Abstract
To investigate the importance of the IE1 p72 regulatory protein during human cytomegalovirus replication, a recombinant virus unable to synthesize IE1 p72 was constructed. The Towne strain mutant CR208 lacked exon 4 of the major immediate-early gene and was isolated and complemented in an IE1-expressing immortalized human fibroblast line (ihfie1.3). Replication of CR208 in primary human fibroblasts was completed after an input multiplicity of 10 PFU/cell but was severely impaired at 0.1 PFU/cell. CR208 formed plaques with lower efficiency on primary fibroblasts than on ihfie1.3 cells, and the relationship between the CR208 inoculum size and the resulting number of undersized plaques was nonlinear, indicating that multiple particles of CR208 were required to initiate lytic replication in a single primary fibroblast. After infection of primary fibroblasts with CR208 at 5 PFU/cell, a normal pattern of viral antigens was detected, although IE1 p72 was absent. During lower-multiplicity infections, IE2 protein was consistently detected at similar levels in a similar proportion of CR208-infected cells relative to the case for a Towne infection, but many fewer CR208-infected cells contained the ppUL44 polymerase accessory protein when evaluated at 24 or 48 h after infection. Furthermore, fibroblasts infected with CR208 at a low multiplicity failed to form viral DNA replication compartments, despite having expressed IE2 p86. These low-multiplicity growth and expression defects were corrected in two rescued derivatives of CR208 able to synthesize IE1 p72. One rescued virus (CR249) carried a deletion removing the large intron between exons 1 and 2 of the ie1-ie2 locus, revealing that this intron was dispensable for growth in cell culture.
Human cytomegalovirus (HCMV) is a widespread herpesvirus, which infects 50 to 100% of the human population. HCMV disease is a significant medical problem, although it is mostly restricted to patients with immature or compromised immune systems (5). HCMV gene expression during productive infection of cultured human fibroblast cells follows an ordered cascade of expression of immediate-early (IE or α) genes, followed by expression of delayed-early (DE or β) genes, followed after viral DNA replication by strong expression of late (L or γ) genes. HCMV also modulates the expression of many cellular genes during the virus life cycle (47).
IE1 p72 (IE1491aa) is the most abundant product of the strongly transcribed major IE locus of HCMV and is detected in the nuclei of infected cells both in culture and in infected individuals. RNA transcripts originating from the major IE enhancer-promoter (MIEP) immediately after infection span five major exons and are alternately spliced and polyadenylated to produce messages for either IE1 p72 (exons 1 to 4) or IE2 p86 (IE2579aa) (exons 1 to 3 and 5) (74, 75). The large exon unique to the IE2 p86 message, originally termed exon 7 (75), is here termed exon 5 (47). Translation of IE1 p72 and IE2 p86 initiates in exon 2, and the proteins share 85 identical residues at their amino termini.
IE2 p86 is thought to be the major specific transcriptional regulator of the lytic cycle of HCMV. Consistent with a such a role, IE2 p86 is a sequence-specific DNA binding protein, which autoregulates by binding adjacent to the transcription start site of the MIEP (12, 32, 40, 41, 44, 53, 83) and also binds to specific sites in other HCMV promoters (3, 66, 67). IE2 p86 interacts with diverse components of the cellular transcription machinery, including TBP, TFIIB, CREB, CBP, and c-Jun (7, 23, 33, 39, 65, 67), and in transient-cotransfection assays IE2 activates transcription from a wide range of HCMV and cellular promoters (15, 28, 35, 45, 54, 70, 73). Transactivation by IE2 p86 may be mediated by upstream promoter elements (39, 65), but promiscuous activation of heterologous promoters is frequently TATA box mediated (23).
In contrast, IE1 p72 acts via discrete promoter elements to stimulate a relatively limited number of viral and cellular promoters. Notably, IE1 p72 transactivates its own promoter, the HCMV MIEP (13, 45, 73), acting via NF-κB sites in the 18-bp repeat sequences of the enhancer (13, 62). IE1 p72 also activates the human immunodeficiency virus type 1 long terminal repeat and the cellular DNA polymerase α, dihydrofolate reductase, and prointerleukin-1β gene promoters (26, 29, 80, 82). IE1 p72 physically interacts with the cellular transcription factors SP-1, E2F-1, and CTF-1 (26, 43, 46), but binding of IE1 to the basal transcription factors TFIIB and TFIID has not been detected (7, 23). As distinct from its promoter-specific transactivator functions, coexpression of IE1 p72 augments the promiscuous transactivating activities of IE2 p86 on a wide variety of viral and cellular promoters (9, 35, 43, 45, 73). However, the accessory viral transactivators TRS1/IRS1, UL36-UL38, and UL112-UL113 are in addition required for the maximal stimulation of a number of HCMV DE promoters (30, 71).
Several other distinct activities have been described for IE1 p72, but considered together, these do not form a clear picture of the probable functions of IE1 in the context of the virus life cycle. IE1 p72 colocalizes in stable cell lines and in infected cells with cellular chromatin, a property shared with the EBNA-1 latent antigen of Epstein-Barr virus (38). IE1 p72 also interacts with the pocket protein p107 (56), inhibits tumor necrosis factor alpha-driven apoptosis in HeLa cells (85), and causes the relocalization of PML protein from nuclear ND10 subdomains (1, 34, 37). Together with IE2 p86, IE1 p72 is one of six auxiliary factors required in trans, alongside core viral DNA replication proteins, for the replication in transfected cells of a plasmid containing the HCMV lytic origin of replication (51). Although this may partly reflect a requirement for IE1 p72 to fully activate core replication gene promoters (30), IE1 p72 was nevertheless required for efficient viral origin replication in permissive human fibroblasts, even when the core replication proteins were constitutively expressed (63).
IE1 p72 and IE2 p86 are widely thought to be the central regulators of the cascade of viral and cellular gene expression during lytic HCMV infection. There is, however, very little direct evidence for this assertion, other than that obtained by the use of antisense phosphorothioate oligonucleotides and antisense RNA expression to inhibit ie1 and ie2 expression and hence viral replication (2, 4, 6). For other herpesviruses, most notably herpes simplex virus type 1 (HSV-1), spontaneous and recombinant viral mutants with defects in gene-regulatory proteins have provided vital experimental information, facilitating a better understanding of the important functions of these proteins in the context of viral infection (59). As HCMV mutants with mutations in ie1 and ie2 had not been described, we set out to generate such mutants through construction of recombinant viruses. We have previously reported the isolation and preliminary characterization of an HCMV ie1 mutant (RC303ΔAcc) by reconstruction from cosmids derived from the Towne and Toledo strains (48). Here, we report the isolation and characterization of an ie1 mutant HCMV in a pure Towne strain background (CR208), derived by recombination between wild-type virus and plasmid DNA containing the selectable marker gene gpt. The two ie1 mutant viruses share a low-multiplicity growth defect in primary fibroblasts; however, viral gene expression during an attenuated low-multiplicity infection by CR208 was more extensive than was previously observed for RC303ΔAcc, indicating that IE1 p72 function may have a strong impact at multiple stages of the virus life cycle.
MATERIALS AND METHODS
Plasmids.
Plasmid DNAs were maintained and propagated in Escherichia coli DH5α, except for gpt-containing plasmids, which were maintained and propagated as previously described in the E. coli gpt mutant strain WB-1 (22). Plasmid pON303G (13) contained the 7.4-kbp SalI IE fragment of HCMV Towne strain (AD169 equivalent nucleotides [nt] 168820 to 176220) (10) ligated into the SalI site of pGEM2 (Promega). pON303G was deleted between two NaeI sites to construct pON2535, which contained a 5.7-kbp viral NaeI-SalI fragment (AD169 equivalent nt 170512 to 176620). Exon 4 was deleted from pON303G by using endonuclease Bst1107I, which cuts at AccI sites in the flanking introns, to produce plasmid pON2551. A 2.6-kbp XhoI-ScaI fragment of pON303G (AD169 equivalent nt 170233 to 172863) was ligated between XhoI and EcoRV sites of pUC21 (79) to make plasmid pON2517. A 2.6-kbp SalI-BglII fragment of pON303G (AD169 equivalent nt 168820 to 171443) was cloned as an XbaI-BglII fragment into pBluescript SK+ (Stratagene) to produce plasmid pON2512. Plasmid pON2523 contained a 3-kbp EcoRI-XbaI fragment of pXbaE (78) ligated between EcoRI and XbaI sites of pUC21. Viral sequences in pON2523 commence at the EcoRI site in UL129 (AD169 equivalent nt 175524) and proceed away from the MIEP into Towne sequences not shared with AD169 (8). Plasmid pON308G (13) contained the 3.9-kbp ClaI IE fragment of HCMV (Towne) (AD169 equivalent nt 170811 to 174751) ligated into the AccI site of pGEM2.
A 1.9-kbp HpaI-PstI fragment of pON308G was ligated between StuI and PstI sites in the E. coli guanosine phosphoribosyl transferase (gpt gene) expression vector pON1101 (22) to produce plasmid pON2518. pON2518 thus contained an HpaI-ClaI viral fragment (AD169 equivalent nt 172892 to 174751), including the MIEP and part of UL127, placed upstream of a gpt expression cassette. A 2.2-kbp EagI-BamHI fragment of pON2518 containing the MIEP and gpt expression cassette was then ligated between EagI and EcoRI sites of pON2535 to construct plasmid pON2540. pON2540 effectively contains a 5.7-kbp NaeI-SalI fragment of the HCMV Towne strain (AD169 equivalent nt 170512 to 176220) with a gpt expression cassette cloned between ClaI and EcoRI sites (AD169 equivalent nt 174751 to 175524), replacing UL127 to UL129. The EagI-HindIII fragment of pON2540 containing the gpt insertion was used to replace the EagI-HindIII fragment of pON2551, producing plasmid pRG208. Plasmid pRG208 effectively contained the 7.4-kbp SalI IE fragment of HCMV Towne, with the following alterations: the deletion of exon 4 sequences between two Bst1107I sites (AD169 equivalent nt 170954 to 172367) and the replacement of the open reading frames (ORFs) UL127 to UL129 between ClaI and EcoRI sites (AD169 equivalent nt 174751 to 175524) with an expression cassette containing the HSV tk promoter, E. coli gpt, and the simian virus 40 early poly(A) signal. pRG208 is represented in Fig. 1a.
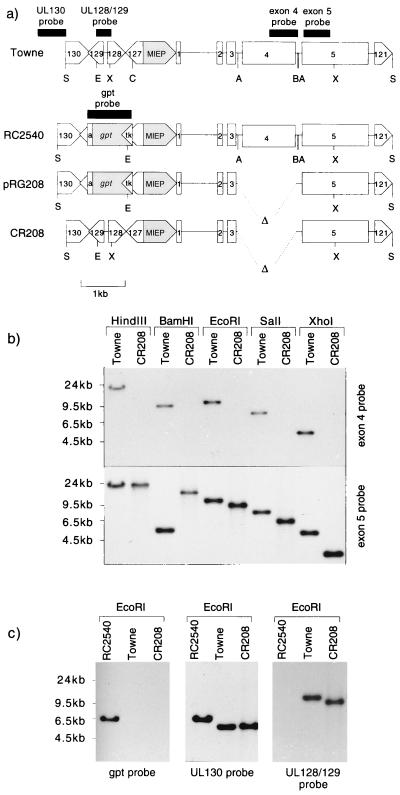
FIG. 1.
(a) Genomic structures of Towne, RC2540, and CR208 viruses in the SalI fragment containing the IE locus, together with the structure of plasmid pRG208. The positions of ORFs UL121 and UL127 to UL130 are shown, together with those of the MIEP (shaded) and exons 1 to 5 of the major IE gene. The gpt expression cassette consists of the HSV-1 thymidine kinase gene promoter (tk), guanosine phosphoribosyl transferase-coding sequences (gpt) (shaded), and the simian virus 40 early polyadenylation signal (a). Δ, HCMV sequences deleted from pRG208 and CR208. Restriction enzyme sites used in Southern blot analysis and for gpt insertion: A, AccI (Bst1107I); B, BamHI; C, ClaI; E, EcoRI; S, SalI; X, XhoI. Probe fragments are shown next to viral sequences as solid blocks. (b) Southern blot analysis of Towne and CR208 DNAs cut with the restriction enzymes shown and hybridized with an exon 4 probe or an exon 5 probe (see panel a). Molecular sizes of standards are shown to the left. (c) Southern blot analysis of Towne, CR208, and RC2540 DNAs cut with EcoRI and hybridized to probes from gpt, UL130, and UL128 and -129 (see panel a). Molecular sizes of standards are shown to the left.
A 933-bp EagI-XhoI fragment from the IE2 p86 cDNA expression vector pON2206 (31) was ligated between the unique EagI and XhoI sites of pRG208 to produce plasmid pRG232. Ligation into pON303G of a 535-bp NdeI-ApaI fragment from pRG232 (containing the proximal MIEP and the spliced exon 1-exon 2 boundary) yielded plasmid pRG249. pRG249 effectively contained the 7.4-kbp SalI IE fragment of the HCMV Towne strain, with the first intron of the major IE gene (AD169 equivalent 172783 to 173609) removed, exactly as occurs in spliced IE mRNAs.
Cell lines and culture.
All cell lines were cultured in Dulbecco modified Eagle medium (GIBCO-BRL) supplemented with penicillin, streptomycin, nonessential amino acids, and 10% (vol/vol) NuSerum I (Collaborative Research). All procedures were carried out with cells cultured at 37°C and with 5% CO2. The human fetal lung fibroblast (HFL) line GM05387 and the human foreskin fibroblast (HFF) line GM03468A were obtained from the National Institute of General Medical Sciences, Camden, N.J.
Approximately 5 × 105 freshly plated early-passage HFFs were treated with 3 ml of filtered (0.45-μm-pore-size filter) culture supernatant from amphotropic PA317-N2-CMVIE packaging cells (60), a gift from Eli Gilboa. After overnight absorption, cells were plated at a low dilution and were cultured in the presence of 400 μg of G418 (GIBCO-BRL) per ml. After 3 weeks of selection, the resulting G418-resistant cells were pooled, and approximately 5 × 105 freshly plated cells were treated as described above with filtered culture supernatant from the amphotropic PA317-LXSN16E6E7 packaging cell line (24), a gift from Denise Galloway. The resulting rapidly growing G418-resistant clones were pooled and were termed ihfie1.3 cells. A G418-resistant polyclonal immortalized cell line was made by treatment of primary HFFs with filtered PA317-LXSN16E6E7 cell supernatant and was called ihf-2.
Virus culture and generation of recombinant HCMV strains.
Cell-free virus stocks were prepared in HFL cells (for Towne, RC2540, CRQ208, and CR249) or ihfie1.3 cells (for CR208). Cells infected with approximately 0.01 PFU/cell were cultured until a cytopathic effect was apparent in 100% of cells and were then refed with fresh culture medium. After 5 days, the culture supernatant was passed through a 0.45-μm-pore-size filter, mixed 1:1 with a 9% (wt/vol) nonfat milk solution (autoclaved), and stored at −70°C.
Detailed procedures for the isolation of recombinant HCMV strains containing the gpt gene have been described previously (22). Viral recombinant RC2540 was isolated by recombination between SalI/NaeI-linearized pON2540 and the HCMV Towne strain and was purified by these standard procedures. To isolate CR208, 8 μg of SalI-linearized pRG208 DNA was transfected (11) into 5 × 105 HFL cells, which were infected 24 h later with 3 PFU of HCMV (Towne strain) per cell. Progeny virus, enriched for gpt expression during three passages on ihfie1.3 cells, was plaque purified twice on ihfie1.3 cells, once with selection for gpt (22) and once without selection. By Southern blot analysis, the first-round viral isolate lacked detectable exon 4 sequences but had heterogenous sequences in the UL127 to -129 region. A homogenous isolate lacking exon 4 and derived from the second round of plaque purification was designated CR208.
To generate CR208 derivatives, 8 μg of SalI-linearized pON303G or pRG249 was transfected into 5 × 105 HFL cells, which were then productively infected at a high multiplicity (approximately 10 PFU/cell) with CR208. A single passage of progeny virus on HFL cells at a low multiplicity (approximately 0.05 PFU/cell) resulted in the outgrowth of a small number of rapidly growing plaques. Viruses from these growth-selected stocks were then plaque purified on HFL cells. A derivative virus from the pON303G recombination was termed CRQ208; a derivative of pRG249 was termed CR249.
Assays of viral growth.
Virus plaque assays were performed on HFLs, HFFs, or ihfie1.3 cells. Virus samples were serially diluted in serum-free medium and absorbed in duplicate onto 105 cells plated 16 to 24 h previously in 12-well dishes, and the inoculum was replaced after 1 h with culture medium supplemented with 0.16 mg of purified human gamma globulin per ml. After growth for 10 days, cells were fixed in 100% methanol (5 min) and stained with 0.04% (wt/vol) Giemsa stain in 10% methanol (20 min), and darkly staining foci or plaques were counted.
For growth curves, 2 × 105 freshly plated cells in six-well dishes were infected with the desired inoculum of virus in serum-free culture medium. After absorption for 1 h, the inoculum was replaced with normal culture medium. Samples of infected-cell medium were removed at each time point and were stored at −70°C. The infected cells were fed with fresh culture medium after each sampling. Duplicate plaque assays of frozen samples were subsequently performed on ihfie1.3 cells.
Analysis of viral DNA.
Viral DNA was isolated from the cytoplasm of the same infected cells from which culture supernatant stocks were harvested. DNA was purified from proteinase K-digested cytoplasmic extracts (69), either by phenol extraction and isopropanol precipitation or by sodium iodide equilibrium density centrifugation (81).
For Southern blot analysis, approximately 0.25 μg of viral DNA was digested with the appropriate restriction endonuclease and was separated by electrophoresis on 0.6% agarose–Tris-borate-EDTA gels. DNA was transferred by capillary blotting overnight to Hybond N+ membranes (Amersham) in 0.2 M NaOH–0.6 M NaCl. Filters were hybridized, as described previously (22), to DNA probes radiolabelled with [α-32P]dCTP (Amersham) by random hexamer priming. Probe fragments for Southern hybridization were generated as follows and were purified from low-melting-point agarose gels: an exon 4-specific probe was isolated as a 474-bp BamHI-BglII fragment of pON2517 (AD169 equivalent nt 170969 to 171443), a 455-bp SacI-NcoI fragment of pON2512 was used to detect exon 5 sequences (AD169 nt 170678 to 170223), a 1-kbp PvuII-BamHI fragment of pON1101 was used to detect gpt sequences, a 626-bp HindIII-SalI fragment of pON2523 detected the UL130 ORF (AD169 nt 176844 to 176218), a 341-bp EcoRI-XhoI fragment of pON303G detected the UL128 and UL129 ORFs (AD169 nt 175524 to 175183), and a 728-bp SacI fragment of pG308 detected intron 1 and exon 1 of the major IE gene (AD169 nt 173016 to 173744). Filters were stripped in 0.1% sodium dodecyl sulfate (SDS) at 100°C and were scanned on an Instant Imager (Hewlett Packard) to assess probe removal prior to subsequent hybridizations with different probes. The results of Southern blotting were visualized by autoradiography.
Direct sequencing of iodide gradient-purified viral DNA was performed by using the Thermo-Sequenase kit (U.S. Biochemicals/Amersham), by direct incorporation of [α-33P]dATP label and according to the manufacturer’s instructions. Viral DNA (2.5 μg) was digested with SalI and ethanol precipitated, and sequence was obtained by using either primer P127 (5′TGGCGATATCAGTTACACAG3′) (AD169 equivalent nt 174683 to 174702) or P129 (5′GAAAACCGCGCGTCATGAGT3′) (AD169 equivalent nt 175636 to 175617).
Analysis of viral antigens and DNA replication.
Hybridoma supernatants of mouse monoclonal antibodies p63-27 (55), SMX (55), and BS510 (64), which recognize IE1 p72, IE2 p86, and ppUL44, respectively, were the gifts of Bodo Plachter. Mouse monoclonal antibody CH160 ascites (16), which recognizes an amino-terminal epitope shared by IE1 p72 and IE2 p86, was the gift of Lenore Pereira. Mouse monoclonal antibody 2A6 ascites, specific for the pp65 virion phosphoprotein (57), was kindly provided by Maria Grazia Revello. Human convalescent serum from a patient recovering from primary HCMV infection was kindly provided by Mark Wills. Antibromodeoxyuridine (anti-BrdU) rat monoclonal antibody MAS-250 hybridoma supernatant and Texas red-linked goat anti-mouse immunoglobulin G (IgG) conjugate were purchased from Sera-Labs. Fluorescein isothiocyanate-conjugated goat anti-mouse IgG antibody was purchased from Cappell.
For Western blot analysis, total proteins from 2 × 105 cells were harvested by a phosphate-buffered saline (PBS) wash and direct lysis into 0.3 ml of SDS-polyacrylamide gel electrophoresis sample buffer. Cells were infected beforehand with appropriate multiplicities of virus, as described for the growth curve experiments. Protein samples were stored at −20°C prior to analysis. Boiled proteins derived from the equivalent of 6 × 103 cells were separated on SDS–8.5% polyacrylamide gels and were electrophoretically blotted onto nitrocellulose filters. Protein transfer was confirmed by Ponceau S staining of filters. Filters were blocked for 1 h in 3% (wt/vol) bovine serum albumin in PBS and were then incubated for 3 h with primary antibody diluted in 10% fetal calf serum in PBS (PBS-FCS). After three 5-min washes in PBS–0.5% Nonidet-P40, the filters were rinsed in PBS and incubated for 1 h with rabbit anti-mouse IgG–horseradish peroxidase (HRP) conjugate (DAKO) or rabbit anti-human Ig–HRP conjugate (DAKO), each diluted 1/1,000 in PBS-FCS. After three further PBS–0.5% Nonidet P-40 washes, bound HRP was visualized by using nickel-enhanced diaminobenzidine-hydrogen peroxide as a substrate (25).
For immunocytochemical staining, 105 cells grown on 19-mm-diameter glass coverslips were infected with virus diluted in serum-free medium, and the culture medium was replaced after a 1-h absorption step. Where indicated, sodium phosphonoformate (PFA) (200 μg/ml) was included in the culture medium. At the appropriate time after infection, cells were washed in PBS and fixed in 1% paraformaldehyde in PBS for 15 min. Cells were then permeabilized by a 5-min incubation in 0.1% Triton X-100 in PBS. PBS-washed fixed monolayers were incubated with primary antibody (diluted as appropriate in PBS-FCS) for 1 h, rinsed three times in PBS, and incubated with fluorescence-labelled secondary antibody diluted 1:100 in PBS-FCS for 1 h. Cell nuclei were counterstained by the inclusion of Hoechst 33258 at 100 ng/ml during the final incubation step. PBS-washed stained cells were mounted in glycerol-PBS (Citifluor) and were visualized by fluorescence microscopy. To double label IE2 protein and viral DNA replication compartments, cells were pulse-labelled by the addition of 10 μM BrdU to the culture medium for 30 min prior to fixation, treated after fixation with 4 N HCl for 10 min, neutralized in PBS, and then incubated for 1 h with a 1:1 mixture of mouse anti-IE2 monoclonal antibody SMX and rat anti-BrdU monoclonal antibody MAS-250, followed by a 1-h incubation with a mixture of non-cross-reactive anti-rat IgG–tetramethyl rhodamine isothiocyanate and anti-mouse IgG–fluorescein isothiocyanate secondary antibodies (Chemicon) (each diluted 1:100 in PBS-FCS). All antibody incubations were carried out at room temperature.
RESULTS
Generation of an ie1 mutant virus.
To complement a viral ie1 mutant, a permissive cell line expressing IE1 p72 was required. As human fibroblasts immortalized by the E6 and E7 genes of human papillomavirus type 16 support HCMV growth to high titers (14), the immortalized IE1-expressing cell line ihfie1.3 (48) was generated by the sequential transduction of an early-passage HFF culture with retroviral vectors expressing IE1 p72 and the human papillomavirus type 16 E6 and E7 products.
As IE1 p72 shares two coding exons with IE2 p86, the disruption of IE1 was accomplished by removing exon 4 of the major IE gene, which encodes all residues unique to IE1 p72. A deletion which removes exon 4 via AccI sites in the flanking introns precludes IE1 p72 expression yet leaves important splicing signals intact and results in the expression of functional IE2 p86 in transient assays (12, 15, 27, 28, 45, 54, 73). The deletion does not disrupt the consensus polyadenylation signal downstream of exon 4. This AccI deletion was linked on a single plasmid to the insertion of a gpt expression cassette into ORFs UL127 to UL129, upstream of the MIEP. The sequences replaced by the gpt insertion overlapped the upstream end of the modulator region of the MIEP (42) but did not include the dyad symmetry element which has been shown to be important for modulator function in undifferentiated teratocarcinoma cells (68). The structure of plasmid pRG208 is depicted in Fig. 1a. Plasmid pON2540, which contained the same gpt cassette insertion into ORFs UL127 to -129 but had a wild-type IE gene region, was also constructed. Recombinant virus RC2540 was readily isolated by recombination between plasmid pON2540 and Towne virus and carried the expected gpt insertion, which was stable upon repeated passage. No major differences between the growth characteristics of RC2540 and the parental Towne strain (21) were detected.
A recombinant between pRG208 and HCMV Towne was generated and purified as described in Materials and Methods and was designated CR208. A Southern blot analysis comparing CR208 genomic DNA to parental Towne DNA is shown in Fig. 1b. A probe to exon 4 sequences detected the expected fragments in Towne DNA (21.3-kbp HindIII, 9.9-kbp BamHI, 10.9-kbp EcoRI, 7.4-kbp SalI, and 5-kbp XhoI fragments) but did not hybridize to CR208 DNA. Hybridization of the same blot to a probe for exon 5 sequences detected similarly sized fragments in Towne DNA, except that a different 5.3-kbp fragment hybridized in the BamHI digest, consistent with the known presence of a BamHI site separating exons 4 and 5. Exon 5 sequences were also detected in CR208 DNA, and in the cases of EcoRI, SalI, and XhoI digests, the fragment sizes (9.5, 6, and 3.6 kbp, respectively) were consistent with a 1.4-kbp deletion of exon 4. Differences in HindIII fragment sizes were barely visible, and the BamHI digest revealed a novel 13.8-kbp fragment, consistent with the loss of a BamHI site in the 1.4-kbp exon 4 deletion and the consequent fusion of adjoining 9.9- and 5.3-kbp BamHI fragments.
As shown in Fig. 1c, a blot of EcoRI-digested viral DNAs from Towne, RC2540, and CR208 was sequentially hybridized to probes representing gpt, UL130, and UL128-UL129 sequences. A 6.2-kbp fragment of RC2540 DNA hybridized to the gpt probe, but gpt sequences were not detected in the Towne control. Surprisingly, gpt sequences were not detected in the CR208 genome. Conversely, 10.9-kbp Towne and 9.5-kbp CR208 fragments hybridized to the UL128-UL129 probe, which did not recognize sequences in the RC2540 genome. All three genomes contained UL130 sequences, but the EcoRI fragment detected in RC2540 (6.4 kbp) was altered relative to that in Towne (5.5 kbp), consistent with the loss of an EcoRI site at the point of the gpt insertion in UL129 and the introduction of a new site in the HSV tk promoter of the gpt expression cassette. The size of the EcoRI fragment detected in CR208 (5.5 kbp) was identical to that in Towne.
Together, these results implied that the UL127- to UL129 region in CR208 was unaltered from the wild type. To establish this fully, genomic DNAs from CR208 and Towne were directly sequenced by using synthetic primers derived from the UL127 and UL129 ORFs, flanking the expected site of gpt insertion, as described in Materials and Methods. The resulting sequence data confirmed that the CR208 genome matched Towne sequences over the boundaries of the missing insertion. Thus, the gpt expression cassette, which presumably enabled the selection of viruses lacking exon 4, was lost from the CR208 genome and was replaced with wild-type sequences. This loss was surprising in the light of the stability of the RC2540 isolate and suggested an influence of the exon 4 deletion upon the stability of the UL127 to -129 gpt insertion. CR208 probably arose by an early homologous recombination event between an exon 4 mutant bearing the gpt insertion and a wild-type virus and became enriched and isolated once selection for gpt was removed during plaque purification. When purified viral DNAs from CR208 and Towne were digested with BamHI, EcoRI, HindIII, or XbaI, separated on agarose gels, and ethidium bromide stained to reveal all DNA fragments, no gross differences besides those expected from the exon 4 deletion were seen (data not shown). The genomic structure of CR208, containing a wild-type UL127 to -129 region, simplified the interpretation of the following experiments, as the intended deletion of exon 4 was the only detectable physical lesion in this virus.
The ie1 cDNA in ihfie1.3 cells did not contain any sequences homologous to regions directly flanking the exon 4 deletion in CR208. ihfie1.3 cells contained exon 4 sequences from the 5′ boundary to the BamHI site in the 3′ noncoding region (AD169 equivalent nt 172221 to 170969), and the deletion in CR208 spanned AD169 equivalent nt 172367 to 170954. Upstream of exon 4, CR208 and ihfie1.3 cells had homology in the enhancer and exons 1, 2, and 3 of the major IE gene. However, homologous recombination through any of these shared sequences to restore exon 4 to the viral genome should have resulted in the loss of the exon 3-intron 3 boundary, the site of differential splicing of the messages for IE1 p72 and IE2 p86 (75). The loss of this potential for alternate splicing would be expected to preclude IE2 p86 expression from such a viral genome. The spontaneous occurrence in CR208 stocks of revertant viruses able to express both IE1 and IE2 was therefore not anticipated, and it has not been observed.
The ie1 mutant was replication deficient after low-multiplicity infection of primary human fibroblasts.
Working stocks of the CR208 deletion mutant were grown and titrated in ihfie1.3 cells. Peak titers reached in the culture medium of infected cells were similar to those generally obtained from Towne virus grown in primary HFL cells, approximately 107 PFU/ml. Prior to all subsequent experiments, CR208 and Towne stocks were titrated in parallel on ihfie1.3 cells. Thus, PFUs in comparative experiments were always defined on an ihfie1.3 background. Towne and CR208 plaques formed on ihfie1.3 cells had similar sizes and morphologies.
Virus replication was assayed after both high (10-PFU/cell)- and low (0.1-PFU/cell)-multiplicity infections of ihfie1.3 cells and primary fibroblasts. The culture medium was sampled at various times postinfection and was titrated on ihfie1.3 cells. In ihfie1.3 cells, the two viruses accumulated to similar levels after infection at high (data not shown) and low (Fig. 2b) input multiplicities. After high-multiplicity infection of primary fibroblasts, the two viruses again accumulated to similar levels (data not shown), consistent with results obtained with a similar ie1 mutant virus generated by cosmid reconstruction (48). As previously observed (48), ie1 mutant progeny from primary HFL cells readily infected fresh primary HFL cultures, and cell-free CR208 was thus passaged over three generations without loss of infectivity and without the appearance of detectable exon 4 sequences in the resulting viral genomes (21). During the first round of infection in HFLs, the assembly of IE1 p72 into CR208 virions released from ihfie1.3 cells could theoretically account for the ability of these virions to undergo a single productive round of infection in the absence of complementation. The demonstration of the serial passage of cell-free CR208 virus in primary HFL cells argued against this explanation and showed that after high-multiplicity input, CR208 could replicate to high levels in the absence of IE1 p72 protein. In contrast, when Towne and CR208 were used to infect primary fibroblasts at a multiplicity of 0.1 PFU/cell, a marked reduction in the subsequent accumulation of replicated virus was seen (Fig. 2a). Replication of CR208 was severely defective compared with that of the parental Towne strain, reaching titers 3 orders of magnitude lower after 9 days of incubation. In addition, the viral cytopathic effect apparent by light microscopy was much reduced, with only a few foci of infection visible in the CR208-infected culture after 10 days.
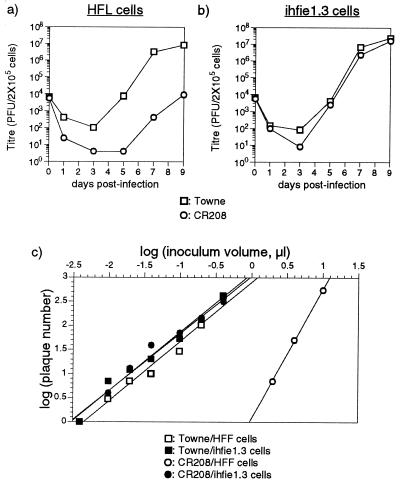
FIG. 2.
(a and b) Multiple-step growth curve analysis of Towne and CR208 viruses in HFL cells (a) and ihfie1.3 cells (b). The results are a time course of the total PFU of infectious virus present in 4 ml of culture supernatant at the indicated sampling times, as measured by plaque formation on ihfie1.3 cells. Experiments were performed with 2 × 105 cells at an input multiplicity of 0.1 PFU/cell. The results of a single experiment are shown; titers are the mean results from two parallel titrations. (c) Plot of titration data taken from a single plaque assay (Table 1), relating the plaque numbers formed on 2 × 105 HFFs or ihfie1.3 cells by various inocula of Towne or CR208 virus stocks. Log plaque number is plotted against log inoculum volume.
Another manifestation of this low-multiplicity growth defect was revealed by plaque assays performed with ihfie1.3 cells, HFLs, HFFs, or the control immortalized ihf-2 cell line. CR208 had a much-reduced plaquing efficiency in each of the three IE1-negative cell lines compared with ihfie1.3 cells. In addition, there was a strongly nonlinear relationship between the input CR208 inoculum volume and the resulting plaque number on HFLs, HFFs, or ihf-2 cells. Plaques formed by CR208 were much smaller than those formed by Towne strain virus on noncomplementing cells and were also much smaller than CR208 plaques formed on the complementing ihfie1.3 cell line. Although there was always considerable variability in HCMV plaque sizes, CR208 plaques on noncomplementing cells would typically consist of 5 to 10 intensely Giemsa-stained cells, at times when Towne plaques comprised 50 to 100 such cells. The results of a representative experiment are shown in Table 1, comparing plaque formation by Towne and CR208 on 2 × 105 ihfie1.3 cells or primary HFFs. The two virus stocks had similar titers when assayed on ihfie1.3 cells and showed reasonably linear relationships between inoculum size and plaque number. In HFFs, Towne virus formed plaques with an efficiency similar to that in ihfie1.3 cells and again showed a reasonably linear relationship between inoculum size and plaque number. In strong contrast, dilution of the CR208 inoculum resulted in a disproportionate drop in the number of small plaques formed on HFF cells. As an example, a 60% reduction in the CR208 inoculum reduced the plaque number 11-fold, from 543 to 50.
TABLE 1.
Comparison of plaque formation by Towne and CR208 viruses on HFF and ihfie1.3 cells
| Virus inoculum (μl) | No. of plaques formed
|
|||
|---|---|---|---|---|
| Towne
|
CR208
|
|||
| HFF cells | ihfie1.3 cells | HFF cells | ihfie1.3 cells | |
| 20 | TNTCa | TNTC | TNTC | TNTC |
| 10 | TNTC | TNTC | 543b | TNTC |
| 4 | TNTC | TNTC | 50b | TNTC |
| 2 | TNTC | TNTC | 7b | TNTC |
| 1 | TNTC | TNTC | 0 | TNTC |
| 0.4 | 412 | 418 | 0 | 318 |
| 0.2 | 102 | 132 | 0 | 145 |
| 0.1 | 29 | 53 | 0 | 70 |
| 0.04 | 10 | 20 | 0 | 39 |
| 0.02 | 7 | 12 | 0 | 13 |
| 0.01 | 3 | 7 | 0 | 4 |
| 0.004 | 1 | 1 | 0 | 1 |
TNTC, too numerous to count.
Small plaques.
These data are consistent with a requirement for multiple CR208 particles to initiate a fully productive viral infection in a single cell. The small size of CR208 plaques might be the consequence of slower virus spread, resulting from the reduced infectivity of single particles. Assuming that the number of particles required to initiate a plaque (n) is relatively constant, at low inputs a likely relationship between inoculum volume (i) and plaque number produced (P) could be approximated by P = ain, where a is a constant for any fixed value of n and depends on n itself, the virus stock titer, and the number of target cells. Therefore, log P = log a + nlog i, so plotting log P against log i should give a straight line with gradient n. The data from Table 1 were treated in this way, and straight lines were fitted to the data points by the least-squares method of linear regression (Fig. 2c). For CR208, n was estimated as 1.22 in ihfie1.3 cells but rose to 2.69 in HFFs. The estimates of n for Towne virus were 1.19 and 1.24 in ihfie1.3 cells and HFFs, respectively. Thus, two or three CR208 particles appeared to be necessary to initiate formation of a plaque.
Growth defects of CR208 were repaired by the restoration of exon 4 sequences: the large intron of the major IE gene and ORFs UL124 and UL125 are dispensable for lytic viral growth.
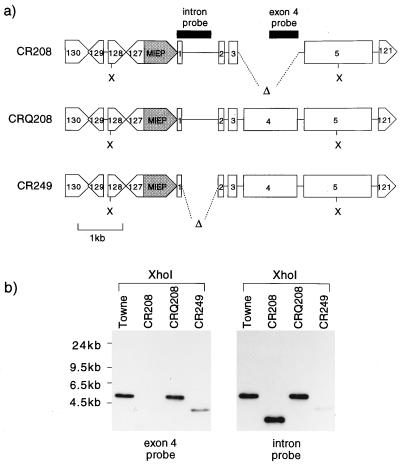
To produce rescued derivatives of CR208, HFLs were transfected with linearized DNAs containing exon 4 and sequences flanking the exon 4 deletion in CR208. pON303G contained a wild-type IE locus. pRG249 lacked the large first intron of the major IE gene but was otherwise identical to pON303G. pGEM3ZF was included as a negative control. Transfected HFLs were then infected at a high multiplicity with CR208 virus. Progeny viruses from these recombinations were then used to infect primary HFL fibroblasts at a low multiplicity. Only stocks derived from the pON303G and pRG249 transfections gave rise to rapidly expanding foci of viral infection in this noncomplementing background. Viruses derived from these rapidly spreading infections were plaque purified and were termed CRQ208 and CR249, respectively. Figure 3a shows the expected genomic structures of CR208, CRQ208, and CR249. Figure 3b shows a Southern blot of XhoI digests of DNAs from these three viruses and the HCMV Towne strain. An exon 4 probe failed to recognize CR208 genomic DNA but detected fragments in Towne (5 kbp), CRQ208 (5 kbp), and CR249 (4.1 kbp). The same blot was hybridized to a probe containing 600 bp of the first intron of the IE gene but also containing 130 bp of exon 1. Fragments were then detected in the genomes of Towne (5 kbp), CR208 (3.6 kbp), and CRQ208 (5 kbp). The 4.1-kbp XhoI fragment of CR249 was recognized only very weakly, due to the deletion of 80% of the sequences homologous to the probe. Thus, the blot confirmed the expected structures of the rescued viruses shown in Fig. 3a. The failure to derive fully growth-competent viruses from the control pGEM3ZF+ recombination showed that the starting CR208 stocks were not contaminated to any detectable level by wild-type virus. The parallel demonstration of a coupled rescue and deletion in the isolation of CR249 provided additional evidence that CRQ208 was a bona fide rescued derivative of CR208.
FIG. 3.
(a) Genomic structures of viruses CR208, CRQ208, and CR249 in the SalI fragment containing the IE locus. The positions of ORFs UL121 and UL127 to UL130 are shown, together with those of the MIEP (shaded) and exons 1 to 5 of the major IE gene. Δ, HCMV sequences deleted from CR208 and CR249. X, XhoI sites used in Southern blot analysis. Probe fragments used are shown next to viral sequences as solid blocks. (b) Southern blot analysis of CR208, CRQ208, and CR249 DNAs cut with XhoI and hybridized sequentially to an exon 4 probe and to an exon 1 plus intron 1 probe (see panel a). CR249 DNA was inadvertently underloaded in this experiment. Molecular sizes of standards are shown to the left.
The growth of CR208 derivatives was evaluated by multiple-step viral growth curve analysis. Working stocks of CRQ208 and CR249 were grown in HFLs, and titers were determined in ihfie1.3 cells. HFLs or ihfie1.3 cells were infected at a low multiplicity (0.1 PFU/cell) with the viruses Towne, CR208, CRQ208, and CR249. The resulting accumulations of cell-free virus were titrated in ihfie1.3 cells, and the results are shown in Fig. 4. Growth of CR208 after a low-multiplicity infection of primary fibroblasts was defective (Fig. 4a), as presented above. The rescued viruses CRQ208 and CR249 grew as well as the Towne strain in HFLs. The equivalent experiment carried out with ihfie1.3 cells demonstrated similar growth kinetics for all four viruses (Fig. 4b). Parallel plaque assays of CRQ208 and CR249 in ihfie1.3 and HFL cells showed that these viruses plated efficiently in either cell type, showing full-sized plaques and a linear relationship between input inoculum size and plaque numbers formed (data not shown).
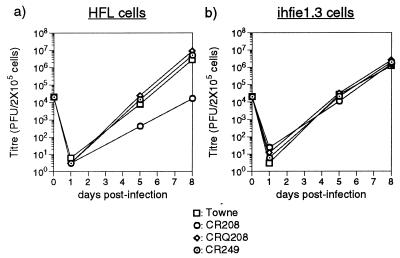
FIG. 4.
Multiple-step growth curve analysis of Towne, CR208, CRQ208, and CR249 viruses in HFL cells (a) and ihfie1.3 cells (b). The results are a time course of the total PFU of infectious virus present in 4 ml of culture supernatant at the indicated sampling times, as measured by plaque formation on ihfie1.3 cells. Experiments were performed with 2 × 105 cells at an input multiplicity of 0.1 PFU/cell. The results of a single experiment are shown; titers are the mean results from two parallel titrations.
Thus, the restoration of exon 4 sequences into two independently derived rescued viral clones repaired the growth defect of CR208. The absence of any appreciable growth defect for CR249 showed that the presence of the 900-bp first intron in the major IE gene was not necessary for lytic viral replication in primary human fibroblasts. ORFs UL124 and UL125 underlie the large first intron and are present in latent transcripts detected in the bone marrow of healthy HCMV carriers (36). Antibodies to the protein product of UL124 have been identified in the sera of some carriers (36), and expression of pUL124 can be detected during the productive infection of cultured HFFs (84). The normal growth of CR249 demonstrated that the protein products of these putative latency genes were not required for productive lytic infection in cultured primary fibroblasts.
Full expression of viral antigens by CR208 after high-multiplicity infection of primary fibroblasts.
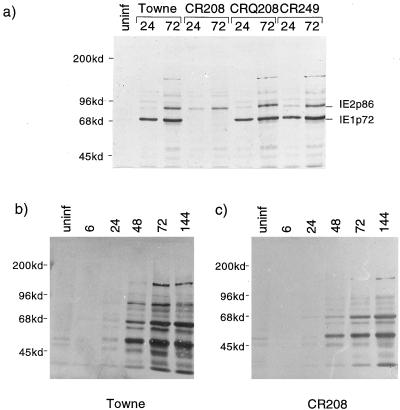
An analysis of viral proteins produced during productive high-multiplicity infections of HFLs was undertaken. Figure 5a shows a Western blot comparing proteins from HFLs infected at 5 PFU/cell with Towne, CR208, CRQ208, and CR249 viruses. IE1 p72 and IE2 p86 were detected in parallel by using antibody CH160, which recognizes a shared epitope encoded in exon 2. CR208 clearly failed to express IE1 p72 at 24 and 72 h postinfection (hpi) but produced IE2 p86 at normal levels. The rescued CRQ208 and CR249 viruses showed patterns of IE antigen expression indistinguishable from that of the wild-type Towne virus. Figure 5b and c show Western blots of total viral antigens expressed during high-multiplicity infections (5 PFU/cell) of HFLs by Towne and CR208 viruses, respectively. Viral antigens were detected by using a human convalescent antiserum. CR208 produced a profile of viral antigens almost identical to that produced by wild-type virus. The accumulation of viral antigens was, however, slower in CR208-infected cells, demonstrating that although CR208 completed the lytic cycle after a high-multiplicity input, the kinetics of total viral protein production were delayed relative to those for wild-type virus. Furthermore, the cytopathic effect observed in cells infected at high multiplicity by CR208 was distinguishable from that in cells infected by Towne, with clearly reduced cell rounding.
FIG. 5.
(a) Western blot analysis of total cellular proteins, detecting IE1 and IE2 proteins produced in HFLs after 24 or 72 h of infection at 5 PFU/cell by Towne, CR208, CRQ208, or CR249 virus. Monoclonal antibody CH160, diluted 1:100, was used. uninf, uninfected-cell proteins. Molecular masses of standards are shown on the left. (b and c) Western blot analysis of time courses of viral proteins produced by Towne (b) and CR208 (c) after high-multiplicity infection (5 PFU/cell) of HFL cells. Viral antigens were detected specifically in total cellular proteins by using human antiserum, diluted 1:50, from a patient convalescing from primary HCMV infection. Molecular masses of standards are shown to the left of each panel.
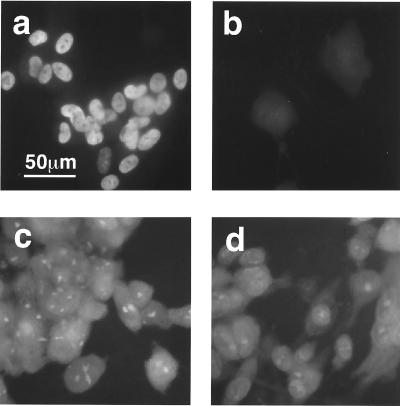
Viral antigens expressed by Towne and CR208 after high-multiplicity infections (5 PFU/cell) were also analyzed by immunofluorescence (Fig. 6). HFFs were used for all cell-staining experiments, as their morphology when grown on glass coverslips provided better resolution. As already described above, CR208 showed defects in plaque formation after low-multiplicity infection of HFFs that were similar to those seen in HFLs. Consistent with the genotype, an IE1 p72-specific monoclonal antibody (p63-27) did not detect nuclear antigen in cells infected by CR208 (Fig. 6b), while Towne-infected cells showed a characteristic diffuse nuclear stain (Fig. 6a). At 72 hpi, the virion structural protein pp65 was detected in the nuclei and cytoplasm of cells infected by either virus by using monoclonal antibody 2A6 (Fig. 6c and d). Immunocytochemical staining has revealed the presence of several other specific viral antigens in cells infected productively with CR208. These include the IE2 protein and the ppUL44 (ICP36) protein (see below).
FIG. 6.
Photomicrographs of immunofluorescence analysis of viral antigens expressed after high-multiplicity infection (5 PFU/cell) of HFFs by Towne (a and c) or CR208 (b and d). (a and b) Staining with IE1-specific antibody p63-27 (undiluted) at 24 hpi. (c and d) Staining with pp65-specific antibody 2A6, diluted 1:100, at 72 hpi. Color positive transparencies were digitized by using a Nikon Coolscan scanner, converted to greyscale using Adobe Photoshop software, and printed as a composite image by using Powerpoint software. Images within a pair were processed identically. The scale bar in panel a applies to all images.
Accumulation of the core viral DNA replication protein ppUL44 was defective after infection of primary fibroblasts at low multiplicity by CR208.
To investigate the nature of the block to viral replication after low-multiplicity infection by CR208, HFLs were infected at various multiplicities and viral proteins were analyzed. The 50-kDa protein product of the UL44 ORF was chosen as a representative protein expressed at DE times. ppUL44 (ICP36) is the processivity factor for the viral DNA polymerase (17) and is necessary in assays of trans-acting factors required for viral DNA replication (52). Expression of antisense RNA to UL44 inhibits virus replication in otherwise permissive U373 astrocytoma cells, apparently by preventing viral DNA replication (58). Thus, although ppUL44 is maximally expressed at late times of infection and UL44 has been described as a late gene (20), DE expression of ppUL44 is very likely required for the progression of infection from the DE to the late phase. The ppUL44 protein is abundant during viral infection of cultured cells, and good specific antibodies are available (49, 64).
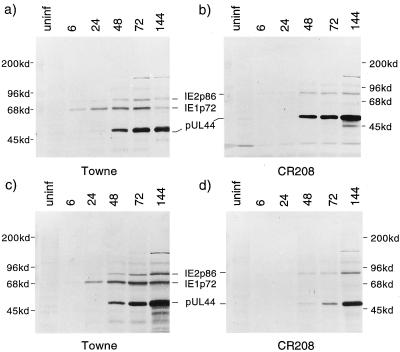
Figure 7 shows a parallel Western blot analysis of IE1 p72, IE2 p86, and ppUL44 proteins. In Fig. 7a and b, HFLs were infected with Towne or CR208 at 5 PFU/cell, and total cell proteins were harvested from parallel samples over a time course. The blots show that IE2 p86 accumulated with similar kinetics during infection by either viral strain, as did the ppUL44 protein. IE1 p72 was detected only in Towne-infected cells. Figure 7c and d show a very similar experiment, conducted in parallel after an input multiplicity of 0.2 PFU/cell. Although IE2 p86 once again accumulated with similar kinetics in both cultures, ppUL44 accumulated at later times, and to lower levels, in the CR208-infected culture. To confirm that this difference between the two viruses was due to the absence of IE1 p72, the experiment shown in Fig. 7c and d was repeated with ihfie1.3 cells. In these complementing cells, IE2 p86 and ppUL44 accumulation by CR208 occurred with kinetics similar to those for the wild type (data not shown). Thus, the defect in ppUL44 accumulation at a low multiplicity was corrected by the provision of IE1 p72 in trans.
FIG. 7.
Western blot analysis of total cellular proteins, detecting IE1 p72, IE2 p86, and ppUL44 proteins produced in HFL cells over an infection time course. A mixture of monoclonal antibodies CH160 (diluted 1:100) and BS510 (diluted 1:25) was used to detect the three viral antigens simultaneously. Molecular masses of standards are shown to the sides. (a and b) Analysis performed after high-multiplicity infection (5 PFU/cell) by Towne (a) or CR208 (b) virus. The death of many Towne-infected cells at 144 h caused reduced protein loading compared to other lanes. (c and d) Analysis performed after low-multiplicity infection (0.2 PFU/cell) by Towne (c) or CR208 (d) virus.
Western blots provide only a semiquantitative measure of steady-state protein levels, and these results must therefore be interpreted with caution. However, this preliminary observation of a discrepancy between IE2 p86 and ppUL44 accumulation after low-multiplicity infection by CR208 was reproducible (21) and was considered worthy of further investigation. In particular, it was necessary to improve sensitivity under genuinely DE conditions, to establish whether any defect in ppUL44 accumulation occurred prior to viral DNA replication or whether it might itself be caused by a failure of viral DNA replication. In addition, our model of a requirement for multiple viral particles to initiate a productive infection predicted that if infection was blocked upstream of ppUL44 expression, the reduction in ppUL44 levels seen by bulk protein analysis should reflect a reduction in the number of infected cells proceeding through the lytic cycle. An analysis of viral antigen accumulation at the single-cell level was therefore undertaken. Monolayers of HFFs infected at various multiplicities with Towne or CR208 were stained in parallel with monoclonal antibodies specific for either IE2 or ppUL44 protein. Antibody SMX, used to detect IE2 antigens in these experiments, is reactive both to IE2 p86 and to the IE2 p40 product, which is made from a late transcript initiating internally within exon 5 (55, 72).
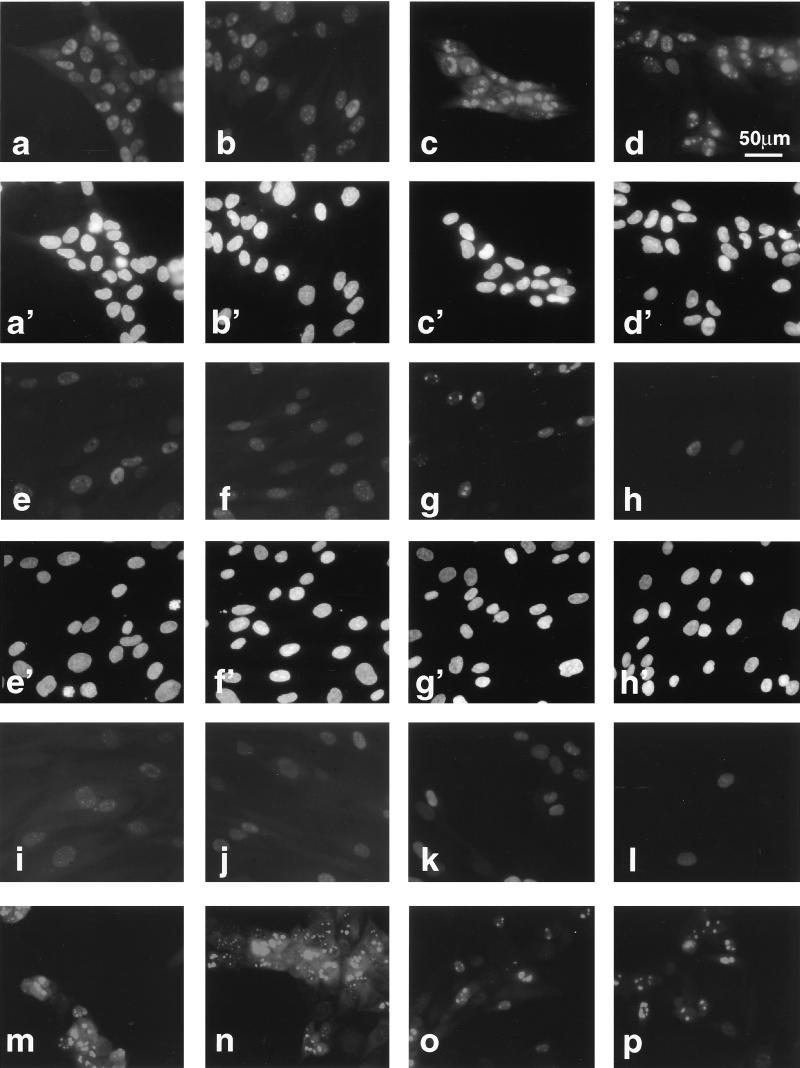
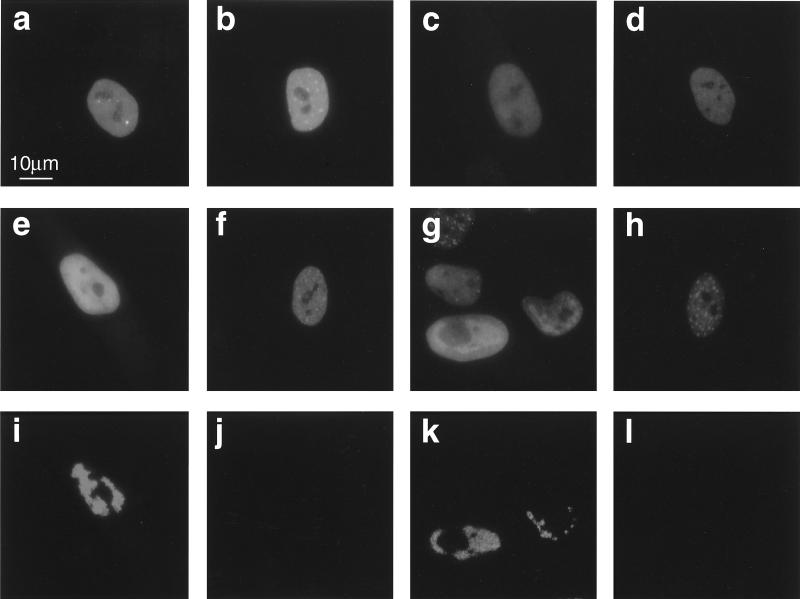
Photomicrographs of cells stained at 48 hpi are shown in Fig. 8. Figure 8a to p show immunostained fields; Fig. 8a′ to h′ show the same respective fields visualized by Hoechst staining. After a high-multiplicity input (5 PFU/cell), high levels of IE2 and ppUL44 were detected in the nuclei of a large percentage of cells infected by Towne or CR208 (Fig. 8a to d). In addition, each protein had a subnuclear distribution which was similar in wild-type- and mutant-infected cells. After a lower-multiplicity input (1 PFU/cell), IE2 was again detected in similar proportions of cells in both Towne- and CR208-infected cultures (Fig. 8e and f). A similar staining pattern was observed in CR208-infected cultures by using antibody CH160, which is reactive to an amino-terminal epitope on IE2 p86 (21), suggesting that full-length IE2 p86 was present in these cells. Similarly to the IE2 staining, approximately 40% of cells in the Towne-infected culture stained for ppUL44 (Fig. 8g). In sharp contrast, ppUL44 staining was seen in many fewer cells in the CR208-infected culture (Fig. 8h), and most of these cells had a diffuse nuclear distribution of antigen, as opposed to the bright patchy pattern characteristic of replication compartment formation during a wild-type infection. At multiplicities of 0.2 PFU/cell and below, cells containing ppUL44 were readily detected in Towne-infected cultures but became extremely scarce in CR208-infected cultures, even though IE2-containing cells were present in numbers similar to those for the wild type in parallel experiments. Thus, many cells infected by CR208 contained IE2 but did not accumulate ppUL44. Similar results were also obtained at 24 hpi (data not shown).
FIG. 8.
Photomicrographs of immunofluorescence analysis of viral antigens expressed after different-multiplicity infections by Towne and CR208 at 48 hpi. (a to p) Immunostained cells; (a′ to h′) same respective fields visualized by Hoechst staining. (a, c, e, g, i, k, m, and o) Towne infection; (b, d, f, h, j, l, n, and p) CR208 infection. (a to d) HFFs infected at 5 PFU/cell and stained either for IE2 protein with antibody SMX diluted 1:2 (a and b) or for ppUL44 with antibody BS510 diluted 1:25 (c and d). (e to h) HFFs infected at 1 PFU/cell and stained either for IE2 (e and f) or for ppUL44 (g and h). (i to l) HFFs infected at 1 PFU/cell in the presence of PFA (200 μg/ml) and stained either for IE2 (i and j) or for ppUL44 (k and l). (m to p) ihfie1.3 cells stained for ppUL44 and infected at 5 PFU/cell (m and n) or 1 PFU/cell (o and p). Color positive transparencies were digitized as for Fig. 6 and printed as a composite image by using Quark Express software. All IE2-stained images were exposed and processed identically, except for panels i and j, which received a doubled exposure time. All ppUL44-stained images were exposed and processed identically, except for panels k and l, which received a doubled exposure time. The scale bar in panel d applies to all images.
To establish that the defect in ppUL44 accumulation was specific to DE phase, the experiments of Fig. 8e to h were performed in the presence of PFA (200 μg/ml), which specifically blocks viral DNA replication. The results are shown in Fig. 8i to l. IE2 staining (Fig. 8i and j) was again approximately equivalent for both viruses and was seen in about 40% of cells, although staining was weaker than in the absence of PFA. Again, many fewer nuclei stained positively for ppUL44 in the CR208 infection than in the Towne infection (Fig. 8k and l). Bright patchy nuclear ppUL44 staining became diffuse in the presence of PFA, probably reflecting the inhibition of the formation of viral DNA replication compartments. Thus, the accumulation of the ppUL44 protein at 24 and 48 h and at 48 h in the presence of PFA was defective during a low-multiplicity infection by CR208. To confirm that the absence of IE1 p72 was responsible for this defect, infections were carried out in ihfie1.3 cells at inputs of 5 and 1 PFU/cell (Fig. 8m to p). In these experiments, no differences in ppUL44 accumulation between Towne and CR208 at either input multiplicity were observed. Most cells contained ppUL44 after infection with 5 PFU of either virus per cell (Fig. 8m and n), and approximately 30% of cells infected at 1 PFU/cell contained ppUL44 (Fig. 8o and p). This equivalence in ihfie1.3 cells was apparent at input multiplicities of as low as 0.008 PFU/cell (21) and demonstrated that the provision of IE1 p72 in trans corrected the low-multiplicity ppUL44 accumulation defect of CR208, just as it corrected the low-multiplicity growth defect of the mutant virus. Rescued viruses CRQ208 and CR249 were also used in similar immunofluorescence experiments and had no demonstrable defects in ppUL44 accumulation relative to a wild-type virus (21).
Two representative fields of cells from HFF monolayers infected at multiplicities of 5, 1, 0.2, and 0.04 PFU/cell and stained after 48 h for either IE2 (antibody SMX) or ppUL44 (antibody BS510) were photographed for specific immunofluorescence and for the Hoechst counterstain. Proportions of cells containing the two antigens were then calculated by comparing numbers of specifically staining nuclei with numbers of Hoechst-staining nuclei. The resulting data (Table 2) are a more quantitative representation of the results of Fig. 8a to h. These data show that IE2 was contained in similar proportions of cells infected with equivalent multiplicities of Towne or CR208 virus, independent of that multiplicity. In Towne infections, accumulation of ppUL44 followed a pattern similar to that for IE2, but in CR208 infections, the effect of reducing the inoculum was much more profound for ppUL44 accumulation, with the percentage of positively staining cells dropping very sharply upon dilution.
TABLE 2.
Frequencies of nuclear staining for IE2 or ppUL44 at 48 hpi of HFF cells
| Virus strain | Multiplicity of infection (PFU/cell) | IE2
|
ppUL44
|
||
|---|---|---|---|---|---|
| Nuclei staining/total cells counted | % Positive cells | Nuclei staining/total cells counted | % Positive cells | ||
| Towne | 5.0 | 172/228 | 75 | 123/177 | 69 |
| 1.0 | 99/244 | 41 | 81/237 | 34 | |
| 0.2 | 27/281 | 9.6 | 21/337 | 6.2 | |
| 0.04 | 4/318 | 1.3 | 3/351 | 0.86 | |
| CR208 | 5.0 | 155/207 | 75 | 93/138 | 67 |
| 1.0 | 110/233 | 47 | 19/318 | 6.0 | |
| 0.2 | 18/265 | 6.8 | 0/296 | <0.34 | |
| 0.04 | 3/227 | 1.3 | 0/277 | <0.36 | |
Viral DNA replication compartments were not formed after low-multiplicity infection by CR208.
Our analysis of the ppUL44 antigen showed that the defect in the CR208 life cycle after a low-multiplicity input correlated with a deficiency in ppUL44 accumulation at DE times. As ppUL44 is a core DNA replication complex protein, which is very likely required for viral DNA replication, our results predict that viral DNA replication may not occur after a low-multiplicity infection by the ie1 mutant virus. To establish this, and to investigate the fate of individual CR208-infected cells over a longer time course, HFFs were infected at a low multiplicity (0.05 PFU/cell) with Towne or CR208 virus. The cells were then analyzed over a time course by combined staining for IE2 protein and for the incorporation of the thymidine analog BrdU into viral DNA replication structures.
Figure 9 shows photomicrographs of representative single infected cells that were fixed and stained at various times after infection. At 24 hpi, similar proportions of infected cells (approximately 2 to 5%) were detected in either culture by staining with an IE2-specific antibody. The staining pattern for IE2 p86, i.e., a diffuse nuclear stain with punctate highlights, was similar in cells infected by either virus (Fig. 9a and b). At 48 hpi, similar proportions of IE2-expressing cells were again detected in each culture (Fig. 9c and d). At 72 hpi, CR208-infected nuclei retained the original IE2 staining pattern, and no DNA replication structures were detected by BrdU incorporation (Fig. 9f and j). In contrast, the majority of Towne-infected cells had a bright patchy configuration of IE2 staining (Fig. 9e), with BrdU incorporation coincident with the brighter patches (Fig. 9i), in the irregular lobed pattern characteristic of CMV DNA replication compartments (63). It is likely that a significant proportion of staining by SMX at this and later stages of productive infection may be due to the accumulation of the late IE2 p40 protein (55, 72). At 144 hpi, cells staining for IE2 were still detectable in the CR208-infected culture, though they had decreased approximately fivefold relative to those at 24 hpi, and the IE2 staining pattern had often become very punctate (Fig. 9h). BrdU incorporation into viral DNA replication structures was not detected in the vast majority of these IE2-containing cells (Fig. 9l), although a careful examination of the culture revealed a very small percentage of the total population (approximately 0.01%) in which viral DNA replication compartments were discernible although retarded relative to the case for a wild-type infection. At 144 hpi, the majority of cells in the Towne-infected culture contained IE2 proteins. The original infected cells were identified by a brighter IE2 stain corresponding to extensive viral DNA replication compartments and were surrounded by other infected cells with earlier staining patterns (Fig. 9g). BrdU incorporation into replication compartments was still strong at 144 hpi and was also detectable in some of the secondarily infected cells (Fig. 9k).
FIG. 9.
Time course immunofluorescence experiment, showing the progression of infection in individual cells after the low-multiplicity inoculation (0.05 PFU/cell) of HFFs with Towne (a, c, e, g, i, and k) or CR208 (b, d, f, h, j, and l). (a and b) Fixation at 24 hpi; IE2 staining (antibody SMX diluted 1:2). (c and d) Fixation at 48 hpi; IE2 staining. (e and f) BrdU pulse and fixation at 72 hpi; IE2 staining. (g and h) BrdU pulse and fixation at 144 hpi; IE2 staining. (i and j) Same fields as panels e and f, respectively, with parallel BrdU uptake staining (antibody MAS-250 diluted 1:2). (k and l) Same fields as panels g and h, respectively, with parallel BrdU uptake staining. Color transparency images were processed as for Fig. 8. All IE2-stained images were exposed and processed identically. All BrdU-stained images were exposed and processed identically. The scale bar in panel a applies to all images.
A parallel experiment was performed by using double staining for ppUL44 and BrdU incorporation (21). In the Towne-infected culture, ppUL44 staining was widespread from 24 hpi onwards, and a large proportion colocalized with BrdU incorporation in cells containing viral DNA replication compartments. Most cells in the Towne-infected culture contained ppUL44 at 144 hpi. In contrast, ppUL44-staining cells were extremely rare in the CR208-infected culture at all time points, but ppUL44 was found to be present in the nuclei of those very rare cells which had formed viral DNA replication compartments at 144 hpi. Similar double-labelling experiments were conducted with the rescued viruses CRQ208 and CR249 (21). Infection with these viruses at low multiplicities resulted in the normal accumulation of IE2 and ppUL44 antigens and the normal establishment of viral DNA replication compartments.
We conclude from these experiments that viral DNA replication compartments were not assembled over 6 days in cells infected at a low multiplicity with CR208 virus, even though IE2 protein was expressed. This result correlated with reduced accumulation of ppUL44 and suggests that viral DNA replication was blocked in the abortively infected cells.
DISCUSSION
We have isolated a recombinant HCMV (CR208) from which all unique coding sequences for the abundant nuclear IE1 p72 protein were deleted. IE1 protein was provided in trans in a complementing immortalized fibroblast line, demonstrating the general applicability of these methods to the isolation of HCMV mutants with deletions in essential viral genes.
IE1 functions were not required for lytic replication upon infection of primary fibroblasts at a high multiplicity, although there were indications that the cascade of viral gene expression was slower in the absence of IE1, and there were also alterations in the viral cytopathic effect. Thus, the most abundant viral IE protein was not absolutely essential for virus replication in cultured fibroblasts. However, after infection at low input multiplicities, replication of the ie1 mutant virus exhibited a marked deficiency. The phenotype of CR208 during plating and growth assays suggested that IE1 p72 function was essential when single virions initiated infection. After low-multiplicity infection, the mutant virus life cycle was apparently stalled between the IE and DE phases, with synthesis of the major regulatory protein IE2 p86 but with defective accumulation of the core DNA replication protein ppUL44 and a failure to establish viral DNA replication compartments. The failure to accumulate ppUL44 in the absence of IE1 p72 was independent of viral DNA replication and occurred in the presence of the inhibitor PFA. The faulty accumulation of ppUL44 observed at 24 and 48 hpi may have contributed to the failure of the formation of viral DNA replication compartments in mutant-infected cells. All of the phenotypic differences observed in viral replication, ppUL44 accumulation, and establishment of DNA replication compartments were corrected in two independently derived rescued derivatives of CR208. The defects were also corrected by the provision of the IE1 protein in trans in the complementing ihfie1.3 cell line. Thus, the observed multiplicity-dependent defects in growth and ppUL44 accumulation by the ie1 mutant CR208 were directly related to the inability to code for IE1 p72.
The previously reported ie1 null mutant RC303ΔAcc (48) exhibited growth properties similar to those of CR208 when growth in HFFs was compared to that in ihfie1.3 cells. Because RC303ΔAcc exhibited a striking multiplicity-dependent expression of IE2 p86, the growth defect was attributed to a defect in IE1 p72-mediated autoregulation (48). This virus also exhibited a reduced expression of other viral antigens, but these effects correlated with the reduced expression of IE2 p86. Expression of IE2 p86 by CR208 appears to be less dependent upon multiplicity (Fig. 7 and 8), a characteristic which might be due to the differences in the genetic backgrounds used to construct RC303ΔAcc and CR208. CR208, characterized here, is purely Towne based, whereas RC303ΔAcc is a chimera of the Towne and Toledo strains. The most relevant differences between these two mutants are probably in the region upstream of the ie1-ie2 enhancer, where the Toledo strain carries a 15-kbp segment that includes a major rearrangement of sequences (8). This difference may account for the different impact of the absence of IE1 p72 on levels of ie2 expression. Irrespective of these differences, or of the possible impact of IE1 p72 on autoregulation in certain strains of virus or under certain circumstances, the phenotype of CR208 demonstrates that IE1 p72 function is important even under conditions when IE2 p86 is synthesized and that the impact of IE1 p72 therefore goes well beyond the IE phase of infection.
A consideration of the characterized activities of IE1 p72 suggests several additional mechanisms which alone or in combination could account for the low-multiplicity growth and expression defects of CR208. First, and most consistent with the data presented here, cooperation between IE1 p72 and IE2 p86 to activate HCMV DE and late promoters has been well documented in transient-assay experiments (9, 35, 43, 45, 73). Further analysis showed that the UL44 promoter and those of other core replication genes require accessory functions IRS1/TRS1, UL112-UL113, and UL36-UL38 in addition to IE2 p86 and IE1 p72 to be fully activated (30, 71). The absence of IE1 p72 during infection could thus lead directly to reduced activation of the UL44 promoter, as observed in these transient assays, or may result in reduced levels of the accessory transactivators. Either mechanism could account for the observed defects in ppUL44 accumulation during CR208 infection. Our data cannot exclude the additional possibility that IE1 p72 up-regulates ppUL44 levels by a posttranscriptional mechanism. Regardless of their exact cause, insufficient levels of ppUL44 or any critical replication protein might result in the attenuation of CR208 infection prior to DNA replication.
Another possibility is that IE1, either alone or acting with other viral functions, sustains the host cell through viral infection. IE1 p72 and IE2 p86 have been attributed antiapoptotic effects (85), and IE1 p72 was the stronger of the two. While we did not directly evaluate apoptosis in this study, it is possible that cells infected nonproductively with CR208 are lost through apoptosis. However, a large population of CR208-infected cells containing IE2 but not ppUL44 was observed at both 24 and 48 hpi. The apparent stability of this abortively infected cell population argues that the observed failure in ppUL44 accumulation occurs prior to any apoptotic events.
Finally, expression of ie2 products mediates a shutoff of ie1-ie2 gene expression via crs (12, 41, 53) that may be accentuated in the absence of IE1 p72. Shutoff of the MIEP may thus occur more readily in cells infected with CR208. However, many CR208-infected cells accumulated significant amounts of the major regulatory protein IE2 p86 without accumulating ppUL44, while wild-type-infected cells containing similar amounts of IE2 p86 had also accumulated ppUL44 (Fig. 8). This observation suggests that even if MIEP shutoff is increased in CR208 infections, the absence of IE1 p72 probably impacts directly on downstream gene expression also.
We do not anticipate that altered ppUL44 levels are the sole mediator of the loss of replicative potential in the absence of IE1 p72. As suggested above, levels of accessory viral transactivators may also be reduced during CR208 infection, as may levels of other viral DE products. The levels during CR208 infection of a wider range of viral IE and DE gene products are currently being investigated. We also do not yet know which features of high-multiplicity infection by CR208 bypass the requirement for IE1 function in ppUL44 accumulation and productive infection. These effects could potentially be mediated either by an increased dosage of the viral genomic template, by an increased dosage of virion transactivator proteins, or by some other stimulatory signal delivered to cells upon infection with HCMV.
The growth phenotype of the ie1 mutants in primary fibroblasts—lytic growth at high multiplicities, a low plating efficiency, small plaques, and a nonlinear relationship between inoculum size and plaque number—is strikingly similar to the growth phenotype of mutants of HSV that are unable to express the IE regulatory protein ICP0 or Vmw110 (61, 77). Other similarities between the two proteins include their synergistic transactivating functions in combination with the major transcriptional transactivators IE2 p86 and ICP4 (18, 45, 50, 73) and the abilities of both proteins to target and modify PML-associated nuclear domain 10 (ND10) bodies (1, 19, 34, 37). It has also been observed that HCMV coinfection can supply functions which complement the low-multiplicity plating defect of an HSV-1 ICP0 mutant (76), although the HCMV gene products responsible have not been identified. It is attractive to speculate that HCMV IE1 and HSV-1 ICP0, although apparently not homologs, may have evolved to perform fundamentally similar functions in these two distantly related herpesviruses.
ACKNOWLEDGMENTS
This work was supported by a Wellcome Career Development Fellowship held by R.F.G. at Cambridge University and by NIH grant PHS RO1 AI33852 to E.S.M.
We thank Denise Galloway, Eli Gilboa, Lenore Pereira, Bodo Plachter, Maria Grazia Revello, and Mark Wills for their generous gifts of reagents. R.F.G. thanks John Sinclair and Patrick Sissons for their active support of his work at Cambridge and Mat Bentham for his technical assistance.
REFERENCES
- 1.Ahn J-H, Hayward G S. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol. 1997;71:4599–4613. doi: 10.1128/jvi.71.6.4599-4613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson K P, Fox M C, Brown-Driver V, Martin M J, Azad R F. Inhibition of human cytomegalovirus immediate-early gene expression by an antisense oligonucleotide complementary to immediate-early RNA. Antimicrob Agents Chemother. 1996;40:2004–2011. doi: 10.1128/aac.40.9.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arlt H, Lang D, Gebert S, Stamminger T. Identification of binding sites for the 86-kilodalton IE2 protein of human cytomegalovirus within an IE2-responsive viral early promoter. J Virol. 1994;68:4117–4125. doi: 10.1128/jvi.68.7.4117-4125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azad R F, Brown-Driver V, Tanaka K, Crooke R M, Anderson K P. Antiviral activity of a phosphorothioate oligonucleotide complementary to RNA of the human cytomegalovirus major immediate-early region. Antimicrob Agents Chemother. 1993;37:1945–1954. doi: 10.1128/aac.37.9.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Britt W J, Alford C A. Cytomegalovirus. In: Fields B N, editor. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2493–2523. [Google Scholar]
- 6.Bryant L A, Sinclair J H. Inhibition of human cytomegalovirus major immediate early gene expression by antisense RNA expression vectors. J Gen Virol. 1993;74:1965–1967. doi: 10.1099/0022-1317-74-9-1965. [DOI] [PubMed] [Google Scholar]
- 7.Caswell R, Hagemeier C, Chiou C J, Hayward G, Kouzarides T, Sinclair J. The human cytomegalovirus 86K immediate early (IE) 2 protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via regions of IE2 required for transcriptional regulation. J Gen Virol. 1993;74:2691–2698. doi: 10.1099/0022-1317-74-12-2691. [DOI] [PubMed] [Google Scholar]
- 8.Cha T A, Tom E, Kemble G W, Duke G M, Mocarski E S, Spaete R R. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol. 1996;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang C P, Malone C L, Stinski M F. A human cytomegalovirus early gene has three inducible promoters that are regulated differentially at various times after infection. J Virol. 1989;63:281–290. doi: 10.1128/jvi.63.1.281-290.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison C A I, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–170. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 11.Chen C A, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cherrington J M, Khoury E L, Mocarski E S. Human cytomegalovirus ie2 negatively regulates α gene expression via a short target sequence near the transcription start site. J Virol. 1991;65:887–896. doi: 10.1128/jvi.65.2.887-896.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cherrington J M, Mocarski E S. Human cytomegalovirus ie1 transactivates the α promoter-enhancer via an 18-base-pair repeat element. J Virol. 1989;63:1435–1440. doi: 10.1128/jvi.63.3.1435-1440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Compton T. An immortalized human fibroblast cell line is permissive for human cytomegalovirus infection. J Virol. 1993;67:3644–3648. doi: 10.1128/jvi.67.6.3644-3648.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Depto A S, Stenberg R M. Regulated expression of the human cytomegalovirus pp65 gene: octamer sequence in the promoter is required for activation by viral gene products. J Virol. 1989;63:1232–1238. doi: 10.1128/jvi.63.3.1232-1238.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dondero D V, Pereira L. Monoclonal antibody production. In: Emmons R, Schmidt N, editors. Diagnostic procedures for viral, rickettsial and chlamydial infections. Washington, D.C: American Public Health Association; 1990. pp. 101–124. [Google Scholar]
- 17.Ertl P F, Powell K L. Physical and functional interaction of human cytomegalovirus DNA polymerase and its accessory protein (ICP36) expressed in insect cells. J Virol. 1992;66:4126–4133. doi: 10.1128/jvi.66.7.4126-4133.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett R D. Trans activation of transcription by herpes virus products: requirement for two HSV-1 immediate-early polypeptides for maximum activity. EMBO J. 1984;3:3135–3141. doi: 10.1002/j.1460-2075.1984.tb02270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Everett R D, Maul G G. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geballe A P, Leach F S, Mocarski E S. Regulation of cytomegalovirus late gene expression: γ genes are controlled by posttranscriptional events. J Virol. 1986;57:864–874. doi: 10.1128/jvi.57.3.864-874.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greaves, R. F. Unpublished data.
- 22.Greaves R F, Brown J M, Vieira J, Mocarski E S. Selectable insertion and deletion mutagenesis of the human cytomegalovirus genome using the Escherichia coli guanosine phosphoribosyl transferase (gpt) gene. J Gen Virol. 1995;76:2151–2160. doi: 10.1099/0022-1317-76-9-2151. [DOI] [PubMed] [Google Scholar]
- 23.Hagemeier C, Walker S, Caswell R, Kouzarides T, Sinclair J. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J Virol. 1992;66:4452–4456. doi: 10.1128/jvi.66.7.4452-4456.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halbert C L, Demers G W, Galloway D A. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J Virol. 1991;65:473–478. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 26.Hayhurst G P, Bryant L A, Caswell R C, Walker S M, Sinclair J H. CCAAT box-dependent activation of the TATA-less human DNA polymerase alpha promoter by the human cytomegalovirus 72-kilodalton major immediate-early protein. J Virol. 1995;69:182–188. doi: 10.1128/jvi.69.1.182-188.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hermiston T W, Malone C L, Stinski M F. Human cytomegalovirus immediate-early two-protein region involved in negative regulation of the major immediate-early promoter. J Virol. 1990;64:3532–3536. doi: 10.1128/jvi.64.7.3532-3536.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hermiston T W, Malone C L, Witte P R, Stinski M F. Identification and characterization of the human cytomegalovirus immediate-early region 2 gene that stimulates gene expression from an inducible promoter. J Virol. 1987;61:3214–3221. doi: 10.1128/jvi.61.10.3214-3221.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunninghake G W, Monks B G, Geist L J, Monick M M, Monroy M A, Stinski M F, Webb A C, Dayer J M, Auron P E, Fenton M J. The functional importance of a cap site-proximal region of the human prointerleukin 1β gene is defined by viral protein transactivation. Mol Cell Biol. 1992;12:3439–3448. doi: 10.1128/mcb.12.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iskenderian A C, Huang L, Reilly A, Stenberg R M, Anders D G. Four of eleven loci required for transient complementation of human cytomegalovirus DNA replication cooperate to activate expression of replication genes. J Virol. 1996;70:383–392. doi: 10.1128/jvi.70.1.383-392.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenkins D E, Martens C L, Mocarski E S. Human cytomegalovirus late protein encoded by ie2: a trans-activator as well as a repressor of gene expression. J Gen Virol. 1994;75:2337–2348. doi: 10.1099/0022-1317-75-9-2337. [DOI] [PubMed] [Google Scholar]
- 32.Jupp R, Hoffmann S, Depto A, Stenberg R M, Ghazal P, Nelson J A. Direct interaction of the human cytomegalovirus IE86 protein with the cis repression signal does not preclude TBP from binding to the TATA box. J Virol. 1993;67:5595–5604. doi: 10.1128/jvi.67.9.5595-5604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jupp R, Hoffmann S, Stenberg R M, Nelson J A, Ghazal P. Human cytomegalovirus IE86 protein interacts with promoter-bound TATA-binding protein via a specific region distinct from the autorepression domain. J Virol. 1993;67:7539–7546. doi: 10.1128/jvi.67.12.7539-7546.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelly C, Van Driel R, Wilkinson G W G. Disruption of PML-associated nuclear bodies during human cytomegalovirus infection. J Gen Virol. 1995;76:2887–2893. doi: 10.1099/0022-1317-76-11-2887. [DOI] [PubMed] [Google Scholar]
- 35.Klucher K M, Sommer M, Kadonaga J T, Spector D H. In vivo and in vitro analysis of transcriptional activation mediated by the human cytomegalovirus major immediate-early proteins. Mol Cell Biol. 1993;13:1238–1250. doi: 10.1128/mcb.13.2.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kondo K, Xu J, Mocarski E S. Human cytomegalovirus latent gene expression in granulocyte-macrophage progenitors in culture and in seropositive individuals. Proc Natl Acad Sci USA. 1996;93:11137–11142. doi: 10.1073/pnas.93.20.11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korioth F, Maul G G, Plachter B, Stamminger T, Frey J. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res. 1996;229:155–158. doi: 10.1006/excr.1996.0353. [DOI] [PubMed] [Google Scholar]
- 38.LaFemina R L, Pizzorno M C, Mosca J D, Hayward G S. Expression of the acidic nuclear immediate-early protein (IE1) of human cytomegalovirus in stable cell lines and its preferential association with metaphase chromosomes. Virology. 1989;172:584–600. doi: 10.1016/0042-6822(89)90201-8. [DOI] [PubMed] [Google Scholar]
- 39.Lang D, Gebert S, Arlt H, Stamminger T. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J Virol. 1995;69:6030–6037. doi: 10.1128/jvi.69.10.6030-6037.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lang D, Stamminger T. The 86-kilodalton IE-2 protein of human cytomegalovirus is a sequence-specific DNA-binding protein that interacts directly with the negative autoregulatory response element located near the cap site of the IE-1/2 enhancer-promoter. J Virol. 1993;67:323–331. doi: 10.1128/jvi.67.1.323-331.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu B, Hermiston T W, Stinski M F. A cis-acting element in the major immediate-early (IE) promoter of human cytomegalovirus is required for negative regulation by IE2. J Virol. 1991;65:897–903. doi: 10.1128/jvi.65.2.897-903.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lubon H, Ghazal P, Hennighausen L, Reynolds-Kohler C, Lockshin C, Nelson J. Cell-specific activity of the modulator region in the human cytomegalovirus major immediate-early gene. Mol Cell Biol. 1989;9:1342–1345. doi: 10.1128/mcb.9.3.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lukac D M, Mannupello J R, Alwine J C. Transcriptional activation by the human cytomegalovirus immediate-early proteins: requirements for simple promoter structures and interactions with multiple components of the transcription complex. J Virol. 1994;68:5184–5193. doi: 10.1128/jvi.68.8.5184-5193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macias M P, Stinski M F. An in vitro system for human cytomegalovirus immediate early 2 protein (IE2)-mediated site-dependent repression of transcription and direct binding of IE2 to the major immediate early promoter. Proc Natl Acad Sci USA. 1993;90:707–711. doi: 10.1073/pnas.90.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malone C L, Vesole D H, Stinski M F. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J Virol. 1990;64:1498–1506. doi: 10.1128/jvi.64.4.1498-1506.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Margolis M J, Pajovic S, Wong E L, Wade M, Jupp R, Nelson J A, Azizkhan J C. Interaction of the 72-kilodalton human cytomegalovirus IE1 gene product with E2F1 coincides with E2F-dependent activation of dihydrofolate reductase transcription. J Virol. 1995;69:7759–7767. doi: 10.1128/jvi.69.12.7759-7767.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mocarski E S. Cytomegaloviruses and their replication. In: Fields B N, editor. Virology. Philadelphia, Pa: Lipincott-Raven; 1996. pp. 2447–2492. [Google Scholar]
- 48.Mocarski E S, Kemble G W, Lyle J M, Greaves R F. A deletion mutant in the human cytomegalovirus gene encoding IE1 (491aa) is replication defective due to a failure in autoregulation. Proc Natl Acad Sci USA. 1996;93:11321–11326. doi: 10.1073/pnas.93.21.11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mocarski E S, Pereira L, Michael N. Precise localization of genes on large animal virus genomes: use of lambda gt11 and monoclonal antibodies to map the gene for a cytomegalovirus protein family. Proc Natl Acad Sci USA. 1985;82:1266–1270. doi: 10.1073/pnas.82.4.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Hare P, Hayward G S. Evidence for a direct role for both the 175,000- and 110,000-molecular-weight immediate-early proteins of herpes simplex virus in the transactivation of delayed-early promoters. J Virol. 1985;53:751–760. doi: 10.1128/jvi.53.3.751-760.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pari G S, Anders D G. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J Virol. 1993;67:6979–6988. doi: 10.1128/jvi.67.12.6979-6988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pari G S, Kacica M A, Anders D G. Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J Virol. 1993;67:2575–2582. doi: 10.1128/jvi.67.5.2575-2582.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pizzorno M C, Hayward G S. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target located near the cap site. J Virol. 1990;64:6154–6165. doi: 10.1128/jvi.64.12.6154-6165.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pizzorno M C, O’Hare P, Sha L, LaFemina R L, Hayward G S. trans activation and autoregulation of gene expression by the immediate-early region 2 gene products of human cytomegalovirus. J Virol. 1988;62:1167–1179. doi: 10.1128/jvi.62.4.1167-1179.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Plachter B, Britt W, Vornhagen R, Stamminger T, Jahn G. Analysis of proteins encoded by IE regions 1 and 2 of human cytomegalovirus using monoclonal antibodies generated against recombinant antigens. Virology. 1993;193:642–652. doi: 10.1006/viro.1993.1172. [DOI] [PubMed] [Google Scholar]
- 56.Poma E E, Kowalik T F, Zhu L, Sinclair J H, Huang E S. The human cytomegalovirus IE1-72 protein interacts with the cellular p107 protein and relieves p107-mediated transcriptional repression of an E2F-responsive promoter. J Virol. 1996;70:7867–7877. doi: 10.1128/jvi.70.11.7867-7877.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Revello M G, Percivalle E, Matteo A D, Morini F, Gerna G. Nuclear expression of the lower matrix protein of human cytomegalovirus in peripheral blood leukocytes of immunocompromised viraemic patients. J Gen Virol. 1992;73:437–442. doi: 10.1099/0022-1317-73-2-437. [DOI] [PubMed] [Google Scholar]
- 58.Ripalti A, Boccuni M C, Campanini F, Landini M P. Cytomegalovirus-mediated induction of antisense mRNA expression to UL44 inhibits virus replication in an astrocytoma cell line: identification of an essential gene. J Virol. 1995;69:2047–2057. doi: 10.1128/jvi.69.4.2047-2057.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roizman B, Sears A. Herpes simplex viruses and their replication. In: Fields B N, editor. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2231–2295. [Google Scholar]
- 60.Roy N. Ph.D. thesis. Ithaca, N.Y: Cornell University; 1993. [Google Scholar]
- 61.Sacks W R, Schaffer P A. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J Virol. 1987;61:829–839. doi: 10.1128/jvi.61.3.829-839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sambucetti L C, Cherrington J M, Wilkinson G W G, Mocarski E S. NF-κB activation of the cytomegalovirus enhancer is mediated by a viral transactivator and by T cell stimulation. EMBO J. 1989;8:4251–4258. doi: 10.1002/j.1460-2075.1989.tb08610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sarisky R T, Hayward G S. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J Virol. 1996;70:7398–7413. doi: 10.1128/jvi.70.11.7398-7413.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmolke S, Kern H F, Drescher P, Jahn G, Plachter B. The dominant phosphoprotein pp65 (UL83) of human cytomegalovirus is dispensable for growth in cell culture. J Virol. 1995;69:5959–5968. doi: 10.1128/jvi.69.10.5959-5968.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwartz R, Helmich B, Spector D H. CREB and CREB-binding proteins play an important role in the IE2 86-kilodalton protein-mediated transactivation of the human cytomegalovirus 2.2-kilobase RNA promoter. J Virol. 1996;70:6955–6966. doi: 10.1128/jvi.70.10.6955-6966.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwartz R, Sommer M H, Scully A, Spector D H. Site-specific binding of the human cytomegalovirus IE2 86-kilodalton protein to an early gene promoter. J Virol. 1994;68:5613–5622. doi: 10.1128/jvi.68.9.5613-5622.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scully A L, Sommer M H, Schwartz R, Spector D H. The human cytomegalovirus IE2 86-kilodalton protein interacts with an early gene promoter via site-specific DNA binding and protein-protein associations. J Virol. 1995;69:6533–6540. doi: 10.1128/jvi.69.10.6533-6540.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shelbourn S L, Kothari S K, Sissons J G, Sinclair J H. Repression of human cytomegalovirus gene expression associated with a novel immediate early regulatory region binding factor. Nucleic Acids Res. 1989;17:9165–9171. doi: 10.1093/nar/17.22.9165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spaete R R, Mocarski E S. Insertion and deletion mutagenesis of the human cytomegalovirus genome. Proc Natl Acad Sci USA. 1987;84:7213–7217. doi: 10.1073/pnas.84.20.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Staprans S I, Rabert D K, Spector D H. Identification of sequence requirements and trans-acting functions necessary for regulated expression of a human cytomegalovirus early gene. J Virol. 1988;62:3463–3473. doi: 10.1128/jvi.62.9.3463-3473.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stasiak P C, Mocarski E S. Transactivation of the cytomegalovirus ICP36 gene promoter requires the α gene product TRS1 in addition to IE1 and IE2. J Virol. 1992;66:1050–1058. doi: 10.1128/jvi.66.2.1050-1058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stenberg R M, Depto A S, Fortney J, Nelson J A. Regulated expression of early and late RNAs and proteins from the human cytomegalovirus immediate-early gene region. J Virol. 1989;63:2699–2708. doi: 10.1128/jvi.63.6.2699-2708.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stenberg R M, Fortney J, Barlow S W, Magrane B P, Nelson J A, Ghazal P. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involve common and unique protein domains. J Virol. 1990;64:1556–1565. doi: 10.1128/jvi.64.4.1556-1565.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stenberg R M, Thomsen D R, Stinski M F. Structural analysis of the major immediate-early gene of human cytomegalovirus. J Virol. 1984;49:190–199. doi: 10.1128/jvi.49.1.190-199.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stenberg R M, Witte P R, Stinski M F. Multiple spliced and unspliced transcripts from human cytomegalovirus immediate-early region 2 and evidence for a common initiation site within immediate-early region 1. J Virol. 1985;56:665–675. doi: 10.1128/jvi.56.3.665-675.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stow E C, Stow N D. Complementation of a herpes simplex virus type 1 Vmw110 deletion mutant by human cytomegalovirus. J Gen Virol. 1989;70:695–704. doi: 10.1099/0022-1317-70-3-695. [DOI] [PubMed] [Google Scholar]
- 77.Stow N D, Stow E C. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J Gen Virol. 1986;67:2751–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 78.Thomsen D R, Stinski M F. Cloning of the human cytomegalovirus genome as endonuclease XbaI fragments. Gene. 1981;16:207–216. doi: 10.1016/0378-1119(81)90077-9. [DOI] [PubMed] [Google Scholar]
- 79.Vieira J, Messing J. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene. 1991;100:189–194. doi: 10.1016/0378-1119(91)90365-i. [DOI] [PubMed] [Google Scholar]
- 80.Wade M, Kowalik T F, Mudryj M, Huang E S, Azizkhan J C. E2F mediates dihydrofolate reductase promoter activation and multiprotein complex formation in human cytomegalovirus infection. Mol Cell Biol. 1992;12:4364–4374. doi: 10.1128/mcb.12.10.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Walboomers J M M, TerSchegget J. A new method for the isolation of herpes simplex virus type 2 DNA. Virology. 1976;74:256–258. doi: 10.1016/0042-6822(76)90151-3. [DOI] [PubMed] [Google Scholar]
- 82.Walker S, Hagemeier C, Sissons J G, Sinclair J H. A 10-base-pair element of the human immunodeficiency virus type 1 long terminal repeat (LTR) is an absolute requirement for transactivation by the human cytomegalovirus 72-kilodalton IE1 protein but can be compensated for by other LTR regions in transactivation by the 80-kilodalton IE2 protein. J Virol. 1992;66:1543–1550. doi: 10.1128/jvi.66.3.1543-1550.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu J, Jupp R, Stenberg R M, Nelson J A, Ghazal P. Site-specific inhibition of RNA polymerase II preinitiation complex assembly by human cytomegalovirus IE86 protein. J Virol. 1993;67:7547–7555. doi: 10.1128/jvi.67.12.7547-7555.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu, J., and E. S. Mocarski. Unpublished data.
- 85.Zhu H, Shen Y, Shenk T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J Virol. 1995;69:7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]