mutation of E2 (in the amino-terminal activation domain which is missing in E2C) has been shown to block replication function in cells (10), and homodimers of this protein are inactive in vitro (28). The heterodimer FE2/WK33-GE2C was inactive in the assays, establishing that the single intact activation domain in the “wild-type” heterodimer is critical for activation.
FIG. 3.
Heterodimer preparations of FE2-GE2C stimulate in vitro replication. (A) Autoradiogram detecting in vitro DNA replication products fractionated by agarose gel electrophoresis. The amount of GE1 in the indicated reactions (+) was 125 ng per standard reaction volume. The amount (in nanograms) of homo- or heterodimer preparation added to each reaction is indicated in the top row. Supercoiled DNA products (arrow, Form 1) and replication intermediates (R.I.) are indicated at the right. Lane 1, background extract synthesis; lane 2, E1-only replication. (B) PhosphorImager quantitation of the data in panel A. The total number of counts for each lane was obtained and converted to picomoles of synthesis by comparison to a standard. dNMP, deoxynucleoside monophosphate.
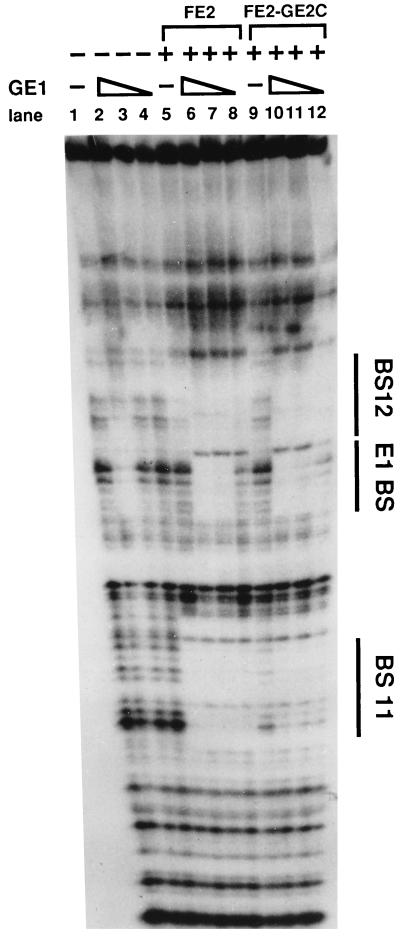
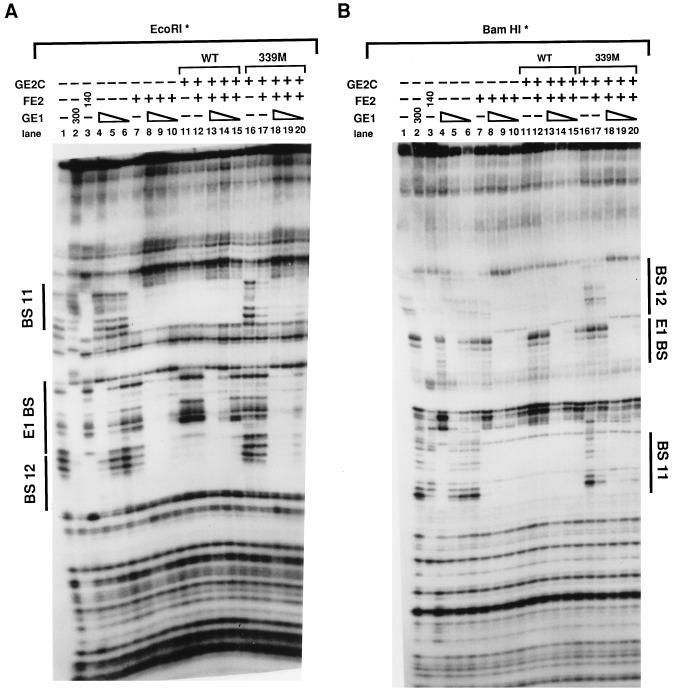
To probe these conclusions further we asked if heterodimers of FE2-GE2C could cooperatively bind with the E1 protein at the origin site as the homodimers of full-length E2 do. Serial dilutions of E1 protein were included in binding buffer with origin DNA fragments either alone or in combination with homodimer or heterodimer E2 protein. The DNA-protein complexes were then analyzed by DNase I footprint analysis. Figure 4 shows that the heterodimers bound slightly more weakly than did the E2 homodimers by themselves but that E1 stimulates binding to a similar extent. There is also a slight difference in the size of the footprint at BS11 (compare lanes 5 and 9). We can only speculate about the reasons why the heterodimers and homodimers display these small differences. One possibility is that the activation domains of the homodimers stabilize each other, resulting in less interference with binding, and that E1 can also stabilize the single E2 activation domain of the heterodimer. In any case, as shown in Fig. 4, the E2-E2C heterodimer and the E2 homodimer show equivalent cooperativity with E1 in DNA binding. With different preparations of E2 proteins we obtained qualitatively similar results via gel shift assays.
FIG. 4.
FE2-GE2C heterodimer preparations enhance E1 DNA binding at the origin. The results of a DNase I protection assay using the BPV-1 origin fragment (nucleotides 7805 to 100) are shown. BS11 and BS12 are the E2 binding sites that flank the E1 binding site (E1BS). The triangles indicate serial dilutions of GE1 (100, 30, and 10 ng). Standard DNase I digestion reactions were carried out in the presence (+) and absence (−) of FE2 (40 ng) or FE2-GE2C (50 ng). A characteristic of the E2-E1 complex on this strand is the DNase I hypersensitive site between BS12 and E1BS; this site is not found in the E1-only footprint.
Homodimers of E2C inhibit E1-E2-dependent replication in vitro.
The implication of the above-described result is that the active form of the repressor is the homodimer of E2C. Liu et al. (29) have previously shown that in vitro DNA replication of HPV-11 origin templates can be inhibited slightly (less than twofold) by HPV-11 E2C protein preparations. To extend and confirm these conclusions we asked if E2-dependent in vitro DNA replication could be repressed by homodimers of the E2C protein and compared this data to a purified homodimer E2C repressor that harbors a mutation (339M) in the alpha helix of the DNA binding domain of E2 (1, 13). This latter protein was created by transferring the 339M mutation previously characterized in the full-length E2 protein (28) to the GE2C gene in a recombinant baculovirus (GE2C-339M). This mutation inactivates DNA binding but leaves dimerization unaltered. As expected neither the GE2C or GE2C-339 protein affected E1-mediated replication at any concentration of E1 or repressor protein tested when only these proteins were present in the reaction (data not shown).
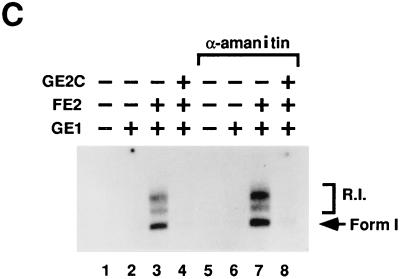
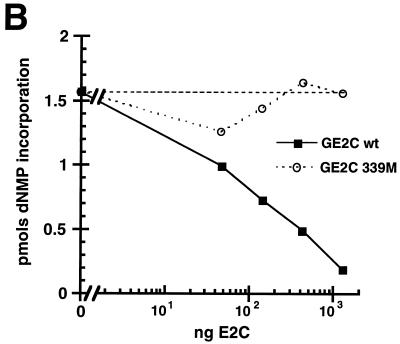
The data presented in Fig. 5 clearly show that the E2C homodimer can repress DNA replication when synthesis is dependent upon the activator form of E2. Moreover, this repression requires the DNA binding domain of the repressor. Interestingly, to achieve a 50% inhibition, a 10-fold molar excess of E2C to E2 is required. This perhaps reflects the differences in cooperativity of binding with the E1 protein between the repressor form and the full-length form of E2 (see below for further discussion). To essentially shut off DNA synthesis in this reaction a 100-fold excess of the short forms is required (Fig. 5B). E2-activated BPV-1 DNA replication in vitro does not require PolII transcription, but E2C has been shown to repress gene expression in vivo. Therefore it was necessary to show that RNA polymerase transcription did not play a role in the repression detected. DNA replication reactions were thus assembled in the absence (Fig. 5C, lanes 1 to 4) and presence (Fig. 5C, lanes 5 to 8) of the transcription inhibitor, α-amanitin. Inhibition of DNA replication by GE2C was found to be independent of the effect of the drug.
FIG. 5.
E2C homodimers inhibit E1-E2-dependent replication in vitro. (A) Autoradiogram detecting in vitro DNA replication products after gel electrophoresis of the products. The amount of GE1 in the indicated reactions (+) was 80 ng per standard reaction volume. The amount of FE2 in the indicated reactions (+) was 10 ng. The titrations of wild-type (WT) E2C and mutant 339M protein are noted at the top of the panel in nanograms. (B) PhosphorImager quantitation of replication data from panel A. The horizontal dashed line indicates the level of E1-E2 replication shown in panel A, lane 3. dNMP, deoxynucleoside monophosphate. (C) E2C inhibition of replication is resistant to α-amanitin. Detection of replication products in vitro was performed as described above. A total of 100 μg of α-amanitin/ml was added to each of the indicated reactions. The amounts of GE1 and FE2 in the indicated reactions (+) were identical to those in panel A; the amount of GE2C in the indicated reactions (+) was the same as that for panel A, lane 7. R.I., replication intermediates; Form 1, supercoiled DNA products.
An important observation made was that inhibition mediated by E2C was critically dependent not only on E2 levels but on E1 concentration. At threefold greater GE1 concentrations than that used in the reactions for Fig. 5, at which point replication becomes E2 independent, the net DNA synthesis was resistant to E2C repression in the range of titration shown for the repressor (data not shown). At threefold lower E1 concentrations no net synthesis was detected even in the absence of E2C. Thus, only at concentrations of E1 at which the replication was dependent upon the presence of E2 protein was inhibition by GE2C observed. These data are thus consistent with the model that E1 and E2 cooperative binding is crucial for efficient DNA replication and that E2C competes with this preinitiation complex by occupying cognate E2 sites. To test this point more directly we asked if E2C could interfere with E1 and E2 interaction on the DNA and if this competition is sensitive to E1 levels.
The experiments shown in Fig. 6 were designed to explore the proposal that E2C could block E1 binding if the latter protein’s DNA occupancy is dependent upon E2. DNase I protection experiments with origin templates were performed by assaying binding as shown by the protection patterns on the bottom strand in Fig. 6A and on the top strand in Fig. 6B. Serial dilution of E1 alone (lanes 4 to 6) compared to a parallel dilution in the presence of E2 (lanes 8 to 10) shows the end point of binding is enhanced by the addition of E2 protein. However, in the presence of E2C the same concentrations of E2 and E1 showed a reduced E1 binding as measured by less E1 protection at the origin palindrome (lanes 13 to 15). Specifically, in lanes 9 and 10, E1 protection of its recognition site was dependent upon E2 proteins; at these threshold concentrations of E1, addition of GE2C at an approximately 10-fold excess to E2 prevented E1 binding (lanes 14 and 15 show no stimulation over lanes 5 and 6, which contain E1 alone). To show this we needed to have GE2C included at a 10-fold excess to E2 to offset the DNA binding cooperativity of E1 and E2. However, at the highest E1 concentration, protection of the E1 binding site was not appreciably affected by GE2C, even at this 10-fold excess of repressor (compare lanes 4, 8, and 13). As a control we showed that the mutant form of the repressor GE2C-339M does not affect the formation of the E1-E2-DNA ternary complex, and at all E1 concentrations tested this protein does not interfere with E1 occupancy at the origin site. In this experiment mixing the mutant E2C with the wild-type E2 has a very small inhibitory effect upon E2 site 12 occupancy (compare lanes 7 and 17) perhaps due to nonspecific protein interactions, but in the presence of E1 this effect is not detectable. These data in sum thus establish that E2C can prevent the binding of E1 to the origin site when the initiator is absolutely dependent upon the E2 enhancer protein for loading.
FIG. 6.
E2C can prevent the assembly of E1-E2 complexes at the origin. E1 and E2 occupancy of cognate sites was assayed by DNase I protection assay of the BPV-1 origin. End-labeled fragments were obtained for the bottom (A) and top (B) strands. Thus, results shown in panel B can be compared to those in Fig. 4. E1 and E2 binding sites are labeled as in the legend for Fig. 4. The triangles indicate serial dilutions of GE1 (100, 30, and 10 ng). Numbers in the rows above the lane numbers indicate nanogram amounts of protein. The amount of FE2 in the indicated reactions (+) was 20 ng; the amount of GE2C in the indicated reactions (+) was 200 ng.
E2 activation of BPV-1 DNA replication in vitro requires E2 DNA binding.
Previous data from our laboratory had shown that the in vitro DNA replication of BPV-1 could be activated by E2 in a manner that did not require flanking viral E2 binding sites in the templates (24, 34). It was found that activation showed little or no dependence upon mutations in these sites. One possible explanation for these results (28) was that the protein-protein interactions between E1 and E2 are dominant contributors to the ΔG driving the ternary complex. Thus E2-DNA interactions would not contribute to, or perturb when mutated, the equilibrium of the ternary complex on naked DNA substrates. While this thought may have some application, it would seem that to achieve repression as described above with the E2C protein, competition for some defined E2 binding sites would be required. This seems particularly true as an intact DNA binding domain of the repressor is required for achieving effective negative regulation. We therefore asked if a consensus E2 binding site present in the vector based on the ColE1 origin of replication might be responsible for this paradoxical set of data. We initially tried to mutate the E2 site in the vector (overlapped by the HgiEII site at nucleotide 1739 of pBluescript), but these attempts were unsuccessful, apparently because the sequences in this region are required for propagation in Escherichia coli (34a). To circumvent this problem we then searched for an E. coli vector that did not contain a consensus E2 binding site, and we found by computer scanning that prokaryote plasmid vectors based upon the p15A origin were suitable for these purposes. We therefore transferred the BPV-1 origin fragments from the templates pKSO and pKSOT to the vector pACYC177 (4), which replicates via this origin. pKSO contains BPV-1 sequence that spans both E2 binding sites 11 and 12, while the 50-bp viral sequences in pKSOT (for origin tiny) do not contain these sites. When these viral sequences were placed in the pACYC vector (resulting in the plasmids pCLO and pCLOT, respectively) the E2 activation effects for E2-positive or E2-negative binding site templates were markedly different. In fact these experiments do not prove that it was indeed the single E2 site at nucleotide 1739 in the pBR-based vectors that explicitly underlies the differences. However, they do show that the choice of recombinant vector backbone does influence the results and that with the appropriate vector E2 activation is indeed dependent upon cis-acting viral E2 binding sites.
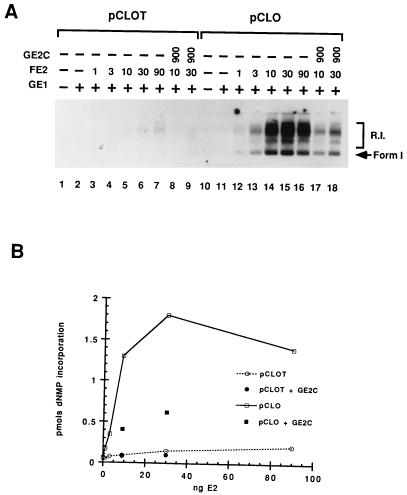
Figure 7 shows that E2 activates the E1-limited in vitro DNA replication of pCLO 10- to 20-fold; in contrast pCLOT shows only marginal activation (less than 2- to 3-fold over the same range of E2 titration). Together with the other results in this report we must conclude that E2 DNA binding to its recognition motifs is important for activation in the in vitro system. In vitro these sites may either be provided by viral-encoded cognate sites (pKSO or pCLO) or vector backbone sites (pKSOT). Consensus sites as in pBR322 or nonconsensus and putatively weak E2 binding sites (those that have, for example, half-sites) contribute as might be anticipated by their affinities. Results shown in Fig. 7 indicate that an excess of E2C can down-regulate replication on the pCLO template (compare lanes 14 and 15 to 17 and 18). Quantitation of these replication products by PhosphorImager analysis shows that deletion of the E2 DNA site lowers E2 activation by 5- to 10-fold and subsequent E2C repression (lanes 5, 6, 8, and 9) is also lower for pCLOT than for pCLO. We should emphasize that where efficient replication is clearly dependent on an E2 site, E2C represses, but that the low level of replication activation detected in the vector without such an E2 site precludes a reliable quantitative measure of repression. We can only infer that nonconsensus E2 sites lead to the marginal activation and repression observed for the pCLOT templates.
FIG. 7.
E2 activation of DNA replication is dependent upon E2 binding sites in BPV or vector DNA. (A) Autoradiogram of fractionated DNA products from in vitro replication assays. The DNA template used in the reactions is indicated above the brackets. pCLOT does not contain the E2 binding sites. The amount of GE1 in the indicated reactions (+) was 80 ng. The amounts (nanograms) of FE2 and GE2C are indicated in their respective rows. R.I., replication intermediates; Form 1, supercoiled DNA products. (B) PhosphorImager quantitation of replication products shown in panel A. dNMP, deoxynucleoside monophosphate.
DISCUSSION
Dimers of the short form of the E2 protein (E2C) can compete with the intact activator for occupancy at cognate E2 binding sites. Our data show that this competition can effectively lead to lower occupancy by E1 at its cognate site because E1 DNA binding by the E1 helicase at low concentrations is dependent upon protein-protein interactions mediated by the activation domain of E2. We do not interpret the repression of DNA replication by E2C or the footprint analysis to mean that E2C activity directly blocks E1 from binding to its separate site, for example, by steric exclusion. We thus postulate that in the in vivo situations where E2C has indeed been shown to possess direct replication repression ability, the E1 levels are low enough such that E2C can interfere with the cooperative binding of activating forms of E2 dimers and the E1 helicase to the origin site. This is consistent with our findings that repression by E2C in the in vitro DNA replication system is only manifest when the E1 levels are low enough to render the reaction absolutely dependent upon E2 and with the results from our in vivo transient-replication experiments. Whereas the repressor can downregulate replication levels almost 10-fold at lower E1-E2 levels, replication driven by fully expressed E1 and E2 genes is less susceptible to even high levels of E2C (see, for example, Fig. 1). E2C is therefore a novel type of DNA binding repressor in the sense that it blocks the binding of an important DNA site-specific mediator of DNA metabolism (E1) by blocking the binding of a required accessory factor (E2) or, alternatively, of a preassembled E1-E2 complex.
These notions are consistent with recent observations by Berg and Stenlund (2), who have shown that E2C can cooperatively bind with E1 to the origin site but with limited cooperativity. These researchers, who used an immunoprecipitation assay to measure binding, showed that E1 and E2 bound with a 30-fold cooperativity but the cooperative interaction between E2C and E1 led only to a 3- to 4-fold higher level of E1 binding. In contrast to Berg and Stenlund we have not been able to ever detect any cooperative binding between E1 and E2C in our equilibrium DNase I footprint analysis (28a, 43). However, this may be due to the lower levels of cooperativity that we detect in general for E1 and E2 complexes. E2C occupancy of DNA templates (compared to E2 binding) therefore leads to a relatively lower occupancy of the initiator and consequently down-modulates replication.
Our data show that heterodimers of E2 and E2C are activators of DNA replication in the in vitro reactions. With multiple preparations, heterodimers activated DNA synthesis in an equivalent manner to that of E2 homodimers. Moreover, the cooperative binding to DNA between E1 and E2 only required a single E2 activation domain. This raises interesting structural possibilities for study of the E1-E2-DNA ternary complexes in that the other unoccupied activation domain of E2 may be able to effectively make contacts with proteins important in other functions of E2. E2C has been shown to repress E2-activated transcription in vivo, but a similar in vitro analysis of heterodimers for transcriptional activation has not been possible. Previously Barsoum et al. (1) speculated that E2C-E2 heterodimers might be repressors of transcription, as mutants of E2C in the DNA binding domain of the short form could still repress transcription while mutations in the dimerization domain were inactive. E2 binding to DNA half-sites of the cognate recognition DNA sequence is known to be severely limited (see, for example, reference 26), and we would speculate that heterodimers in which one subunit contains a mutated DNA binding domain would also possess reduced affinity for DNA. However, we have found that a heterodimer containing only one nonmutated DNA binding domain still activated replication (data not shown). Thus, interpretations of in vivo studies with specifically mutated E2C vectors may not reflect the activity of wild-type heterodimers. The ratio of heterodimers to dimers of E2 in virally transformed cells is not known and has not been addressed. This number is difficult to obtain given the low abundance of all forms, and it is reasonable to expect that a single protease cleavage in the hinge region of E2 in the homodimer could create a heterogeneous population of pseudoheterodimers. Nevertheless in the baculovirus overexpression system all forms are produced as predicted by mass action and random assortment. Clearly the in vivo half-lives of the different forms in transformed cells could be substantially different. In any case the structural and functional implications of our in vitro data are that a single E2 activation domain is sufficient for replication functions.
In the steady state, transformed cells have a 7- to 10-fold excess of repressor forms to the E2 enhancer protein as measured by SDS-PAGE analysis (16), and even in S phase when the viral DNA replicates there is a twofold abundance of E2C to E2 (44). Assuming that dimer formation is equally efficient for the various forms of E2, this would result in only ∼45% of the total E2 dimers being the repressive form of E2 (i.e., E2C homodimers). Taking these points into consideration and the low levels of E2C homodimers and the cooperativity of E1 and E2 as well as extrapolating from our in vitro results that E2-E2C heterodimers are activators for DNA replication, it is likely that E2C only acts as a poor repressor, serving to modulate or to set the copy number rather than to shut off DNA synthesis. This is consistent with the finding that transient replication mediated by viral genomes containing mutated repressors is found to be higher than wild-type replication. Curiously, mutations in both repressor forms create viral genomes that do not replicate in the cell to higher levels than do genomes harboring a singly mutated repressor (23a, 32). Yet high-copy-number plasmids cannot be stably maintained in the double-mutant case. Such “overreplication” could lead to cell death or apoptosis. Overexpression of E2 has been found to be cytostatic (8, 17), and we have observed similar and synergistic effects with overexpression of E1 and E2 in the C127 cells usually employed for transformation assays (11a). Alternatively, heterodimers or repressor forms may have unknown and pivotal roles in stable plasmid maintenance in oncogenically transformed cells.
The observations that E2C could repress in vitro DNA replication and that this repression was dependent upon the DNA binding domain of the repressor led us to reexamine the role of E2 DNA binding sites for the cell-free system. This is because we could not understand in any simple way how competition between E2 and E2C in an E1-concentration-dependent manner could lead to repression unless specific and defined E2 DNA binding sites were involved. The data presented in Fig. 7 indicate but do not prove that a consensus E2 sequence (ACCACCGCTGGT) in the plasmid DNA backbone is sufficient to account for most of the E2 activation previously reported by our group when we utilized BPV-1 templates mutated or deleted for cognate viral E2 sites. We posit that when such ColE1 vectors are transfected into cells, histone octamers occupy those distal vector-encoded E2 sites. The binding energy provided by the E1-E2-DNA ternary complex with the large concomitant loop structure would not be sufficient for competition with histones. In contrast even with E2 half-sites appropriately placed close to the cognate E1 site the binding energy is sufficient for such occupancy. Such a model is also consistent with previous in vivo studies from Ustav et al. (41) who showed that multiple tandem distal E2 sites are required to provide replication activation; in contrast single weak sites located closer to the E1 docking position are sufficient for replication activation. Alternatively, many more subtle differences in the plasmid backbones may account for the disparate results obtained in vitro with regard to the requirements for a cis-acting viral E2 binding site. In any case our data show that with the appropriate vector apparent differences between in vivo and in vitro results disappear.
Li and Botchan (25) showed that when BPV-1 templates were coated with histone octamers a proximal E2 DNA binding site was indeed required for in vitro viral DNA replication. Those data led us to suggest that E2 might play an additional role in viral replication in that it allows for E1 occupancy on chromatin templates. We did not conclude that this chromatin function of E2 was sufficient for replication enhancement but indicated that this antirepressor activity was a simple consequence of protein-protein interaction between E1 and E2. This notion remains a viable hypothesis and could indeed be mediated by one of the E2 dimer’s two activation domains not essential for association with E1.
We believe that our present in vitro studies also shed some light on the mechanism of E2 activation. Sedman and Stenlund (37) have previously challenged the notion that E2 activation of BPV-1 DNA replication is due solely (or mainly) to the cooperative DNA binding interactions of E1 and E2 at the origin. They argued that though E2 could activate DNA replication at limiting E1 concentrations in vitro, in the cell E2 was absolutely required and the cooperativity in DNA binding was only 10- to 30-fold in any case. They proposed that E2 serves as a specificity factor for E1. This was perhaps supported by the observation that E2 became absolutely required for DNA replication in vitro over a range of E1 concentrations when competitor DNA was added to the reaction. We, however, would suggest that adding competitor to the reaction is a very effective way of lowering the E1 concentration as the protein binds to nonproductive sites in proportion to the concentration of such sequences. Therefore, the point derived from earlier studies (43) is that the effective concentration of E1 in the cell or in extracts in which competitor is added is low. These different points of view are not merely semantic, as one might presuppose that E2 could act to specifically decrease the binding of E1 to noncognate sites. Thus E2 could help transfer E1 from nonproductive sites, either by intra- or interstrand transfer, to the DNA origin site where oligomerization to an active helicase could occur. To discuss specificity as a thermodynamic parameter one must consider the ratio Kequilibrium specific/ Kequilibrium nonspecific, where both the numerator and denominator are independent terms. E2 has been shown to clearly increase the DNA binding of E1 at the origin site, and thus, the numerator of the ternary complex is greater than the numerator in the parallel ratio for the “E1-only” complex. We point out that comparing the Kequilibrium specific/Kequilibrium nonspecific ratios for these two different DNA protein structures is still the way to describe and compare the specificities of the complexes even if the precise protein compositions are different. No direct evidence has been provided to show that E2 actually decreases the E1 equilibrium for binding at nonconsensus E1 sites. In fact data show that E2 does not influence measurable E1 helicase activity on templates that do not contain cognate E1 sites (38). Furthermore, as we have shown here, E2C does not repress replication by increasing E1’s affinity for nonproductive association. For if it did, E2C would repress E1-only activity. Thus we are driven to propose, in conclusion, that the special “specificity” role of E2 in its activation function for DNA replication works by enhancing E1 occupancy at the origin site and that E2C blocks this cooperative interaction.
ACKNOWLEDGMENTS
We appreciate technical assistance from Jeff Lee and Sun Y. Kim. Thanks also go to members of the Botchan lab for helpful discussions.
M.G. was supported by EMBO postdoctoral fellowship ALTF644-1993; C.W.L. was supported in part by NIH postdoctoral fellowship CA09041 to the Cancer Research Lab, University of California, Berkeley. This work was supported by Public Health Service grants CA42414 and CA30490 from the NIH to M.R.B.
REFERENCES
- 1.Barsoum J, Prakash S S, Han P, Androphy E J. Mechanism of action of the papillomavirus E2 repressor: repression in the absence of DNA binding. J Virol. 1992;66:3941–3945. doi: 10.1128/jvi.66.6.3941-3945.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg M, Stenlund A. Functional interactions between papillomavirus E1 and E2 proteins. J Virol. 1997;71:3853–3863. doi: 10.1128/jvi.71.5.3853-3863.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonne-Andrea C, Santucci S, Clertant P, Tillier F. Bovine papillomavirus E1 protein binds specifically DNA polymerase α but not replication protein A. J Virol. 1995;69:2341–2350. doi: 10.1128/jvi.69.4.2341-2350.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng S, Schmidt-Grimminger D C, Mumat T, Broker T R, Chow L T. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 1995;9:2335–2349. doi: 10.1101/gad.9.19.2335. [DOI] [PubMed] [Google Scholar]
- 6.Chiang C M, Dong G, Broker T R, Chow L T. Control of human papillomavirus type 11 origin of DNA replication by the E2 family of transcriptional regulatory proteins. J Virol. 1992;66:5224–5231. doi: 10.1128/jvi.66.9.5224-5231.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choe J, Vaillancourt P, Stenlund A, Botchan M. Bovine papillomavirus type 1 encodes two forms of a transcriptional repressor: structural and functional analysis of new viral cDNAs. J Virol. 1989;63:1743–1755. doi: 10.1128/jvi.63.4.1743-1755.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dowhanick J J, McBride A A, Howley P M. Suppression of cellular proliferation by the papillomavirus E2 protein. J Virol. 1995;69:7791–7799. doi: 10.1128/jvi.69.12.7791-7799.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dvoretzky I, Shober R, Chattopadhyay S K, Lowy D. A quantitative in vitro focus assay for bovine papilloma virus. Virology. 1980;103:369–375. doi: 10.1016/0042-6822(80)90195-6. [DOI] [PubMed] [Google Scholar]
- 10.Ferguson M K, Botchan M R. Genetic analysis of the activation domain of bovine papillomavirus protein E2: its role in transcription and replication. J Virol. 1996;70:4193–4199. doi: 10.1128/jvi.70.7.4193-4199.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Gossen, M., J. Lee, and M. R. Botchan. Unpublished observations.
- 12.Grussenmeyer T, Scheidtmann K, Hutchinson M A, Eckhart W, Walter G. Complexes of polyoma virus medium T antigen and cellular proteins. Proc Natl Acad Sci USA. 1985;82:7952–7954. doi: 10.1073/pnas.82.23.7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hedge R S, Grossman S R, Laimins L A, Sigler P B. Crystal structure at 1.7 A of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target. Nature. 1992;359:505–512. doi: 10.1038/359505a0. [DOI] [PubMed] [Google Scholar]
- 14.Hopp T P, Prickett K S, Price V L, Libby R T, March C J, Cerretti D P, Urdal D L, Conlon P J. A short polypeptide marker sequence useful for recombinant protein identification and purification. Bio/Technology. 1988;6:1204–1210. [Google Scholar]
- 15.Howley P M. Papillomavirinae: the viruses and the replication. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 947–978. [Google Scholar]
- 16.Hubbert N L, Schiller J T, Lowy D R, Androphy E J. Bovine papilloma virus-transformed cells contain multiple E2 proteins. Proc Natl Acad Sci USA. 1988;85:5864–5868. doi: 10.1073/pnas.85.16.5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang E S, Naeger L K, DiMaio D. Activation of the endogenous p53 inhibitory pathway in HeLa cervical carcinoma cells by expression of the bovine papillomavirus E2 gene. Oncogene. 1996;12:795–803. [PubMed] [Google Scholar]
- 18.Knight J D. Biochemistry of transcription activation by the E2 protein of bovine papillomavirus type 1. Ph.D. thesis. University of California at Berkeley; 1991. [Google Scholar]
- 19.Lacey M, Alpert S, Hanahan D. Bovine papillomavirus genome elicits skin tumours in transgenic mice. Nature. 1986;322:609–612. doi: 10.1038/322609a0. [DOI] [PubMed] [Google Scholar]
- 20.Lambert P F, Hubbert N L, Howley P M, Schiller J T. Genetic assignment of multiple E2 gene products in bovine papillomavirus-transformed cells. J Virol. 1989;63:3151–3154. doi: 10.1128/jvi.63.7.3151-3154.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambert P F, Spalholz B A, Howley P M. A transcriptional repressor encoded by BPV-1 shares a common carboxy terminal domain with the E2 transactivator. Cell. 1987;50:68–78. doi: 10.1016/0092-8674(87)90663-5. [DOI] [PubMed] [Google Scholar]
- 22.Lambert P L, Monk B C, Howley P M. Phenotypic analysis of bovine papillomavirus type 1 E2 repressor mutants. J Virol. 1990;64:950–956. doi: 10.1128/jvi.64.2.950-956.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehman C W, King D S, Botchan M R. A papillomavirus E2 phosphorylation mutant exhibits normal transient replication and transcription but is defective in transformation and plasmid retention. J Virol. 1997;71:3652–3665. doi: 10.1128/jvi.71.5.3652-3665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Lehman, C. W. Unpublished data.
- 24.Li R, Botchan M R. The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell. 1993;73:1207–1221. doi: 10.1016/0092-8674(93)90649-b. [DOI] [PubMed] [Google Scholar]
- 25.Li R, Botchan M R. Acidic transcription factors alleviate nucleosome-mediated repression of BPV-1 DNA replication. Proc Natl Acad Sci USA. 1994;91:7051–7055. doi: 10.1073/pnas.91.15.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li R, Knight J, Bream G, Stenlund A, Botchan M. Specific recognition nucleotides and their DNA context determine the affinity of E2 protein for 17 binding sites in the BPV-1 genome. Genes Dev. 1989;3:510–526. doi: 10.1101/gad.3.4.510. [DOI] [PubMed] [Google Scholar]
- 27.Li R, Knight J D, Jackson S P, Tjian R, Botchan M R. Direct interaction between Sp1 and the BPV enhancer E2 protein mediates synergistic activation of transcription. Cell. 1991;65:493–505. doi: 10.1016/0092-8674(91)90467-d. [DOI] [PubMed] [Google Scholar]
- 28.Li R, Yang L, Fouts E, Botchan M R. Site specific DNA binding proteins important for replication and transcription have multiple activities. Cold Spring Harbor Symp. 1993;58:403–413. doi: 10.1101/sqb.1993.058.01.047. [DOI] [PubMed] [Google Scholar]
- 28a.Lim, D. A., and M. R. Botchan. Unpublished data.
- 29.Liu J-S, Kuo S-R, Broker T R, Chow L T. The functions of human papillomavirus type 11 E1, E2, and E2C proteins in cell-free DNA replication. J Biol Chem. 1995;270:27283–27291. doi: 10.1074/jbc.270.45.27283. [DOI] [PubMed] [Google Scholar]
- 30.McBride A A, Byrne J C, Howley P M. E2 polypeptides encoded by bovine papillomavirus 1 form dimers through the carboxy-terminal DNA-binding domain: transactivation is mediated through the amino-terminal domain. Proc Natl Acad Sci USA. 1989;86:510–514. doi: 10.1073/pnas.86.2.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melendy T, Sedman J, Stenlund A. Cellular factors required for papillomavirus DNA replication. J Virol. 1995;69:7857–7867. doi: 10.1128/jvi.69.12.7857-7867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendoza R B. Viral cis and trans elements that regulate replication of the bovine papillomavirus. Ph.D. thesis. University of California at Berkeley; 1993. [Google Scholar]
- 33.Muller F, Seo Y-S, Hurwitz J. Replication of bovine papillomavirus type 1 origin containing DNA in crude extracts and with purified factors. J Biol Chem. 1994;269:17086–17094. [PubMed] [Google Scholar]
- 34.Park P, Yang L, Wang T, Botchan M R, Mohr I J. The cellular DNA polymerase α-primase is required for papillomavirus DNA replication and associates with the viral E1 helicase. Proc Natl Acad Sci USA. 1994;91:8700–8704. doi: 10.1073/pnas.91.18.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Polisky, B. Personal communication.
- 35.Prakash S S, Grossman S R, Pepinsky R B, Laimins L A, Androphy E J. Amino acids necessary for DNA contact and dimerization imply novel motifs in the papillomavirus E2 trans-activator. Genes Dev. 1992;6:105–116. doi: 10.1101/gad.6.1.105. [DOI] [PubMed] [Google Scholar]
- 36.Riese D J, Settleman J, Neary K, DiMaio D. Bovine papillomavirus E2 repressor mutant displays a high-copy-number phenotype and enhanced transforming activity. J Virol. 1990;64:944–949. doi: 10.1128/jvi.64.2.944-949.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sedman J, Stenlund A. Cooperative interaction between the initiator E1 and the transcriptional activator E2 is required for replicator specific DNA replication of bovine papillomavirus in vivo and in vitro. EMBO J. 1995;14:6218–6228. doi: 10.1002/j.1460-2075.1995.tb00312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seo Y S, Muller F, Lusky M, Gibbs E, Kim H-Y, Phillips B, Hurwitz J. Bovine papillomavirus (BPV)-encoded E2 protein enhances binding of E1 protein to the BPV replication origin. Proc Natl Acad Sci USA. 1993;90:2865–2869. doi: 10.1073/pnas.90.7.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stenlund A. Papillomavirus DNA replication. In: DePamphilis M L, editor. DNA replication in eukaryote cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 679–697. [Google Scholar]
- 40.Stoler M H, Wolinsky S M, Whitbeck A, Broker T R. Differentiation-linked human papillomavirus types 6 and 11 transcription in genital condylomata revealed by in situ hybridization with message-specific RNA probes. Virology. 1989;172:331–340. doi: 10.1016/0042-6822(89)90135-9. [DOI] [PubMed] [Google Scholar]
- 41.Ustav E, Ustav M, Szymanski P, Stenlund A. The bovine papillomavirus origin of replication requires a binding site for the E2 transcriptional activator. Proc Natl Acad Sci USA. 1993;90:898–902. doi: 10.1073/pnas.90.3.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaillancourt P, Nottoli T, Choe J, Botchan M R. The E2 transactivator of bovine papillomavirus type 1 is expressed from multiple promoters. J Virol. 1990;64:3927–3937. doi: 10.1128/jvi.64.8.3927-3937.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang L, Li R, Mohr I J, Clark R, Botchan M R. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature. 1991;353:628–633. doi: 10.1038/353628a0. [DOI] [PubMed] [Google Scholar]
- 44.Yang L, Mohr I, Li R, Nottoli T, Sun S, Botchan M. The transcription factor E2 regulates BPV-1 DNA replication in vitro by direct protein-protein interaction. Cold Spring Harbor Symp Quant Biol. 1991;56:335–346. doi: 10.1101/sqb.1991.056.01.040. [DOI] [PubMed] [Google Scholar]