Abstract
We have used immunofluorescent staining and confocal microscopy to examine the subcellular localization of structural and nonstructural bovine papillomavirus (BPV) proteins in cultured cells that produce infectious virions. When expressed separately, L1, the major capsid protein, showed a diffuse nuclear distribution while L2, the minor capsid protein, was found to localize to punctate nuclear regions identified as promonocytic leukemia protein (PML) oncogenic domains (PODs). Coexpression of L1 and L2 induced a relocation of L1 into the PODs, leading to the colocalization of L1 and L2. The effect of L2 expression on the distribution of the nonstructural viral proteins E1 and E2, which are required for maintenance of the genome and viral DNA synthesis, was also examined. The localization of the E1 protein was unaffected by L2 expression. However, the pattern of anti-E2 staining was dramatically altered in L2-expressing cells. Similar to L1, E2 was shifted from a dispersed nuclear locality into the PODs and colocalized with L2. The recruitment of full-length E2 by L2 occurred in the absence of other viral components. L2 was shown previously to be essential for the generation of infectious BPV. Our present results provide evidence for a role for L2 in the organization of virion components by recruiting them to a distinct nuclear domain. This L2-dependent colocalization probably serves as a mechanism to promote the assembly of papillomaviruses either by increasing the local concentration of virion constituents or by providing the physical architecture necessary for efficient packaging and assembly. The data also suggest a role for a nonstructural viral protein, E2, in virion assembly, specifically the recruitment of the viral genome to the sites of assembly, through its high-affinity interaction with specific sequences in the viral DNA.
Papillomaviruses are nonenveloped, icosahedral DNA viruses that persistently infect stratified squamous epithelia from a wide spectrum of animals. The 8-kb double-stranded genome is maintained nonproductively in low copy number as an autonomous nuclear replicon in the basal layers of the epithelium, while productive viral replication occurs in the differentiating cells located in the more superficial layers of the epithelium. Productive viral replication cannot occur in the lower layers because the structural viral proteins are not expressed. Differentiation of the epithelium triggers a coordinate increase in the replication of the viral genome and expression of the L1 major and L2 minor structural viral proteins, leading to the assembly of infectious viral particles in the nucleus (for a review, see reference 53).
The process of viral genome encapsidation is poorly understood, particularly for the small DNA tumor viruses such as papillomaviruses. Technical difficulties in reproducing the normal pattern of differentiation in cultured epithelial cells have hampered efforts to produce infectious papillomavirus in vitro (23). Consequently, little is known about the cellular and viral factors that control the switch to the productive phase or the process by which the papillomavirus genome and virion capsid proteins assemble into infectious virions. It is unclear how the viral genome is preferentially packaged into virions, since neither L1 nor L2 binds DNA in a sequence-specific manner (38, 59). Furthermore, it is not known where in the nucleus virion assembly occurs.
We have recently reported a nonepithelial culture system for making infectious papillomavirus in which the contribution of certain viral genes to this process can be assessed (45). Infection of rodent fibroblasts with BPV virions leads to steady-state autonomous replication of the viral genome and transcription of only the nonstructural viral proteins, as is seen in the basal layers of the stratified squamous epithelium (35, 48, 57). Replication of the viral genome is dependent on two nonstructural proteins, E1 and E2, which specifically bind the viral DNA (1, 8, 42, 54, 56). Although E1 and E2 are expressed in BPV-infected fibroblasts, the structural viral genes are not (48). Therefore, no virus is produced. However, expression of the structural viral genes L1 and L2 via recombinant defective SFV vectors leads to the production of infectious BPV (45). This finding indicates that epithelial cell-specific factors are not required to generate infectious papillomavirus. Furthermore, this system provides a model for studying aspects of the latent and productive phases of the virus life cycle. With this system, preliminary genetic analysis has already shown that L2 is required for encapsidation of the viral genome into particles, although expression of L1 alone leads to nuclear assembly of empty VLPs (30, 45).
In the present study, we sought to characterize the subnuclear localization of the viral proteins involved in autonomous replication of the viral genome and in the assembly of infectious virus. To express the viral genes, we have used the SFV expression system, in part because the only vector protein expressed from the recombinant SFV RNA is the NSP1-4 polyprotein, which produces cytoplasmic RNAs for translation of the recombinant proteins (37). Therefore, this expression vector is unlikely to have a direct effect on the intranuclear events of papillomavirus assembly. Our results indicate that there is a major redistribution of viral components when latently infected cells are induced to produce infectious virus. Genetic dissection of this process has led us to propose a model for papillomavirus virion assembly in which L2 mediates the colocalization of L1 and an E2-viral genome complex at distinct nuclear structures previously identified as PODs (6, 14). Thus, this model implicates a nonstructural viral protein (E2), in addition to the structural viral proteins, in the assembly of infectious virus, as well as a particular subnuclear structure in which assembly occurs.
MATERIALS AND METHODS
Abbreviations used in this paper.
Ad5, adenovirus type 5; BPV, bovine papillomavirus; FITC, fluorescein isothiocyanate; HPV, human papillomavirus; HSV, herpes simplex virus; IgG, immunoglobulin G; NIH, National Institutes of Health; PBS, phosphate-buffered saline; PML, promyelocytic leukemia protein; POD, PML oncogenic domain; SFV, Semliki Forest virus; SV40, simian virus 40; VLP, virus-like particle.
Antibodies.
Monoclonal antibody B201, directed against the BPV E2 protein, and polyclonal antiserum 150-1, which recognizes the BPV E1 protein, were provided by Elliot Androphy (New England Medical Center, Tufts University School of Medicine). Monoclonal antibody 5B6, which recognizes the BPV L1 capsid protein, and rabbit polyclonal antiserum 17/28, raised against the full-length BPV L2 capsid protein, were generated in this laboratory and have been described previously (46). Monoclonal antibody 6A8, directed against the BPV L2 protein, was provided by A. Bennett Jenson (Georgetown University) (28). The antibody against SC35 was purchased from Sigma Chemicals (St. Louis, Mo.). The anti-PML antibody, 5E10, was generated by R. van Driel (University of Amsterdam) and was a kind gift of Louis Staudt (National Cancer Institute) (50). FITC-conjugated goat anti-mouse IgG and Texas red-conjugated goat anti-rabbit IgG were purchased from Jackson Immunoresearch (West Grove, Pa.).
Cell lines.
BPHE-1 cells, obtained from Andrew Lewis Jr. (NIH), were grown in Dulbecco’s modified Eagle’s medium supplemented with antibiotics and 10% fetal calf serum (57). BHK-21 cells were grown in Glascow’s medium supplemented with 10% tryptose phosphate broth, antibiotics, nonessential amino acids, HEPES, and 5% fetal calf serum. For microscopic analyses, the cells were seeded onto acid-washed no. 01 coverslips in 24-well plates at a density of 105 cells/well and cultured overnight.
Recombinant SFV expression system.
The production of recombinant SFV RNAs and replication-defective virus expressing the BPV L1 or L2 capsid protein and the SFV infection protocols have been described previously (45). BPV E1 and E2 were cloned into the BamHI site of pSFV-1 as PCR products amplified from the BPV genome (primers for E1, 5′ ccgctggatccgcaccatggcaaacgataaaggtagc and 3′ gcggtggatccgatcttgcaacttatcactac; primers for E2, 5′ ccgctggatccgcaccatggagacagcatgcgaacg and 3′ gcggtggatccgaagaaaaggcaatggcagtg [boldface indicates restriction sites and start sites]), and recombinant viruses expressing each gene were generated as described for L1 and L2. For infection of cells, high-titer recombinant SFV stock was treated with 500 μg of chymotrypsin A4 per ml on ice for 30 min and then aprotinin was added to 500 μg/ml for an additional 10 min. The activated virus was diluted 100-fold in Dulbecco’s PBS with calcium and magnesium and added to cells in 24-well plates. After 60 min at 37°C, virus-containing medium was removed and replaced with the normal growth medium supplemented with 100 mM KCl for the remainder of the infection to maintain cellular protein expression. Infections were allowed to continue for 5 to 6 h prior to cell fixation and immunolocalization. Although SFV infection will induce cell death in 48 h, the morphology of the infected cells was not visibly altered at this early time point.
Immunofluorescence staining.
The cells were washed three times with cold PBS (pH 7.4), fixed by a 10-min incubation at room temperature with 1.0% paraformaldehyde diluted in PBS, and washed three times with PBS–200 mM glycine. They were then incubated with primary antibody diluted in PBS–0.1% polyoxyethylene 20 cetyl ether (Brij 58; Sigma Chemicals) and incubated at 4°C. Polyclonal antisera were used at a dilution of 1/1,000. Monoclonal antibodies used as hybridoma supernatants were diluted 1/100. Purified antibodies were used at a concentration of 5 μg/ml. For double immunofluorescence staining, the primary antibodies were incubated in unison. After incubation, the coverslips were washed three times with PBS–0.1% Brij. Secondary antibodies were diluted to 5 μg/ml in PBS–0.1% Brij, and incubation was performed at 4°C. After this incubation, the cells were washed thoroughly in PBS–0.1% Brij and inverted onto Fluoromount-G mounting solution (Southern Biotechnology Associates, Birmingham, Ala.) on a glass slide. Fluorescence was examined with a Bio-Rad MRC 1024 laser scanning confocal system attached to a Zeiss Axioplan microscope. All the images were acquired with a Zeiss 63x N.A. 1.4 Planapo objective, using the photon-counting mode. The use of control coverslips established that fluorescence in the green and red channels was not overlapping and that antibody binding was specific for the intended antigen. The images were collaged and subjected to scale adjustment with Adobe Photoshop software.
RESULTS
Subnuclear localization of BPV capsid proteins.
BPHE-1 is a hamster fibroblast cell line that is latently infected with multiple copies of autonomously replicating BPV genomes and expresses the nonstructural viral proteins (57). We used the SFV expression system to introduce the L2 minor capsid protein into BPHE-1 cells and localized the L2 protein by immunofluorescence staining and laser scanning confocal microscopy. Figure 1A and D, which shows the typical distribution of L2 6 h after SFV infection, indicates that the protein was displayed in a distinct intranuclear punctate pattern.
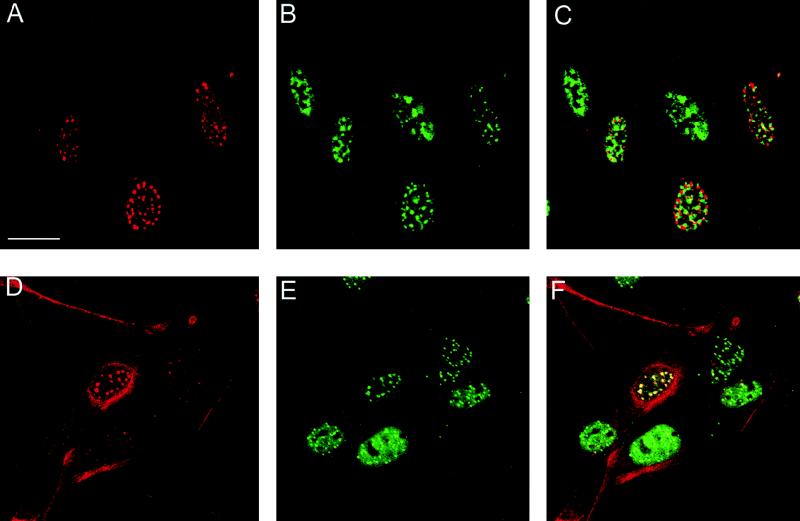
FIG. 1.
Colocalization of L2 with PML protein. BPHE-1 cells were infected with the L2-SFV recombinant. The cells were fixed, and double-staining immunolocalization against the L2 protein and either SC35 or PML was performed. (A and D) The L2 protein was detected with rabbit polyclonal antiserum 17/28 and Texas red-conjugated goat anti-rabbit IgG. (B) SC35 was detected with a mouse monoclonal antibody (Sigma) and FITC-conjugated goat anti-mouse IgG. The same field is shown in panels A and B. (C) Digital superimposition of the two images. Coincidence of staining would be depicted in yellow in the merged panel. (E) PML localization. PML was detected with mouse monoclonal antibody 5E10 and FITC-conjugated goat anti-mouse IgG. Panel E shows the same field as the anti-L2 staining in panel D. (F) The overlap in the distribution of the patterns is evident in the merged image. Note that in panels D to F, the L2-PML colocalization is independent of the level of expression of L2. Bar, 10 mm.
To rule out the possibility that this distribution depended on the BPV components in the BPHE-1 cells, L2 was expressed, via the SFV vector, in cells that did not harbor papillomavirus sequences. A similar punctate nuclear pattern of L2 staining was also observed in other cell types, including COS-7, BHK-21, and the human fibroblast cell line 1634 (see Fig. 5A; data not shown). Therefore, this distinct L2 localization is dependent only upon cellular factors and appears to be independent of cell lineage. To determine if this localization was a common feature of papillomavirus L2, the distribution of the HPV16 L2 protein, expressed via an SFV vector, in these cell lines was also examined. The pattern with HPV16 L2 protein was similar to that seen with BPV L2 (data not shown), strongly suggesting that this localization is characteristic of papillomavirus L2.
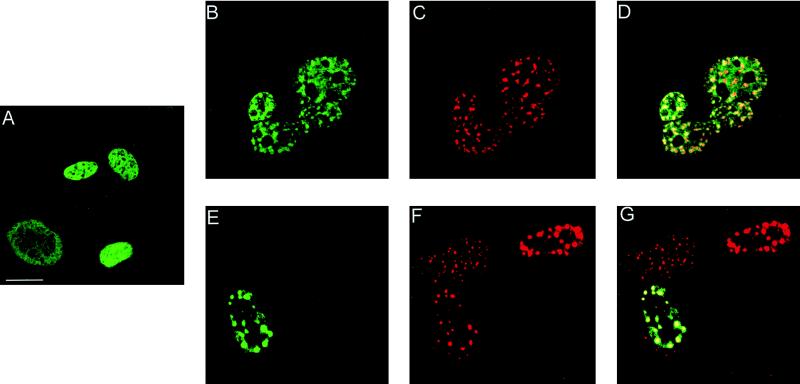
FIG. 5.
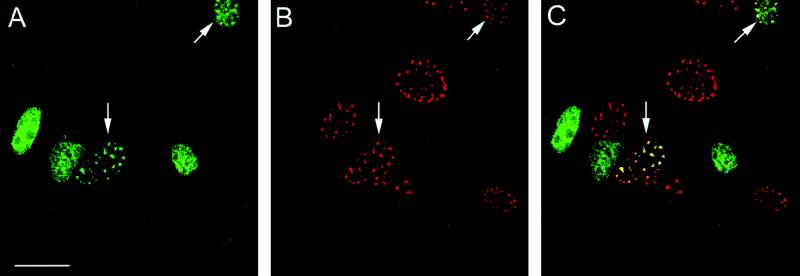
The full-length form of E2 relocates to PODs. BHK-21 cells were infected with either the L2-SFV recombinant or the E2-SFV recombinant or coinfected to simultaneously express both proteins. (A) In cells that were infected with L2-SFV, the L2 protein was detected with 17/28 and Texas red-conjugated goat anti-rabbit IgG. (B) Cells that were infected with E2-SFV were stained with anti-E2 monoclonal antibody B201 and FITC conjugated goat anti-mouse IgG. The proteins in the coinfected cells were detected by double staining with the same primary and secondary antibodies. (C) Anti-L2 staining, detected in the red channel. (D) Anti-E2 staining of the same field, detected in the green channel. A cell that is coinfected is indicated by the arrows in these panels. Bar, 20 μm.
L2-containing punctate structures are PODs.
To identify the nuclear domains in which the BPV L2 protein localized, we performed double-staining experiments with a number of described nuclear proteins and L2. We found no colocalization of the L2 protein with coiled bodies, the retinoblastoma protein p53, or the splicing factor SC35. Although the staining pattern seen with the anti-SC35 antibody was similar to that seen with the anti-L2 antibody (Fig. 1A and B), it was evident from the merged image that these regions were exclusive (Fig. 1C). However, when we compared the distribution of the L2 protein with that of anti-PML protein staining, we observed a nearly complete overlap in protein distribution (Fig. 1D to F).
The PML protein is a putative growth suppressor gene product that localizes in subnuclear organelles termed PODs (6, 14). The PML distribution appeared to be unaffected by the expression of the L2 protein, and the localization of L2 in the PODs was unrelated to the level of L2 in the cell. This is seen in Fig. 1D, where cells expressing high, intermediate, and low levels of L2 can be compared. All the cells expressing L2 showed a similar punctate distribution, in which L2 colocalized with PML in every cell. Therefore, it is unlikely that this colocalization is an artifact of overexpression. It is also unlikely that localization of L2 to PODs is due to expression via defective SFV, since neither morphological changes nor apoptosis (as evidenced by DNA fragmentation) was detected at the 5- to 6-h postinfection time point at which the analysis was conducted (data not shown). In addition, SFV infection did not induce POD localization of other proteins, independent of L2 (see below).
L2 redirects L1 to PODs.
Since L1 and L2 coassemble into capsids, we sought to determine whether L1 might display a nuclear staining pattern similar to that of L2. However, when L1 was expressed in BPHE-1 cells, its distribution protein differed markedly from that of L2. L1 was present in a nuclear pattern that varied from a diffuse to a slightly speckled arrangement with nucleolar exclusion (Fig. 2A).
FIG. 2.
L2 expression affects the cellular distribution of L1. BPHE-1 cells were either infected with the L1-SFV recombinant (A) or coinfected with the L1-SFV recombinant and the L2-SFV recombinant, and the proteins were detected by double immunofluorescence (B to G). (A) Staining with the anti-L1 antibody, 5B6, detected with FITC-conjugated goat anti-mouse IgG. Panels B to D show the same field of cells as do panels E to G. (B and E) Optically gated images showing the unique fluorescence of the 5B6 antibody; (C and F) optically gated images showing the unique fluorescence of the anti-L2 antiserum, 17/28, detected with Texas-red conjugated goat anti-rabbit IgG. The digital merge of the images is shown in panels D and G. Coincidence of staining appears yellow in the merged image. Bar, 10 μm.
This result led us to explore the possibility that the subcellular distribution of L1 protein is affected by coexpression of L2. Therefore, we coinfected BPHE-1 cells with recombinant L1-SFV and recombinant L2-SFV, which are the conditions that lead to the formation of infectious BPV in BPHE-1 cells. The L1 staining, which was collected in the green channel, is shown in Fig. 2B and E. This pattern was dramatically altered from the diffuse nuclear pattern seen after L1 SFV infection alone. The L2 staining pattern in the coinfected cells was consistent with the distribution of L2 observed in the absence of L1 (Fig. 2C and F). The distributions of L1 and L2 overlapped substantially in the merge of the two images (Fig. 2D and G). In general, L1 did not appear as tightly coalesced as L2 (Fig. 2D). However in some cells, L1 almost completely colocalized with L2 (Fig. 2G), while in other cells, we have observed L1 mostly surrounding rather than overlapping the L2 domain. This variability may be due to differences in the kinetics of the infection of individual cells or may reflect different stages in L1 relocation or virion assembly. We conclude that L2 induced the redirection of a substantial proportion of L1 to PODs.
L2 induces colocalization of E2.
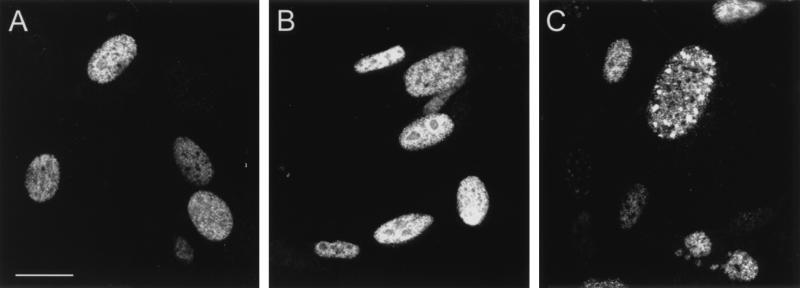
We next examined the effect of expression of the BPV capsid proteins on the distribution of the nonstructural viral protein E2, which is involved in viral genome replication and viral transcription (8, 47, 54). In BPHE-1 cells, E2 was detected as a nuclear protein with a diffuse distribution (Fig. 3A). There was no apparent effect on the localization of this protein when the L1 capsid protein was expressed in these cells (Fig. 3B). In contrast, L2 expression shifted E2 into punctate regions similar to those observed with the anti-L2 staining shown in Fig. 1. Figure 3C shows representative E2 immunostaining in BPHE-1 cells that have been infected with recombinant L2-SFV. The levels of E2 often decreased substantially during recombinant SFV infection, presumably due to the well-documented inhibition of host protein synthesis by SFV, although this did not interfere with determining the localization of E2 (49). This effect is partially due to interference with the Na+K+ transporter by SFV (4, 18). A decrease in E2 was also observed in control infections with unrelated SFV recombinants (data not shown). Infection in the presence of 100 mM KCl helped counteract this problem, but nevertheless, as seen in Fig. 4B and C, the level of immunoreactive E2 was low in a number of the SFV-infected cells.
FIG. 3.
E2 distribution in BPHE-1 cells that are uninfected or infected with either the L1-SFV or L2-SFV recombinant. The E2 protein was detected with monoclonal antibody B201 and FITC-conjugated goat anti-mouse IgG. (A) E2 distribution in uninfected cells. (B) E2 distribution in L1-SFV-infected cells. (C) E2 distribution in L2-SFV-infected cells. All panels were collected with identical image settings. Bar, 10 μm.
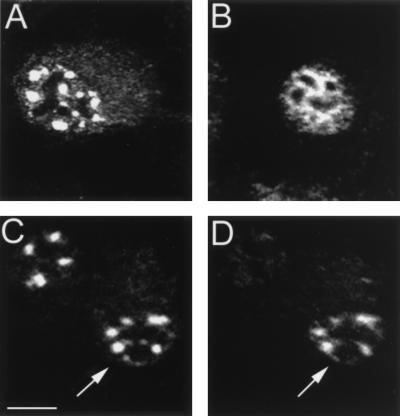
FIG. 4.
L2 recruits the E2 protein into PODs. BPHE-1 cells were infected with the L2-SFV recombinant, and the L2 and E2 proteins were detected by double immunofluorescence. Panels A and B show the same field of cells. (A) Localization of E2 detected with B201 and FITC-conjugated goat anti-mouse IgG. (B) Localization of L2 detected with 17/28 and Texas red-conjugated goat anti-rabbit IgG. (C) Digital merge of the images in panels A and B. Yellow areas indicate regions of colocalized staining. Bar, 10 μm.
To determine if L2 induced the redistribution of the E2 protein into the L2-staining PODs, we performed double staining of the BPHE-1 cells after infection with L2-SFV. Figure 4A shows the E2 staining in these cells as detected by fluorescence in the green channel. Most of the cells show the diffusely distributed nuclear pattern. However, the cells indicated by the arrows demonstrate the relocation of E2 into the punctate pattern. Figure 4B shows the anti-L2 staining of these cells as detected in the red channel. All of the L2-expressing cells showed the punctate pattern of staining. Figure 4C is the merge of the red and green channels shown in Fig. 4A and B. The coincidence of the E2 and L2 staining is striking in the infected cells that maintain detectable levels of E2.
L2 is sufficient to redistribute full-length E2.
BPV-transformed cells with autonomously replicating genomes express three forms of the E2 protein: a full-length 48-kDa form that functions in genome replication and transcriptional transactivation and two smaller forms which act as repressors of viral transcription (25, 32, 40). The antibody used in the immunofluorescence studies recognizes an epitope in the C-terminal DNA binding domain common to all three proteins and would not distinguish among them. Another feature of the BPHE-1 cells is that an unknown proportion of E2 molecules are bound to the viral genome. Therefore, it was unclear whether the L2-dependent redistribution of E2 in the BPHE-1 might depend on the presence of the viral genome in the cells.
To determine if the L2-dependent redistribution of E2 observed in the BPHE-1 cells could occur between L2 and the full-length E2 protein, independently of the viral genome, we infected BHK-21 cells (which do not contain the papillomavirus genome) with both the L2-SFV recombinant and an SFV recombinant expressing the full-length E2. Since the RNA for E2 was produced entirely by the SFV RNA-dependent polymerase in the cytoplasm, production of the alternative E2 mRNAs was precluded. As expected, only the 48-kDa form was detected on Western blots of SFV-E2-infected cell extracts (data not shown). As noted above, the L2 distribution in BHK-21 cells was similar to that observed with the BPHE-1 cells (Fig. 5A). When E2 was expressed in BHK-21 cells, in the absence of L2, most of the protein was present in a diffuse nuclear distribution (Fig. 5B). When the cells were coinfected with L2 and E2, the L2 pattern was unaltered but E2 assumed the punctate staining pattern of L2 in the cells that coexpressed the two proteins (Fig. 5C and D). These results indicate that L2-dependent localization of the full-length E2 to PODs is independent of the viral genome and viral gene products other than L2.
L2 does not induce the redistribution of E1.
We also examined the localization of E1, which participates in viral DNA replication and so is presumably expressed in BPHE-1 cells. The immunostaining with an anti-E1 antibody in BPHE-1 cells was weak. This result was expected, since only low levels of E1 expression from steady-state autonomously replicating BPV genomes have been reported (51). No change in the speckled staining pattern was observed after SFV-mediated expression of either capsid protein. Because the intensity of the staining was so low and the parental line of BPHE-1 was not available as a control, no firm conclusions could be drawn from the E1 analysis in these cells.
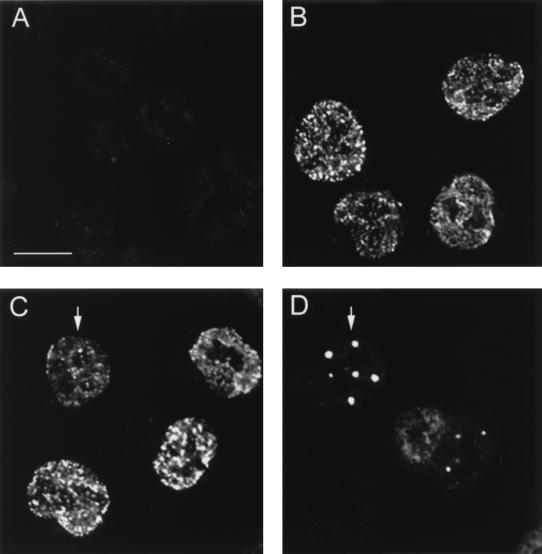
To overcome these problems, BHK-21 cells were infected with an E1-SFV recombinant, which resulted in clear immunostaining in a speckled nuclear pattern, while uninfected cells were negative (Fig. 6A and B). Coinfection with the L2 and E1 recombinant SFVs resulted in the typical punctate L2 staining pattern (Fig. 6D), but this expression did not alter the E1 pattern in the coinfected cells (Fig. 6C). Therefore, the L2 protein does not directly induce a redistribution of E1. However, these results do not preclude the possibility that E1 localizes to PODs indirectly through its well-documented interaction with E2 (42, 56).
FIG. 6.
E1 localization is unaffected by L2 expression. BHK-21 cells were coinfected with the E1-SFV and L2-SFV recombinants. E1 was detected with rabbit polyclonal antiserum 150-1 and Texas-red conjugated goat anti-rabbit IgG. (A) Binding of these antisera to uninfected cells. (B) 150-1 staining of cells infected with E1-SFV. (C) E1 localization after coinfection with E1-SFV and L2-SFV (arrow). Panels A through C were collected with identical image settings. (D) The L2 protein was detected in the coinfected cells by staining with monoclonal antibody 6A8-E6H6 and FITC-conjugated goat anti-mouse IgG. Panels C and D show identical fields; a coexpressing cell is indicated by the arrow. Bar, 20 μm.
DISCUSSION
In this study, we have found that the minor capsid protein L2 has the intrinsic capacity to localize to PODs in the absence of other viral components. Further, the presence of L2 in PODs is associated with the recruitment of the major capsid protein L1 and the nonstructural protein E2, which binds the genome with high affinity at multiple sequence-specific sites (1, 36). It is therefore attractive to speculate that PODs are the main structures in which papillomaviruses assemble.
PODs are interchromatinic matrix-bound nuclear bodies with an average diameter of 0.3 to 0.5 μm in most cells. The cellular function(s) of PODs is largely unknown (2, 19). They have also been designated Kr bodies or nuclear domain 10 (ND10) based on the average number of bodies per cell, although their number actually varies and may be greater in transformed cells (2, 33, 55). PODs may be required for normal maturation of myeloid cells, since their fragmentation is often seen in acute promyelocytic leukemia (PML) (14). Disruption of PODs in this leukemia is associated with heterodimer formation between the normal PML protein and a PML-retinoic acid receptor α fusion protein that results from a characteristic t(15;17) chromosomal translocation (10, 11, 29, 55). In addition to PML, PODs contain several other proteins. These include the SP100 protein, which was originally identified as an autoantigen in patients with primary biliary cirrhosis, Int-6, the PIC-1 protein, and 55-kDa (NDP55) and 65-kDa proteins (2, 3, 9, 15, 52).
Some associations have been reported between PODs and the replication of other DNA viruses. Productive viral replication appears to commence in association with PODs for HSV-1, Ad5, and SV40 (5, 13, 16, 27, 39, 44). Despite the remarkable convergence to this structure for these three genetically unrelated viruses, the role that this localization plays in the virus-cell interaction has remained unclear.
A number of potential roles in viral replication have been suggested for the association of viral components with PODs. It has been proposed that POD association may be a cellular mechanism that has evolved to limit initial virus replication (26). The fact that Ad5 E4-ORF3 and HSV-1 ICP0 encode proteins that disrupt PODs as infection proceeds has been taken as evidence supporting this possibility (13, 16, 39, 44). Also, the observation that interferon upregulates the expression of POD proteins is consistent with PODs acting as an antiviral defense mechanism (7, 22, 34).
Alternatively, POD association may play a positive role in viral replication. This localization might (i) increase the local concentration of viral products and so promote assembly, (ii) interfere with normal differentiation and/or apoptotic responses to the viruses in the epithelial cells that are their usual sites of initial replication, (iii) facilitate access to cellular transcription and/or replication factors (although there is little evidence that PODs possess these functions), and (iv) promote essential processing of viral products. With regard to the last of these items, it is interesting that a ubiquitin-dependent protease has recently been shown to be POD associated (3). Our finding that the conversion from latent to productive papillomavirus infection in our in vitro system is associated with a redistribution of the relevant viral products to PODs supports to the view that PODs play a positive role in the replication of papillomaviruses.
While studies of Ad5, HSV, SV40, and Epstein-Barr virus have identified early-gene products that interact with, and in some cases disassemble, PODs, these proteins have not been implicated in virion assembly and the gene products responsible for POD localization of the virion components have not been determined (13, 16, 27, 39, 44). In this study, we have demonstrated that the association of other papillomavirus proteins with PODs during productive infection depends upon the L2 minor capsid protein. In the absence of L2, which is essential for the generation of infectious virus, the other viral components display indistinct, heterogeneous distributions. Our results suggest that L2 may function to facilitate virion production by inducing the colocalization of the other components required for virion assembly. The recruitment to PODs is likely to represent an important feature that distinguishes productive from latent papillomavirus infection. It is possible that the POD-binding proteins HSV-1 ICP0 and Epstein-Barr virus EBNA-5, which have been implicated in the escape from latency, serve an analogous function for their respective viruses.
The results of this study suggest the following model for the productive phase of the papillomavirus life cycle (Fig. 7). The productive cycle begins when L1 and L2 expression is induced by differentiation-specific signals in the infected epithelial cells (12, 41). SFV-mediated expression of these two genes substitutes for this in our system and demonstrates that differentiation per se is not required for virus production. Virus assembly appears to be triggered by the association of L2 with PODs and the colocalization of L1. It is likely that the association of L1 with the PODs is the result of a direct interaction of L1 with L2, since stable L1-L2 complexes form in both fully assembled VLPs in vivo and also in partially assembled viral capsid structures, including L1 pentamers, in vitro (42a). Although L1 can self-assemble into VLPs in the absence of L2 (30, 31), L2 increases VLP production 4-fold in insect cells and 100-fold in mammalian cells (24, 31, 58). This greater efficiency could be the result of an increased rate of capsid assembly as a consequence of the L2-mediated concentration of L1 at the PODs.
FIG. 7.
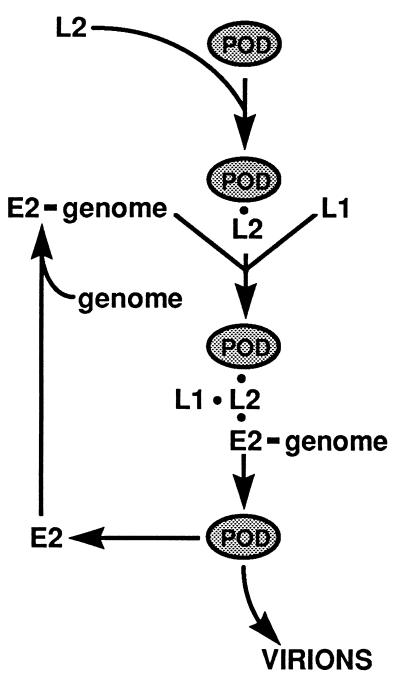
Model for L2-mediated assembly of papillomavirus virions. It is proposed that L2 acts to mediate papillomavirus assembly by causing the concentration of the virion components within the PODs. L2 localizes to PODs independently of other viral proteins. The L2 localization will cause the subsequent recruitment of E2 with the bound genome and L1. These events are independent of each other. This L2-L1-E2-genome association within the PODs confers an appropriate environment and/or concentration for virion assembly. See the text for details.
It appeared that in some cells containing L2, the L1 protein was located predominantly around the perimeter of the L2 domains rather than overlapping them. These variations may reflect temporal differences in the SFV infection of individual cells, since all infections appeared to show a mixture of the two patterns. It is likely that we detected a variety of L1 assembly states with the anti-L1 antibody used here. In vitro, the antibody recognizes pentameric L1 as well as in intact virions (our unpublished results). It is possible that the L1 detected around the POD perimeter is due to mature virions that have been released from the sites of assembly and show a diminished reactivity with the anti-L2 antibody. Alternatively, the peripheral anti-L1 staining could be due to L1 pentamers in the process of assembling with L2 or to L1-only VLPs in instances where L1 is in excess.
L2 also induced the redistribution of E2. The experiments with the BHK-21 cells clearly demonstrated that E2 association with the PODs is dependent on L2 but is independent of L1, other early papillomavirus gene products, or the viral genome. However, we have no evidence that E2 interacts directly with L2. Despite considerable efforts, including coimmunoprecipitation experiments and cosedimentation in sucrose gradients, we have not detected soluble E2-L2 complexes in vivo or in vitro. At present, we cannot distinguish between the possibilities that L2 and E2 bind with relatively low affinity, that E2 binds to a component of the PODs that has been altered by L2, or that E2, L2 and a POD component form a trimolecular complex.
It is unclear how papillomaviruses preferentially package their genomes over cellular DNA, since neither the individual capsid proteins nor the assembled VLPs bind the genome in a sequence-specific manner (38, 59). We speculate that the specificity of viral genome encapsidation is due to the L2-dependent translocation of an E2-genome complex to the PODs, since E2 avidly binds multiple sites on the viral genome (1, 36). Preliminary fluorescent in situ hybridization experiments with BPHE-1 cells suggest that L2 can induce a change in the nuclear distribution of the autonomously replicating viral genomes from diffuse to punctate (our unpublished observation). Since we have been unable to detect E2 in infectious BPV virions extracted from cattle warts (unpublished observation), E2 is depicted in Fig. 7 as acting catalytically in the process of virion assembly. SV40 T antigen, which is a nonstructural protein of that virus, may be functionally analogous to E2 in this regard. T antigen is a viral genome binding transcription/replication factor that associates with PODs but does not cause their disruption (27). A signal on the SV40 genome that is required for its packaging into virions has been mapped to a segment on the viral DNA that includes the T antigen binding sites (43).
Translocation of the viral products to PODs could potentially promote virus production in additional ways. It is possible that overreplication of the viral genome is induced by redistribution of the papillomavirus genome to this location, since lytic DNA viruses commence their replicative cycles at PODs. There has been no model system to study the induction of vegetative DNA replication of papillomaviruses, and nothing is known about what triggers the process or which viral gene products are required. It will be interesting to examine the temporal relationship between POD localization of the viral proteins and replication of the viral genome. Most of the analyses in this study were done approximately 6 h after SFV infection. However, we routinely wait until at least 30 h postinfection to maximize the harvest of infectious papillomavirus (45). Therefore, accumulation of viral genomes might be more evident at time points later than those analyzed here.
Disruption of PODs caused by the PML-retinoic acid receptor α fusion protein is associated with the inhibition of terminal differentiation of promyelocytes (14, 17, 20, 21). This finding raises the possibility that association of the papillomavirus gene products with these structures influences the differentiation program of normal keratinocytes. Although induction of epithelial differentiation is normally required for capsid gene expression, it may be advantageous for the virus to delay or prevent the final stages of terminal differentiation. For instance, indiscriminate peptide cross-linking by transglutaminase and DNA-RNA degradation by terminal differentiation-specific nucleases would seemingly be at odds with the production of infectious virions. Examination of keratinocyte-specific differentiation markers in normal keratinocytes induced to differentiate after the introduction of L2 (or L2 plus the viral proteins that depend on L2 for POD localization) might reveal interesting relationships among PODs, papillomavirus proteins, and epithelial differentiation.
ACKNOWLEDGMENTS
We thank Andrew M. Lewis (NIH, Bethesda, Md.) for the BPHE-1 cells, Louis Staudt (NIH) for the 5E10 antibody, Elliot Androphy for the 150-1 antiserum and the B201 antibody, and A. Bennett Jenson for antibody 6A8. We are especially grateful to Jon Yewdell, Jack Bennink, and the Laboratory of Viral Diseases (NIH) for the use of their confocal microscope.
REFERENCES
- 1.Androphy A J, Lowy D R, Schiller J T. Bovine papillomavirus E2 trans activating gene product binds to specific sites in papillomavirus DNA. Nature. 1987;325:70–73. doi: 10.1038/325070a0. [DOI] [PubMed] [Google Scholar]
- 2.Ascoli C, Maul G J. Identification of a novel nuclear domain. J Cell Biol. 1991;112:785–795. doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boddy M N, Howe K, Etkin L D, Solomon E, Freemont P S. Pic 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 4.Carrasco L. The inhibition of cell functions after viral infection: a proposed general mechanism. FEBS Lett. 1977;76:11–15. doi: 10.1016/0014-5793(77)80110-5. [DOI] [PubMed] [Google Scholar]
- 5.Carvalho T, Seeler J S, Ohman K, Jordan P, Petterson U, Akusjarvi G, Carmo-Fonseca M, Dejean A. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J Cell Biol. 1995;131:45–56. doi: 10.1083/jcb.131.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang K S, Fan Y H, Andreef M, Liu J, Mu Z M. The PML gene encodes a phosphoprotein associated with the nuclear matrix. Blood. 1995;85:3646–3653. [PubMed] [Google Scholar]
- 7.Chelbi-Alix M K, Pelicano L, Quignon F, Koken M H M, Venturini L, Stadler M, Pavlovic J, Degos L, de The H. Induction of PML protein by interferons in normal and APL cells. Leukemia. 1995;9:2027–2033. [PubMed] [Google Scholar]
- 8.Chiang C-M, Ustav M, Stenlund A, Ho T, Broker T R, Chow L T. Viral E1 and E2 proteins support replication of homologous and heterologous papillomavirus origins. Proc Natl Acad Sci USA. 1992;89:5799–5803. doi: 10.1073/pnas.89.13.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desbois C, Rousset R, Bantignies F, Jalinot P. Exclusion of Int-6 from PML nuclear bodies by binding to the HTLV-I Tax oncoprotein. Science. 1996;273:951–3. doi: 10.1126/science.273.5277.951. [DOI] [PubMed] [Google Scholar]
- 10.de The H, Chomienne C, Lanotte M, Degos L, Dejean A. The t(15;17) translocation of the acute promyelocytic leukemia fuses the retinoic acid receptor α gene to a novel transcribed locus. Nature. 1990;347:558–561. doi: 10.1038/347558a0. [DOI] [PubMed] [Google Scholar]
- 11.de The H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 12.Dollard S C, Wilson J L, Demeter L M, Bonnez W, Reichman R C, Broker T R, Chow L T. Production of human papillomavirus and modulation of the infectious program in epithelial raft cultures. Genes Dev. 1992;6:1131–1142. doi: 10.1101/gad.6.7.1131. [DOI] [PubMed] [Google Scholar]
- 13.Doucas V, Ishov A M, Romo A, Juguilon H, Weitzman M D, Evans R M, Maul G G. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 14.Dyck J A, Maul G G, Miller W H, Jr, Chen J D, Kakizuka A, Evans R M. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 15.Epstein A L. Immunobiochemical characterization with monoclonal antibodies of Epstein-Barr virus-associated early antigens in chemically induced cells. J Virol. 1984;50:372–379. doi: 10.1128/jvi.50.2.372-379.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett R D, Maul G G. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fagioli M, Grignani F, Ferrucci P F, Alcalay M, Mencarelli A, Nicoletti I, Grignani F, Pelicci P G. Effect of the acute promyelocytic leukemia PML/RAR alpha protein on differentiation and survival of myeloid precursors. Leukemia. 1994;8:S7–S11. [PubMed] [Google Scholar]
- 18.Garry R F, Bishop J M, Parker S, Westbrook K, Lewis G, Waite M R F. Na+ and K+ concentrations and the regulation of protein synthesis in Sindbis virus-infected chick cells. Virology. 1979;96:108–120. doi: 10.1016/0042-6822(79)90177-6. [DOI] [PubMed] [Google Scholar]
- 19.Grande M A, van der Kraan I, van Steensel B, Schul W, de The H, van der Voort H T, de Jong L, van Driel R. PML-containing nuclear bodies: their spatial distribution in relation to other nuclear components. J Cell Biochem. 1996;63:280–291. doi: 10.1002/(sici)1097-4644(19961201)63:3<280::aid-jcb3>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 20.Grignani F, Testa U, Fagioli M, Barberi T, Masciulli R, Mariani G, Peschle C, Pelicci P G. Promyelocytic leukemia-specific PML-retinoic acid receptor alpha fusion protein interferes with erythroid differentiation of human erythroleukemia K562 cells. Cancer Res. 1995;55:440–443. [PubMed] [Google Scholar]
- 21.Grignani F, Testa U, Rogaia D, Ferrucci P F, Samoggia P, Pinto A, Aldinucci D, Gelmetti V, Fagioli M, Alcalay M, Seeler J, Grignani F, Nicoletti I, Peschle C, Pelicci P G. Effects on differentiation by the promyelocytic leukemia PML/RARalpha protein depend on the fusion of the PML protein dimerization and RARalpha fusion domains. EMBO J. 1996;15:4949–4958. [PMC free article] [PubMed] [Google Scholar]
- 22.Grotzinger T, Jensen K, Guldner H H, Sternsdorf T, Szostecki C, Schwab M, Savelyeva L, Reich B, Will H. A highly amplified mouse gene is homologous to the human interferon-responsive Sp100 gene encoding an autoantigen associated with nuclear dots. Mol Cell Biol. 1996;16:1150–1156. doi: 10.1128/mcb.16.3.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagensee M, Galloway D. Growing human papillomaviruses and virus-like particles in the laboratory. Papillomavirus Rep. 1993;4:121–124. [Google Scholar]
- 24.Hagensee M E, Yaegashi N, Galloway D A. Self-assembly of human papillomavirus type 1 capsids by expression of the L1 protein alone or by coexpression of the L1 and L2 capsid proteins. J Virol. 1993;67:315–22. doi: 10.1128/jvi.67.1.315-322.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hubbert N L, Schiller J T, Lowy D R, Androphy E J. Bovine papillomavirus transformed cells contain multiple E2 proteins. Proc Natl Acad Sci USA. 1988;85:5864–5868. doi: 10.1073/pnas.85.16.5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishov A M, Maul G G. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang W Q, Szekely L, Klein G, Ringertz N. Intranuclear redistribution of SV40T, p53, and PML in a conditionally SV40T-immortalized cell line. Exp Cell Res. 1996;229:289–300. doi: 10.1006/excr.1996.0374. [DOI] [PubMed] [Google Scholar]
- 28.Jin X W, Cowsert L M, Pilacinski W P, Jenson A B. Identification of L2 open reading frame gene products of bovine papillomavirus type 1 using monoclonal antibodies. J Gen Virol. 1989;70:1133–1140. doi: 10.1099/0022-1317-70-5-1133. [DOI] [PubMed] [Google Scholar]
- 29.Kakizuka A, Miller W H J, Umesono K, Warrel R P J, Frankel S R, Murty V V V S, Dmitrovsky E, Evans R M. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RARa with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 30.Kirnbauer R, Booy F, Cheng N, Lowy D R, Schiller J T. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc Natl Acad Sci USA. 1992;89:12180–12184. doi: 10.1073/pnas.89.24.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirnbauer R, Taub J, Greenstone H, Roden R B S, Durst M, Gissmann L, Lowy D R, Schiller J T. Efficient self-assembly of human papillomavirus type 16 L1 and L1-L2 into virus-like particles. J Virol. 1993;67:6929–6936. doi: 10.1128/jvi.67.12.6929-6936.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lambert P F, Hubbert N L, Howley P M, Schiller J T. A transcriptional repressor encoded by BPV1 shares a common carboxy-terminal domain with the E2 transactivator. J Virol. 1989;63:3151–3154. [Google Scholar]
- 33.Lamond A I, Carmo-Fonseca M. The coiled body. Trends Cell Biol. 1993;3:198–204. doi: 10.1016/0962-8924(93)90214-l. [DOI] [PubMed] [Google Scholar]
- 34.Lavau C, Marchio A, Fagioli M, Jansen J, Falini B, Lebon P, Grosveld F, Pandolfi P P, Pelicci P G, Dejean A. The PML gene is a primary target for interferon. Oncogene. 1995;11:871–876. [PubMed] [Google Scholar]
- 35.Law M F, Lowy D R, Dvoretzky I, Howley P M. Mouse cells transformed by bovine papillomavirus contain only extrachromosomal viral DNA sequences. Proc Natl Acad Sci USA. 1981;78:2727–2731. doi: 10.1073/pnas.78.5.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li R, Knight J, Bream G, Stenlund A, Botchan M. Specific recognition nucleotides and their context determine the affinity of E2 protein for 17 binding sites in the BPV-1 genome. Genes Dev. 1989;3:510–526. doi: 10.1101/gad.3.4.510. [DOI] [PubMed] [Google Scholar]
- 37.Liljestrom P, Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Bio/Technology. 1991;9:1356–1361. doi: 10.1038/nbt1291-1356. [DOI] [PubMed] [Google Scholar]
- 38.Mallon R G, Wojciechowicz D, Defendi V. DNA binding activity of papillomavirus proteins. J Virol. 1987;61:1655–1660. doi: 10.1128/jvi.61.5.1655-1660.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maul G G, Ishov A M, Everett R D. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- 40.McBride A A, Byrne J C, Howley P M. The papillomavirus E2 regulatory proteins. J Biol Chem. 1991;266:18411–18414. [PubMed] [Google Scholar]
- 41.Meyers C, Frattini M G, Hudson J B, Laimins L A. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science. 1992;257:971–973. doi: 10.1126/science.1323879. [DOI] [PubMed] [Google Scholar]
- 42.Mohr I J, Clark R, Sun S, Androphy E J, MacPherson P, Botchan M R. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science. 1990;250:1694–1699. doi: 10.1126/science.2176744. [DOI] [PubMed] [Google Scholar]
- 42a.Okun, M., et al. Unpublished data.
- 43.Oppenheim A, Sandalon Z, Peleg A, Shaul O, Nicolis S, Ottolenghi S. A cis-acting DNA signal for encapsidation of simian virus 40. J Virol. 1992;66:5320–5328. doi: 10.1128/jvi.66.9.5320-5328.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puvion-Dutilleul F, Chelbi-Alix M K, Koken M, Quignon F, Puvion E, de The H. Adenovirus infection induces rearrangements in the intranuclear distribution of the nuclear body-associated PML protein. Exp Cell Res. 1995;218:9–16. doi: 10.1006/excr.1995.1125. [DOI] [PubMed] [Google Scholar]
- 45.Roden R B S, Greenstone H L, Kirnbauer R, Booy F P, Jessie J, Lowy D R, Schiller J T. In vitro generation and type-specific neutralization of a human papillomavirus type 16 virion pseudotype. J Virol. 1996;70:5875–5883. doi: 10.1128/jvi.70.9.5875-5883.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roden R B S, Weissinger E M, Henderson D W, Booy F, Kirnbauer R, Mushinski J F, Lowy D R, Schiller J T. Neutralization of bovine papillomavirus by antibodies to L1 and L2 capsid proteins. J Virol. 1994;68:7570–7574. doi: 10.1128/jvi.68.11.7570-7574.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spalholz B A, Yang Y C, Howley P M. Transactivation of a bovine papilloma virus transcriptional regulatory element by the E2 gene product. Cell. 1985;42:183–191. doi: 10.1016/s0092-8674(85)80114-8. [DOI] [PubMed] [Google Scholar]
- 48.Stenlund A, Zabielski J, Ahola H, Moreno-Lopez J, Pettersson U. Messenger RNAs from the transforming region of bovine papillomavirus type 1. J Mol Biol. 1985;182:541–554. doi: 10.1016/0022-2836(85)90240-2. [DOI] [PubMed] [Google Scholar]
- 49.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stuurman N, de Graaf A, Floore A, Josso A, Humbel B, de Jong L, van Driel R. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J Cell Sci. 1992;101:773–784. doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- 51.Sun S, Thorner L, Lentz M, MacPherson P, Botchan M. Identification of a 68-kilodalton nuclear ATP-binding phosphoprotein encoded by bovine papillomavirus type 1. J Virol. 1990;64:5093–5105. doi: 10.1128/jvi.64.10.5093-5105.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szostecki D, Guldner H H, Netter H J, Will H. Isolation and characterization of a cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with primary biliary cirrhosis. J Immunol. 1990;145:4338–4347. [PubMed] [Google Scholar]
- 53.Taichman L B, LaPorta R F. The expression of papillomaviruses in human epithelial cells. In: Salzman N P, Howley P M, editors. The Papovaviridae. 2. The papillomaviruses. New York, N.Y: Plenum Press; 1987. pp. 109–139. [Google Scholar]
- 54.Ustav M, Stenlund A. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 1991;10:449–457. doi: 10.1002/j.1460-2075.1991.tb07967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RARα in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 56.Wilson V G, Ludes-Meyer J. A bovine papillomavirus E1-related protein binds specifically to bovine papillomavirus DNA. J Virol. 1991;65:5314–5322. doi: 10.1128/jvi.65.10.5314-5322.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y-L, Lewis A, Wade-Glass M, Schlegel R. Levels of bovine papillomavirus RNA and protein expression correlate with variations in the tumorigenic phenotype of hamster cells. J Virol. 1987;61:2924–2928. doi: 10.1128/jvi.61.9.2924-2928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou J, Stenzel D J, Sun X Y, Frazer I H. Synthesis and assembly of infectious bovine papillomavirus particles in vitro. J Gen Virol. 1993;74:763–768. doi: 10.1099/0022-1317-74-4-763. [DOI] [PubMed] [Google Scholar]
- 59.Zhou J, Sun X Y, Louis K, Frazer I H. Interaction of human papillomavirus (HPV) type 16 capsid proteins with HPV DNA requires an intact L2 N-terminal sequence. J Virol. 1994;68:619–625. doi: 10.1128/jvi.68.2.619-625.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]