Abstract
Mutations in Drosophila ISWI, a member of the SWI2/SNF2 family of chromatin remodeling ATPases, alter the global architecture of the male X chromosome. The transcription of genes on this chromosome is increased 2-fold relative to females due to dosage compensation, a process involving the acetylation of histone H4 at lysine 16 (H4K16). Here we show that blocking H4K16 acetylation suppresses the X chromosome defects resulting from loss of ISWI function in males. In contrast, the forced acetylation of H4K16 in ISWI mutant females causes X chromosome defects indistinguishable from those seen in ISWI mutant males. Increased expression of MOF, the histone acetyltransferase that acetylates H4K16, strongly enhances phenotypes resulting from the partial loss of ISWI function. Peptide competition assays revealed that H4K16 acetylation reduces the ability of ISWI to interact productively with its substrate. These findings suggest that H4K16 acetylation directly counteracts chromatin compaction mediated by the ISWI ATPase.
INTRODUCTION
The Drosophila ISWI protein functions as the ATPase subunit of several nucleosome remodeling complexes, including CHRAC, ACF and NURF (Längst and Becker, 2001). Genetic and biochemical studies have revealed important roles for ISWI in transcriptional activation, transcriptional repression and chromatin assembly (Längst and Becker, 2001). ISWI participates in these processes by catalyzing ATP-dependent alterations in chromatin structure (Längst and Becker, 2001). One ISWI complex, NURF, disrupts the periodicity of nucleosomal arrays (Wu et al., 1998), while others, including CHRAC and ACF, create chromatin with more regularly spaced arrays of nucleosomes (Fyodorov and Kadonaga, 2001; Längst and Becker, 2001). By altering the structure or spacing of nucleosomes, ISWI appears to modulate interactions between regulatory proteins and DNA in the context of chromatin.
ISWI is also required for the maintenance of higher-order chromatin structure. Although ISWI is an essential gene in Drosophila, individuals homozygous for ISWI null mutations survive until late larval development due to the high maternal contribution of ISWI (Deuring et al., 2000). The severe reduction in ISWI levels in the salivary glands of ISWI mutant larvae causes the male X chromosome, but not the autosomes or female X chromosome, to appear much less compact than normal (Deuring et al., 2000). The aberrant morphology of the male X chromosome in ISWI mutant larvae may reflect the ability of some ISWI-containing remodeling complexes to improve the regularity of nucleosomal arrays and hence promote chromatin folding.
The unusual chromosome defects observed in ISWI mutants suggests that some unique feature of the male X chromosome makes it particularly sensitive to the partial loss of ISWI function. One good candidate for such a feature is the acetylation of lysine 16 of histone H4 (H4K16), which leads to a 2-fold increase in the transcription of X-linked genes in males relative to females (Park and Kuroda, 2001; Smith et al., 2001). This covalent modification of chromatin is dependent on the activity of the dosage compensation complex, which contains the Male-specific lethal proteins MSL-1, MSL-2 and MSL-3, Maleless (MLE), the histone acetyltransferase Males-absent on the first (MOF) and the non-coding roX1 and roX2 RNAs (Park and Kuroda, 2001; Smith et al., 2001).
Histone acetylation is causally involved in transcriptional activation, possibly through partial decompaction of higher-order structure (Hansen et al., 1998; Tse et al., 1998; Akhtar and Becker, 2000). We therefore suspected that acetylation of H4K16 by the dosage-compensation machinery might directly or indirectly counteract compaction mediated by the ISWI ATPase. Here we report that the process of dosage compensation is both necessary and sufficient for the X chromosome defects observed in ISWI mutant larvae. Furthermore, we show that the acetylation of H4K16 antagonizes ISWI function both in vivo and in vitro. These findings demonstrate that the activity of a chromatin remodeling ATPase can be directly regulated by the site-specific acetylation of its chromatin substrate.
RESULTS AND DISCUSSION
The dosage compensation complex is necessary and sufficient for the chromosome defects observed in ISWI mutant larvae
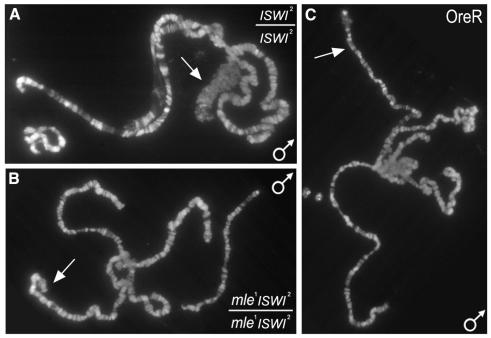
To investigate a functional antagonism between ISWI and histone H4 acetylation, we examined the epistatic relationship between ISWI and mle. Because mle encodes an essential subunit of the dosage compensation complex, the X chromosome is not acetylated on H4K16 in homozygous mle males (Kuroda et al., 1991). As shown in Figure 1A, the X chromosome of homozygous ISWI2 males is grossly abnormal, exhibiting a bloated appearance and an almost complete loss of its characteristic banding pattern (compare Figure 1A and C). In contrast, this phenotype is not observed in male larvae homozygous for both ISWI2 and mle1 (Figure 1B). The activity of the dosage compensation complex is therefore necessary for the altered appearance of the male X chromosome in ISWI mutant larvae.
Fig. 1. The dosage compensation complex is necessary for the X chromosome defects observed in ISWI2 males. (A) Salivary gland polytene chromosomes from homozygous ISWI2 male larvae. Note the bloated appearance of the X chromosome. (B) Salivary gland polytene chromosomes from homozygous mle1 ISWI2 male larvae. Loss of mle function completely suppresses X chromosome defects associated with loss of ISWI function. No staining of the male X chromosome was detected in these larvae using antibodies against acetylated H4K16 (data not shown). (C) Salivary gland polytene chromosomes from OreR male larvae. In all panels, the X chromosome is marked by an arrow.
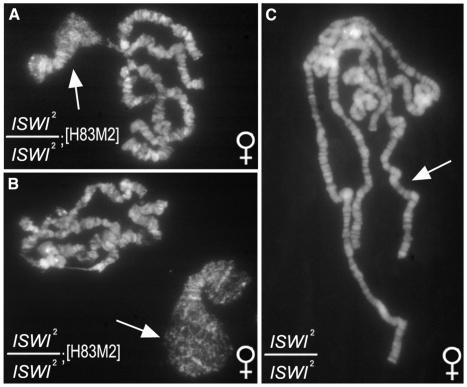
As an alternative approach for investigating whether the dosage compensation complex alters the ability of chromatin to be remodeled by ISWI in vivo, we examined the salivary gland polytene chromosomes of homozygous ISWI2 females expressing the MSL-2 protein. In wild-type females, the X chromosome is not acetylated on H4K16 due to the absence of the MSL-2 subunit of the dosage compensation complex (Bashaw and Baker, 1995). The acetylation of the female X chromosome on H4K16 can be artificially induced using a transgene (H83M2) that expresses MSL-2 in females under the control of the hsp83 promoter (Kelley et al., 1995). The expression of MSL-2 in female ISWI2 (but not wild-type) larvae causes the X chromosome to exhibit the bloated morphology that is typically seen only in ISWI mutant males (compare Figure 2A and C). In ∼30% of the salivary gland nuclei from these females, this phenotype is so extreme that the X chromosome is almost completely dispersed and detaches from the chromocenter (Figure 2B). Thus, the activity of the dosage compensation complex is sufficient for the altered appearance of the X chromosome in ISWI2 mutant larvae.
Fig. 2. The activity of the dosage compensation complex is sufficient for the X chromosome defects resulting from loss of ISWI function. (A) Salivary gland chromosomes from female ISWI2/ISWI2; [H83M2]/+ larvae. Note the bloated appearance of the X chromosome (marked by an arrow) resulting from the expression of the MSL-2 protein in ISWI2 females. An extreme example of this phenotype is shown in (B).The X chromosome was recognized in these individuals by staining with an antibody against acetylated H4K16 (data not shown). (C) Salivary gland polytene chromosomes from homozygous ISWI2 female larvae. Note that all chromosomes appear normal.
H4K16 acetylation antagonizes ISWI function in vivo
We next sought to determine whether the acetylation of H4K16, as opposed to some other activity of the dosage compensation complex, makes chromatin more sensitive to the loss of ISWI function. This was not a trivial concern since the dosage compensation complex physically interacts with the JIL-1 kinase, thus increasing the phosphorylation of serine 10 of histone H3 (H3S10) on the male X chromosome (Jin et al., 2000; Wang et al., 2001). The relative contributions of H4K16 acetylation and elevated H3S10 phosphorylation to the ISWI mutant phenotype could not be resolved simply by examining the chromosomes of mof ISWI double mutants, since MOF is required for the spreading of the dosage compensation complex along the X chromosome (Gu et al., 2000).
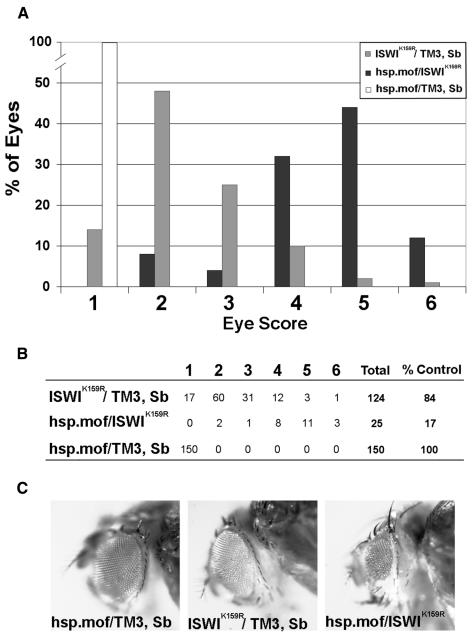
MOF does not require components of the dosage compensation complex or other proteins to acetylate H4K16 in vitro (Akhtar and Becker, 2000), and the expression of high levels of MOF leads to widespread acetylation of H4K16 on all salivary gland polytene chromosomes (data not shown). As an alternative approach for investigating a potential functional antagonism between MOF and ISWI, we examined whether the increased expression of MOF enhances phenotypes resulting from the partial loss of ISWI function. The expression of dominant-negative ISWI protein (ISWIK159R) in the eye-antennal imaginal disc leads to the development of adults with slightly smaller or rough eyes (Deuring et al., 2000; Papoulas et al., 2001). A transgene expressing Mof under the control of a heat-inducible promoter (Scott et al., 2000) strongly enhances these phenotypes (Figure 3), but not similar phenotypes resulting from the expression of a dominant-negative BRM protein, BRMK804R (Papoulas et al., 2001; data not shown). This strong genetic interaction is observed in both sexes, even though females lack functional dosage compensation complexes. Furthermore, JIL-1 mutations have no effect on eye phenotypes resulting from the expression of ISWIK159R in the eye-antennal imaginal disc (data not shown). These data suggest that the acetylation of H4K16, as opposed to some other activity of the dosage compensation complex, antagonizes ISWI function in vivo.
Fig. 3. Elevated MOF expression enhances phenotypes resulting from partial loss of ISWI function in the developing eye. (A) hsp.mof/TM3, Sb males were crossed to ey-Gal4, UAS-ISWIK159R/TM3, Sb females at 29°C and female progeny were scored for eye defects as follows: 1, wild-type eye; 2, roughness comprising <50% of normal eye area; 3, roughness comprising >50% of normal eye area; 4, eye rough and <50% reduced in size; 5, eye rough and >50% reduced in size; 6, eye absent. Note that the hsp.mof transgene dramatically enhances eye defects associated with ISWIK159R expression. No enhancement was observed in progeny raised at 18°C. (B) Table of data used to generate graph shown in (A). % Control refers to the number of progeny recovered relative to hsp.mof/TM3, Sb siblings. Note the decreased viability resulting from the expression of both MOF and ISWIK159R. (C) Representative eyes of hsp.mof/TM3, Sb (class 1), ISWIK159R/TM3, Sb (class 2) and hsp.mof/ISWIK159R (class 5).
H4K16 acetylation interferes with the interaction between ISWI and its nucleosomal substrate in vitro
How might histone acetylation render chromatin more sensitive to the loss of ISWI function? One possibility is that the acetylation of H4K16 counteracts chromatin compaction mediated by the ISWI ATPase. Luger et al. (1997) have proposed that H4K16 interacts with an acidic patch of the H2A–H2B dimer of an adjacent nucleosome. Acetylation of H4K16 might disrupt this interaction, creating less densely packaged chromatin. It is also possible that the acetylation of the H4 tail directly alters the ability of ISWI to remodel chromatin structure. We favored this possibility, since the H4 tail has been shown to be critical for the nucleosome spacing and other nucleosome-stimulated ATPase activities of ISWI in vitro (Clapier et al., 2001). The critical determinant of ISWI function within the H4 tail is a DNA-bound basic patch including amino acids R17H18R19 (Clapier et al., 2002). This region is also critical for the nucleosome sliding and ATPase activities of NURF (Hamiche et al., 2001). It is possible that acetylation of the adjacent lysine 16 modulates the structure of this patch and hence interferes with ISWI function.
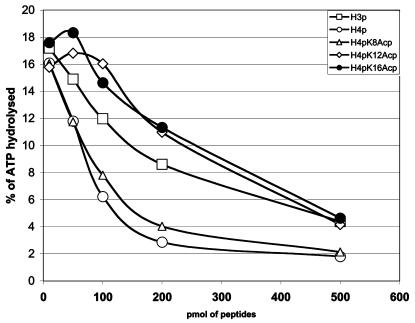
To determine whether the acetylation of the histone H4 tail directly affects the interaction of ISWI with its substrate, we tested the ability of synthetic, modified peptides corresponding to specific histone tails to inhibit the nucleosome-stimulated ATPase activity of ISWI in vitro. Isolated histone N-termini by themselves do not stimulate the ATPase of ISWI, but effectively inhibit its activity through competition with the nucleosomal substrate (Georgel et al., 1997; Corona et al., 1999). Consistent with its critical role in mediating interactions between ISWI and the nucleosome, peptides corresponding to the unacetylated histone H4 N-terminus inhibit ISWI ATPase activity 5-fold at a concentration of 200 pmol (Figure 4). At this concentration the corresponding H3 tail peptide is much less effective. Acetylation of the H4 peptide at lysine 16 reduces its competition properties to levels even below the H3 peptide (Figure 4). H4 tail peptides acetylated at lysine 8 or methylated at lysine 20 show competition properties very similar to the unmodified histone H4 tail, indicating that the interaction between ISWI and the H4 tail in vitro is weakened by site-specific acetylation and not by non-specific charge reduction (Figure 4; C. Clapier and P.B. Becker, unpublished data). Interestingly, acetylation at lysine 12 also abolishes the functional interaction of the H4 N-terminus with ISWI (Figure 4). The H4K12Ac isoform is present at many euchromatic sites of polytene chromosomes and is highly enriched in heterochromatin (Turner, 2000). This suggests that the ability of ISWI to sense the modification state of chromatin and to modulate its activity accordingly may be of more general significance.
Fig. 4. The ATPase activity of ISWI is modulated by site-specific acetylation of the H4 N-terminal tail. The ATPase of ISWI is stimulated strongly by the presence of nucleosomes. Synthetic peptides corresponding to the H3 and H4 N-terminus were added to ATPase reactions to determine their potential to compete with the nucleosomal substrate. The competition properties of peptides acetylated at H4K16 (filled circles), H4K12 (open diamonds), H4K8 (open triangles) are best compared to wild-type H4 (open circles) and H3 (open squares) N-termini in reactions containing 200 pmol of peptide. The graph reflects the average of two independent experiments with similar competition profiles. The result has been qualitatively reproduced under a variety of conditions, albeit with different absolute values.
In support of the histone code hypothesis (Turner et al., 1992; Jenuwein and Allis, 2001), our data suggest that the acetylation of specific residues of histone H4, including lysine 16 and lysine 12, may directly counteract repressive chromatin compaction mediated by the ISWI ATPase. Functional synergism between energy-dependent nucleosome remodeling factors and histone acetyltransferases has been documented previously (Cosma et al., 1999; Wu and Grunstein, 2000; Gregory et al., 2001). In contrast, our findings demonstrate that the activity of a chromatin remodeling ATPase can be regulated directly by the site-specific acetylation of its chromatin substrate.
Speculation
Our data suggest that the degree of compaction of chromosomal domains may be governed by a complex interplay between ATP-dependent chromatin remodeling factors and targeted site-specific histone acetylation. Two ISWI-containing complexes, CHRAC and ACF, promote the formation of chromatin with regularly spaced nucleosomes in vitro (Längst and Becker, 2001). Regular nucleosomal arrays are a hallmark of folded chromatin in vitro (Blank and Becker, 1995) and of heterochromatin in vivo (Wallrath and Elgin, 1995). ISWI may therefore contribute to the assembly and maintenance of repressive higher-order chromatin structure. Indeed, a growing body of evidence suggests that ISWI is involved in transcriptional repression in both yeast and flies (Deuring et al., 2000; Goldmark et al., 2000). By reducing the ability of ISWI to promote the formation of regular nucleosomal arrays, MOF and other acetyltransferases may derepress genes silenced by higher-order chromatin structure. In the case of MOF, this could alter the expression of large numbers of genes within broad chromosomal domains. Although it would be difficult to detect cytologically, we suspect that the modification of the H4 tail in the vicinity of specific genes may cause local changes in chromatin structure and transcription via a similar mechanism.
METHODS
Fly strains and genetic crosses. Flies were raised at 25°C using standard agar/cornmeal/molasses media. All mutations are described in FlyBase (http://www.flybase.org). Homozygous mle1 ISWI2 male larvae were generated by crossing w/Y; mle1 ISWI2/T(2:3) CyO;TM6B Tb males to y w; mle1 ISWI2/CyO y+ females and recognized by their yellow mouthhooks and the absence of the dominant Tb mutation. ISWI2/ISWI2; [H83M2] female larvae were generated by crossing w/Y; ISWI2; [H83M2]/T(2:3) CyO;TM6B Tb males to y w; ISWI2/T(2:3) CyO;TM6B Tb virgin females and recognized by their darkly pigmented mouthhooks and the lack of the dominant Tb mutation. To examine the effect of increased MOF expression on the rough-eye phenotype caused by partial loss of ISWI function, y w; P[w+, hsp70:mof] M38/TM3, Sb males were crossed to w, P[w+, ey-Gal4], P[w+, UASGALhsp70:ISWIK159R] 11-4/TM3, Sb females at 29°C. Progeny were recognized based on their bristle morphology and eye color. Eye defects were scored using a 1 to 6 scale, which permits the unbiased representation of the range of eye defects present in each progeny class (Papoulas et al., 2001). No genetic interaction between hsp.mof and ISWIK159R was observed when the above cross was conducted at 18°C.
Polytene chromosomes squashes. Late third-instar larvae were grown at 18°C, identified using the larval markers y and Tb as described above, and allowed to age for up to 1 week. Salivary glands were dissected in 0.7% NaCl and fixed for 15 min in 45% acetic acid. Chromosome spreads were prepared and stained with DAPI.
ATPase competition assays. ATPase assays were performed at 26°C with 150 µM ATP and 0.7 µCi [γ-32P]ATP under ISWI remodeling conditions, as described previously (Corona et al., 1999). Unhydrolyzed ATP and free phosphate were separated after 1 h by thin-layer chromatography using TLC PEI-Cellulose foils (Merck). Spots were quantified by FluoroImager using Tina software. Peptide competition assays contained 300 ng of DNA, 250 ng of histones, 35 ng of yNAP-1, 1 pmol of ISWI and were incubated in the presence of 10, 50, 100, 200 and 500 pmol of the indicated peptide (Upstate Biotech).
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to J. Armstrong and G. Hartzog for comments and critical reading of the manuscript, and R. Kelley, M. Kuroda, J. Lucchesi and M.J. Scott for providing stocks and advice. Antibody against H4K16Ac was generously provided by Dr B. Turner. This work was supported by grants from the NIH (GM49883) to J.W.T. and the Deutsche Forschungsgemeinschaft (SFB 594) to P.B.B. D.F.V.C. was supported by EMBO and HFSP postdoctoral fellowships. C.R.C. is a fellow of the EMBL international PhD program.
REFERENCES
- Akhtar A. and Becker, P.B. (2000) Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell, 5, 367–375. [DOI] [PubMed] [Google Scholar]
- Bashaw G.J. and Baker, B.S. (1995) The msl-2 dosage compensation gene of Drosophila encodes a putative DNA-binding protein whose expression is sex specifically regulated by Sex-lethal. Development, 121, 3245–3258. [DOI] [PubMed] [Google Scholar]
- Blank T.A. and Becker, P.B. (1995) Electrostatic mechanism of nucleosome spacing. J. Mol. Biol., 252, 305–313. [DOI] [PubMed] [Google Scholar]
- Clapier C.R., Längst, G., Corona, D.F., Becker, P.B. and Nightingale, K.P. (2001) Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol. Cell. Biol., 21, 875–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier C.R., Nightingale, K.P. and Becker, P.B. (2002) A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI. Nucleic Acids Res., 30, 649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona D.F., Längst, G., Clapier, C.R., Bonte, E.J., Ferrari, S., Tamkun, J.W. and Becker, P.B. (1999) ISWI is an ATP-dependent nucleosome remodeling factor. Mol. Cell, 3, 239–245. [DOI] [PubMed] [Google Scholar]
- Cosma M.P., Tanaka, T. and Nasmyth, K. (1999) Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle and developmentally regulated promoter. Cell, 97, 299–311. [DOI] [PubMed] [Google Scholar]
- Deuring R. et al. (2000) The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell, 5, 355–365. [DOI] [PubMed] [Google Scholar]
- Fyodorov D.V. and Kadonaga, J.T. (2001) The many faces of chromatin remodeling: SWItching beyond transcription. Cell, 106, 523–525. [DOI] [PubMed] [Google Scholar]
- Georgel P.T., Tsukiyama, T. and Wu, C. (1997) Role of histone tails in nucleosome remodeling by Drosophila NURF. EMBO J., 16, 4717–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmark J.P., Fazzio, T.G., Estep, P.W., Church, G.M. and Tsukiyama, T. (2000) The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell, 103, 423–433. [DOI] [PubMed] [Google Scholar]
- Gregory P.D., Wagner, K. and Horz, W. (2001) Histone acetylation and chromatin remodeling. Exp. Cell. Res., 265, 195–202. [DOI] [PubMed] [Google Scholar]
- Gu W., Wei, X., Pannuti, A. and Lucchesi, J.C. (2000) Targeting the chromatin-remodeling MSL complex of Drosophila to its sites of action on the X chromosome requires both acetyl transferase and ATPase activities. EMBO J., 19, 5202–5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiche A., Kang, J.G., Dennis, C., Xiao, H. and Wu, C. (2001) Histone tails modulate nucleosome mobility and regulate ATP-dependent nucleosome sliding by NURF. Proc. Natl Acad. Sci. USA, 98, 14316–14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J.C., Tse, C. and Wolffe, A.P. (1998) Structure and function of the core histone N-termini: more than meets the eye. Biochemistry, 37, 17637–17641. [DOI] [PubMed] [Google Scholar]
- Jenuwein T. and Allis, C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Jin Y., Wang, Y., Johansen, J. and Johansen, K.M. (2000) JIL-1, a chromosomal kinase implicated in regulation of chromatin structure, associates with the male specific lethal (MSL) dosage compensation complex. J. Cell Biol., 149, 1005–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R.L., Solovyeva, I., Lyman, L.M., Richman, R., Solovyev, V. and Kuroda, M.I. (1995) Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell, 81, 867–877. [DOI] [PubMed] [Google Scholar]
- Kuroda M.I., Kernan, M.J., Kreber, R., Ganetzky, B. and Baker, B.S. (1991) The maleless protein associates with the X chromosome to regulate dosage compensation in Drosophila. Cell, 66, 935–947. [DOI] [PubMed] [Google Scholar]
- Längst G. and Becker, P.B. (2001) Nucleosome mobilization and positioning by ISWI-containing chromatin remodeling factors. J. Cell Sci., 114, 2561–2568. [DOI] [PubMed] [Google Scholar]
- Luger K., Mader, A.W., Richmond, R.K., Sargent, D.F. and Richmond, T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- Papoulas O., Daubresse, G., Armstrong, J.A., Jin, J., Scott, M.P. and Tamkun, J.W. (2001) The HMG-domain protein BAP111 is important for the function of the BRM chromatin-remodeling complex in vivo. Proc. Natl Acad. Sci. USA, 98, 5728–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y. and Kuroda, M.I. (2001) Epigenetic aspects of X-chromosome dosage compensation. Science, 293, 1083–1085. [DOI] [PubMed] [Google Scholar]
- Scott M.J., Pan, L.L., Cleland, S.B., Knox, A.L. and Heinrich, J. (2000) MSL1 plays a central role in assembly of the MSL complex, essential for dosage compensation in Drosophila. EMBO J., 19, 144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.R., Allis, C.D. and Lucchesi, J.C. (2001) Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J. Biol. Chem., 276, 31483–31486. [DOI] [PubMed] [Google Scholar]
- Tse C., Sera, T., Wolffe, A.P. and Hansen, J.C. (1998) Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol., 18, 4629–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B.M. (2000) Histone acetylation and an epigenetic code. BioEssays, 22, 836–845. [DOI] [PubMed] [Google Scholar]
- Turner B.M., Birley, A.J. and Lavender, J. (1992) Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell, 69, 375–384. [DOI] [PubMed] [Google Scholar]
- Wallrath L.L. and Elgin, S.C. (1995) Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev., 9, 1263–1277. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang, W., Jin, Y., Johansen, J. and Johansen, K.M. (2001) The JIL-1 tandem kinase mediates histone H3 phosphorylation and is required for maintenance of chromatin structure in Drosophila. Cell, 105, 433–443. [DOI] [PubMed] [Google Scholar]
- Wu C., Tsukiyama, T., Gdula, D., Georgel, P., Martinez-Balbas, M., Mizuguchi, G., Ossipow, V., Sandaltzopoulos, R. and Wang, H.M. (1998) ATP-dependent remodeling of chromatin. Cold Spring Harb. Symp. Quant. Biol., 63, 525–534. [DOI] [PubMed] [Google Scholar]
- Wu J. and Grunstein, M. (2000) 25 years after the nucleosome model: chromatin modifications. Trends Biochem. Sci., 25, 619–623. [DOI] [PubMed] [Google Scholar]