Abstract
Diverse cellular processes have been observed or predicted to occur in biomolecular condensates, which are comprised of proteins and nucleic acids that undergo liquid-liquid phase separation (LLPS). Protein-driven LLPS often involves weak, multivalent interactions between intrinsically disordered regions (IDRs). Due to their inherent lack of defined tertiary structures, NMR has been a powerful resource for studying the behavior and interactions of IDRs in condensates. While IDRs in proteins are necessary for phase separation, core proteins enriched in condensates often contain structured domains that are essential for their function and contribute to phase separation. How phase separation can affect the structure and conformational dynamics of structured domains is critical for understanding how biochemical reactions can be effectively regulated in cellular condensates. In this perspective, we discuss the consequences phase separation can have on structured domains and outline NMR observables we believe are useful for assessing protein structure and dynamics in condensates.
Graphical Abstract
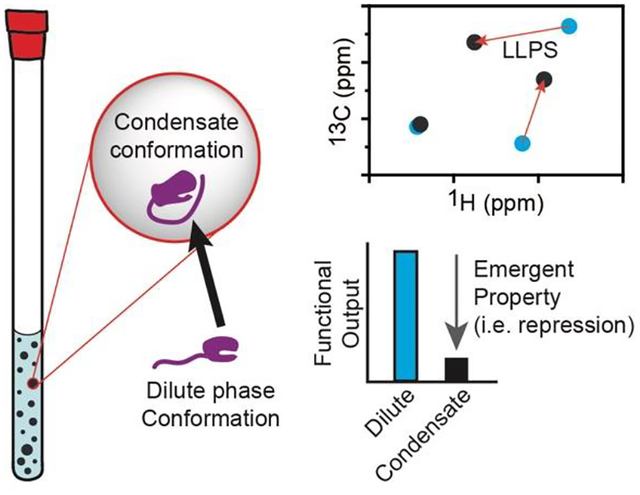
Condensates in biology
Cellular processes require the coordination of multiple steps in order to achieve a desired functional outcome. The evolution of multisubunit complexes that carry out sequential steps in a biochemical reaction has been essential for the spatial organization of these processes key to ensuring rapid, efficient responses to cell stimuli that maintain homeostasis. The relatively recent observation of liquid-like membraneless organelles (MLOs) in C. elegans ushered in a new field of cellular organization in biology1. The formation of the MLOs, or biomolecular condensates, results from the spontaneous partitioning of biomolecules into discrete compartments with a greater concentration relative to the surrounding solution2,3. Since their initial observation, numerous previously annotated cellular puncta have been shown to exhibit properties consistent with condensate formation. Together, the processes encapsulated within these puncta account for many essential aspects of cellular biochemistry. Among them includes the organization of heterochromatin and ribosome biogenesis in the nucleus, translational regulation and mRNA decay in the cytoplasm, as well as the organization of signaling cascades4–9.
The molecular basis for condensate formation draws extensively from seminal studies in polymer chemistry10,11. Liquid-liquid phase separation (LLPS) and percolation are two predominant thermodynamic driving forces of condensate formation12. Briefly, phase separation of a single macromolecular system arises from unfavorable interactions between the macromolecule and surrounding solvent to cause concentrated sites enriched in homotypic intermolecular interactions (Figure 1A, top). Percolation occurs when macromolecules with a high degree of multivalency lead to the formation of extended networks through oligomerization, protein-protein interactions or association with nucleic acids (Figure 1A, bottom)13. Moreover, phase separation and percolation can be coupled to promote condensate formation. In this perspective we collectively refer to these processes as phase separation for simplicity.
Figure 1:
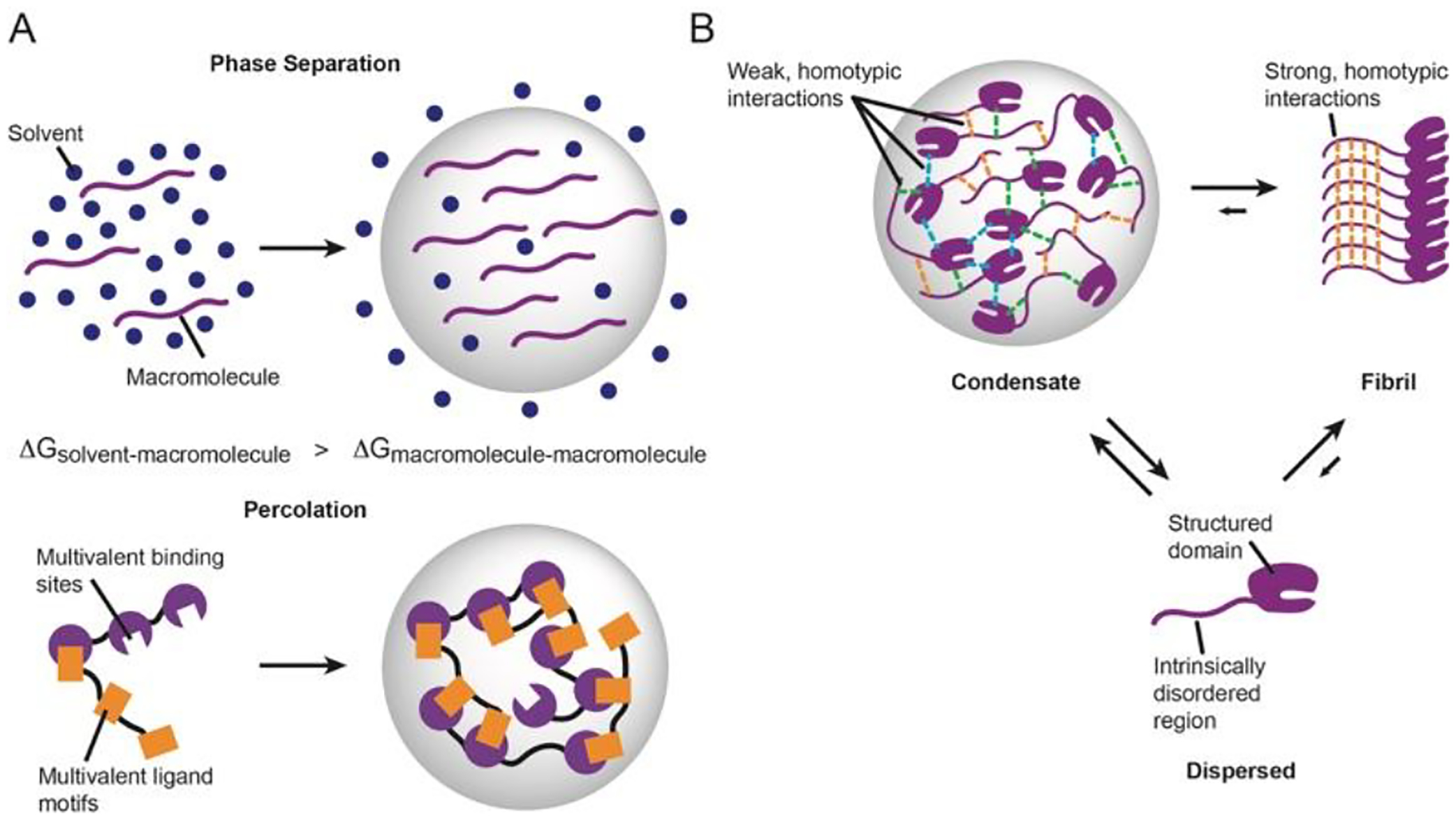
Interactions control condensate formation and liquid-like properties. A, Condensate formation results from phase separation (top) and percolation (bottom). B, Intrinsically disordered regions often flank structured regions and promote liquid-liquid phase separation through weak, multivalent interactions. Disordered regions can also promote stable fibril formation through strong interactions following maturation of condensates.
Since a variety of cellular processes occur in condensates, the list of proteins observed and hypothesized to undergo phase separation in vitro and in vivo is also diverse and constantly growing. Thus, it has been difficult to devise a consensus sequence or structural feature of the types of interactions promoting phase separation, but a few notable patterns have emerged14. Namely, many proteins that are essential for or accelerate condensate formation are intrinsically disordered (IDPs) or contain structured domains flanked by intrinsically disordered regions (IDRs) (Figure 1B). Atomistically, both electrostatic and hydrophobic interactions between amino acids have been shown to drive phase separation and alterations in these interactions can change the macroscopic behavior of droplets. For example, changing lysine:RNA interactions to arginine:RNA led to less dynamic condensates and decreasing the uniform distribution of aromatic residues caused hRNPA1 condensates to lose their liquid-like properties15,16. These and other observations underscore how the underlying molecular grammar driving phase separation can have important implications for tuning the microenvironment in condensates.
Protein and protein-RNA aggregates represent an extreme and often pathogenic form of MLOs. These stable aggregates result from the formation of amyloid-like fibrils that are seeded by interactions between glutamine and asparagine rich repeats within proteins17,18. Interestingly, several amyloidogenic proteins including FUS, Tau, hnRNPA1/2, and TDP-43 localize in various biomolecular condensates and undergo LLPS19–22. Moreover, mutations associated with neurodegenerative disease in TDP-43 inhibit its propensity to undergo phase separation while disease-associated mutations in tau enhance its phase separation and gelation14,17. The localization of FUS and TDP-43 to stress granules, for example, has been demonstrated to promote fibrilization and disease mutations associated with amyotrophic lateral sclerosis (ALS) prevented stress granule clearance and accelerated fibril formation21,22. Thus, the highly concentrated environment within condensates might represent a precarious precipice between homeostasis and pathogenesis whereby the liquid-like properties in condensates are an important checkpoint for clearing seeding intermediates to prevent nonreversible fibril formation.
Studying phase separation by NMR
Because proteins that undergo phase separation contain extensive disordered regions, exhibit dynamic properties and are formed through weak, transient interactions, they are refractory to traditional structural methods like X-ray crystallography and Cryo-electron microscopy. This has made solution- and solid-state NMR particularly well-suited for gaining atomic information on systems undergoing LLPS (reviewed in23–25). In vitro droplets containing the IDR of DDX4 were observed to have a translational diffusion constant similar to an E. coli cell and similar results were obtained for FUS26,27. Despite such slow translational diffusion, 2D HSQC spectra could be recorded and interpreted due to significant flexibility of the IDR in the condensed phase, giving rise to relatively fast local rotational diffusion, and consequently, dephasing that is slow enough to allow resolution of multiple crosspeaks. At the other extreme, magic-angle spinning solid-state NMR (MAS-SSNMR) is a powerful approach to characterize the structure of fibrils and condensate maturation that are refractory to solution NMR methods28–33. Thus, the combination of solution and solid-state NMR experiments allows for interrogation of phase separation from initial seeding to aggregate formation.
Several studies have utilized solution-state NMR to elucidate the molecular interactions important for phase separation by IDPs and IDRs23,24,26,34. These studies have mainly combined analysis of proteins in dilute phase to make informed mutations that can be assayed biochemically to assess their effects on phase separation. Spectral crowding arising from the poor chemical shift dispersion of unstructured proteins can confound analysis. To overcome this problem, an array of 13C-detected pulse schemes in conjunction with cryogenically cooled probes have been developed to allow assignment of IDPs35–44. The recent implementation of proton-detected haCONHA experiments have helped provide enhanced sensitivity to resolve structural changes that occur in proteins upon phase separation45.
Reconstituted membraneless organelles can exist in liquid or hydrogel like states, much like lipid bilayers, which can exist in fluid or gel phases46. Hybrid solution and solid-state NMR approaches in conjunction with MAS can be used to characterize, on a per residue basis, whether protein interactions in condensates are more liquid or gel-like. For example, both Methyl-TROSY and solid-state NMR studies of the enhancer of decapping protein Edc3 demonstrated its IDR remains largely flexible in condensates except for a small region that contacts the structured C-terminal YfeF N domain, which had not been previously observed47. Furthermore, hybrid NMR studies of hydrogels were used to characterize the behavior of FG repeats in phase separation of the nuclear pore complex protein Nup98 and to identify specific serine residues that contribute to the liquid-to-gel transition of human HP1a48,49. Finally, the study of squid beak proteins revealed that histidine deprotonation is an early step in their phase separation followed by tyrosine-tyrosine interactions and hydrophobic interactions50. The above studies highlight the power of NMR for determining how interactions change during LLPS, gelation and fibril formation, which can inform on biological function and disease.
Consequences of phase separation on molecular structure
The demixing of biomolecules from solution has several consequences for controlling both enzymes and scaffolding proteins3,51. The simplest view is that there is a division of labor between the IDR and structured enzymatic core domains, with the former promoting LLPS and the latter carrying out a specific function-such as scaffolding or an enzymatic reaction. Thus, the simplest direct consequence of phase separation is an increase of protein concentration in the condensed phase, which can accelerate enzyme catalyzed reactions52–54. However, there are several examples whereby condensate formation leads to inhibition or activation of functional processes beyond simple concentration effects, suggesting the unique chemical environment of the condensed phase can bias structured domains to achieve a switch like response in output55–59. In this case, we imagine the IDRs are poised to respond to these changes and alter the physical interactions to influence structured domain conformation. For example, the scaffolding of proteins within condensates can increase enzymatic rates by enhancing the association of reactants enzyme (decreased KM) or preventing nonproductive interactions compared to the unscaffolded proteins2,60. Alternatively, the condensed phase may promote nonproductive interactions that inhibit function by masking allosteric sites or substrate binding, for example. Finally, the unique solvation properties within condensates could affect reaction rates by biasing interactions between cofactors and enzymes or altering the free energy of the transition state51.
Given that multi-step processes can be enriched into a single condensate, there is the potential for numerous types of interactions to occur. Altering these interactions can have consequences for favoring the types of reactions that take place in condensates. For example, the nucleolus has been shown to organize into a condensate with distinct molecular composition that thermodynamically promotes the release of matured ribosomal RNA because of decreased multivalent interactions61. In addition, arginine-phosphate interactions were important for phase separation and mRNA partitioning to influence whether FMRP/CAPRIN1 condensates promote translation or deadenylation (repression)62. We also recently showed that mRNA decapping activity in is regulated in a composition dependent manner because a structured domain of the activator Edc3 interacts with motifs in the IDR of the decapping enzyme Dcp2 to change the molecular network promoting phase separation56. Thus, favoring certain interactions between structured and disordered regions in condensates is a robust mechanism to control cellular biochemistry.
Phase separation is regulated by molecular conformation and we envision the unique environment in condensates can perturb molecular conformation. G3BP1, an essential component of stress granule formation, must undergo a conformational change from an autoinhibited to open state that reveals an IDR to interact with RNA and promote phase separation63,64. The heterochromatin protein HP1 changes conformation because of phosphorylation of its N-terminal disordered region or ligand binding to cause phase separation and the fission yeast protein Swi6 promotes heterochromatin phase separation by distorting the nucleosome core4,65. As these examples illustrate, molecular conformation represents an additional mechanism to control condensate formation, composition, and function.
Considerations for sample preparation
Using NMR to studying macromolecules that undergo phase separation leads to several challenges and considerations that need to be assessed to ensure monitoring of the appropriate observable. For instance, studying interactions in the dilute phase provides valuable insights into the initial events in phase separation but are an indirect readout of the interactions in condensates66,67. Direct observation of the condensed phase is difficult because it usually requires large quantities to create a homogenous monophasic sample across the entire NMR coil to avoid the effects of settling and higher viscosity that reduce signal intensity23. In addition, condensate-glass interactions can alter the magnetic susceptibility of the sample to cause additional crosspeaks that confound analysis68. This latter challenge was overcome by the suspension of condensates in an agarose hydrogel that mimics the cytoplasm and maintains a more physiologic surface area-to-volume ratio68. Gaining a complete understanding of the regulatory mechanisms imparted by phase separation will likely require a combination of the sample types described here and presents an opportunity for the advancement of spectroscopic hardware and molecular labeling schemes.
Approaches for studying structured domains in phase-separated systems
So far, most NMR analyses of condensates have focused on the IDRs and these studies have been crucial for illuminating the interactions important for phase separation23. While IDRs are replete in the proteome, they are often found adjacent to structured domains that are necessary for proper protein function69. In addition, IDRs can regulate the function of structured domains through direct and allosteric mechanisms70. In the context of biomolecular condensates, structured domains can promote phase separation and alter the material properties of condensates in vitro and in vivo71,72. Thus, examining the behavior of structured domains in condensates is important to understanding how biochemical processes are regulated in condensates. However, how phase separation may influence structured domains of proteins has remained poorly studied, likely due to the challenges in expressing and purifying these proteins as well as the additional spectral complexity arising from additional crosspeaks and slower rates of molecular tumbling.
It is our view that studying the impact of IDRs on the dynamics of structured domains in solution is a good prelude for understanding how biological condensates can achieve additional control of function. This necessitates a combination of liquid, solid-state NMR and pulsed EPR approaches such as DEER68,73. A major goal in the field will be to answer the fundamental question of how conformational landscapes can be impacted by phase separation to impart additional regulation.
Systems with globular domains and IDRs will have a great degree of spectral complexity. One workaround is to introduce spectroscopically active labels at specific positions throughout the protein sequence, a now common practice for NMR studies large proteins and complexes74–77. The labeling of terminal methyl groups in isoleucine, leucine, valine, methionine, and alanine (ILVMA) residues has greatly enhanced the sensitivity of NMR signals for the study of large protein assemblies, and we have outlined some of its applications for studying phase separation above (Figure 2)74,78.
Figure 2:
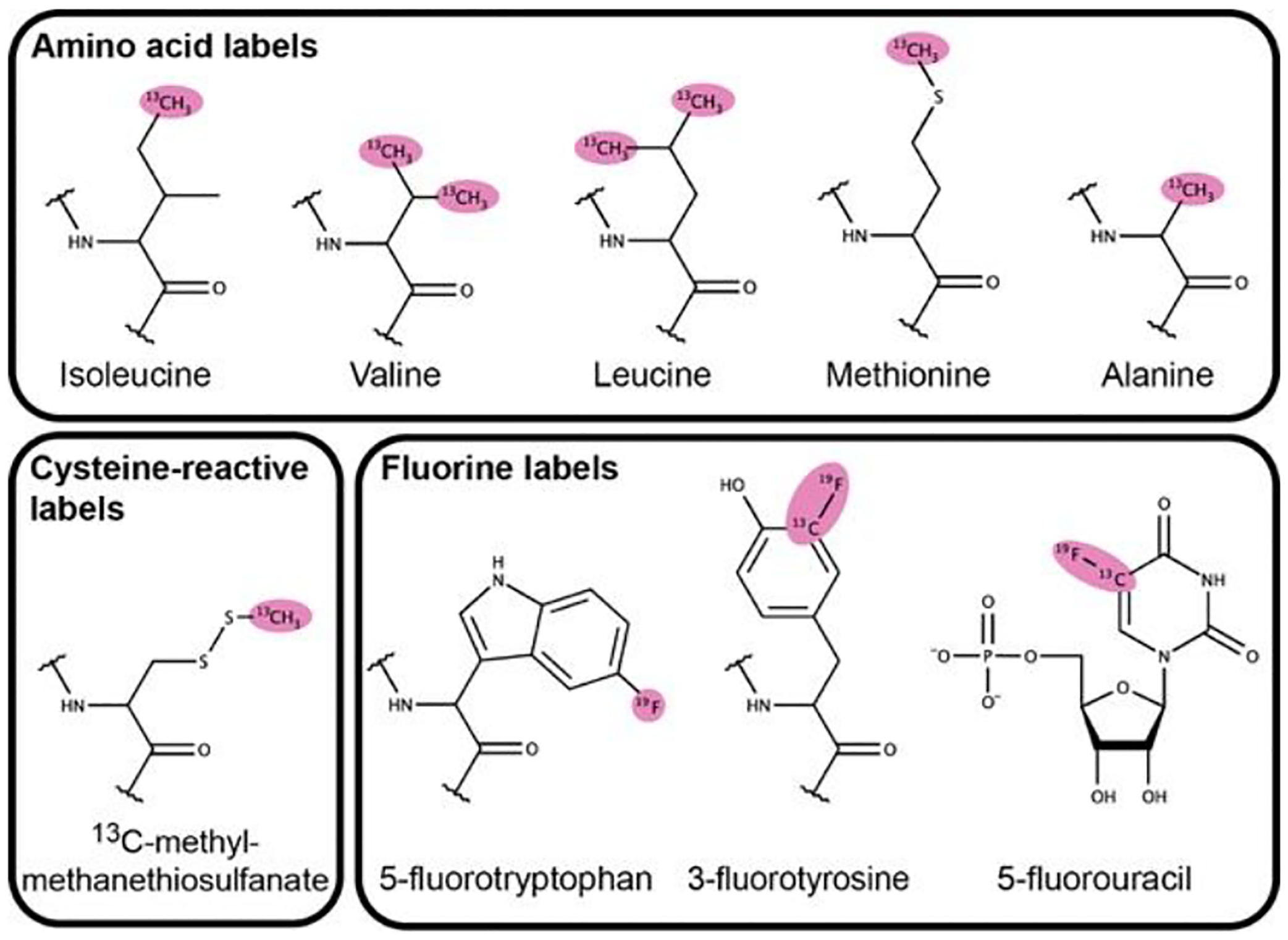
Amino and nucleic acid derivatives used in biomolecular NMR. These labels have advantageous NMR relaxation properties useful for studying large macromolecular assemblies such as condensates. Observable signals are highlighted in pink.
In addition, the conjugation of NMR reporter groups such as 13C-methyl-methanethiosulfonate at reactive amino acids (i.e. cysteine) following purification allows for expression of proteins of interest in insect and mammalian cultures, which may be required to obtain NMR quantities of proteins that are recalcitrant to E. coli expression (Figure 2)79. Furthermore, the incorporation of fluorine into biomolecules at aromatic residues and through chemical conjugation has been applied to the study of biomolecular interactions and dynamics80–82. In particular, the development of aromatic 19F-13C TROSY pulse sequences has extended its application to larger biomolecules, including RNA (Figure 2)83. Given the favorable spectroscopic properties, sensitivity to chemical environment, and lack of natural incorporation into biomolecules, fluorine may be a powerful tool to studying molecules within condensates using solution or SSNMR.
Another means for minimizing spectral overlap is through the segmental labeling of proteins wherein only one region of interest is NMR-active. Segmental labeling has the advantage of maintaining the protein in a near-native state. Two protein-based methods for ligation have been the most extensively used for NMR studies: intein and Sortase - mediated ligation84,85. In the former, two components of interest (exteins) are ligated through the formation of an intermediate thioester that is resolved by a N-terminal cysteine on the second extein (Figure 3A). The N-terminal cysteine is required for ligation and either needs to be synthesized or exposed upon proteolytic cleavage during purification. Sortase-mediated ligation makes use of the protein encoded by the S. aureus SortaseA gene, a member of the transpeptidase family of enzymes that anchor proteins to the cell wall of gram-positive bacteria (Figure 3B)86. A recognition sequence LPXTG (X = any amino acid) is appended to the C-terminus of one fragment and ligated to an N-terminal glycine residue of the second protein fragment. Thus, the mutational scar LPXTGn (n= number of glycine residues) is present in the final ligated protein. We recently used SortaseA ligation to study how the disordered region in the decapping enzyme Dcp2 influences the conformation of the structured catalytic domains (Figure 3C,D)56. In addition, the labeling and ligation methods outlined here can be used in combination to produce differential labeling schemes within a single polypeptide that would allow for greater coverage of the total protein structure87. Moreover, these strategies can also be combined to label multiple proteins that together undergo phase separation.
Figure 3:
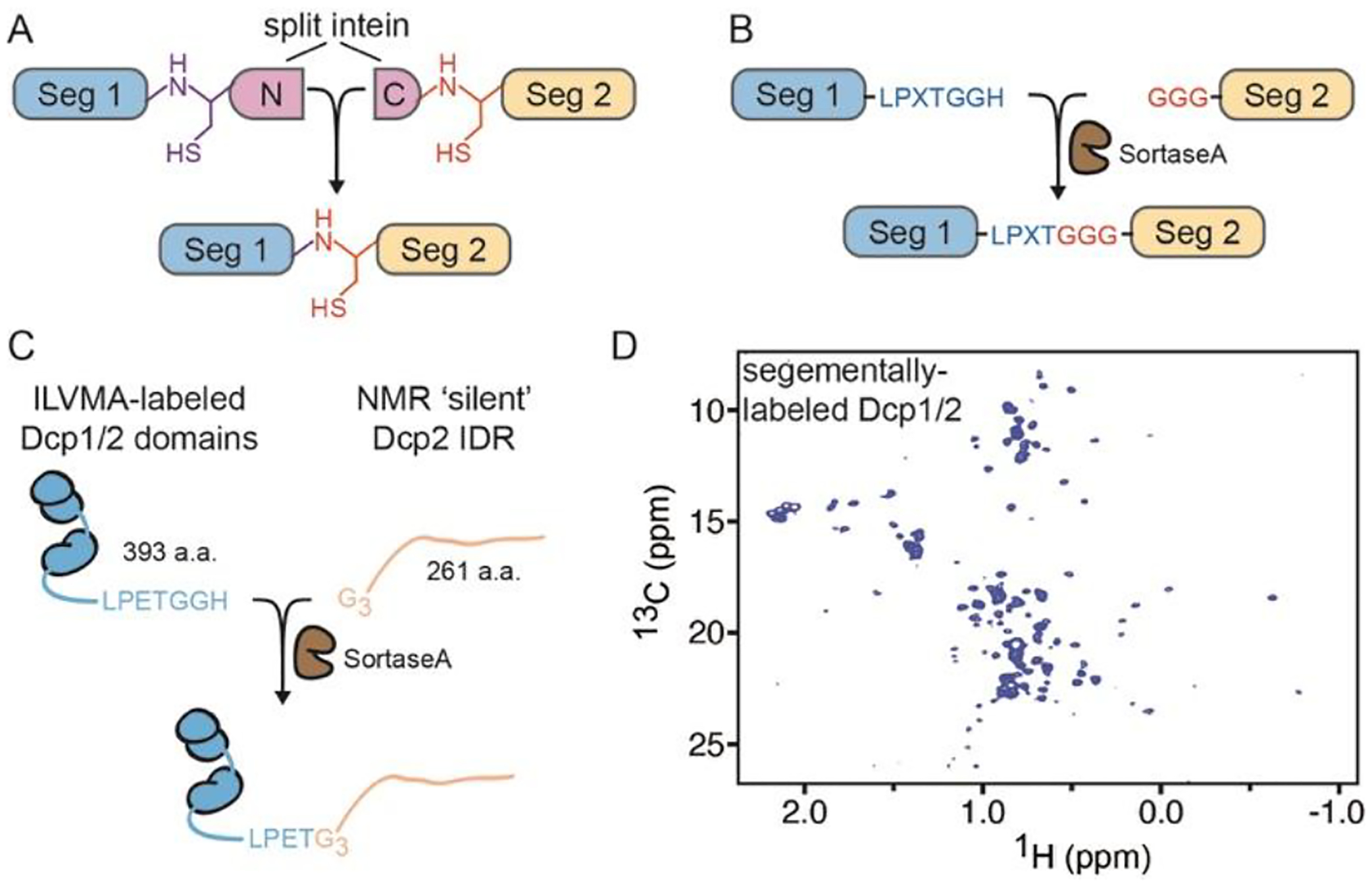
Protein conjugation strategies for segmental labeling of proteins. A, Split intein can be fused to two polypeptide fragments (Seg1 and Seg2) that are subsequently ligated to form a single chain. B, SortaseA enzyme ligates a protein fragment (Seg2) with N-terminal glycine residues to its C-terminal recognition sequence in a second fragment (Seg1). C, SortaseA ligation applied to S. pombe decapping complex Dcp1/2 D, 1H-13C HSQC ILVMA spectrum of structured domains in segmentally-labeled Dcp1/2.
Conformational landscapes of structured core domains may be remodelled by interactions with IDRs88,89. Paramagnetic relaxation enhancement (PRE) NMR experiments can also be employed to observe transient, lowly-populated conformations that may be present in solution87. With known structures of core domains, the ability to back calculate PREs allows one to ask if and how the IDR would influence conformation of dynamic core domains90. This approach is quite complementary to pulsed EPR approaches such as DEER which are carried out on frozen solutions73,91. PRE-NMR has been extensively used to characterize the intra- and intermolecular interactions important for controlling phase separation of FUS, TDP-43, and hnRNPA2 in the dilute phase and how post-translational modifications influence these interactions92,93. In addition, NMR and pulsed EPR studies employing DEER to generate distance distribution functions from nitroxide spin pairs incorporated into FUS demonstrated it undergoes compaction upon phase separation68. Cross-linking mass spectrometry (XL-MS) corroborated these results and demonstrated more extensive interactions between and within FUS molecules in condensates94. The combination of these methods and the development of new ones would greatly enhance our ability to understand how structured regions are influenced by phase separation.
Consequences of phase separation on structured domains
What interactions emerge in condensates?
Protein IDRs are often crucial for condensate formation through multivalent interactions. In addition, short linear motifs (SLiMs) embedded within IDRs can promote phase separation and regulate protein function. In dilute solution, interactions between a regulatory SLiM and structured domain may be transient but their partitioning into condensates creates an environment that stabilizes the interaction and exerts emergent regulatory control (Figure 4A). We have observed that the C-terminal IDR of the decapping protein Dcp2 mediates its phase separation with the essential cofactor Dcp1, which leads to enhanced repression of activity56. Our study did not characterize the interactions between the inhibitory IDR of Dcp2 and structure core domains of Dcp1/Dcp2 in condensates directly, but we observed by solution state NMR that the Dcp2 C-terminal IDR stabilizes an inactive conformation of the Dcp1/Dcp2 complex. We hypothesize that the high local concentrations and unique solvation properties present in condensates may stabilize interactions between the C-terminal IDR and the structured core domains of the Dcp1/Dcp2 complex. In addition, as we describe in the next section, phase separation may allow proteins to access lowly-populated states through alterations in their energetic landscape.
Figure 4:
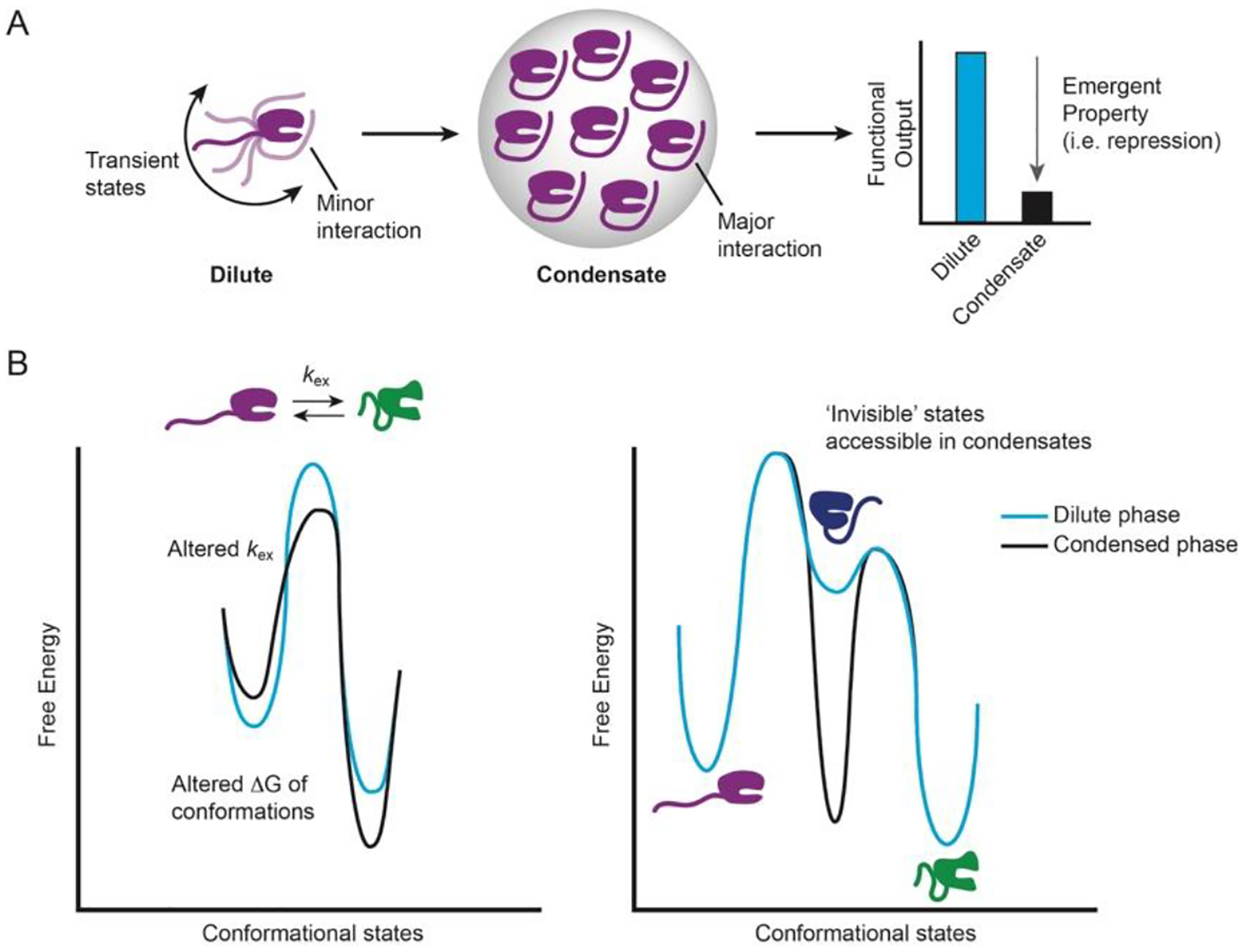
Potential effects of phase separation on structured domains. A, In dispersed solution, an IDR samples many conformational states, but one minor interaction may become stabilized in condensates and cause an emergent functional outcome for activity (i.e. repression, activation, etc.) B, The condensate environment can alter the conformational landscape of proteins in multiple ways, including changes to the rate of conformational exchange or the free energy of a given state.
How are conformational landscapes of multidomain proteins affected by LLPS?
Proteins can explore a wide conformational landscape with motions spanning from picoseconds to hours or longer and these conformational fluctuations are often important for proper function95. The unique properties of biomolecular condensates, including increased viscosity, high molar concentrations, and altered dielectric constants have the potential to reshape a protein’s conformational landscape (Figure 4B). A recent cross-linking mass spectrometry (XL-MS) demonstrated that a RRM domain in FUS undergoes a partial denaturing upon LLPS that is not readily observed in dilute solution94. In addition, the heterochromatin protein HP1 ortholog from S. pombe, Swi6, causes distortions in buried regions of the nucleosome and disrupting the distortions abrogates phase separation in vitro and leads to improper heterochromatic centers in vivo65. Thus, it seems probable previously ‘invisible’ states can be accessed and stabilized in the condensed phase. It is also intriguing to consider that structured oligomeric states not readily detectable in dilute solution become accessible in condensates to increase cooperativity and avidity in reactions. In sum, these ‘new’ states could impart additional regulation, enhance function, or represent unstable intermediates on pathway to amyloid formation. However, the population and structural features of these states has not been studied, but we believe NMR is well-suited to rigorously address these questions.
Concluding Remarks
The work of many research groups has advanced our knowledge of how intrinsic disorder contributes to protein function and, more recently, its fundamental role in liquid-liquid phase separation. The increasing suite of biophysical and computational methods is a promising integrated framework to address the questions presented here regarding emergent interactions and altered conformational landscapes in biomolecular condensates. The ability to apply and expand existing NMR approaches to study the interplay between disorder and structured regions in condensates presents an exciting opportunity to uncover new modes of regulation. These contributions will further our understanding of why phase separation is a common feature of diverse cellular processes.
Highlights.
NMR has been extensively used to study proteins undergoing phase separation
There is an interplay between IDRs and structured domains in condensates
Structured domains undergoing phase separation are poorly studied
Conformational landscapes in condensates may reveal new regulatory mechanisms
Acknowledgements
This article is dedicated to Gerhard Wagner for his foundational and ongoing contributions to studying biomolecules by NMR. JDG greatly appreciates his support over the years, fondly remembering that with his gentle question “How are things going?”, much can be revealed. The lesson learned is that it is not only what is asked, but how it is asked that makes space for true insight. The authors thank Haribabu Arthanari, Lewis Kay, and Gaetano Montelione for the invitation to contribute to this special issue. This work is supported by the U.S. National Institutes of Health (R01 GM078360 to J.D.G.) and the Sandler Program for Breakthrough Biomedical Research.
Footnotes
Declaration of Competing Interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Brangwynne CP et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Banani SF, Lee HO, Hyman AA & Rosen MK Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeynaems S et al. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol. 28, 420–435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larson AG et al. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strom AR et al. Phase separation drives heterochromatin domain formation. Nature 547, 241–245 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su X et al. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352, 595–599 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheth U & Parker R Decapping and Decay of Messenger RNA Occur in Cytoplasmic Processing Bodies. Science 300, 805–808 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Decker CJ & Parker R P-Bodies and Stress Granules: Possible Roles in the Control of Translation and mRNA Degradation. Cold Spring Harb. Perspect. Biol 4, a012286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tulpule A et al. Kinase-mediated RAS signaling via membraneless cytoplasmic protein granules. Cell 184, 2649–2664.e18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flory PJ Thermodynamics of High Polymer Solutions. J. Chem. Phys 10, 51–61 (1942). [Google Scholar]
- 11.Huggins ML Solutions of Long Chain Compounds. J. Chem. Phys 9, 440–440 (1941). [Google Scholar]
- 12.Mittag T & Pappu RV A conceptual framework for understanding phase separation and addressing open questions and challenges. Mol. Cell 82, 2201–2214 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi J-M, Hyman AA & Pappu RV Generalized models for bond percolation transitions of associative polymers. Phys. Rev. E 102, 042403 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vernon RM & Forman-Kay JD First-generation predictors of biological protein phase separation. Curr. Opin. Struct. Biol 58, 88–96 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Ukmar-Godec T et al. Lysine/RNA-interactions drive and regulate biomolecular condensation. Nat. Commun 10, 2909 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin EW et al. Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 367, 694–699 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derkatch IL et al. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc. Natl. Acad. Sci 101, 12934–12939 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cascarina SM & Ross ED Yeast prions and human prion-like proteins: sequence features and prediction methods. Cell. Mol. Life Sci. CMLS 71, 2047–2063 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambadipudi S, Biernat J, Riedel D, Mandelkow E & Zweckstetter M Liquid–liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nat. Commun 8, 275 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conicella AE, Zerze GH, Mittal J & Fawzi NL ALS Mutations Disrupt Phase Separation Mediated by α-Helical Structure in the TDP-43 Low-Complexity C-Terminal Domain. Struct. Lond. Engl. 1993 24, 1537–1549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molliex A et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim HJ et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murthy AC & Fawzi NL The (un)structural biology of biomolecular liquid-liquid phase separation using NMR spectroscopy. J. Biol. Chem 295, 2375–2384 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibbs E, Perrone B, Hassan A, Kümmerle R & Kriwacki R NPM1 exhibits structural and dynamic heterogeneity upon phase separation with the p14ARF tumor suppressor. J. Magn. Reson 310, 106646 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fawzi NL, Parekh SH & Mittal J Biophysical studies of phase separation integrating experimental and computational methods. Curr. Opin. Struct. Biol 70, 78–86 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brady JP et al. Structural and hydrodynamic properties of an intrinsically disordered region of a germ cell-specific protein on phase separation. Proc. Natl. Acad. Sci 114, E8194–E8203 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burke KA, Janke AM, Rhine CL & Fawzi NL Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II. Mol. Cell 60, 231–241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petkova AT et al. A structural model for Alzheimer’s β-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci 99, 16742–16747 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tycko R Solid-state NMR studies of amyloid fibril structure. Annu. Rev. Phys. Chem 62, 279–299 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berkeley RF, Kashefi M & Debelouchina GT Real-time observation of structure and dynamics during the liquid-to-solid transition of FUS LC. Biophys. J 120, 1276–1287 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karamanos TK, Kalverda AP, Thompson GS & Radford SE Mechanisms of amyloid formation revealed by solution NMR. Prog. Nucl. Magn. Reson. Spectrosc 88–89, 86–104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fawzi NL, Ying J, Ghirlando R, Torchia DA & Clore GM Atomic-resolution dynamics on the surface of amyloid-β protofibrils probed by solution NMR. Nature 480, 268–272 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fonda BD, Jami KM, Boulos NR & Murray DT Identification of the Rigid Core for Aged Liquid Droplets of an RNA-Binding Protein Low Complexity Domain. J. Am. Chem. Soc 143, 6657–6668 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Murthy AC et al. Molecular interactions underlying liquid−liquid phase separation of the FUS low-complexity domain. Nat. Struct. Mol. Biol 26, 637–648 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pantoja-Uceda D & Santoro J New 13C-detected experiments for the assignment of intrinsically disordered proteins. J. Biomol. NMR 59, 43–50 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Murrali MG et al. 13C APSY-NMR for sequential assignment of intrinsically disordered proteins. J. Biomol. NMR 70, 167–175 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Piai A et al. “CON-CON” assignment strategy for highly flexible intrinsically disordered proteins. J. Biomol. NMR 60, 209–218 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Felli IC & Pierattelli R Novel methods based on 13C detection to study intrinsically disordered proteins. J. Magn. Reson 241, 115–125 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Bermel W et al. High-dimensionality 13C direct-detected NMR experiments for the automatic assignment of intrinsically disordered proteins. J. Biomol. NMR 57, 353–361 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Gibbs EB & Kriwacki RW Direct detection of carbon and nitrogen nuclei for high-resolution analysis of intrinsically disordered proteins using NMR spectroscopy. Methods 138–139, 39–46 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brutscher B et al. NMR Methods for the Study of Instrinsically Disordered Proteins Structure, Dynamics, and Interactions: General Overview and Practical Guidelines. in Intrinsically Disordered Proteins Studied by NMR Spectroscopy (eds. Felli IC & Pierattelli R) 49–122 (Springer International Publishing, 2015). doi: 10.1007/978-3-319-20164-1_3. [DOI] [PubMed] [Google Scholar]
- 42.Ambadipudi S, Reddy GJ, Biernat J, Mandelkow E & Zweckstetter M Residue-specific identification of phase separation hot spots of Alzheimer’s-related protein tau. Chem. Sci 10, 6503–6507 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeuchi K, Heffron G, Sun Z-YJ, Frueh DP & Wagner G Nitrogen-detected CAN and CON experiments as alternative experiments for main chain NMR resonance assignments. J. Biomol. NMR 47, 271–282 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bermel W et al. Improving the chemical shift dispersion of multidimensional NMR spectra of intrinsically disordered proteins. J. Biomol. NMR 55, 231–237 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Wong LE, Kim TH, Muhandiram DR, Forman-Kay JD & Kay LE NMR Experiments for Studies of Dilute and Condensed Protein Phases: Application to the Phase-Separating Protein CAPRIN1. J. Am. Chem. Soc 142, 2471–2489 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Tristram-Nagle S & Nagle JF Lipid bilayers: thermodynamics, structure, fluctuations, and interactions. Chem. Phys. Lipids 127, 3–14 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Damman R et al. Atomic-level insight into mRNA processing bodies by combining solid and solution-state NMR spectroscopy. Nat. Commun 10, 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ackermann BE & Debelouchina GT Heterochromatin Protein HP1α Gelation Dynamics Revealed by Solid-State NMR Spectroscopy. Angew. Chem. Int. Ed 58, 6300–6305 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Najbauer EE, Ng SC, Griesinger C, Görlich D & Andreas LB Atomic resolution dynamics of cohesive interactions in phase-separated Nup98 FG domains. Nat. Commun 13, 1494 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gabryelczyk B et al. Hydrogen bond guidance and aromatic stacking drive liquid-liquid phase separation of intrinsically disordered histidine-rich peptides. Nat. Commun 10, 5465 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Narlikar GJ & Kutateladze TG Enzymatic Reactions inside Biological Condensates. J. Mol. Biol 433, 166624 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sheu-Gruttadauria J & MacRae IJ Phase Transitions in the Assembly and Function of Human miRISC. Cell 173, 946–957.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gallego LD et al. Phase separation directs ubiquitination of gene-body nucleosomes. Nature 579, 592–597 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao H et al. Quantitative analysis of purine nucleotides indicates that purinosomes increase de novo purine biosynthesis. J. Biol. Chem 290, 6705–6713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pitchiaya S et al. Dynamic Recruitment of Single RNAs to Processing Bodies Depends on RNA Functionality. Mol. Cell 74, 521–533.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tibble RW, Depaix A, Kowalska J, Jemielity J & Gross JD Biomolecular condensates amplify mRNA decapping by biasing enzyme conformation. Nat. Chem. Biol 17, 615–623 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y et al. Nuclear condensates of p300 formed though the structured catalytic core can act as a storage pool of p300 with reduced HAT activity. Nat. Commun 12, 4618 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horvathova I et al. The Dynamics of mRNA Turnover Revealed by Single-Molecule Imaging in Single Cells. Mol. Cell 68, 615–625.e9 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Moon SL et al. Multicolour single-molecule tracking of mRNA interactions with RNP granules. Nat. Cell Biol 21, 162–168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peeples W & Rosen MK Mechanistic dissection of increased enzymatic rate in a phase-separated compartment. Nat. Chem. Biol 17, 693–702 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Riback JA et al. Composition-dependent thermodynamics of intracellular phase separation. Nature 1–6 (2020) doi: 10.1038/s41586-020-2256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim TH et al. Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science 365, 825–829 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Guillén-Boixet J et al. RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell 181, 346–361.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang P et al. G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell 181, 325–345.e28 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanulli S et al. HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 575, 390–394 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Conicella AE et al. TDP-43 α-helical structure tunes liquid–liquid phase separation and function. Proc. Natl. Acad. Sci 117, 5883–5894 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reichheld SE, Muiznieks LD, Keeley FW & Sharpe S Direct observation of structure and dynamics during phase separation of an elastomeric protein. Proc. Natl. Acad. Sci 114, E4408–E4415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Emmanouilidis L et al. NMR and EPR reveal a compaction of the RNA-binding protein FUS upon droplet formation. Nat. Chem. Biol 17, 608–614 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pentony MM & Jones DT Modularity of intrinsic disorder in the human proteome. Proteins 78, 212–221 (2010). [DOI] [PubMed] [Google Scholar]
- 70.Wright PE & Dyson HJ Intrinsically Disordered Proteins in Cellular Signaling and Regulation. Nat. Rev. Mol. Cell Biol 16, 18–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dignon GL, Best RB & Mittal J Biomolecular Phase Separation: From Molecular Driving Forces to Macroscopic Properties. Annu. Rev. Phys. Chem 71, 53–75 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martin EW et al. Interplay of folded domains and the disordered low-complexity domain in mediating hnRNPA1 phase separation. Nucleic Acids Res. 49, 2931–2945 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Prisner T, Rohrer M & MacMillan F Pulsed EPR spectroscopy: biological applications. Annu. Rev. Phys. Chem 52, 279–313 (2001). [DOI] [PubMed] [Google Scholar]
- 74.Schütz S & Sprangers R Methyl TROSY spectroscopy: A versatile NMR approach to study challenging biological systems. Prog. Nucl. Magn. Reson. Spectrosc 116, 56–84 (2020). [DOI] [PubMed] [Google Scholar]
- 75.Gardner KH & Kay LE Production and Incorporation of 15N, 13C, 2H (1H-δ1 Methyl) Isoleucine into Proteins for Multidimensional NMR Studies. J. Am. Chem. Soc 119, 7599–7600 (1997). [Google Scholar]
- 76.Tugarinov V, Kanelis V & Kay LE Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat. Protoc 1, 749–754 (2006). [DOI] [PubMed] [Google Scholar]
- 77.Debelouchina GT & Muir TW A molecular engineering toolbox for the structural biologist. Q. Rev. Biophys 50, e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goto NK & Kay LE New developments in isotope labeling strategies for protein solution NMR spectroscopy. Curr. Opin. Struct. Biol 10, 585–592 (2000). [DOI] [PubMed] [Google Scholar]
- 79.Religa TL, Ruschak AM, Rosenzweig R & Kay LE Site-Directed Methyl Group Labeling as an NMR Probe of Structure and Dynamics in Supramolecular Protein Systems: Applications to the Proteasome and to the ClpP Protease. J. Am. Chem. Soc 133, 9063–9068 (2011). [DOI] [PubMed] [Google Scholar]
- 80.Didenko T, Liu JJ, Horst R, Stevens RC & Wüthrich K Fluorine-19 NMR of integral membrane proteins illustrated with studies of GPCRs. Curr. Opin. Struct. Biol 23, 740–747 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boeszoermenyi A, Ogórek B, Jain A, Arthanari H & Wagner G The precious fluorine on the ring: fluorine NMR for biological systems. J. Biomol. NMR 74, 365–379 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Overbeck JH, Kremer W & Sprangers R A suite of 19F based relaxation dispersion experiments to assess biomolecular motions. J. Biomol. NMR 74, 753–766 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boeszoermenyi A et al. Aromatic 19F-13C TROSY: a background-free approach to probe biomolecular structure, function, and dynamics. Nat. Methods 16, 333–340 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Muralidharan V & Muir TW Protein ligation: an enabling technology for the biophysical analysis of proteins. Nat. Methods 3, 429–438 (2006). [DOI] [PubMed] [Google Scholar]
- 85.Refaei MA et al. Observing selected domains in multi-domain proteins via sortase-mediated ligation and NMR spectroscopy. J. Biomol. NMR 49, 3–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mazmanian SK, Liu G, Ton-That H & Schneewind O Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285, 760–763 (1999). [DOI] [PubMed] [Google Scholar]
- 87.Clore GM & Iwahara J Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem. Rev 109, 4108–4139 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim Y, Furman CM, Manhart CM, Alani E & Finkelstein IJ Intrinsically disordered regions regulate both catalytic and non-catalytic activities of the MutLα mismatch repair complex. Nucleic Acids Res. 47, 1823–1835 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Keul ND et al. The entropic force generated by intrinsically disordered segments tunes protein function. Nature 563, 584–588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Iwahara J, Schwieters CD & Clore GM Ensemble approach for NMR structure refinement against (1)H paramagnetic relaxation enhancement data arising from a flexible paramagnetic group attached to a macromolecule. J. Am. Chem. Soc 126, 5879–5896 (2004). [DOI] [PubMed] [Google Scholar]
- 91.Chiang Y-W, Borbat PP & Freed JH The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J. Magn. Reson. San Diego Calif 1997 172, 279–295 (2005). [DOI] [PubMed] [Google Scholar]
- 92.Monahan Z et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 36, 2951–2967 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ryan VH et al. Mechanistic View of hnRNPA2 Low-Complexity Domain Structure, Interactions, and Phase Separation Altered by Mutation and Arginine Methylation. Mol. Cell 69, 465–479.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Boczek EE et al. HspB8 prevents aberrant phase transitions of FUS by chaperoning its folded RNA-binding domain. eLife 10, e69377 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Henzler-Wildman K & Kern D Dynamic personalities of proteins. Nature 450, 964–972 (2007). [DOI] [PubMed] [Google Scholar]


