Abstract
The human cytomegalovirus (HCMV) UL94 gene product is a herpesvirus-common virion protein that is expressed with true late kinetics. To identify the important cis- and trans-acting factors which contribute to UL94 transcriptional regulation, we have cloned, sequenced, and analyzed UL94 promoter function by transient transfection analysis. Transfection of UL94 promoter-reporter gene constructs into permissive human fibroblasts or U373(MG) cells indicated that promoter activity was detected following infection with HCMV. Point mutations within a TATA-like element located upstream of the RNA start site significantly reduced UL94 promoter activity. Deletion mutagenesis of the promoter indicated that a positive regulatory element (PRE) was likely to exist downstream of the UL94 mRNA start site, while a negative regulatory element (NRE) was present upstream of the TATA box. At late times of infection, the PRE appeared to have a dominant effect over the NRE to stimulate maximum levels of UL94 promoter activity, while at earlier times of infection, no activity associated with the PRE could be detected. The NRE, however, appeared to cause constitutive down-regulation of UL94 promoter activity. Binding sites for the cellular p53 protein located within the NRE appeared to contribute to NRE function, and NRE function could be recapitulated in cotransfection assays by concomitant expression of p53 and HCMV IE2-86 protein. Our results suggest a novel mechanism by which the cellular protein p53, which is involved in both transcriptional regulation and progression of cellular DNA synthesis, plays a central role in the regulation of a viral promoter which is not activated prior the onset of viral DNA replication.
Human cytomegalovirus (HCMV) is a member of the beta class of human herpesviruses and is associated with a number of serious medical conditions, particularly in individuals with compromised immune systems (3). Its genome is the largest of the human herpesviruses (240 kb) and has the capacity to encode over 200 distinct viral gene products (7). Results from a number of different laboratories have indicated that two of these gene products, the major immediate-early (IE) gene products of HCMV (IE1-72 and IE2-86), are essential viral proteins that regulate expression of a variety of viral and heterologous genes at the level of transcription (4, 9, 31, 32, 43, 47, 54, 58, 59). IE1-72 and IE2-86 have been demonstrated to act synergistically to mediate transcriptional activation of several viral genes of subsequent kinetic classes, including viral early genes, such as the viral DNA polymerase gene (and other early genes), as well as early-late genes, such as pp65 (1, 37, 38). Both IE1-72 and IE2-86 have the capacity to regulate transcription from viral and cellular promoters through interactions with cellular activator proteins, such as E2F, p53, CAAT-box-binding factor, CREB-binding factor, and probably others (10, 24, 31, 44, 48, 73). In vitro evidence also implicates IE2-86 interactions with basal transcription factors, such as TATA-box-binding protein and TFIIB, in regulation of viral and cellular promoters, including IE2-86 autoregulation of its own promoter (4, 9, 32, 35, 36, 43, 46, 47, 54, 61, 78).
Despite mounting evidence that the HCMV IE gene products can stimulate expression of a wide variety of viral and heterologous promoters, evidence also suggests that not all HCMV promoters are stimulated by the viral IE proteins. The HCMV true late gene, pp28 (or UL99), is transcribed by a mechanism that does not appear to require the HCMV IE1-72 or IE2-86 proteins (15). While it is not clear whether UL99 can be activated by IE1-72 and/or IE2-86 in conjunction with other viral proteins, cotransfection experiments indicated that IE1-72 and IE2-86 do not stimulate expression of the promoter upstream of the 1.6-kb UL99 RNA start site. Because of the relative promiscuity of the HCMV IE proteins, it is of particular interest to understand (i) why this or other late promoters would be refractory to stimulation by the IE gene products and (ii) what, if any, role is played by the IE proteins in regulating late promoter expression.
Much of our understanding of the transcription of herpesvirus late promoters comes from work with the alphaherpesvirus herpes simplex virus (HSV) (22, 27, 30). In the HSV system, transient transfection experiments using late promoter-reporter constructs have indicated that the structure of viral late promoters is fairly simple. They contain TATA boxes and/or initiator elements which direct the transcription start site. However, in contrast to promoters of other kinetic classes, very few sequence elements upstream of the TATA box or RNA start site appear to significantly enhance late promoter activation. Rather, there appear to be specific sequence elements located downstream of the RNA start site which interact with both viral and cellular proteins and act to both enhance late gene transcription and limit transcription from these promoters to late times of infection. Viral factors such as the ICP4 protein, which appear to enhance HSV late gene expression, and cellular factors such as the DAS protein, which interact with sequence elements more proximal to the RNA start site, appear to play a more significant role in the temporal regulation of late gene expression (22). In addition to these observations, it has also been suggested that other HSV late promoters are subject to negative regulation at earlier times of infection. Still other experiments suggest that the simple structure of late promoters makes them relatively weak with respect to their ability to compete for viral and cellular transcription factors. Thus, it is only through increased concentration of the promoter sequences, via viral DNA replication, that late promoters are capable of successfully competing for factors necessary to initiate transcription. This model may explain why late promoters are often activated inappropriately at early times of infection when expressed from bacterial plasmids. It has also been suggested that late promoters are not accessible to transcription factors prior to viral DNA replication due to local chromatin structure; however, recombination of late promoter-reporter gene constructs into regions of the viral genome that are normally expressed at early times of infection indicates that late kinetics are maintained; thus, it is not clear whether local chromatin structure actually plays a role in late promoter regulation. The available evidence suggests that several of these factors—negative regulation at early times of infection, enhancement at late times of infection, and increased template concentration—are all likely to contribute to the strict temporal activity of late promoters.
Even less information is available concerning regulation of the HCMV true late promoters. As mentioned above, the pp28 promoter cannot be stimulated by the HCMV IE1-72 and IE2-86 proteins (15). Mutagenesis of the pp28 promoter and analysis by transient transfection or recombinant virus experiments indicate that a region from −40 to +103 of the promoter (relative to the RNA start site) is capable of recapitulating true late promoter activity (15, 39). The important sequence elements within this region (other than a likely TATA box) have not, however, been extensively analyzed. Analysis of the ICP36 late promoter suggests that a combination of HCMV activators IE1-72, IE2-86, and TRS-1 stimulates expression of this promoter; however, the significance of this observation is unclear since the ICP36 promoter also contains two start sites, both of which are also stimulated by a combination of IE1-72, IE2-86, and TRS-1, that are activated at early times of infection (42, 63). Indeed, recent evidence suggests that a variety of HCMV early promoters are stimulated to a greater extent by cotransfection of other viral factors—including TRS-1—with the HCMV major IE proteins (33, 37). Thus, it is unlikely that this is a late-specific regulatory mechanism (15).
Because current data do not allow any general conclusions regarding HCMV true late promoter regulation to be drawn, we have investigated the regulation of expression of the HCMV UL94 gene product. Previous experiments indicated that UL94-specific transcripts were detected only at late times of a productive HCMV infection and were sensitive to treatment with ganciclovir, establishing UL94 as a true late gene product (76, 77). In this study, we provide additional evidence that UL94 transcription is restricted to late times of infection and that a region encompassing −120 to +48 of the UL94 promoter is likely to contain the cis-acting sequences necessary to impart true late specificity on the UL94 promoter. We also demonstrate that three important sequence elements within this region—a TATA box, an upstream negative regulatory element (NRE), and a downstream positive regulatory element (PRE)—contribute to regulation of the UL94 promoter. Cotransfection experiments indicate that the repressive effect of the NRE can be recapitulated by expression of cellular p53 and HCMV IE2-86 proteins. Furthermore, this recapitulation requires p53, suggesting that this cellular protein plays a central role in regulation of the UL94 promoter.
MATERIALS AND METHODS
Cell culture and virus.
Human embryonic lung (HEL) cells, human astrocytoma cell line U373(MG), and human osteosarcoma cell line Saos-2 were propagated in Dulbecco modified Eagle medium (DMEM) supplemented with 10% [HEL and U373(MG) cells] or 15% (Saos-2 cells) fetal bovine serum (FBS) as well as antibiotics. Towne strain HCMV [HCMV (Towne)] was propagated in HEL cells as previously described (40, 80).
DNA cloning, sequencing, and plasmids.
UL94 promoter fragments were cloned by PCR amplification of the appropriate regions by using Vent DNA polymerase (New England Biolabs) and a Perkin-Elmer Cetus DNA thermocycler. The 9412 region was generated by amplification from HindIII-digested HCMV (Towne) genomic DNA. The remaining constructs were amplified from plasmid pCAT9412. Oligonucleotides were synthesized at the UNC-Chapel Hill Lineberger Comprehensive Cancer Center Oligonucleotide Facility. All primers were engineered to contain either HindIII sites (upstream primers) or PstI sites (downstream primers) by inclusion of the following sequence at the 5′ end of each primer: (NNNN)2AAGCTT for generation of HindIII sites or (NNNN)2CTGCAG for generation of PstI sites, where the restriction site is underlined and NNNN corresponds to either ACGT (primers 1 and 2), CGAT (primers 3 to 8), or GCAT (primers 9 to 15). Each construct was designated by “94” followed by the numbers of the primer pair used in its amplification. The HCMV-specific region of each primer and the constructs generated using each primer are listed in Table 1.
TABLE 1.
Primers and constructs used
| Primer | HCMV-specific region (5′-3′) | Used for amplification of: |
|---|---|---|
| Upstream (HindIII ends) | ||
| 1 | GCTGATTCGCGCCGTGCGCGACGAGATC | 9412–9415, 94115 |
| 6 | GTTTGGGAACTTTGCCGTC | 9436 |
| 7 | GCTGTATTATTAAGGCGC | 9437, 9447 |
| 8 | GCCTTGGCGCTCTGGATGG | 9438 |
| 9 | GAACATGTCCTGGCGC | 9439 |
| 10 | GCTGTAGGAGTAAGGCGCTAACGCCTCG | 94310 |
| 13 | TTTCATGTCCTGGCGC | 94313 |
| Downstream (PstI ends) | ||
| 2 | GAAGAGTGACGCGCGAAGAG | 9412 |
| 3 | GCACCCCGGCTCAGACGAGG | 9413, 9436–9439, 94310, 94313 |
| 4 | GCGCACCACGTCAGCGTACC | 9414, 9447 |
| 5 | GAGAGCCAACGTCGCAGGCG | 9415 |
| 15 | GCGCCGAGCGCTC | 94115 |
Following amplification, fragments were digested with HindIII-PstI (Promega) and ligated into HindIII-PstI-digested pCATBasic (Promega). Ligations were transformed into Escherichia coli DH5α, and ampicillin-resistant clones were screened by colony hybridization using nick-translated, [32P]dATP-labeled PCR fragments as described previously (76). Positive clones were grown, and plasmid DNA was purified by using Qiagen-tip 500 columns (Qiagen). Some promoter-chloramphenical acetyltransferase (CAT) constructs were analyzed by DNA sequence analysis using a Sequenase kit (United States Biochemical) as suggested by the manufacturer, while others were analyzed by fluorescence tagging at the UNC-Chapel Hill Automated DNA Sequencing Facility.
Expression plasmids for HCMV IE2-86 (pcDNA3-IE86) and IE1-72 (pcDNA-IE72) proteins have been previously described (80). Expression plasmids for wild-type (pC53-SN3) and mutant (pC53-SCX3) p53 were supplied by Bert Vogelstein and have also been previously described (5).
Primer extension analysis.
UL94 transcripts were analyzed by primer extension using whole-cell RNA isolated from HCMV-infected HEL cells as described previously (76). The first UL94-specific primer, UL94-3 (5′ CACCACGTCAGCGTACCAAGTCTGTTC 3′), used in these assays has also been previously described (76). The second primer, UL94-2, which overlaps the UL94 open reading frame (ORF), has the sequence 5′ ATGGCTTGGCGCAGCGGTAT 3′.
CAT assays.
For infection-transfection experiments, cells were seeded into 35-mm-diameter six-well plates at 3 × 106 cells/well. The following day, cells were transfected via liposome-mediated transfection using 1,3-dioleoyloxy-2-(6-carboxyspermyl)propylamide (DOSPER; Boehringer Mannheim). For each 35-mm-diameter well, 0.5 μg of reporter plasmid along with 0.5 μg of Rous Sarcoma virus (RSV)–β-galactosidase (β-Gal) or simian virus 40 (SV40)–β-Gal plasmid was mixed with 4 μl of DOSPER in a final volume of 100 μl of HEPES-buffered saline (20 mM HEPES, 150 mM NaCl [pH 7.4]). Sixty microliters of the DNA-liposome complexes was added dropwise to cells cultured with 1 ml of medium. All transfections were done in triplicate and were allowed to proceed overnight. The next day, the transfectant was removed; cells were washed once with 2 ml of DMEM and subsequently infected with HCMV at a multiplicity of infection of approximately 2 to 5 PFU/cell. Following a 2-h absorption period, 1 ml of DMEM supplemented with 4% heat-inactivated FBS was added to each 35-mm-diameter well. For drug block experiments, the medium was supplemented with 10 μM ganciclovir [9-(1,3-dihydroxy-2-propoxymethyl)guanine (DHPG); Syntex] and was changed daily. Cells were harvested at the appropriate time point postinfection in 250 μl of 1× cell lysis buffer (Promega). For CAT assays, cell extract was mixed with acetylenzyme coenzyme A (Boehringer Mannheim) and [14C]chloramphenicol (New England Nuclear), and CAT assays were performed as described previously (83). Samples were standardized by using the Promega β-Gal enzyme assay system. Assays were carried out as suggested by the manufacturer, and absorbance at 420 nm for each sample was determined with a Beckman DU-70 spectrophotometer.
For cotransfection experiments, 0.1 to 0.5 μg of each effector plasmid was added to the transfection mixture along with 0.5 μg of each of the reporter and standardization plasmids. DNA amounts were standardized by inclusion of the appropriate amount of plasmid pGEM-7zf(+) (Promega). Transfections were carried out as described above except that following the transfection, the medium was replaced with 2 ml of DMEM supplemented with 10% (HEL and U373(MG) cells) or 15% (Saos-2 cells) FBS. At 72 h posttransfection, cells were harvested and CAT assays were performed as described above.
EMSA.
For p53 electrophoretic mobility shift assays (EMSA), we used purified baculovirus-expressed p53 protein with a six-histidine tag (57). Complementary oligonucleotides containing either wild-type or mutated p53-binding sites from the UL94 promoter were annealed to generate double-stranded probes. Sequences of oligonucleotides pairs (5′ to 3′) are as follows: 94p53W2, TCACGGAACATGTCCTGGCGC; 94p53C2, GCGCCAGGACATGTTCCGTGA; 94p53W3, TCACGGAACATGTCCTGGCGCGTTGTTTGGGAACTTTGCCGTCAT; 94p53C3, ATGACGGCAAAGTTCCCAAACAACGCGCCAGGACATGTTCCGTGA; 94p53m1, TCACGGAATCGCTCCTGGCGCGTTGTTTGGGAATCGCGCCGTCAT; and 94p53m2, ATGACGGCGCGATTCCCAAACAACGCGCCAGGAGCGATTCCGTGA.
EMSA were performed as previously described (40) except that 1× binding buffer consisted of 10% glycerol, 25 mM HEPES (pH 7.6), 50 mM NaCl, 1 mM dithiothreitol, 0.5 μg of bovine serum albumin/μl, 0.1% Triton X-100, and 0.1 μg of poly(dI-dC)/μl. For antibody supershift experiments, reactions were performed with 1 μl of antibody per 15 of μl reaction mixture for 30 min at room temperature prior to addition of the probe. Anti-p53 antibodies 421 and DO-1 were obtained from Calbiochem Oncogene Research Products.
RESULTS
Late-specific RNA start site usage in UL94 transcription.
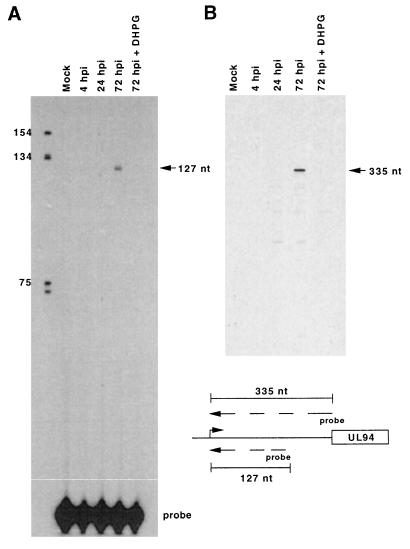
We previously reported that UL94-specific DNA probes detected two classes of transcripts of approximately 9.1 and 2.0 kb in Northern blot analysis of HCMV-infected cell RNA (76, 77). Both transcript classes could be detected only at late times of infection and were sensitive to treatment with ganciclovir, suggesting that UL94 was a member of the true late kinetic class. We also mapped a putative RNA start site upstream of the UL94 ORF (76). This start site, located 336 nucleotides (nt) upstream of the UL94 initiation codon, was positioned 30 bp downstream of a TATA-box-like sequence and was detected by using RNA isolated from HCMV-infected cells at 72 h postinfection (hpi). To ascertain whether this start site was utilized exclusively at late times of infection, we performed primer extension analysis on RNA isolated from HCMV-infected cells at IE, early, and late times of infection, as well as RNA from mock-infected cells and HCMV-infected cells treated with ganciclovir, using a UL94-specific primer that was utilized in our previous RNA mapping experiments. As demonstrated in Fig. 1, extension of the primer to the putative RNA start site could be detected only at 72 hpi in the infection time course; in addition, no initiation of transcription at this site was detected at 72 hpi in the presence of ganciclovir or in mock-infected cells. Results consistent with those shown in Fig. 1 were also obtained in analyses using a second primer (UL94-2) which overlapped the UL94 ORF. These results confirm our previous observations that UL94-specific mRNA can be detected only at late times of infection and also demonstrate that the previously mapped UL94 transcript start site is utilized exclusively at late times of infection.
FIG. 1.
Late-specific utilization of the mRNA start site immediately upstream of the UL94 ORF, determined by primer extension analysis of UL94 mRNA from HCMV (Towne)-infected fibroblasts using primers UL94-3 (A), located 208 nt upstream of the UL94 ORF, and primer UL94-2 (B), which overlaps the ATG for UL94 ORF. The relative positions of the two probes with respect to the UL94 ORF are shown. The concentration of DHPG was 10 μM.
UL94 promoter sequence analysis.
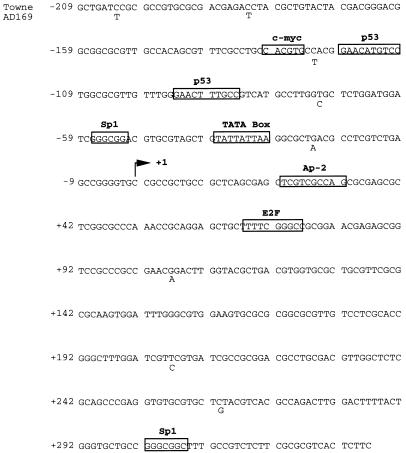
To begin to understand how UL94 transcription is regulated, we sequenced a 525-bp region proximal to the TATA-box-like element located upstream of the UL94 ORF. The sequence analyzed included approximately 210 bp upstream of the UL94 RNA start site as well as 335 bp of sequence located between the RNA start site and the predicted UL94 initiation codon. The results (Fig. 2) are presented in comparison to the published sequence for HCMV strain Ad169 in this region (7). We observed only minor differences in nucleotide sequence between Towne and Ad169 in the putative UL94 promoter region. The likely TATA box at −30 is nonconsensus but is consistent in sequence with TATA-like elements found in other HCMV promoter regions. As in other HCMV early and late promoters, single consensus binding sites for several cellular transcription factors, including Sp1 and c-Myc, are located immediately upstream of the TATA box (15, 31). In addition, a possible 20-nt p53 binding element (with the two 10-nt half-sites separated by 13 nt of intervening sequence) is located in the upstream region. The upstream-most p53 half-site is a perfect match with the consensus p53-binding sequence WWWCATGRRR, while the downstream half-site has a 2-nt mismatch. The sequence and arrangement of these elements is consistent with that observed for cellular promoters which have been demonstrated to be regulated by p53 (18, 20).
FIG. 2.
DNA sequence analysis of the UL94 promoter region from HCMV (Towne). All bases are identical to the reported sequence for the same region of HCMV (Ad169), except where indicated, below the Towne sequence. Also shown are the putative UL94 TATA box and RNA start site (+1), as well as consensus binding sites (boxed) for cellular transcription factors.
Analysis of sequences located downstream of the UL94 RNA start site showed no additional TATA-like elements were found in this region, although we did detect several consensus initiator elements (YYAN[T/A]YY). We have thus far, however, been unable to detect transcription initiation at any of these sites, suggesting that the presence of these elements is completely fortuitous. No cis-regulatory sequence (crs)-like or other binding sites for the HCMV IE2-86 viral transactivator could be found in the UL94 promoter region. However, sequence elements for cellular transcription factors E2F, Sp1, CREB, and the TATA box have been demonstrated to mediate virus-induced transcription of viral and heterologous promoters, suggesting that these elements, or others, could play a role in regulation of UL94 transcription (41, 48, 74, 80).
Functional analysis of UL94 promoter sequences.
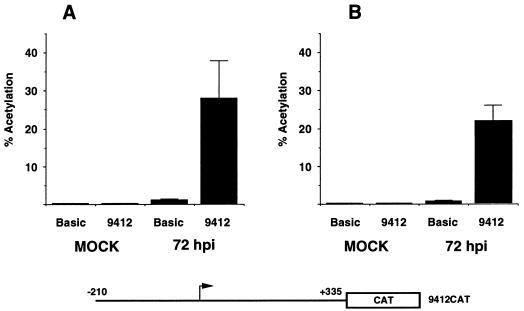
To examine the ability of the putative UL94 promoter sequences to direct transcription, we subcloned the 525-bp fragment sequenced as described above into reporter vector pCATBasic. Human fibroblasts were transfected with the resultant plasmid (9412CAT) and subsequently infected or mock infected with HCMV. As shown in Fig. 3A, extracts from 9412CAT-transfected fibroblasts contained significantly higher CAT activity following a 72 h superinfection with HCMV relative to mock-infected cells or infected cells which were transfected with the parent vector lacking promoter sequences. These results indicated that the 525-bp UL94 promoter fragment contains a functional promoter that is activated in a virus-dependent manner.
FIG. 3.
Transient transfection analysis of UL94 promoter activity from reporter construct 9412CAT, containing the full-length UL94 promoter region. UL94 promoter-CAT (9412CAT) or the parent vector (pCATBasic), lacking enhancer-promoter sequences, was transfected into HEL (A) or U373(MG) (B) cells. Twenty-four hours later, cells were infected (72 hpi) or mock-infected (mock) with HCMV (Towne). Cell lysates were prepared at 72 hpi and assayed for CAT activity. Shown is the mean percent acetylation from triplicate samples for each time point and plasmid. Samples were standardized for transfection efficiency by determining β-Gal activity from a cotransfected RSV–β-Gal plasmid.
As a comparison to UL94 promoter activity in fibroblasts, we repeated the experiment with the HCMV permissive human astrocytoma cell line U373(MG) and obtained results analogous to the results obtained with fibroblasts (Fig. 3B) (13, 49, 56). It is worth noting that in quantitative comparisons between fibroblasts and U373(MG) cells, we generally observed higher levels of UL94 promoter activity in U373(MG) cells. A comparison of the transfection efficiencies of HEL and U373(MG) cells by in situ staining for β-Gal activity, following transfection with an RSV–β-Gal vector, indicated that U373(MG) cells were transfected at a significantly higher frequency than the HEL cells and with much less variability. Since U373(MG) cells are fully permissive for HCMV infection and are more efficiently transfected, we often used these cells preferentially in subsequent transfection-infection experiments. However, many experiments were also reproduced in HEL cells to ensure that the results were consistent within the context of normal human cells.
Deletion analysis of the UL94 promoter.
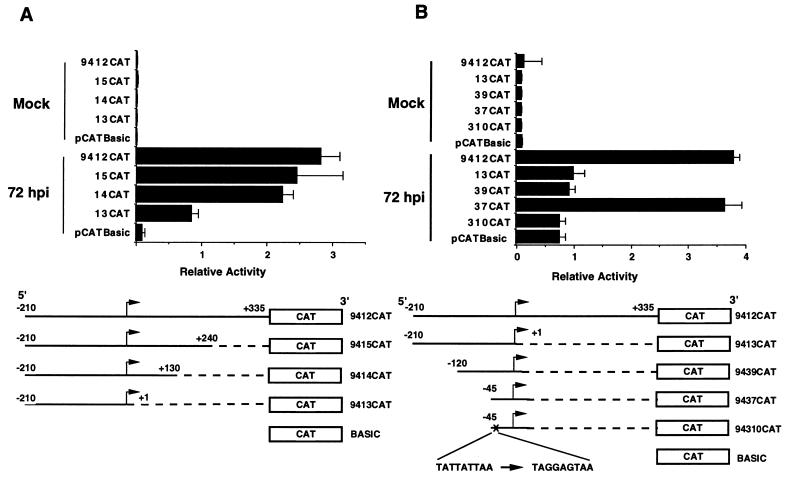
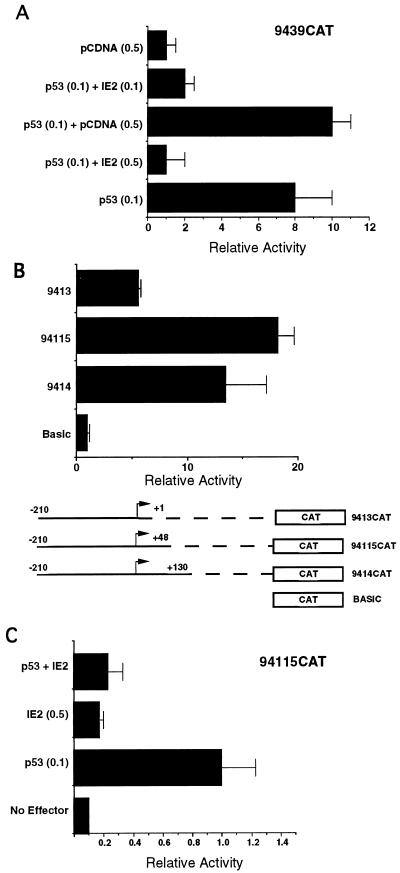
To more precisely define the important functional regions of the UL94 promoter, we generated a series of deletion mutants of the original 525-bp construct and analyzed the ability of these constructs to support expression of CAT in transfection-infection experiments. As can be seen in Fig. 4A, deletion of sequences between +130 and +335 resulted in no substantial loss in CAT activity as measured at 72 hpi in U373(MG) cells. In contrast, deletion of sequences between +2 and +130 resulted in at least a 60% reduction in CAT activity relative to the full-length construct. The relative decrease in activity observed between constructs 9414CAT and 9413CAT (i.e., 3-fold) is identical to the reported decrease in activity of HCMV DNA polymerase promoter-reporter constructs following deletion of the essential IR1 sequence element and analysis by transient transfection-infection (38). Thus, these results suggest that while sequences between +130 and +335 of the UL94 promoter region do not contribute significantly to the activity of the UL94 promoter, a putative positive regulatory region is located between +2 and +130.
FIG. 4.
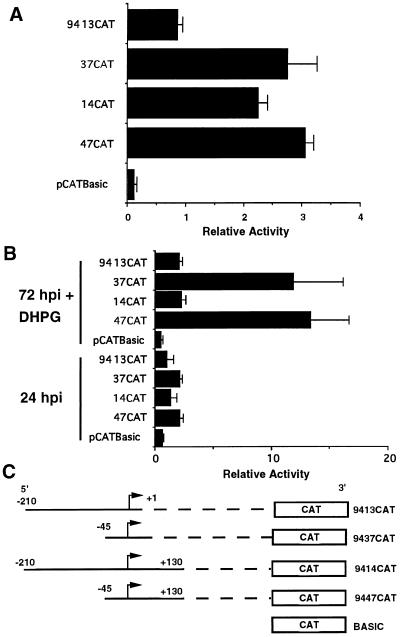
Deletion and point mutation analysis suggests that three important regulatory elements located within 130 bp of the UL94 mRNA start site affect UL94 promoter activity: a 3′ PRE, a 5′ NRE, and a TATA box. (A) Progressive deletions in downstream sequences (with respect to the RNA start site); (B) progressive deletions in the upstream region, as well as point mutations in the TATA box. All constructs were generated by subcloning the regions shown in the diagram into pCATBasic. Plasmids were transfected into U373(MG) cells and subsequently infected with HCMV (Towne) or mock infected. The relative activity determined for triplicate samples of each construct following standardization by β-Gal activity is shown.
To examine the influence of sequences upstream of the TATA box on UL94 promoter activity, we also created a series of 5′-deletion mutants, as shown in Fig. 4B. To minimize any confounding effects of the downstream positive regulatory region mapped above, we initially constructed a series of 5′-deletion constructs which extended only to the RNA start site and thus did not include the downstream regulatory region. As shown in Fig. 4B, when these constructs were analyzed by transfection-infection of U373(MG) cells, no significant CAT activity was observed in mock-infected cells; in addition, we observed a relatively low CAT activity from constructs containing 210 or 120 bp of upstream sequence. However, deletion of all sequences upstream of the TATA box (9137CAT) resulted in CAT activity at least threefold greater than for constructs which contained 120 bp or more of upstream sequence. The CAT activity from this minimal construct was similar to that observed with the 9414CAT construct, which contained 210 bp of upstream sequence as well as the putative downstream regulatory region. Since addition of 120 bp of upstream sequence resulted in diminished CAT activity relative to the minimal construct containing only the region immediately proximal to the TATA box and RNA start site, this experiment suggested that a possible negative regulatory region is located upstream of the TATA box in the UL94 promoter.
To demonstrate that the TATA sequence found in our promoter-CAT constructs was indeed a TATA box, we created a minimal promoter construct (94310CAT) in which specific point mutations were introduced into the TATA sequence (TATTATTAA to TAGGAGTAA). As shown in Fig. 4B, the level of CAT activity from this construct was threefold lower than that of the otherwise identical wild-type construct (compare 94310CAT and 9437CAT). This reduction in CAT activity is similar to that observed in constructs which contain a wild-type TATA box and the putative 5′ NRE, indicating that the wild-type TATA sequence is essential to maximum promoter activity.
To determine the effect of the putative 3′ PRE on promoter activity, with or without the presence of the 5′ NRE, we created an additional construct which contained no sequences upstream of the TATA box but extended 3 to +130 (i.e., into the PRE). As shown in Fig. 5, there was no significant difference in CAT activity between constructs 9447CAT and 9437CAT at 72 hpi. This finding suggested that the PRE region did not significantly add to the promoter activity that was observed with the TATA box alone at late times of infection. In contrast to these results, however, CAT activity from construct 9414CAT, containing the TATA box, PRE, and NRE, was significantly higher at 72 hpi than the activity of construct 9413CAT, which contains only the TATA box and NRE. These results suggested that the positive effect of the PRE is dominant over the negative effect of the NRE at late times of infection.
FIG. 5.
(A) The putative downstream PRE is dominant to the upstream NRE at late times of infection. (B) The dominant effect of the PRE over the NRE is not obviously observed prior to the onset of viral DNA replication (24 hpi) or if viral DNA replication is inhibited (72 hpi + DHPG [10 μM]). Shown are the relative CAT activities of triplicate samples for each plasmid construct standardized by β-Gal activity. Transfection-infection was carried out in U373(MG) cells, and cells were harvested at 24 or 72 hpi as indicated. (C) Constructs containing the UL94 TATA box as well as the PRE (9447CAT), the NRE (9413CAT), both elements (9414CAT), or neither element (9437CAT).
Taken together, the results of this series of 5′- and 3′-deletion constructs, as well as the TATA-box point mutants, suggests the existence of three important regulatory elements within a region encompassing −120 to +130 of the UL94 promoter: a 5′ NRE, a 3′ PRE, and a TATA box. At late times of infection, the repressive effect of the 5′ NRE is observed only when the 3′ PRE is absent, indicating that PRE function is dominant to NRE function at late times of infection. However, constructs which contain only the TATA box and the 5′ NRE exhibit only minimal activity in comparison to the TATA-only construct or constructs containing the 3′ PRE. Thus, the PRE appears to function as a derepression element, as it has the capacity to overcome the negative effect exerted by the NRE on the TATA box region of UL94 promoter at late times of infection.
UL94 promoter activity at 24 hpi and in the presence of ganciclovir.
True late viral promoters are active only after the onset of viral DNA replication. Deletion mutagenesis of late promoters from HSV has suggested that in addition to the requirement for viral DNA replication, cis-regulatory sequences located in late promoters influence promoter activity by either enhancing activity exclusively at late times of infection or by preventing late promoter activity at earlier times of infection (19, 21, 22, 27, 29, 30). Indeed, it is likely that both positive and negative regulatory mechanisms are necessary for precise regulation of late promoters. To address this question, we performed another series of transfection-infection experiments in which CAT activity was measured following a 24-h infection with HCMV or a 72-h infection in the presence of ganciclovir. Specifically, we compared the activity of our promoter construct containing only the TATA box (9437CAT) with those of constructs containing the TATA box and either the NRE (9413CAT), the PRE (9447CAT), or both (9414CAT) at both 24 hpi and at 72 hpi in the presence of ganciclovir. As can be seen in Fig. 5B, none of the UL94 promoter constructs had significant CAT activity at 24 hpi, as directly compared to the activity of the promoterless-vector control. There was a small but measurable increase in activity with constructs 9437CAT and 9447CAT, in comparison to the other constructs, suggesting that lack of the NRE resulted in higher activity of the UL94 promoter at early times of infection. This conclusion was substantiated by the results obtained at 72 hpi with ganciclovir: constructs 9437CAT and 9447CAT had significantly higher activity under these conditions than either 9413CAT or 9414CAT; indeed, the activity of 9413CAT and 9414CAT was no higher at 72 hpi with ganciclovir than at 24 hpi. Interestingly, whereas at 72 hpi the activity of 9414CAT was equivalent to that seen with 9447CAT or 9437CAT (Fig. 5A), its activity at 24 hpi and at 72 hpi with ganciclovir was extremely low. This suggests that the dominant effect of the PRE is a late-phase-specific event, since we do not observe the derepression at 24 hpi or at 72 hpi with ganciclovir. In addition, since both 9437CAT and 9447CAT, which lack the NRE, are active both at 24 hpi and at 72 hpi with ganciclovir, it is likely that the UL94 promoter TATA box can be activated at early times of infection in the absence of the NRE.
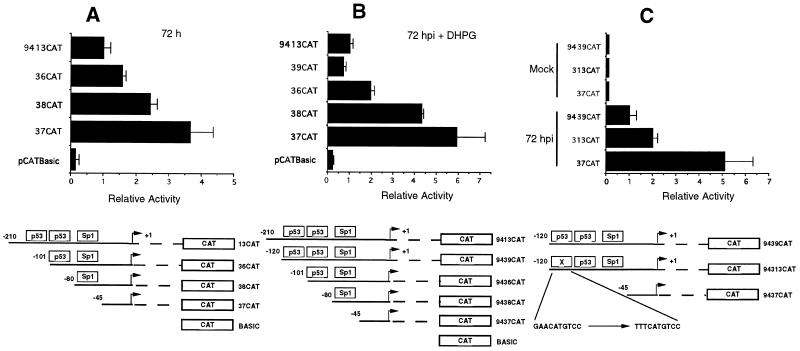
Additional deletion mapping of the 5′ NRE.
Sequence analysis of the UL94 promoter demonstrated that putative binding sites for several cellular transcription factors, including p53, were located within the NRE region (Fig. 2). It has previously been reported that whereas p53 generally activates transcription in a DNA-binding-dependent manner, in the presence of viral proteins such as HCMV IE2-86, adenovirus E1A or E1B, hepatitis B virus (HBV) X protein, or SV40 large T antigen, transcription from promoters with p53-binding sites is repressed (12, 28, 48, 55, 57, 62, 64, 71, 72). Since the NRE appears to be involved in repression of the UL94 promoter, we tested the effect of deletion of either one or both p53 half-sites on UL94 promoter activity in transfection-infection experiments. There was an additive effect of deletion of either one or both p53 sites: when both sites were deleted, there was an approximately 2.5-fold increase in UL94 promoter activity at 72 hpi in comparison to constructs containing the p53-binding-site region (Fig. 6A). The effect was even more pronounced at 72 hpi with ganciclovir: as much as a fivefold increase in UL94 promoter activity was seen when the region containing the p53 sites was deleted (Fig. 6B). In addition, we detected a close to twofold increase in promoter activity at late times of infection when point mutations were introduced into the upstream p53-binding site (Fig. 6C). These results suggest that the region containing the p53-binding sites contributes to the repression activity observed with the UL94 promoter NRE. Since deletion of the p53-binding sites did not restore promoter activity to that observed with deletion of the entire NRE, we conclude that other sequences besides the p53-binding-site region play a role in full NRE activity.
FIG. 6.
Deletion or mutation of the consensus p53-binding sites in the putative NRE region leads to increased UL94 promoter activity at late times of infection or when viral DNA replication is inhibited. (A) CAT activities at 72 hpi for UL94 promoter constructs containing both p53 sites (9413CAT) or with deletions in the perfect consensus p53 site (9438CAT), both (9436CAT) p53 sites, or all sequences upstream of the TATA box; (B) CAT activities of the same constructs at 72 hpi with DHPG (10 μM); (C) CAT activities at 72 hpi from constructs containing both (9439CAT) or neither (9437CAT) p53 site, or point mutations in the first perfect consensus p53 site (94313CAT). The mean relative activity of triplicate samples of each construct, standardized by β-Gal activity, is shown. All constructs were transfected into U373(MG) cells, infected with HCMV 24 h later, and harvested for CAT activity at 72 hpi.
Effect of p53 on UL94 promoter activity in cotransfection assays.
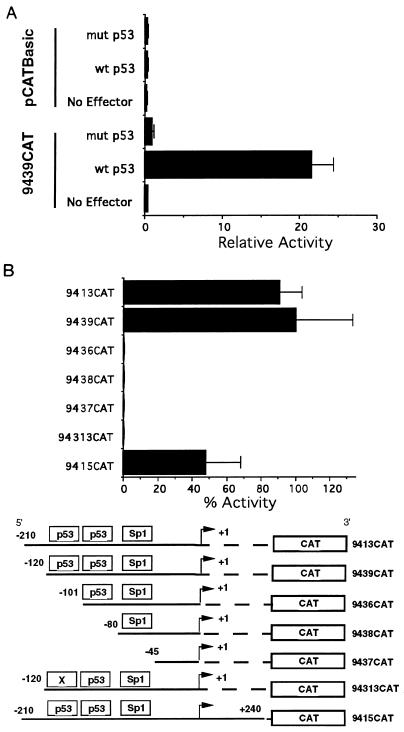
To determine the effect of p53 on UL94 promoter activity, we cotransfected p53-negative Saos-2 cells with plasmids expressing either wild-type or mutant p53 and UL94 promoter construct 9439CAT, which contains the putative p53-binding sites. Cotransfection of 9439CAT with wild-type p53 resulted in an approximately 20-fold increase in promoter activity compared to cotransfection with mutant p53 or vector alone controls (Fig. 7A). Likewise, wild-type p53 had no effect on the promoterless vector control. In addition to promoter construct 9439CAT, we tested several other UL94 promoter constructs for responsiveness to p53 in Saos-2 cells (Fig. 7B). All of the constructs containing both p53-binding sites were significantly activated by cotransfection with wild-type p53, whereas constructs lacking the p53-binding sites were not activated. Interestingly, construct 9436CAT, which contains only the downstream, nonconsensus p53-binding site, and construct 94313CAT, which contains point mutations in the upstream p53-binding site, were also not activated by p53 in Saos-2 cells. These results imply that both p53-binding sites are necessary for responsiveness to p53 or, alternatively, that the downstream p53 site is simply unresponsive to p53. These two possibilities are addressed below.
FIG. 7.
Stimulation of UL94 promoter activity by p53 in Saos-2 cells. (A) Saos-2 cells were cotransfected with UL94 promoter construct 9439CAT, containing both p53 sites, or pCATBasic, along with expression plasmids for wild-type (wt) or mutant (mut) p53. (B) UL94 promoter constructs containing both p53 sites (9415CAT, 9439CAT, and 9413CAT), neither p53 site (9437CAT and 9436CAT), the downstream nonconsensus p53 site (9438CAT), or point mutations in the upstream perfect consensus p53 site (94313CAT) were cotransfected with expression plasmid for wild-type p53. For both panels, the relative mean activity for triplicate samples of each construct and/or condition is shown.
Binding of p53 to the UL94 promoter.
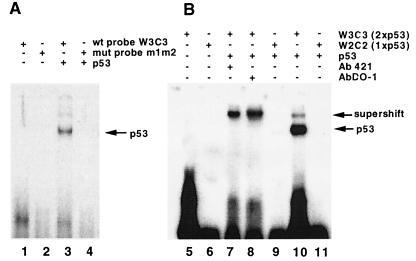
To demonstrate binding of p53 to the UL94 promoter, we used, in EMSA, oligonucleotides containing either one or both UL94 promoter p53-binding sites, or oligonucleotides containing mutated p53 sites, and purified baculovirus-expressed p53. As shown in Fig. 8, p53 bound to the wild-type but not the mutated UL94 p53 sites (lanes 3 and 4). This binding could be specifically competed by excess unlabeled wild-type (W3C3) but not mutant (m1/m2) UL94 p53 site probe (data not shown). We could also supershift the p53-UL94 promoter complex with p53-specific antibodies (lanes 7 and 8). In addition, p53 protein bound at a much higher affinity to probes containing both p53 sites (lane 10 [with W3C3 probe]) than a probe containing only the upstream consensus p53-binding site (lane 9 [with W2C2]). This result suggests that binding of p53 to the UL94 promoter requires both p53-binding sites and is consistent with previous reports which indicate that binding of tetrameric p53 to responsive promoters requires more than one 10-bp p53 consensus sequence. These results also suggest that the downstream p53 site in the UL94 promoter is likely to be necessary for stabilization of the p53-DNA interaction, despite the fact that this element alone cannot support p53-mediated transactivation. It should be noted that while the gel shift and CAT assay data demonstrate that the two 10-bp p53-binding sites in the NRE are essential to obtain p53 binding to the NRE (EMSA) and transactivation (p53 expression alone) or suppression (coexpression of p53 and IE2-86 [see below]) of NRE-containing promoters (CAT assays), we have not yet determined which NRE nucleotides are bound to p53.
FIG. 8.
Binding of baculovirus-expressed p53 to the UL94 promoter p53 sites by EMSA. (A) Free, labeled, wild-type (wt) (lane 1) or mutant (mut) (lane 2) oligonucleotide probes, or wild-type (lane 3) and mutant (lane 4) probes incubated with purified, histidine-tagged p53; (B) p53 binding to labeled wild-type p53 probe containing both p53 sites of UL94 promoter (lane 10, W3C3) compared to only the upstream consensus p53 site (lane 9 and 11, W2C2) as well as both probes alone (lane 5 and 6, respectively), and supershift/stabilization of p53 binding to UL94 promoter p53 sites with p53-specific antibodies 421 (lanes 7) and DO-1 (lane 8), respectively.
Effect of HCMV IE2-86 on the UL94 promoter.
Since the p53-binding sites in the UL94 promoter appear to contribute to repression of the UL94 promoter during infection, we wondered why they mediated p53-dependent activation in Saos-2 cells. As mentioned earlier, the HCMV IE2-86 protein has been reported to tether a transcriptional repression domain to p53 through protein-protein interaction (71). The effect of this interaction is a p53-binding-site-dependent IE2-mediated repression of transcription by p53. To examine the effect of IE2-86 on p53-mediated activation of the UL94 promoter, we cotransfected Saos-2 cells with UL94 promoter construct 9439CAT and expression plasmids for both p53 and IE2. As can be seen in Fig. 9A, IE2-86 alone had no effect on UL94 promoter activity; in contrast, when cotransfected with p53, IE2-86 abrogated p53-mediated activation of the UL94 promoter. The level of repression that we observed is consistent with previously published data showing repression of p53-mediated transcription by IE2-86. Furthermore, Western blot analysis showed this repression was not due to a decrease in p53 levels in the presence of IE2-86 (data not shown). These results demonstrate that IE2-86-mediated repression of p53 can recapitulate the transcriptional repression observed during infection by the UL94 promoter NRE region.
FIG. 9.
IE2-mediated inhibition of p53 transactivation of UL94 promoter constructs, with or without the PRE region, recapitulates NRE function. (A) Cotransfection of UL94 promoter construct 9439CAT, which lacks the PRE, with expression vectors for p53, p53 plus IE2-86, or the empty expression vector (pCDNA); (B) transfection-infection experiment demonstrating that UL94 promoter construct 94115CAT, which contains an additional deletion in the PRE region, has levels of activity comparable to those of 9414CAT at late times of infection; (C) IE2-86-mediated inhibition of p53 transactivation of UL94 promoter construct 94115CAT, containing the reduced PRE region. The relative mean activity of triplicate samples for each condition is shown in each case.
Of the functionally important UL94 promoter regions, construct 9439CAT contains only the TATA box and NRE; however, we also wanted to measure the effect of p53 on promoter constructs containing the PRE region, since our transfection-infection data suggested that the PRE had the capacity to disrupt NRE function at late times of infection. In addition, we were interested in more precisely localizing the PRE region. To address the second point, we generated an additional deletion mutant, 94115CAT, which extends to +48 with respect to the RNA initiation site. As can be seen in Fig. 9B, when this construct was tested for functionality in infection-transfection assays, its activity was similar to that of 9414CAT, which contained an additional 82 bp of downstream sequence. We conclude from this experiment that PRE function resides in the first 48 bp downstream of the RNA initiation site. To determine the effect of p53 and IE2 on 94115CAT, we cotransfected Saos-2 cells with 94115CAT and expression plasmids for p53 and IE2-86, as before. The results (Fig. 9C) indicated that 94115CAT is also responsive to p53 and, further, that IE2-86 could abrogate this stimulation. Thus, in the absence of any other viral gene products, the PRE region of UL94 promoter has no obvious role in the repression of the UL94 promoter activity by IE2-86 or p53.
DISCUSSION
In this report, we provide a characterization of the promoter region regulating expression of the HCMV UL94 protein. We show that, consistent with our previous data, transcription from the putative UL94 promoter occurs only at late times of infection and is sensitive to treatment with the viral DNA replication inhibitor ganciclovir (77). DNA sequence analysis of the UL94 promoter indicated the presence of a TATA-box-like element at −30 with respect to the RNA start site. Analysis of the UL94 promoter in transient transfection assays indicated that it could support transcription of CAT during a productive infection with HCMV. Mutagenesis of the UL94 promoter indicated that maximum activity was dependent on the TATA box-like element, since point mutations in this element significantly reduced UL94 promoter activity. In the absence of any other UL94 promoter regulatory sequences, this basic TATA box can activate transcription at early and late times of infection.
Deletion analysis suggested that regions both upstream and downstream of the RNA start site are important for regulation of the UL94 promoter. The downstream PRE region appears to be necessary for stimulation of UL94 promoter activity at late times of infection, since in context of the entire promoter, its absence leads to diminished promoter activity. In contrast, the upstream NRE appears to prevent activation of the UL94 promoter, since its presence curtails TATA-box-mediated activation. The NRE appears to confer constitutive repression on the UL94 TATA box, since promoter constructs containing only the NRE and TATA box have diminished activity throughout infection. In contrast, the PRE appears to enhance activation of the UL94 promoter exclusively at late times of infection, since constructs containing both the NRE and PRE have diminished activity at 24 hpi and at 72 hpi with ganciclovir but are fully active at 72 hpi. Given these observations, the primary role of the PRE appears to overcome the repressive effect of the NRE at late times of infection, since it is only at late times of infection in the presence of the NRE that any effect mediated through the PRE can be demonstrated.
Mutagenesis of the PRE region indicated that the functionally important sequences were located within the first 48 bp downstream of the RNA start site. Mutagenesis of the NRE suggests that its activity is mediated, in part, through two 10-bp binding sites for the cellular protein p53. Deletion of the p53-binding sites increased UL94 promoter activity; point mutations in the upstream p53 site had the same effect. Purified p53 protein bound to wild-type but not mutant UL94 promoter p53 binding sites in EMSA. This binding occurred at a much higher affinity when a probe containing both p53 sites was used.
In addition to these experiments, the biological effect of the NRE region was recapitulated in cotransfection experiments using expression plasmids for p53 and HCMV IE2-86 protein in conjunction with UL94 promoter-CAT constructs. We also demonstrated that p53 stimulated UL94 promoter activity in cotransfection assays in a manner dependent on the presence of both p53-binding sites. This stimulation was abrogated by cotransfection with the HCMV IE2-86 protein, suggesting that a p53-IE2 interaction is important for repression of the UL94 promoter. This effect is consistent with the observed role of the NRE region in the transfection-infection experiments.
The exact nature of the PRE and NRE regions is unclear, although it appears that the NRE functions, in part, via interaction with cellular p53. As has been seen with other HCMV viral promoters that contain NRE and PRE regions, constructs containing only the PRE were constitutively active (31). In addition, as has been shown with at least one other HCMV viral promoter (UL4), cellular factors bind to and are important for the NRE function, as we have demonstrated with p53 in this study. However, in the UL4 promoter, IE2 expression could activate gene expression (31), whereas IE2-86 is required for the NRE region to prevent UL94 promoter activation. Clearly, there are other important sequences within the NRE, since deletion of the p53 sites does not result in complete NRE inactivation. We are currently investigating the role of other sequences within the NRE, including the Sp1 site. Since p53 has been reported to physically interact with Sp1, it is possible that a larger complex containing both of these proteins enhances NRE function (20).
Both IE2-86 and p53 are rapidly induced and/or their levels are stabilized throughout HCMV infection (34, 51, 52, 62, 65); thus, it is likely that an interaction between the two proteins results in a constitutive inactivation of promoters containing certain p53 sites during HCMV infection. While no data addressing this possibility have been published, it has been suggested that levels of the p53-regulated protein p21/WafI/CipI are not altered during HCMV infection (38). Since other stimuli which increase intracellular concentrations of p53 often lead to a p53-mediated induction of p21 (6, 14, 23), it is likely that p53, through its interactions with IE2-86, is inactivated during HCMV infection with respect to its ability to transactivate certain promoters. This effect of IE2-86 on p53 has been documented in other studies via transient assays using p53-reporter constructs in cotransfections with p53 and IE2 (65, 74). It has also been reported that p53 and IE2-86 physically interact to mediate this effect (74). Active repression of p53-responsive promoters has also been observed in several other viral systems, including adenovirus, SV40, and HBV; in each case, the repressive effect is mediated by important viral transcriptional regulators, including adenoviruses E1A/E1B, SV40 large T antigen, and the HBV X protein (28, 55, 64, 67, 68, 70, 75). Since the HCMV IE2-86 protein shares many functional characteristics with these proteins, it is reasonable to speculate that it has a similar effect on p53 activity during the viral replication cycle. In fact, it is likely that induction or stabilization of p53 during viral infection ensures that p53-responsive promoters remain actively repressed during the replication cycle, in a manner dependent on viral transcriptional regulators. p53 has been well documented to be involved in both cell cycle arrest and apoptosis (1, 2, 6, 8, 11, 17, 25, 26, 45, 60, 66, 79). Whether the primary goal of the HCMV-mediated inactivation of p53 function is to prevent p53-mediated cell cycle arrest or, alternatively, p53-mediated apoptosis is not yet clear. It has been reported that IE2-86 can inhibit apoptosis in certain cell types and that HCMV infection results in cell cycle arrest in G2/M (36, 84); both of these effects are possibly mediated through p53. Regardless of this distinction, our results suggest that p53 is involved in regulating the UL94 promoter, providing functional relevance, in the context of HCMV infection, for the interaction between IE2-86 and p53.
While we have identified important cis-regulatory signals and trans-acting factors which have the capacity to regulate the UL94 promoter, we have not yet addressed the role of viral DNA replication. Since the UL94 promoter is not active at early times of infection or at 72 hpi with ganciclovir in its normal genomic context, it is clear that viral DNA replication plays an important role in UL94 promoter regulation. As has been demonstrated for other herpesvirus late promoters, we have observed that even those UL94 promoter constructs that have reduced activity due to mutation are not completely inactivated in transient transfection experiments. This is likely to be due to the fact that our promoter constructs are presented in the context of bacterial plasmids instead of the viral genome: such a context has been demonstrated to abrogate some aspects of viral late gene regulation. The mechanism responsible for this effect, however, is not known. Now that we have defined a minimal UL94 promoter region that appears to contain all of the important cis-regulatory regions, we would like to recombine a UL94 promoter-reporter gene construct into the viral genome in order to study its regulation in a more appropriate context. Such an approach has been used to study HSV late promoters as well as the HCMV UL99 true late promoter (20, 22, 39). We anticipate that in the context of the viral genome, the effects of mutations within the NRE and/or PRE will be substantially greater than what has been observed in transient assays and that we will be able to more precisely define the role of these cis-regulatory regions as well as viral and cellular proteins in the regulation of UL94 expression.
It is particularly intriguing that a late promoter—whose activity requires the onset of viral DNA replication—would be regulated, in part, through p53. Aside from its role in regulating transcription of important cell cycle regulators such as p21 and mdm2, p53 can also actively inhibit cellular DNA replication through protein-protein and protein-DNA interactions (49). While the role for p53 in HCMV viral DNA replication has not been demonstrated, regulation of the UL94 promoter by a cellular protein that can regulate both transcription and DNA replication is consistent with the association between viral late gene transcription and viral DNA replication. Thus, our results suggest a possibly novel mechanism by which viral DNA replication and late gene expression are concomitantly regulated.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
We thank Eric Poma and Andrew Yurochko for critical reading of the manuscript and Shu-mei Huong and Ke-Wu Lei for excellent technical assistance.
This work was supported by grant AI12712 and CA 21773 from the National Institutes of Health to E.-S.H.
REFERENCES
- 1.Abrahamson J L, Lee J M, Bernstein A. Regulation of p53-mediated apoptosis and cell cycle arrest by Steel factor. Mol Cell Biol. 1995;15:6953–6960. doi: 10.1128/mcb.15.12.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agarwal M L, Agarwal A, Taylor W R, Stark G R. p53 controls both the G2/M and the G1 cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc Natl Acad Sci USA. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alford C A, Britt W J. Cytomegalovirus. In: Fields B N, Knipe D M, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. Fields virology. New York, N.Y: Raven Press; 1990. pp. 1981–2010. [Google Scholar]
- 4.Arlt H, Lang D, Gebert S, Stamminger T. Identification of binding sites for the 86-kilodalton IE2 protein of human cytomegalovirus within an IE2-responsive viral early promoter. J Virol. 1994;68:4117–4125. doi: 10.1128/jvi.68.7.4117-4125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker S J, Markowitz S, Fearon E R, Willson J K V, Vogelstein B. Supression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 6.Brugarolas J, Chandrasekaran C, Gordon J I, Beach D, Jacks T, Hannon G J. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 7.Chee M S, Bankier A T, Beck S, Bonhi R, Brown C M, Cerny R, Horsnell T, Hutchison III C A, Kouzarides T, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microb Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Wu X, Lin J, Levine A J. mdm-2 inhibits the G1 arrest and apoptosis functions of the p53 tumor suppressor protein. Mol Cell Biol. 1996;16:2445–2452. doi: 10.1128/mcb.16.5.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherrington J M, Khoury E L, Mocarski E S. Human cytomegalovirus ie2 negatively regulates alpha gene expression via a short target sequence near the transcription start site. J Virol. 1991;65:887–896. doi: 10.1128/jvi.65.2.887-896.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherrington J M, Mocarski E S. Human cytomegalovirus ie1 transactivates the alpha promoter-enhancer via an 18-base-pair repeat element. J Virol. 1989;63:1435–1440. doi: 10.1128/jvi.63.3.1435-1440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiou S K, Rao L, White E. Bcl-2 blocks p53-dependent apoptosis. Mol Cell Biol. 1994;14:2556–2563. doi: 10.1128/mcb.14.4.2556. . (Erratum, 14:4333.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiou S K, Tseng C C, Rao L, White E. Functional complementation of the adenovirus E1B 19-kilodalton protein with Bcl-2 in the inhibition of apoptosis in infected cells. J Virol. 1994;68:6553–6566. doi: 10.1128/jvi.68.10.6553-6566.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dal Monte P, Bessia C, Ripalti A, Landini M P, Topilko A, Plachter B, Virelizier J L, Michelson S. Stably expressed antisense RNA to cytomegalovirus UL83 inhibits viral replication. J Virol. 1996;70:2086–2094. doi: 10.1128/jvi.70.4.2086-2094.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datto M B, Li Y, Panus J F, Howe D J, Xiong Y, Wang X F. Transforming growth factor beta induces the cyclin-dependent kinase inhibitor p21 through a p53-independent mechanism. Proc Natl Acad Sci USA. 1995;92:5545–5549. doi: 10.1073/pnas.92.12.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Depto A S, Stenberg R M. Functional analysis of the true late human cytomegalovirus pp28 upstream promoter: cis-acting elements and viral trans-acting proteins necessary for promoter activation. J Virol. 1992;66:3241–3246. doi: 10.1128/jvi.66.5.3241-3246.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Depto A S, Stenberg R M. Regulated expression of the human cytomegalovirus pp65 gene: octamer sequence in the promoter is required for activation by viral gene products. J Virol. 1989;63:1232–1238. doi: 10.1128/jvi.63.3.1232-1238.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dulic V, Kaufmann W K, Wilson S J, Tlsty T D, Lees E, Harper J W, Elledge S J, Reed S I. p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell. 1994;76:1013–1023. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- 18.Friedman P N, Chen X, Bargonetti J, Prives C. The p53 protein is an unusually shaped tetramer that binds directly to DNA. Proc Natl Acad Sci USA. 1993;90:3319–3323. doi: 10.1073/pnas.90.8.3319. . (Erratum, 90:5878.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodart S A, Guzowski J F, Rice M K, Wagner E K. Effect of genomic location on expression of beta-galactosidase mRNA controlled by the herpes simplex virus type 1 UL38 promoter. J Virol. 1992;66:2973–2981. doi: 10.1128/jvi.66.5.2973-2981.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gualberto A, Hixon M L, Finco T S, Perkins N D, Nabel G J, Baldwin A S., Jr A proliferative p53-responsive element mediates tumor necrosis factor alpha induction of the human immunodeficiency virus type 1 long terminal repeat. Mol Cell Biol. 1995;15:3450–3459. doi: 10.1128/mcb.15.6.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guzowski J F, Singh J, Wagner E K. Transcriptional activation of the herpes simplex virus type 1 UL38 promoter conferred by the cis-acting downstream activation sequence is mediated by a cellular transcription factor. J Virol. 1994;68:7774–7789. doi: 10.1128/jvi.68.12.7774-7789.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guzowski J F, Wagner E K. Mutational analysis of the herpes simplex virus type 1 strict late UL38 promoter/leader reveals two regions critical in transcriptional regulation. J Virol. 1993;67:5098–5108. doi: 10.1128/jvi.67.9.5098-5108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halevy O, Novitch B G, Spicer D B, Skapek S X, Rhee J, Hannon G J, Beach D, Lassar A B. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- 24.Hayhurst G P, Bryant L A, Caswell R C, Walker S M, Sinclair J H. CCAAT box-dependent activation of the TATA-less human DNA polymerase alpha promoter by the human cytomegalovirus 72-kilodalton major immediate-early protein. J Virol. 1995;69:182–188. doi: 10.1128/jvi.69.1.182-188.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hermeking H, Eick D. Mediation of c-Myc-induced apoptosis by p53. Science. 1994;265:2091–2093. doi: 10.1126/science.8091232. [DOI] [PubMed] [Google Scholar]
- 26.Hiebert S W, Packham G, Strom D K, Haffner R, Oren M, Zambetti G, Cleveland J L. E2F-1:DP-1 induces p53 and overrides survival factors to trigger apoptosis. Mol Cell Biol. 1995;15:6864–6874. doi: 10.1128/mcb.15.12.6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Homa F, Krikos A, Glorioso J C, Levine M. Functional analysis of regulatory regions controlling strict late HSV gene expression. In: Wagner E K, editor. Herpesvirus transcription and its regulation. Boca Raton, Fla: CRC Press; 1991. pp. 207–222. [Google Scholar]
- 28.Horikoshi N, Usheva A, Chen J, Levine A J, Weinmann R, Shenk T. Two domains of p53 interact with the TATA-binding protein, and the adenovirus 13S E1A protein disrupts the association, relieving p53-mediated transcriptional repression. Mol Cell Biol. 1995;15:227–234. doi: 10.1128/mcb.15.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang C J, Petroski M D, Pande N T, Rice M K, Wagner E K. The herpes simplex virus type 1 VP5 promoter contains a cis-acting element near the cap site which interacts with a cellular protein. J Virol. 1996;70:1898–1904. doi: 10.1128/jvi.70.3.1898-1904.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang C J, Wagner E K. The herpes simplex virus type 1 major capsid protein (VP5-UL19) promoter contains two cis-acting elements influencing late expression. J Virol. 1994;68:5738–5747. doi: 10.1128/jvi.68.9.5738-5747.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang L, Malone C L, Stinski M F. A human cytomegalovirus early promoter with upstream negative and positive cis-acting elements: IE2 negates the effect of the negative element, and NF-Y binds to the positive element. J Virol. 1994;68:2108–2117. doi: 10.1128/jvi.68.4.2108-2117.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang L, Stinski M F. Binding of cellular repressor protein or the IE2 protein to a cis-acting negative regulatory element upstream of a human cytomegalovirus early promoter. J Virol. 1995;69:7612–7621. doi: 10.1128/jvi.69.12.7612-7621.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iskenderian A C, Huang L, Reilly A, Stenberg R M, Anders D G. Four of eleven loci required for transient complementation of human cytomegalovirus DNA replication cooperate to activate expression of replication genes. J Virol. 1996;70:383–392. doi: 10.1128/jvi.70.1.383-392.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jault F M, Jault J M, Ruchti F, Fortunato E A, Clark C, Corbeil J, Richman D D, Spector D H. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J Virol. 1995;69:6697–6704. doi: 10.1128/jvi.69.11.6697-6704.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jupp R, Hoffmann S, Depto A, Stenberg R M, Ghazal P, Nelson J A. Direct interaction of the human cytomegalovirus IE86 protein with the cis repression signal does not preclude TBP from binding to the TATA box. J Virol. 1993;67:5595–5604. doi: 10.1128/jvi.67.9.5595-5604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jupp R, Hoffmann S, Stenberg R M, Nelson J A, Ghazal P. Human cytomegalovirus IE86 protein interacts with promoter-bound TATA-binding protein via a specific region distinct from the autorepression domain. J Virol. 1993;67:7539–7546. doi: 10.1128/jvi.67.12.7539-7546.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kerry J A, Priddy M A, Jervey T Y, Kohler C P, Staley T L, Vanson C D, Jones T R, Iskenderian A C, Anders D G, Stenberg R M. Multiple regulatory events influence human cytomegalovirus DNA polymerase (UL54) expression during viral infection. J Virol. 1996;70:373–382. doi: 10.1128/jvi.70.1.373-382.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kerry J A, Priddy M A, Stenberg R M. Identification of sequence elements in the human cytomegalovirus DNA polymerase gene promoter required for activation by viral gene products. J Virol. 1994;68:4167–4176. doi: 10.1128/jvi.68.7.4167-4176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohler C P, Kerry J A, Carter M, Muzithras V P, Jones T R, Stenberg R M. Use of recombinant virus to assess human cytomegalovirus early and late promoters in the context of the viral genome. J Virol. 1994;68:6589–6597. doi: 10.1128/jvi.68.10.6589-6597.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kowalik T F, Wing B, Haskill J S, Azizkhan J C, Baldwin A S, Jr, Huang E S. Multiple mechanisms are implicated in the regulation of NF-kappa B activity during human cytomegalovirus infection. Proc Natl Acad Sci USA. 1993;90:1107–1111. doi: 10.1073/pnas.90.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lang D, Gebert S, Arlt H, Stamminger T. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J Virol. 1995;69:6030–6037. doi: 10.1128/jvi.69.10.6030-6037.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leach F S, Mocarski E S. Regulation of cytomegalovirus late-gene expression: differential use of three start sites in the transcriptional activation of ICP36 gene expression. J Virol. 1989;63:1783–1791. doi: 10.1128/jvi.63.4.1783-1791.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu B, Hermiston T W, Stinski M F. A cis-acting element in the major immediate-early (IE) promoter of human cytomegalovirus is required for negative regulation by IE2. J Virol. 1991;65:897–903. doi: 10.1128/jvi.65.2.897-903.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu B, Stinski M F. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J Virol. 1992;66:4434–4444. doi: 10.1128/jvi.66.7.4434-4444.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Livingstone L R, White A, Sprouse J, Livanos E, Jacks T, Tlsty T D. Altered cell cycle arrest and gene amplification potential accompany loss of wild-type p53. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 46.Macias M P, Stinski M F. An in vitro system for human cytomegalovirus immediate early 2 protein (IE2)-mediated site-dependent repression of transcription and direct binding of IE2 to the major immediate early promoter. Proc Natl Acad Sci USA. 1993;90:707–711. doi: 10.1073/pnas.90.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malone C L, Vesole D H, Stinski M F. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J Virol. 1990;64:1498–1506. doi: 10.1128/jvi.64.4.1498-1506.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Margolis M J, Pajovic S, Wong E L, Wade M, Jupp R, Nelson J A, Azizkhan J C. Interaction of the 72-kilodalton human cytomegalovirus IE1 gene product with E2F1 coincides with E2F-dependent activation of dihydrofolate reductase transcription. J Virol. 1995;69:7759–7767. doi: 10.1128/jvi.69.12.7759-7767.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Michelson S, Turowski P, Picard L, Goris J, Landini M P, Topilko A, Hemmings B, Bessia C, Garcia A, Virelizier J L. Human cytomegalovirus carries serine/threonine protein phosphatases PP1 and a host-cell derived PP2A. J Virol. 1996;70:1415–1423. doi: 10.1128/jvi.70.3.1415-1423.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller S D, Farmer G, Prives C. p53 inhibits DNA replication in vitro in a DNA-binding-dependent manner. Mol Cell Biol. 1995;15:6554–6560. doi: 10.1128/mcb.15.12.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mocarski E S. Cytomegalovirus biology and replication. In: Roizman B, Whitely R J, Lopez C, editors. The human herpesviruses. New York, N.Y: Raven Press; 1991. pp. 173–226. [Google Scholar]
- 52.Muganda P, Mendoza O, Hernandez J, Qian Q. Human cytomegalovirus elevates levels of the cellular protein p53 in infected fibroblasts. J Virol. 1994;68:8028–8034. doi: 10.1128/jvi.68.12.8028-8034.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pari G S, Kacica M A, Anders D G. Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J Virol. 1993;67:2575–2582. doi: 10.1128/jvi.67.5.2575-2582.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pizzorno M C, Hayward G S. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target sequence located near the cap site. J Virol. 1990;64:6154–6165. doi: 10.1128/jvi.64.12.6154-6165.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quartin R S, Cole C N, Pipas J M, Levine A J. The amino-terminal functions of the simian virus 40 large T antigen are required to overcome wild-type p53-mediated growth arrest of cells. J Virol. 1994;68:1334–1341. doi: 10.1128/jvi.68.3.1334-1341.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ripalti A, Boccuni M C, Campanini F, Landini M P. Cytomegalovirus-mediated induction of antisense mRNA expression to UL44 inhibits virus replication in an astrocytoma cell line: identification of an essential gene. J Virol. 1995;69:2047–2057. doi: 10.1128/jvi.69.4.2047-2057.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sabbatini P, Chiou S K, Rao L, White E. Modulation of p53-mediated transcriptional repression and apoptosis by the adenovirus E1B 19K protein. Mol Cell Biol. 1995;15:1060–1070. doi: 10.1128/mcb.15.2.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwartz R, Sommer M H, Scully A, Spector D H. Site-specific binding of the human cytomegalovirus IE2 86-kilodalton protein to an early gene promoter. J Virol. 1994;68:5613–5622. doi: 10.1128/jvi.68.9.5613-5622.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scully A L, Sommer M H, Schwartz R, Spector D H. The human cytomegalovirus IE2 86-kilodalton protein interacts with an early gene promoter via site-specific DNA binding and protein-protein associations. J Virol. 1995;69:6533–6540. doi: 10.1128/jvi.69.10.6533-6540.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Slebos R J, Lee M H, Plunkett B S, Kessis T D, Williams B O, Jacks T, Hedrick L, Kastan M B, Cho K R. p53-dependent G1 arrest involves pRb-related proteins and is disrupted by the human papillomavirus 16 E7 oncoprotein. Proc Natl Acad Sci USA. 1994;91:5320–5324. doi: 10.1073/pnas.91.12.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sommer M H, Scully A L, Spector D H. Transactivation by the human cytomegalovirus IE2 86-kilodalton protein requires a domain that binds to both the TATA box-binding protein and the retinoblastoma protein. J Virol. 1994;68:6223–6231. doi: 10.1128/jvi.68.10.6223-6231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Speir E, Modali R, Huang E S, Leon M B, Shawl F, Finkel T, Epstein S E. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science. 1994;265:391–394. doi: 10.1126/science.8023160. [DOI] [PubMed] [Google Scholar]
- 63.Stasiak P C, Mocarski E S. Transactivation of the cytomegalovirus ICP36 gene promoter requires the alpha gene product TRS1 in addition to IE1 and IE2. J Virol. 1992;66:1050–1058. doi: 10.1128/jvi.66.2.1050-1058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Steegenga W T, van Laar T, Riteco N, Mandarino A, Shvarts A, van der Eb A J, Jochemsen A G. Adenovirus E1A proteins inhibit activation of transcription by p53. Mol Cell Biol. 1996;16:2101–2109. doi: 10.1128/mcb.16.5.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stenberg R M, Depto A S, Fortney J, Nelson J A. Regulated expression of early and late RNAs and proteins from the human cytomegalovirus immediate-early gene region. J Virol. 1989;63:2699–2708. doi: 10.1128/jvi.63.6.2699-2708.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Symonds H, Krall L, Remington L, Saenz-Robles M, Lowe S, Jacks T, Van Dyke T. p53-dependent apoptosis suppresses tumor growth and progression in vivo. Cell. 1994;78:703–711. doi: 10.1016/0092-8674(94)90534-7. [DOI] [PubMed] [Google Scholar]
- 67.Tack L C, Wright J H. Altered phosphorylation of free and bound forms of monkey p53 and simian virus 40 large T antigen during lytic infection. J Virol. 1992;66:1312–1320. doi: 10.1128/jvi.66.3.1312-1320.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thukral S K, Blain G C, Chang K K, Fields S. Distinct residues of human p53 implicated in binding to DNA, simian virus 40 large T antigen, 53BP1, and 53BP2. Mol Cell Biol. 1994;14:8315–8321. doi: 10.1128/mcb.14.12.8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tiemann F, Zerrahn J, Deppert W. Cooperation of simian virus 40 large and small T antigens in metabolic stabilization of tumor suppressor p53 during cellular transformation. J Virol. 1995;69:6115–6121. doi: 10.1128/jvi.69.10.6115-6121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Truant R, Antunovic J, Greenblatt J, Prives C, Cromlish J A. Direct interaction of the hepatitis B virus HBx protein with p53 leads to inhibition by HBx of p53 response element-directed transactivation. J Virol. 1995;69:1851–1859. doi: 10.1128/jvi.69.3.1851-1859.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsai H-L, Kou G-H, Chen S-C, Wu C-W, Lin Y-S. Human cytomegalovirus immediate-early protein IE2 tethers a transcriptional repression domain to p53. J Biol Chem. 1996;271:3534–3540. [PubMed] [Google Scholar]
- 72.van den Heuvel S J, van Laar T, The I, van der Eb A J. Large E1B proteins of adenovirus types 5 and 12 have different effects on p53 and distinct roles in cell transformation. J Virol. 1993;67:5226–5234. doi: 10.1128/jvi.67.9.5226-5234.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wade E J, Klucher K M, Spector D H. An AP-1 binding site is the predominant cis-acting regulatory element in the 1.2-kilobase early RNA promoter of human cytomegalovirus. J Virol. 1992;66:2407–2417. doi: 10.1128/jvi.66.4.2407-2417.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wade M, Kowalik T F, Mudryj M, Huang E S, Azizkhan J C. E2F mediates dihydrofolate reductase promoter activation and multiprotein complex formation in human cytomegalovirus infection. Mol Cell Biol. 1992;12:4364–4374. doi: 10.1128/mcb.12.10.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.White E, Cipriani R, Sabbatini P, Denton A. Adenovirus E1B 19-kilodalton protein overcomes the cytotoxicity of E1A proteins. J Virol. 1991;65:2968–2978. doi: 10.1128/jvi.65.6.2968-2978.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wing B A, Huang E S. Analysis and mapping of a family of 3′-coterminal transcripts containing coding sequences for human cytomegalovirus open reading frames UL93 through UL99. J Virol. 1995;69:1521–1531. doi: 10.1128/jvi.69.3.1521-1531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wing B A, Lee G C, Huang E S. The human cytomegalovirus UL94 open reading frame encodes a conserved herpesvirus capsid/tegument-associated virion protein that is expressed with true late kinetics. J Virol. 1996;70:3339–3345. doi: 10.1128/jvi.70.6.3339-3345.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu J, Jupp R, Stenberg R M, Nelson J A, Ghazal P. Site-specific inhibition of RNA polymerase II preinitiation complex assembly by human cytomegalovirus IE86 protein. J Virol. 1993;67:7547–7555. doi: 10.1128/jvi.67.12.7547-7555.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu X, Levine A J. p53 and E2f-1 cooperate to mediate apoptosis. Proc Natl Acad Sci USA. 1994;91:3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yurochko A D, Kowalik T F, Huong S M, Huang E S. Human cytomegalovirus upregulates NF-κ B activity by transactivating the NF-κ B p105/p50 and p65 promoters. J Virol. 1995;69:5391–5400. doi: 10.1128/jvi.69.9.5391-5400.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhu H, Shen Y, Shenk T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J Virol. 1995;69:7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]