Abstract
TOM22 is an essential mitochondrial outer membrane protein required for the import of precursor proteins into the organelles. The amino-terminal 84 amino acids of TOM22 extend into the cytosol and include 19 negatively and 6 positively charged residues. This region of the protein is thought to interact with positively charged presequences on mitochondrial preproteins, presumably via electrostatic interactions. We constructed a series of mutant derivatives of TOM22 in which 2 to 15 of the negatively charged residues in the cytosolic domain were changed to their corresponding amido forms. The mutant constructs were transformed into a sheltered Neurospora crassa heterokaryon bearing a tom22::hygromycin R disruption in one nucleus. All constructs restored viability to the disruption-carrying nucleus and gave rise to homokaryotic strains containing mutant tom22 alleles. Isolated mitochondria from three representative mutant strains, including the mutant carrying 15 neutralized residues (strain 861), imported precursor proteins at efficiencies comparable to those for wild-type organelles. Precursor binding studies with mitochondrial outer membrane vesicles from several of the mutant strains, including strain 861, revealed only slight differences from binding to wild-type vesicles. Deletion mutants lacking portions of the negatively charged region of TOM22 can also restore viability to the disruption-containing nucleus, but mutants lacking the entire region cannot. Taken together, these data suggest that an abundance of negative charges in the cytosolic domain of TOM22 is not essential for the binding or import of mitochondrial precursor proteins; however, other features in the domain are required.
Nucleus-encoded proteins destined to reside in mitochondria are synthesized as precursors on cytosolic ribosomes. Most precursors contain an N-terminal mitochondrial targeting signal, or presequence, which is ultimately removed from the protein. The process of translocation into mitochondria (reviewed in references 28, 42, 44, 53, 56) is initiated by binding of precursors to the cis site on the surface of the organelle by interaction with receptors of the outer membrane translocase (the TOM complex). The major components of the cis site are the cytosolic domains of the TOM20 and TOM22 proteins, which act in tandem as the receptor for the majority of precursors (32, 50). A small subset of precursors, most of which lack cleavable presequences, first interact with the TOM70-TOM37 receptor system prior to being passed to the TOM22-TOM20 complex (15, 18, 24, 26, 40).
Following cis site binding, precursors are routed through the TOM complex translocation channel. For precursors with cleavable targeting signals, the presequence interacts with the trans site on the intermembrane space side of the outer membrane as it emerges from the translocation channel (33, 49, 50). Precursors in this position are poised for interaction with the TIM23-containing (4, 54) inner membrane translocase (the TIM complex) and ΔΨ- and ATP-dependent movement through the inner membrane. One family of precursors utilizes a different TIM complex for insertion directly into the mitochondrial inner membrane (23, 25, 59, 60).
Several studies have shown that the presequence is both necessary and sufficient to direct proteins to the organelle (19–21). Presequences have no common primary structure but share certain properties (42, 45, 52, 62, 63): most are 20 to 80 amino acid residues long, they are enriched for both positively charged (mostly Arg) and hydroxylated (mostly Ser) residues, they are deficient in negatively charged residues, and they have the potential to form an amphipathic α-helix. These characteristics have suggested various mechanisms by which presequences could facilitate targeting and/or import into mitochondria. First, their distinct features could be recognized by surface receptors. Second, they might penetrate lipid bilayers due to their amphipathic nature. Third, their positively charged nature might allow them to be drawn into the matrix by electrophoretic forces imparted by the membrane potential (ΔΨ) across the mitochondrial inner membrane (30).
The N-terminal, cytosolic domains of both Neurospora crassa and Saccharomyces cerevisiae TOM22 are highly negatively charged. This led to the suggestion that targeting of mitochondrial precursors to the organelle might be achieved via electrostatic interaction of these charges with the positive residues in the presequence (24). Similar suggestions were made for the action of TOM20, since it contains clusters of acidic residues even though its overall charge is nearly neutral (6, 17, 35). Recently, several reports have provided support for the notion of electrostatic interaction between precursors and receptors. For example, cis site binding of presequence-containing precursors to either mitochondria (17) or outer membrane vesicles (OMV [32]) is salt sensitive and can be competed by presequence peptides. In vitro assays of the binding of various precursors to the cytosolic domains of either TOM20 or TOM22 have shown variable influences of salt and competition by presequence peptides depending on the combination of precursor and receptor examined (8, 26, 57). Two studies have addressed the role of the negative charges on receptor proteins in vivo. In the first, it was found that changing one neutral and one negatively charged residue to positively charged residues in yeast TOM22 affected growth on nonfermentable carbon sources (6). Mitochondria isolated from the mutant strain were strongly impaired in their ability to import preproteins. Removal of the acidic residues at the C terminus of yeast TOM20 resulted in a reduction of import in isolated mitochondria but did not affect growth (6). A second study utilized a mutant form of the rat TOM20 protein lacking the acidic C terminus. When this truncated form of the rat protein was expressed in a yeast tom20 deletion strain, it complemented both the growth and import defects of the cells (22).
We decided to systematically evaluate the role of the negative charges in the cytosolic domain of TOM22. To achieve this, we neutralized various numbers of the acidic residues and expressed the mutant constructs in N. crassa cells lacking a wild-type tom22 gene. In addition, we constructed deletion mutants lacking large portions of the cytosolic domain. Our analysis suggests that most of the negative charges in the domain can be removed without drastic effects on either the growth of cells or on the import of precursors into mitochondria. Furthermore, large portions of the negatively charged region of the TOM22 cytosolic domain can be deleted without severe effects on the function of the protein.
MATERIALS AND METHODS
Strains, media, and growth.
Growth and handling of N. crassa strains were as described elsewhere (11). Strain ND-113-1 is a sheltered heterokaryon composed of two nuclei with the genotypes a his-3 mtr tom22::hygR and a pan-2 bml (40). The mutation in the mtr gene provides resistance to both 4-methyltryptophan (MTR) and p-fluorophenylalanine (FPA). Strain 76-26 (a his-3 mtr) is the parent of the ND-113-1 nucleus that carries the tom22 disruption in the sheltered heterokaryon. Two different classes of mutants were used in this study. The first class contains mutations which alter negatively charged residues in the cytosolic domain of the TOM22 protein. These were derived by transformation of ND-113-1 with plasmids carrying bleomycin resistance (2), ampicillin resistance, and tom22 genes that had been altered by site-directed mutagenesis of a genomic tom22 clone. These strains (designated 96, 40, 104, 98, 006, 068, 008, 861, 95, 100) and their isolation are described in Results. A single strain containing a frameshift mutation at codon 29 (FS29) was also derived by this method. The second class contained deletions in the tom22 gene. These strains were isolated by cotransforming ND-113-1 with a bleomycin resistance plasmid and different plasmids containing deleted forms of the genomic tom22 gene created by site-directed mutagenesis. These strains (Δ2-28, Δ32-44, Δ2-28+Δ32-44, and Δ95-104) are also described in Results.
As a simple method to quantify growth, we measured the rate of mycelial elongation in race tubes (11). These were constructed by laying sterile disposable 25-ml pipettes flat on a bench top and filling them approximately one-third full of molten solid growth medium (64). Strains were inoculated at one end of the tube, and the extent of mycelial growth was recorded each day.
Isolation of mitochondria.
Mitochondria were isolated by the procedure of Pfanner and Neupert (46), except that the phenylmethylsulfonyl fluoride concentration during the grinding step was increased to 1 mM. Further modifications were introduced because the outer membranes of mitochondria from strain 861 were found to be unusually fragile. Therefore, isolation of mitochondria was carried out with isolation buffer containing 500 or 750 mM sucrose, and the duration of manual grinding with quartz sand was reduced to about 30 s.
Protein import into isolated mitochondria.
The procedure described by Harkness et al. (16) was utilized to assay import of precursors into mitochondria. Import conditions were modified so that the sucrose concentration during all phases of the reaction was adjusted to equal the concentration used for isolation of mitochondria. Trypsin pretreatment was done according to the method described by Mayer et al. (31).
Whole-cell PCR.
A small sample of conidia (approximately one inoculating loop full) from a fresh N. crassa culture was suspended in 100 μl of 1 M sorbitol–20 mM EDTA (pH 8.0) containing lysing enzymes (Sigma, St. Louis, Mo.) at 3 mg/ml, and the suspension was incubated for 10 min at 37°C. The sample was centrifuged at 15,000 rpm for 1 min in a microcentrifuge, and the supernatant was discarded. The pellet was washed by resuspending in 500 μl of the sorbitol-EDTA solution without lysing enzymes, and the centrifugation was repeated. The pellet was resuspended in 100 μl of sterile distilled water and boiled for 10 min. After boiling, the sample was thoroughly vortexed and 300 μl of 6 M NaI and 5 μl of glassmilk (61) were added. The sample was then gently mixed for 30 min. The glassmilk, with bound DNA, was pelleted in a microcentrifuge and washed three times with 10 mM Tris-HCl (pH 7.2)–100 mM NaCl–1 mM EDTA–50% ethanol. The DNA was removed from the glassmilk with 20 μl of sterile distilled water, and 2-μl samples were used in PCRs.
Other techniques.
The standard techniques of agarose gel electrophoresis, transformation of Escherichia coli, and isolation of bacterial plasmid DNA were according to the methods described by Ausubel et al. (3). DNA sequencing with the Thermo Sequence radiolabeled terminator cycle sequencing kit (Amersham, Cleveland, Ohio) and site-directed mutagenesis with the Muta-Gene system (Bio-Rad, Hercules, Calif.) were done according to the suppliers’ instructions. The following procedures were performed by previously published procedures: separation of mitochondrial proteins by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (27), Western blotting (14), determination of mitochondrial protein concentrations (7), transformation of N. crassa spheroplasts (58) with the modifications described by Akins and Lambowitz (1), isolation of mitochondrial OMV (31), and assays of preprotein binding to OMV (33).
RESULTS
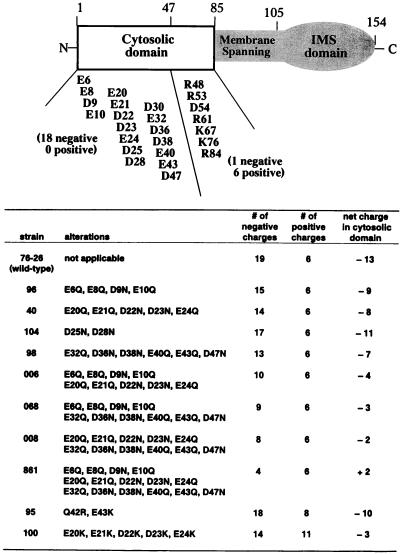
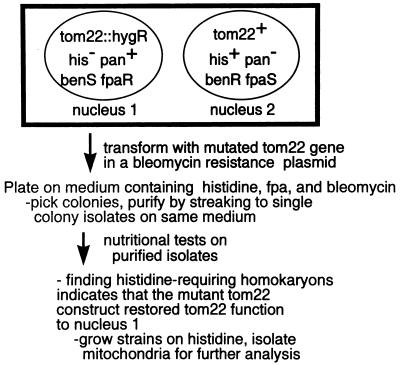
To investigate the role of the negative charges in TOM22, we changed various numbers of the 19 acidic residues in the cytosolic domain to their corresponding amido forms using site-directed mutagenesis (Fig. 1). The mutant versions of the gene were inserted into a vector carrying a bleomycin resistance gene. These plasmids were introduced into spheroplasts of a sheltered heterokaryon (strain ND-113-1) carrying a tom22::hygromycin R disruption in one nucleus that prevents synthesis of TOM22 and renders that nucleus inviable as a homokaryon (Fig. 2) (40). Transformed spheroplasts were plated on selective medium containing bleomycin, p-fluorophenylalanine, and histidine. In principle, this medium could allow the formation of two possible types of colonies following ectopic integration of the tom22 constructs. If the disruption-carrying nucleus is transformed with both the bleomycin resistance gene and a functional tom22, then histidine-requiring homokaryons should be found. If the mutations introduced to the tom22 gene render it nonfunctional or if integration occurs in the tom22+ nucleus of the heterokaryon, then only bleomycin-resistant heterokaryons should grow. However, since the presence of p-fluorophenylalanine and histidine in the medium should strongly favor the growth of colonies with a high ratio of the nucleus bearing p-fluorophenylalanine resistance (nucleus 1; Fig. 2), it was unlikely that heterokaryons would appear on the selective medium.
FIG. 1.
Structure of the TOM22 protein and mutants derived by site-directed mutagenesis of acidic residues. (Top) Putative structure of N. crassa TOM22 showing the cytosolic domain, the membrane-spanning region, and the intermembrane space (IMS) domain of the protein. The positions of all acidic and basic residues in the cytosolic domain are indicated. (Bottom) Charge neutralization mutant strains used in this study. These were derived by site-directed mutagenesis of various numbers of acidic residues in the cytosolic domain. The numbers of negative and positive charges present in each mutant as well as the net charge in the cytosolic domain are indicated.
FIG. 2.
Scheme for isolation of tom22 mutant strains. Strain ND-113-1 (40) (see Materials and Methods) is the sheltered heterokaryon represented by the box at the top. Nucleus 1 contains a tom22 gene disrupted by a gene encoding hyrgromycin resistance (hygR). In addition, nucleus 1 carries auxotrophy for histidine and a resistance gene for p-fluorophenylalanine (fpaR). It is possible to select for rescue of the tom22 deficiency in nucleus 1 when that nucleus is transformed with both a bleomycin resistance gene and a mutant tom22 gene when that mutant gene gives rise to a functional protein. The presence of histidine in the selective medium frees nucleus 1 from its dependence on nucleus 2 for growth, while the presence of FPA forces transformants to be greatly enriched for nucleus 1. Purification and nutritional testing of the colonies derived from transformation with any of the mutant forms of tom22 described in the legend to Fig. 1 revealed that all bleomycin-resistant transformants were incapable of growth on medium lacking histidine. Thus, all of the transformants were homokaryotic for nucleus 1 and all of the mutant forms of tom22 were able to restore TOM22 function.
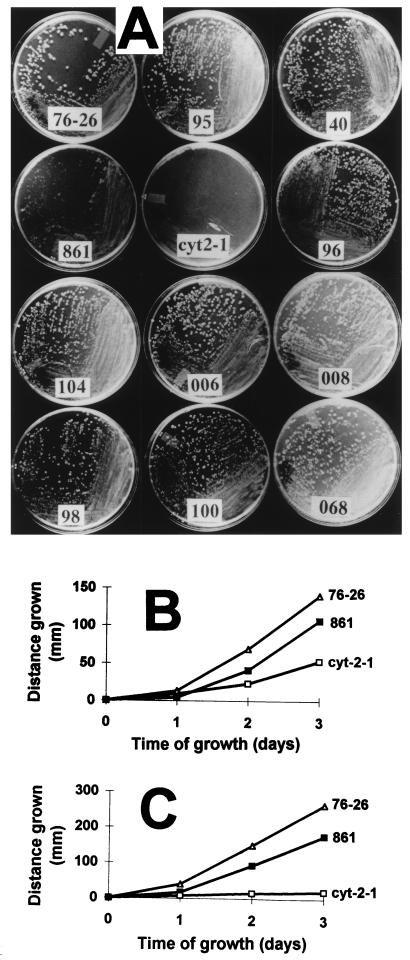
Transformants were isolated, allowed to conidiate, purified by streaking to single-colony isolates on the medium used for selection, and tested for biochemical requirements (Fig. 2). Transformants obtained with any of the mutant constructs shown in Fig. 1 were found to be exclusively histidine-requiring homokaryons and, therefore, must contain only the disruption-bearing nucleus transformed with a tom22 allele supplying a functional version of the TOM22 protein. To exclude the possibility of contamination or alterations in the mutated tom22 sequences, PCR-amplified DNA from each homokaryotic strain was sequenced. All strains were found to carry the predicted mutations. Taken together, the above results indicate that all of the mutant constructs were able to restore TOM22 function. Furthermore, the inclusion of p-fluorophenylalanine and histidine in the medium provided efficient selection against the growth of sheltered heterokaryons transformed to bleomycin resistance. It is noteworthy that transformation with the bleomycin resistance plasmid alone gave no transformants on this medium, indicating that spontaneous suppressors of the TOM22 deficiency in the disruption nucleus did not arise. Strains containing any of the altered versions of TOM22 shown in Fig. 1 formed colonies at 30°C at a rate comparable to that for the wild type, with the exception of mutant 861, in which 15 of the negative charges were neutralized (Fig. 3A). To provide a comparison for the severity of the growth defect of strain 861, we included the previously described cytochrome c heme lyase-deficient cyt-2-1 mutant (13, 39) in our analysis. The cyt-2-1 mutant provides an example of one of the most slowly growing N. crassa mutants known that is affected in mitochondrial function (38). Strain 861 grew at a rate intermediate between those of the wild type and the cyt-2-1 mutant, so that the effects of the alterations in strain 861 on growth may be judged as relatively mild. A more quantitative assessment of the growth defect in strain 861 was obtained by measuring the rate of mycelial elongation at different temperatures in race tubes (Fig. 3B and C). Growth of strain 861 was slightly slower than that of wild-type strain 76-26 at 22 and 37°C. The behaviors of mutants 95, 100, 006, 008, and 068 in race tubes at either 22 or 37°C were indistinguishable from that of the wild type (not shown).
FIG. 3.
Growth of wild-type strain 76-26 and mutant strains. (A) Colonies formed on sorbose-containing medium after 48 h of growth at 30°C are shown. The previously described CCHL-deficient strain (the cyt-2-1 strain) is shown as an example of a slowly growing mutant. (B) Mycelial elongation of strains 76-26 and 861 and the cyt-2-1 strain as measured in race tubes (see Materials and Methods) at 22°C. (C) Same as panel B, but with growth at 37°C.
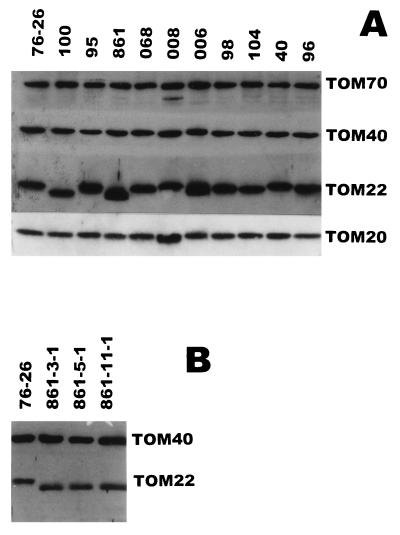
The levels of TOM22 and other TOM complex proteins in the mitochondria of the homokaryotic transformants did not differ dramatically from wild-type levels (Fig. 4A). Thus, the information required for the uptake of TOM22 into mitochondria is not contained in the negatively charged amino acid residues. Furthermore, we assume that the altered TOM22 proteins correctly assemble with the remaining TOM complex components, since none of the strains had a severe growth phenotype.
FIG. 4.
Immunostaining of mitochondrial proteins in wild-type (76-26) and tom22 mutant strains. Equal amounts of mitochondrial protein were loaded in each lane. Proteins were separated by SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and decorated with antiserum to the indicated proteins. (A) Levels of TOM complex components in the wild type and all charge neutralization mutant strains generated in this study. Alterations in many of the TOM22 mutants result in altered electrophoretic mobilities relative to that of wild-type TOM22. (B) Levels of TOM40 and TOM22 in three different isolates obtained following transformation of strain ND-113-1 with the 861 mutant form of tom22.
Since integration of transforming DNA in N. crassa usually occurs at ectopic sites, we also wished to ensure that there were no drastic differences in the level of TOM22 protein that arose from locus-specific effects on the expression of integrated sequences. Therefore, the levels of TOM22 were examined in three separate isolates obtained by transformation with the 861 tom22 construct, in which 15 negative charges had been neutralized (Fig. 4B [861 series]). No significant variation in TOM22 levels was observed between the strains examined.
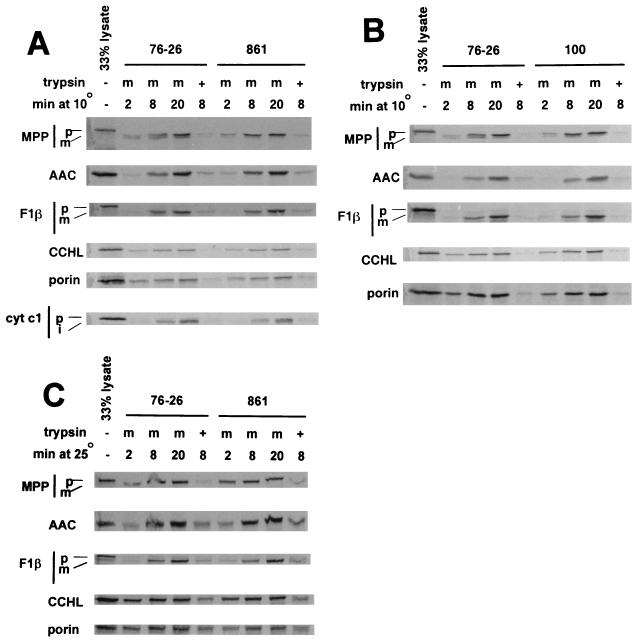
In vitro import of mitochondrial precursors was measured by using mitochondria isolated from wild-type strain 76-26 and two strains chosen for the severity and differing nature of the alterations in TOM22 (Fig. 1), i.e., strains 861 and 100. We used precursors representing mitochondrial proteins found in the matrix (the β-subunit of the F1 ATPase [F1β] and the α-subunit of mitochondrial processing peptidase [MPP]), the inner membrane (the ATP-ADP carrier [AAC]), the intermembrane space (cytochrome c heme lyase [CCHL] and cytochrome c1), and the outer membrane (porin). The precursors for F1β, MPP, and cytochrome c1 contain N-terminal presequences, while the other precursors contain internal targeting signals. All of the precursors tested have been previously shown to depend strictly on TOM22 function for their import (24, 40). Import experiments were conducted at 10°C, at which the uptake of all precursors was in the linear range. No significant differences in the import of any of the precursors tested between the wild type and mutant strains 861 and 100 were observed (Fig. 5A and B). To ensure that import rates were not different at a more physiological temperature, we also measured import at 25°C for strain 861. Again, no dramatic differences from the wild type were observed (Fig. 5C).
FIG. 5.
Import of [35S]methionine-labeled precursor proteins into isolated mitochondria. Mitochondria isolated from strain 861 (A), strain 100 (B), and wild-type control strain 76-26 were either pretreated with trypsin (+) or mock treated (m). Import was conducted at 10°C for the time periods indicated. Following import reactions, mitochondria were reisolated and subjected to SDS-PAGE. The gels were blotted to a polyvinylidene difluoride membrane and subjected to autoradiography. The precursors used in the import reactions are indicated on the left. The leftmost lane for each precursor contained 33% of the input lysate used in each import reaction. cyt c1, cytochrome c1; p, m, and i, positions of the precursor, mature, and intermediate forms of the preproteins, respectively. (C) Same as for panel A, except with import performed at 25°C.
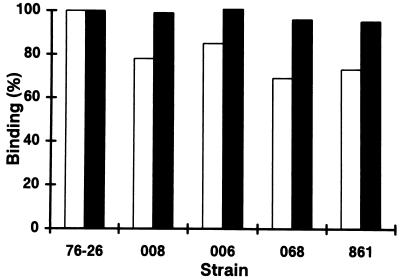
To examine directly the ability of a presequence-containing precursor to interact with receptors on the surface of mitochondria, OMV were prepared from mitochondria of various mutant strains and compared to OMV from wild-type strain 76-26 for their abilities to bind a fusion protein containing the presequence of subunit 9 of the F0-ATPase fused to mouse dihydrofolate reductase (pSu9-DHFR [47]). pSu9-DHFR was incubated with OMV under conditions which result in exclusive binding to either the cis or the trans site of the outer membrane (49). trans site binding levels were virtually identical in mutant and wild-type OMV, whereas minor reductions in cis site binding were observed in the mutant OMV (Fig. 6). Control experiments with all strains showed that treatment with high salt buffer reduced the amount of material bound at the cis site by 90%. Removal of the OMV surface receptors by pretreatment with trypsin reduced cis and trans site binding to about 5 and 10%, respectively, of the levels shown in Fig. 6 (see reference 49).
FIG. 6.
Charges in TOM22 are not required for preprotein binding at the cis site or for presequence translocation to the trans site of the outer membrane. OMV (5 μg of protein per sample) were isolated from the indicated tom22 mutant strains. Binding of pSu9-DHFR was performed in 100 μl of binding buffer (0.25-mg/ml bovine serum albumin, 2.5 mM MgCl2, 20 mM KCl, 10 mM morpholinepropanesulfonic acid [MOPS]-KOH [pH 7.2]) in the absence (trans site binding) or presence (cis site binding) of 1 mM NADPH and 1 μM methotrexate for 15 min at 25 or 0°C, respectively. Following incubation, samples were diluted with 700 μl of buffer (10 mM MOPS-KOH, 1 mM EDTA [pH 7.2]) containing 120 mM KCl (trans site binding) or 20 mM KCl (cis site binding). OMV were reisolated by centrifugation for 20 min at 125,000 × g, and the pellets were subjected to SDS-PAGE. Bound pSu9-DHFR was quantitated by PhosphorImager analysis. The amount of pSu9-DHFR bound to the wild-type strain (76-26) was set to 100%. Data shown are the averages of seven trials. Standard deviations of the different data sets ranged from 15 to 25%. White bars, cis site binding; black bars, trans site binding.
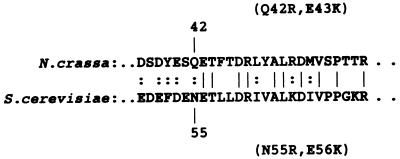
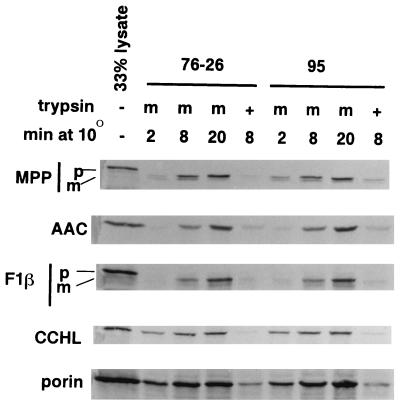
In a previous report on the role of the negative charges in the cytosolic domain of TOM22, one neutral and one negatively charged residue were simultaneously changed to positively charged residues in yeast TOM22. It was observed that these changes resulted in a decreased growth rate on nonfermentable carbon sources and a reduction in the import of various mitochondrial precursors into isolated mitochondria (6). Since we were unable to detect significant effects on TOM22 function in our charge mutants, we wished to determine if the particular changes made in the previous study may have affected a crucial region of the protein. Changes at the corresponding positions were introduced into the N. crassa version of the protein, which shows reasonable similarity to yeast TOM22 in this region (Fig. 7). Transformation of the sheltered heterokaryon ND-113-1 with this version of N. crassa tom22 gave rise to homokaryotic strain 95. The strain was found to grow at a rate indistinguishable from that for wild-type strain 76-26 (Fig. 3 and race tube data at 22 and 37°C [not shown]). Measurement of preprotein import into mitochondria isolated from strain 95 also revealed no differences from the wild type (Fig. 8).
FIG. 7.
Alignment of N. crassa and S. cerevisiae TOM22 proteins. Only the region altered in mutant 95 and the corresponding yeast mutant (6) is shown. The mutations generated in each of the two species are indicated in parentheses. |, identical amino acids; :, chemically similar amino acids.
FIG. 8.
Import of preproteins into mitochondria isolated from strain 95 and wild-type strain 76-26. Details are as described in the legend to Fig. 5.
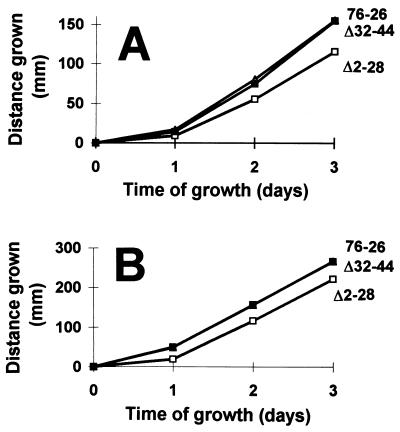
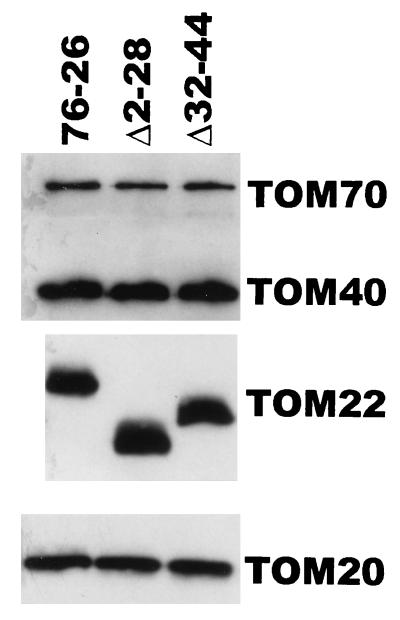
The surprising nature of our observations with the charge mutants led us to examine the ability of more grossly altered TOM22 proteins to restore the essential function of the protein. We constructed a series of tom22 deletion mutants and a frameshift mutation at codon 29 (FS29). These mutant forms were used in transformation experiments to determine if they were capable of giving rise to viable homokaryotic strains carrying only the disruption nucleus from sheltered heterokaryon ND-113-1. As shown in Table 1, deletion of different portions (Δ2-28 or Δ32-44) of the negatively charged region of the cytosolic domain of TOM22 resulted in mutant proteins that were able to restore viability to the disruption nucleus, giving rise to histidine-requiring homokaryons. Strains containing the deletion Δ2-28 grew slightly more slowly than wild-type controls, but Δ32-44 strains could not be distinguished from the wild type (Fig. 9). The content of TOM complex components in these strains was indistinguishable from that of the wild type (Fig. 10). In contrast, mutant tom22 alleles with a deletion of almost the entire negatively charged domain (Δ2-28 plus Δ32-44) or a deletion of residues 95 to 104 in the membrane-spanning domain or containing the frameshift F529 destroyed the ability of the transforming DNA to rescue the disruption nucleus. In summary, no particular region of the negatively charged cytosolic domain of TOM22 is essential for function; however, deletion of the entire domain results in a protein unable to support cell viability. Since the negative charges in this domain can be neutralized without severe effects, these data suggest that other features of the region are required for TOM22 function.
TABLE 1.
Deletions in tom22 and their effects
| Deleted residues | Region affected by deletiona | Restores viability to tom22 disruption nucleus | Growth of deletion strainb |
|---|---|---|---|
| Δ2-28 | Part of the negatively charged region of the cytosolic domain | Yes | Slow |
| Δ32-44 | Part of the negatively charged region of the cytosolic domain | Yes | Wild type |
| Δ2-28 + Δ32-44 | Almost entire negatively charged region of the cytosolic domain | No | Inviable |
| Δ95-104 | Membrane-spanning domain | No | Inviable |
FIG. 9.
Growth of tom22 deletion strains. Mycelial elongation of the wild-type strain (76-26) and the Δ2-28 and Δ32-44 mutant strains was measured in race tubes at 22°C (A) and at 37°C (B). The curves for 76-26 and Δ32-44 in panel B are entirely overlapping.
FIG. 10.
Immunostaining of TOM complex proteins in mitochondria isolated from tom22 deletion strains. Details are as described in the legend to Fig. 4.
DISCUSSION
The initial determination of the sequence of TOM22 led to the suggestion that its receptor function might be achieved via interaction of its negatively charged cytosolic domain with positively charged presequences (24). Based on an analogy with the methionine bristles of the signal recognition particle, which interact with hydrophobic regions on the signal sequence of proteins targeted for the endoplasmic reticulum (5), it was subsequently proposed that “acid bristles” on mitochondrial receptors interact with positively charged presequences on mitochondrial precursors (6). Negatively charged regions in proteins involved in almost the entire import pathway, have now been identified, including Mft52, a protein found in the yeast cytosol required for efficient delivery of precursors to mitochondria (9); the cytosolic domains of the TOM22 and TOM20 receptors (6); TOM5 at the entrance of the putative outer membrane channel (12); the intermembrane space domain of TOM22 (6); and, in TIM23, the gate of the inner membrane pore (4). These observations have suggested an acid chain that electrostatically binds the positively charged presequence along each step of the import pathway (55).
However, certain aspects of the acid chain model and the general interaction of presequences and receptors remain controversial. For example, Mayer et al. (32) showed that TOM20 and TOM22 must act in tandem to form an effective binding site for precursors on OMV, even though studies with the purified cytosolic domains of these receptors show binding of precursors to the individual proteins (8, 26, 57). Furthermore, one study demonstrated salt-enhanced binding of presequence-containing precursors to the purified cytosolic domain of TOM20 (8), while analogous studies showed salt-sensitive binding (26, 57). There is also conflicting evidence regarding the role of the negatively charged intermembrane space domain of TOM22 in the trans binding site (6, 10, 34, 37). Thus, it is apparent that the mechanisms of precursor-receptor interaction and the role of negative charges on proteins of the import apparatus are not yet entirely understood.
The data presented in this report address the role of the negative charges in the cytosolic domain of TOM22 by using both in vivo and in vitro studies. The protein is known to be essential for viability of both yeast and N. crassa (18, 29, 36, 40) and is the more negatively charged of the two proteins (TOM20 and TOM22) that form the major receptor for mitochondrial precursor proteins. It seems reasonable to assume that TOM22 would play a major role in any scheme utilizing acid binding sites during the movement of precursors along the import pathway. However, the majority of charges can be removed from the protein without dramatic effects. Even in strain 861, in which TOM22 lacks 15 of the 19 negative charges in the cytosolic domain, no major alterations in the ability to import or bind mitochondrial precursors were observed. We assume that the reduced growth rate of this mutant is the result of a decreased ability to import one or more specific precursors in vivo. Given the number of changes in the protein in strain 861, it cannot simply be inferred that the growth defect is due to the lack of negative charges rather than an overall change in the structure of the protein. In fact, we have preliminary evidence supporting the idea that some of the mutant TOM22 proteins, including the 861 version, may have an aberrant structure, since they exhibit altered sensitivity to proteases (48). We also show that the negative charges are not required for the import and assembly of TOM22 itself into the outer membrane, in agreement with our recent in vitro studies of TOM22 assembly (51). The negative charges also seem unnecessary for efficient assembly of other members of the TOM complex.
It was previously determined that the binding of pSu9-DHFR to the cis site of the outer membrane was salt sensitive, dependent on TOM22, and competed by a synthetic presequence peptide (32). Since we observe little reduction of this binding in OMV isolated from several of our mutant strains, including strain 861, it appears that the binding does not require an overall negative charge on the cytosolic domain of TOM22. An earlier investigation of yeast TOM22 in vivo demonstrated rather severe effects from two relatively minor changes in the protein (6). Although the mutations in one of our N. crassa mutants (strain 95) were analagous to those in the yeast study, we did not observe any effects in N. crassa. There is no obvious explanation for this discrepancy, but perhaps the changes in yeast TOM22 provoke a severe change in protein structure not observed in the N. crassa homolog.
Our findings with the deletion mutants suggest that no specific portion of the negatively charged region of the cytosolic domain of TOM22 is required for its function. A minor effect on growth rate is seen in the Δ2-28 strain, while Δ32-44 is indistinguishable from the wild type. On the other hand, deletion of most of the negatively charged region (Δ2-28 plus Δ32-44) prevents the mutant protein from rescuing the disruption nucleus. This deletion would result in a TOM22 protein with an overall charge of +3 in the cytosolic domain, which is quite similar to charge mutant 861 (with an overall charge of +2). The observation that the Δ2-28-plus-Δ32-44 deletion fails to rescue the disruption nucleus while the 861 form of the protein does rescue the nucleus strongly suggests that features in this region (other than the negative charges) must be important for TOM22 function in mitochondrial protein import.
We have recently shown that the positively charged region of TOM22 preceding the membrane-spanning domain (Fig. 1) is essential for the targeting-assembly of TOM22 itself to the mitochondrial outer membrane (51). It is well documented that the intermembrane space domain of TOM22 is not required for cell viability (6, 10, 34, 37). Our data show that the transmembrane domain of TOM22 performs an essential function, since a deletion in the region results in an inactive form of the protein. This might reflect an inability of the mutant protein to assemble into the membrane rather than demonstrate a role for the membrane-spanning domain in postassembly function. Still, it seems likely that the membrane domain plays a role in the formation of the outer membrane pore and/or in providing a binding site for preproteins. This idea is supported by the observation that import of almost all precursors is virtually eliminated in mitochondria from which TOM22 has been depleted (40, 41). On the other hand, bypass import of precursors is still observed when only the cytosolic domains of the surface receptors are removed (43).
Not all steps of the import pathway are mediated by electrostatic interaction of precursors with negatively charged residues, since binding of precursors at the trans site is known to be salt resistant (34, 49, 50). As shown in this paper, the negative charges in the cytosolic domain of TOM22 appear not to play a major role in precursor binding at the cis site, although TOM22 function is dependent on the presence of the region containing those negative charges. Thus, forces other than ionic interactions may be important for recognition of preproteins and their translocation across the mitochondrial outer membrane.
ACKNOWLEDGMENTS
This work was supported by grants from the Medical Research Council of Canada (M.R.C.) and from the Sonderforschungsbereich 184 of the Deutsche Forschungsgemeinschaft. D.R. was supported by a fellowship from the European Molecular Biology Organization.
We are grateful to Bonnie Crowther, Albert Ussher, and Petra Heckmeyer for excellent technical assistance.
REFERENCES
- 1.Akins R A, Lambowitz A M. General method for cloning Neurospora crassa nuclear genes by complementation of mutants. Mol Cell Biol. 1985;5:2272–2278. doi: 10.1128/mcb.5.9.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Austin B, Hall R M, Tyler B M. Optimized vectors for transformation of Neurospora crassa and Aspergillus nidulans to bleomycin and phleomycin resistance. Gene. 1990;93:157–162. doi: 10.1016/0378-1119(90)90152-h. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel R A, Brent R, Kingston R E, Moore D D, Seidman J G. Current protocols in molecular biology. New York, N.Y: Greene and Wiley Interscience; 1992. [Google Scholar]
- 4.Bauer M F, Sirrenberg C, Neupert W, Brunner M. Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell. 1996;87:33–41. doi: 10.1016/s0092-8674(00)81320-3. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein H D, Poritz M A, Strub K, Hoben P J, Brenner S, Walter P. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature. 1989;340:482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- 6.Bolliger L, Junne T, Schatz G, Lithgow T. Acidic receptor domains on both sides of the outer membrane mediate translocation of precursor proteins into yeast mitochondria. EMBO J. 1995;14:6318–6326. doi: 10.1002/j.1460-2075.1995.tb00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Brix J, Dietmeier K, Pfanner N. Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J Biol Chem. 1997;272:20730–20735. doi: 10.1074/jbc.272.33.20730. [DOI] [PubMed] [Google Scholar]
- 9.Cartwright P, Beilharz T, Hansen P, Garrett J, Lithgow T. Mft52, and acid-bristle protein in the cytosol that delivers precursor proteins to yeast mitochondria. J Biol Chem. 1997;272:5320–5325. doi: 10.1074/jbc.272.8.5320. [DOI] [PubMed] [Google Scholar]
- 10.Court D A, Nargang F E, Steiner H, Hodges R S, Neupert W, Lill R. Role of the intermembrane space domain of the preprotein receptor Tom22 in protein import into mitochondria. Mol Cell Biol. 1996;16:4034–4042. doi: 10.1128/mcb.16.8.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis R H, De Serres F J. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 1970;17:79–143. [Google Scholar]
- 12.Dietmeier K, Hönlinger A, Bömer U, Dekker P J T, Eckerskorn C, Lottspeich F, Kübrich M, Pfanner N. Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nature. 1997;388:195–200. doi: 10.1038/40663. [DOI] [PubMed] [Google Scholar]
- 13.Drygas M E, Lambowitz A M, Nargang F E. Cloning and analysis of the Neurospora crassa gene for cytochrome c heme lyase. J Biol Chem. 1989;264:17897–17907. [PubMed] [Google Scholar]
- 14.Good A G, Crosby W L. Anaerobic induction of alanine aminotransferase in barley root tissue. Plant Physiol. 1989;90:1305–1309. doi: 10.1104/pp.90.4.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gratzer S, Lithgow T, Bauer R E, Lamping E, Paltauf F, Kohlwein S D, Haucke V, Junne T, Schatz G, Horst M. Mas37p, a novel receptor subunit for protein import into mitochondria. J Cell Biol. 1995;129:25–34. doi: 10.1083/jcb.129.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harkness T A A, Nargang F E, Van der Klei I, Neupert W, Lill R. A crucial role of the mitochondrial protein import receptor MOM19 for the biogenesis of mitochondria. J Cell Biol. 1994;124:637–648. doi: 10.1083/jcb.124.5.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haucke V, Lithgow T, Rospert S, Hahne K, Schatz G. The yeast mitochondrial protein import receptor Mas20p binds precursor proteins through electrostatic interaction with the positively charged presequence. J Biol Chem. 1995;270:5565–5570. doi: 10.1074/jbc.270.10.5565. [DOI] [PubMed] [Google Scholar]
- 18.Hönlinger A, Kübrich M, Moczko M, Gärtner F, Mallet L, Bussereau F, Eckerskorn C, Lottspeich F, Dietmeier K, Jacquet M, Pfanner N. The mitochondrial receptor complex: MOM22 is essential for cell viability and directly interacts with preproteins. Mol Cell Biol. 1995;15:3382–3389. doi: 10.1128/mcb.15.6.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horwich A L, Kalousek F, Mellman I, Rosenberg L E. A leader peptide is sufficient to direct mitochondrial import of a chimeric protein. EMBO J. 1985;4:1129–1135. doi: 10.1002/j.1460-2075.1985.tb03750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurt E C, Pesold-Hurt B, Schatz G. The cleavable prepiece of an imported mitochondrial protein is sufficient to direct cytosolic dihydrofolate reductase into the mitochondrial matrix. FEBS Lett. 1984;178:306–310. doi: 10.1016/0014-5793(84)80622-5. [DOI] [PubMed] [Google Scholar]
- 21.Hurt E C, Pesold-Hurt B, Suda K, Oppliger W, Schatz G. The first twelve amino acids (less than half of the presequence) can direct mouse cytosolic dihydrofolate reductase into the yeast mitochondrial matrix. EMBO J. 1985;4:2061–2068. doi: 10.1002/j.1460-2075.1985.tb03892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwahashi J, Yamazaki S, Komiya T, Nomura N, Nishikawa S, Endo T, Mihara K. Analysis of the functional domain of the rat liver mitochondrial import receptor Tom20. J Biol Chem. 1997;272:18467–18472. doi: 10.1074/jbc.272.29.18467. [DOI] [PubMed] [Google Scholar]
- 23.Kerscher O, Holder J, Srinivasan M, Leung R S, Jensen R E. The Tim54p-Tim22p complex mediates insertion of proteins into the mitochondrial inner membrane. J Cell Biol. 1997;139:1663–1675. doi: 10.1083/jcb.139.7.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiebler M, Keil P, Schneider H, van der Klei I, Pfanner N, Neupert W. The mitochondrial receptor complex: a central role of MOM22 in mediating transfer of preproteins from receptors to the general insertion pore. Cell. 1993;74:483–492. doi: 10.1016/0092-8674(93)80050-o. [DOI] [PubMed] [Google Scholar]
- 25.Koehler C M, Jarosch E, Tokatlidis K, Schmid D, Schweyen R J, Schatz G. Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science. 1998;279:369–373. doi: 10.1126/science.279.5349.369. [DOI] [PubMed] [Google Scholar]
- 26.Komiya T, Rospert S, Schatz G, Mihara K. Binding of mitochondrial precursor proteins to the cytoplasmic domains of the import receptors Tom70 and Tom20 is determined by cytoplasmic chaperones. EMBO J. 1997;16:4267–4275. doi: 10.1093/emboj/16.14.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Lill R, Neupert W. Mechanisms of protein import across the mitochondrial outer membrane. Trends Cell Biol. 1996;6:56–61. doi: 10.1016/0962-8924(96)81015-4. [DOI] [PubMed] [Google Scholar]
- 29.Lithgow T, Junne T, Suda K, Gratzer S, Schatz G. The mitochondrial outer membrane protein Mas22p is essential for protein import and viability of yeast. Proc Natl Acad Sci USA. 1994;91:11973–11977. doi: 10.1073/pnas.91.25.11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin J, Mahlke K, Pfanner N. Role of an energized inner membrane in mitochondrial protein import. J Biol Chem. 1991;266:18051–18057. [PubMed] [Google Scholar]
- 31.Mayer A, Lill R, Neupert W. Translocation and insertion of precursor proteins into isolated outer membranes of mitochondria. J Cell Biol. 1993;121:1233–1243. doi: 10.1083/jcb.121.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayer A, Nargang F E, Neupert W, Lill R. MOM22 is a receptor for mitochondrial targeting sequences and cooperates with MOM19. EMBO J. 1995;14:4204–4211. doi: 10.1002/j.1460-2075.1995.tb00094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayer A, Neupert W, Lill R. Mitochondrial protein import: Reversible binding of the presequence at the trans side of the outer membrane drives partial translocation and unfolding. Cell. 1995;80:127–137. doi: 10.1016/0092-8674(95)90457-3. [DOI] [PubMed] [Google Scholar]
- 34.Moczko M, Bömer U, Kübrich M, Zufall N, Hönlinger A, Pfanner N. The intermembrane space domain of mitochondrial Tom22 functions as a trans binding site for preproteins with N-terminal targeting sequences. Mol Cell Biol. 1997;17:6574–6584. doi: 10.1128/mcb.17.11.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moczko M, Ehmann B, Gärtner F, Hönlinger A, Schäfer E, Pfanner N. Deletion of the receptor MOM19 strongly impairs import of cleavable preproteins into Saccharomyces cerevisiae mitochondria. J Biol Chem. 1994;269:9045–9051. [PubMed] [Google Scholar]
- 36.Nakai M, Endo T. Identification of yeast MAS17 encoding the functional counterpart of the mitochondrial receptor complex protein MOM22 of Neurospora crassa. FEBS Lett. 1995;357:202–206. doi: 10.1016/0014-5793(94)01362-5. [DOI] [PubMed] [Google Scholar]
- 37.Nakai M, Kinoshita K, Endo T. Mitochondrial receptor complex protein. The intermembrane space domain of yeast Mas17 is not essential for its targeting or function. J Biol Chem. 1995;270:30571–30575. doi: 10.1074/jbc.270.51.30571. [DOI] [PubMed] [Google Scholar]
- 38.Nargang, F. E. Unpublished data.
- 39.Nargang F E, Drygas M E, Kwong P L, Nicholson D W, Neupert W. A mutant of Neurospora crassa deficient in cytochrome c heme lyase activity cannot import cytochrome c into mitochondria. J Biol Chem. 1988;263:9388–9394. [PubMed] [Google Scholar]
- 40.Nargang F E, Künkele K-P, Mayer A, Ritzel R G, Neupert W, Lill R. “Sheltered disruption” of Neurospora crassa MOM22, an essential component of the mitochondrial protein import complex. EMBO J. 1995;14:1099–1108. doi: 10.1002/j.1460-2075.1995.tb07093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nargang F E, Lill R. Import of proteins into mitochondria. In: Brambl R, Marzluf G, editors. The Mycota III. Berlin, Germany: Springer-Verlag; 1996. pp. 85–107. [Google Scholar]
- 42.Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 43.Pfaller R, Pfanner N, Neupert W. Mitochondrial protein import: bypass of proteinaceous surface receptors can occur with low specificity and efficiency. J Biol Chem. 1989;264:34–39. [PubMed] [Google Scholar]
- 44.Pfanner N, Craig E A, Hönlinger A. Mitochondrial preprotein translocase. Annu Rev Cell Dev Biol. 1997;13:25–51. doi: 10.1146/annurev.cellbio.13.1.25. [DOI] [PubMed] [Google Scholar]
- 45.Pfanner N, Neupert W. The mitochondrial protein import apparatus. Annu Rev Biochem. 1990;59:331–353. doi: 10.1146/annurev.bi.59.070190.001555. [DOI] [PubMed] [Google Scholar]
- 46.Pfanner N, Neupert W. Transport of proteins into mitochondria: a potassium diffusion potential is able to drive the import of ADP/ATP carrier. EMBO J. 1985;4:2819–2825. doi: 10.1002/j.1460-2075.1985.tb04009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pfanner N, Tropschug M, Neupert W. Mitochondrial protein import: nucleoside triphosphates are involved in conferring import-competence to precursors. Cell. 1987;49:815–823. doi: 10.1016/0092-8674(87)90619-2. [DOI] [PubMed] [Google Scholar]
- 48.Rapaport, D., K.-P. Künkele, M. Dembowski, U. Ahting, F. E. Nargang, W. Neupert, and R. Lill. Dynamics of the TOM complex of mitochondria during binding and translocation of preproteins. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 49.Rapaport D, Mayer A, Neupert W, Lill R. cis and trans sites of the TOM complex of mitochondria in unfolding and initial translocation of preproteins. J Biol Chem. 1998;273:8806–8813. doi: 10.1074/jbc.273.15.8806. [DOI] [PubMed] [Google Scholar]
- 50.Rapaport D, Neupert W, Lill R. Mitochondrial protein import: Tom40 plays a major role in targeting and translocation of preproteins by forming a specific binding site for the presequence. J Biol Chem. 1997;272:18725–18731. doi: 10.1074/jbc.272.30.18725. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez-Cousino, N., F. E. Nargang, R. Baardman, W. Neupert, R. Lill, and D. A. Court. An import signal in the cytosolic domain of the Neurospora mitochondrial outer membrane protein TOM22. J. Biol. Chem., in press. [DOI] [PubMed]
- 52.Roise D, Schatz G. Mitochondrial presequences. J Biol Chem. 1988;263:4509–4511. [PubMed] [Google Scholar]
- 53.Ryan K R, Jensen R E. Protein translocation across mitochondrial membranes: What a long, strange trip it is. Cell. 1995;83:517–519. doi: 10.1016/0092-8674(95)90089-6. [DOI] [PubMed] [Google Scholar]
- 54.Ryan K R, Leung R S, Jensen R E. Characterization of the mitochondrial inner membrane translocase complex: the Tim23p hydrophobic domain interacts with Tim17p but not other Tim23p molecules. Mol Cell Biol. 1998;18:178–187. doi: 10.1128/mcb.18.1.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schatz G. Just follow the acid chain. Nature. 1997;388:121–122. doi: 10.1038/40510. [DOI] [PubMed] [Google Scholar]
- 56.Schatz G. The protein import system of mitochondria. J Biol Chem. 1996;271:31763–31766. doi: 10.1074/jbc.271.50.31763. [DOI] [PubMed] [Google Scholar]
- 57.Schleiff E, Shore G C, Goping I S. Interactions of the human mitochondrial protein import receptor hTOM20 with precursor proteins in vitro reveal pleiotropic specificities and different receptor domain requirements. J Biol Chem. 1997;272:17784–17789. doi: 10.1074/jbc.272.28.17784. [DOI] [PubMed] [Google Scholar]
- 58.Schweizer M, Case M E, Dykstra C C, Giles N H, Kushner S R. Identification and characterization of recombinant plasmids carrying the complete qa gene cluster from Neurospora crassa including the qa-1+ regulatory gene. Proc Natl Acad Sci USA. 1981;78:5086–5090. doi: 10.1073/pnas.78.8.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sirrenberg C, Bauer M, Guiard B, Neupert W, Brunner M. Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22. Nature. 1996;384:582–585. doi: 10.1038/384582a0. [DOI] [PubMed] [Google Scholar]
- 60.Sirrenberg C, Endres M, Fölsch H, Stuart R A, Neupert W, Brunner M. Carrier protein import into mitochondria mediated by the intermembrane space proteins Tim10/Mrs11p and Tim12/Mrs10p. Nature. 1998;391:912–915. doi: 10.1038/36136. [DOI] [PubMed] [Google Scholar]
- 61.Vogelstein B, Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci USA. 1979;73:615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.von Heijne G. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 1986;5:1335–1342. doi: 10.1002/j.1460-2075.1986.tb04364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.von Heijne G. Targeting signals for protein import into mitochondria and other subcellular organelles. Adv Mol Cell Biol. 1996;17:1–12. [Google Scholar]
- 64.White B, Woodward D. A simple method for making disposable race tubes. Fungal Genet Newsl. 1995;42:79. [Google Scholar]