Abstract
A variety of nuclear localization signals (NLSs) are experimentally known although only one motif was available for database searches through PROSITE. We initially collected a set of 91 experimentally verified NLSs from the literature. Through iterated ‘in silico mutagenesis’ we then extended the set to 214 potential NLSs. This final set matched in 43% of all known nuclear proteins and in no known non-nuclear protein. We estimated that >17% of all eukaryotic proteins may be imported into the nucleus. Finally, we found an overlap between the NLS and DNA-binding region for 90% of the proteins for which both the NLS and DNA-binding regions were known. Thus, evolution seemed to have used part of the existing DNA-binding mechanism when compartmentalizing DNA-binding proteins into the nucleus. However, only 56 of our 214 NLS motifs overlapped with DNA-binding regions. These 56 NLSs enabled a de novo prediction of partial DNA-binding regions for ∼800 proteins in human, fly, worm and yeast.
INTRODUCTION
Simplification of nuclear import
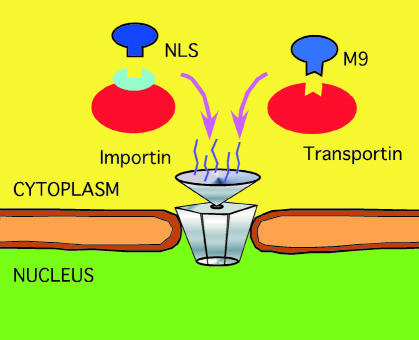
A nuclear localization signal (NLS) is a short stretch of amino acids that mediates the transport of nuclear proteins into the nucleus (Figure 1). NLS motifs play a key role in this mechanism; (i) typically, deletion of the NLS disrupts nuclear import; and (ii) frequently, a non-nuclear protein will be imported into the nucleus if fused to an NLS. Both facts have been used routinely to unravel NLS motifs experimentally (Tinland et al., 1992; Moede et al., 1999).
Fig. 1. Simplified scheme for nuclear import. Upon synthesis of nuclear proteins in the cytoplasm, e.g. the family of importins or transportins bind to the NLS. The complex importin/NLS protein (or transportin/protein) is then actively transported into the nucleus through nuclear pores involving the Ran GTPase cycle. Currently, this is the only known mechanism for nuclear import (Mattaj and Englmeier, 1998; Weis, 1998).
Variety of NLS motifs
Do experimentally known NLS motifs have a consensus? Positively charged residues are abundant in NLSs, in general, since some of these positive residues bind to e.g. importins (Conti et al., 1998). Mutating positive charges is often the simplest way to disrupt nuclear import; however, there are glycine-rich NLS motifs with few positive charges (Bonifaci et al., 1997). The best described experimentally are monopartite and bipartite motifs (Boulikas, 1993). Typically, the monopartite motif is characterized by a cluster of basic residues preceded by a helix-breaking residue. Similarly, the bipartite motif consists of two clusters of basic residues separated by 9–12 residues. However, not all experimentally known NLSs comply with the above ‘rules’ (Hsieh et al., 1998; Truant and Cullen, 1999; Irie et al., 2000). Furthermore, many non-nuclear proteins match such simplified ‘consensus rules’.
Finding an NLS in silico?
A wealth of experimental data about NLSs has been accumulated. How can you find a known NLS in your protein? If a standard database search reveals a ‘significant similarity’ between your protein and a protein of experimentally known and annotated NLS, you can infer the NLS from the homologue. If not, can you find most experimental motifs in PROSITE (Hofmann et al., 1999)? The negative answer was the starting point for this work: build an ‘expert database’ of experimentally known NLSs. Another motivation was the observation that NLSs defined by experiments often appeared too specific. Theoretical generalizations for NLSs have been suggested: ‘NLS cores are hexapeptides with at least four basic residue and neither acidic nor bulky residues’ (Boulikas, 1994); however, this motif matches only few nuclear and many non-nuclear proteins.
Do homologues have similar NLSs?
Two naturally evolved proteins with >30% identical residues have similar three-dimensional structures (Rost, 1999). The sequence similarity required to infer function is much higher (Devos and Valencia, 2000). Structural thresholds depend on alignment length, e.g. two identical 11-residue peptides can adopt different structures (Minor and Kim, 1996). NLSs are short stretches of residues. Thus, at which levels of sequence similarity can we infer that two proteins will have a similar NLS? A lack of data prevented us from thoroughly answering this question; however, we found some upper boundaries.
Here we present an extended expert database of experimentally known and potential NLS motifs. We evaluate the validity of the set by a rigorous test against known nuclear and non-nuclear proteins. Our method comprised two steps: (i) data collection—collect experimental NLS motifs from literature, extend motifs through close homologues; (ii) generalization—refine motifs found by shortening (too specific) or lengthening (not specific enough), and test new motifs conceptually similar to known motifs found in many families of nuclear proteins. The crucial component of both steps was to accept motifs if not found in non-nuclear proteins.
RESULTS AND DISCUSSION
Improved accuracy and coverage of the NLS database
Inferring NLSs based on very limited sequence
We found ∼30 protein pairs with >80% sequence identity and different annotations (nuclear and cytoplasmic) in our subset of SWISS-PROT (see Methods, e.g. the nuclear elongation factor 1α2 in mouse and the cytoplasmic transcription elongation factor 1α in Zebra fish had 91% identity over 460 residues). At 50–65% sequence identity, we found many pairs aligned over a substantial length, and annotated in different localizations (e.g. 60% nuclear and extracellular: fbrl_rat/ndl_drome; 63% nuclear and mitochondrial: hmgt_mouse/mtt1_human; 51% nuclear and chloroplast: grp1_sinal/ro30_nicpl). Thus, we can infer that a protein is nuclear only if it is almost identical to a known nuclear protein. However, for all the experimental NLSs we extracted we succeeded to infer correctly the nuclear localization knowing the NLS. Note, this failed for all NLSs from previously published theoretical generalizations (Boulikas, 1994).
Raising coverage from 9 to 43%
Before we started, we had three ways to find an NLS in protein A. (i) We could memorize NLSs published and visually detect one (or several) of these in A. Obviously, this requires time and ample expertise. Furthermore, all experimental NLSs covered only 10% of the known nuclear proteins (too specific, Table I). (ii) We could automatically detect the NLS in PROSITE (Hofmann et al., 1999); however, this covered only ∼3% of all known proteins, and was not always correct (Table I). (iii) We could find a significant level of sequence similarity to a protein for which the NLS was annotated in SWISS-PROT (Bairoch and Apweiler, 1999). This covered ∼9% of all known nuclear proteins (Table I). Furthermore, standard database searches starting with the proteins known to be nuclear yielded <25% of the known nuclear families at a generous BLAST cut-off of 10–3. In contrast, our final expert set of potential NLSs matched 43% of all nuclear proteins without any false positive (Table I).
Table I. Accuracy and coverage of NLS motifs.
| Seta | N NLSb | Nprot nucc | Nfam nucd | Accuracy (%)e | Coverage (%)f |
|---|---|---|---|---|---|
| PROSITE | 1 | 96 | 31 | 90 | 3 |
| SWISS-PROT | 322 | 290 | n.a. | 9 | |
| NLS-lit cleaned | 91 | 309 | 35 | 100 | 10 |
| NLS-lit consensus | 91 | 537 | 35 | 100 | 17 |
| PredictNLS_DB | 214 | 1354 | 186 | 100 | 43 |
aPROSITE, motifs annotated in the PROSITE database of functional motifs (Hofmann et al., 1999); SWISS-PROT, subset of SWISS-PROT database (Bairoch and Apweiler, 1999) annotating NLSs (note that a few proteins had more than one NLS annotated); NLS-lit cleaned, subset of motifs from literature with 100% accuracy; NLS-lit consensus, motifs refined by consensus of close homologues; PredictNLS_DB, final data set after in silico mutagenesis.
bNumber of NLS motifs in set.
cNumber of proteins matching any of the NLSs and known to be nuclear.
dNumber of unique protein families matching any of the NLSs and known to be nuclear (Methods, data set).
ePercentage of nuclear proteins in set of proteins matching any of the NLSs.
fPercentage of known nuclear proteins (Methods, data set) matching any of the motifs in the set (total number of known nuclear proteins 3142).
Limitations and error margin of method
Proteins often contain more than one NLS. Thus, our method might fail to propose the functional NLS. Furthermore, a few of our potential NLSs might just be motifs common to nuclear proteins such as DNA-binding motifs. Examples for motifs common to nuclear proteins we found with the motif-detection programs PRATT (Jonassen, 1997) and the Gibbs-sampler (Hertz and Stormo, 1999) were long repeats of glycines, glutamic acids and glutamine, and zinc-finger type II motifs. Most importantly, we found possible NLSs in 54 Escherichia coli proteins, only 26 of which could be explained by DNA-binding motifs. Assuming that the remaining 28 comprised errors, we estimated the error margin of our method as <1% (28/4286).
Lessons learned from ‘in silico mutagenesis’
(i) As expected, amino acids with similar physico-chemical properties could often be exchanged (leucine/isoleucine). (ii) Unexpectedly, positive amino acids (arginine and lysine) often could not be inter-changed. (iii) None of the NLSs previously proposed by theory passed our criterion of 100% accuracy. (iv) We found that proteins may have similar structure and function and yet may utilize different NLSs. (v) Very peculiar motifs we added to our final list were (a) GGGxGGGxxSSS, e.g. found by generalization of the M9 domain motif (human RNP A1 protein), and (b) SGxxG{3,}?xG{3,}?xG{3,}?S (any number of more than three consecutive Gs), e.g. found in the transcriptional activator protein of mouse.
More than 17% of eukaryotic proteins are nuclear
Extrapolating from the SWISS-PROT coverage, we could estimate a lower limit (SWISS-PROT biased towards known NLSs) for the fraction of nuclear proteins in eukaryotes. We detected potential NLSs in 4187 proteins from human, fly, yeast and worm (Table II). Thus, >17% of all eukaryotic proteins appeared to be imported into the nucleus. All entire genomes investigated had a similar percentage of nuclear proteins, although they clearly differed in the content of extracellular, helical membrane and coiled-coil proteins (J. Liu and B. Rost, submitted).
Table II. Nuclear proteins in genomes.
| Genomea | No. of ORFsb | No. of proteins with NLSc | Estimated nuclear content (%)d |
|---|---|---|---|
| Human | 13 933 | 1311 | >22e |
| Drosophila | 14 219 | 1256 | >21 |
| C. elegans | 16 232 | 1141 | >17 |
| Yeast | 6307 | 479 | >18 |
| E. coli | 4286 | 54 | 0 |
aWe obtained the incomplete set of human sequences from the latest releases of SWISS-PROT and TrEMBL (Bairoch and Apweiler, 1999), and the complete lists of proteins for the genomes of Drosophila melangoster, Caenorhabditis elegans, Saccharomyces cerevisiae and Escherichia coli from the respective web sites (Liu and Rost, 2000).
bNumber of open reading frames (proteins) in entire genome.
cNumber of proteins for which the set PredictNLS_DB found an NLS in that genome.
dGiven that our data set of NLS covers ∼43% of all known nuclear proteins (Table I), we estimated the content of nuclear proteins in the entire genome based on the number of proteins for which we found NLSs; supposedly, these estimates provided a lower boundary (Results).
eSince our current data set for human contains only ∼10% of all the proteins expected in the human genome, and since most of these are strongly biased by ‘experimental focus’, we could not estimate whether or not the coverage for human will be similar for the remaining 90% of all human proteins.
Specific NLS motifs used to bind DNA
20% of NLS motifs co-localized with the DNA-binding region
Too few complexes of DNA–protein were solved by X-ray crystallography to conclude that the NLS and DNA-binding motifs were co-localized. Instead, we used 1115 proteins with SWISS-PROT annotations about DNA-binding regions; 736 of these had a known NLS (66%), and for 664 the NLS overlapped with the DNA-binding region. Thus, for 90% of all proteins, for which we knew both the NLS and the DNA-binding region, both motifs overlapped. For 10% of the proteins, we could establish that the NLS and the DNA-binding region did not overlap. Furthermore, the NLS motifs co-localizing with DNA binding constituted about one fourth (56 of 214) of our final NLS set. The very observation that DNA binding and the NLS overlap frequently was not novel. In fact, based on a 20 times larger data set, we verified the original results from LaCasse and Lefebvre (1995). We also corrected their estimate upwards: where they found that 67% of the DNA-binding regions co-localized with the NLS, we found this number to be 90%. In contrast, our results suggested that most NLS motifs were not used to bind DNA.
RNA-binding regions typically not overlapping with NL
Contrary to LaCasse and Lefebvre (1995), we found that only 33 of the 99 regions annotated in SWISS-PROT as RNA binding in nuclear proteins overlapped with an NLS. The difference resulted largely from their definition of ‘RNA-binding region’ as the entire region between two consecutive RNA-binding sites. In contrast, SWISS-PROT—correctly—annotated only regions experimentally shown to bind RNA.
Structures for DNA binding and NLS
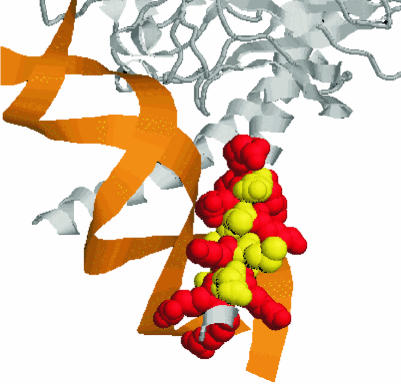
For 20 of the investigated 22 proteins of known structure, we found the known NLS to overlap with the DNA-binding region (Figure 2). The only exceptions were rap1 from yeast and the segmentation protein fushi tarazu from fly (PDB codes: 1ign and 1ftz, respectively) for which we did not find the respective NLS in the known DNA-binding regions. However, these two exceptions did not have any of the 56 NLSs found to co-localize with DNA binding. As expected, we found all NLSs on the protein surface.
Fig. 2. NLS motif also used for DNA binding. Zoom into the interface between DNA and P55-C-fos proto-oncogene protein [note, the other parts of the amazing crystal structure of the complex with PDB code 1a02 (Chen et al., 1998) are not shown]. The coloured region corresponds to the residues RRERNKMAAAKSRNRRR. In fact, this motif is also contained in our data set of potential NLS motifs. Colouring scheme: basic residues shown in red, others in yellow. Graph created with RASMOL (Sayle and Milner-White, 1995).
Speculation about evolution
The co-localization of NLSs and DNA-binding regions suggested that DNA and shuttle proteins like importins and transportins utilized similar binding residues. Protein–DNA interactions may have preceded the ‘invention’ of a nucleus used by eukaryotes to compartmentalize all processes involving DNA. How are proteins to import into this compartment recognized? Common to many nuclear proteins are DNA-binding regions. Thus, it seems likely to utilize fragments of these regions to manage nuclear import. Consequently, we expect to find importin-like proteins and NLS-like sequences in prokaryotic organisms. In fact, we did find such motifs in E. coli protein (Table II); many of these appeared to be involved in DNA binding. Obviously, evolution created other NLS motifs (only 56 of 214 of the NLSs co-localized with DNA binding) over time. NLSs are often also used to target nuclear export (Mattaj and Englmeier, 1998). Could we thus perceive the co-localization of DNA binding and NLS as an elegant mechanism to also prevent export for some of the proteins? And did evolution in fact have to create novel NLS motifs to manage export rather than import? Our data did not falsify such speculations.
De novo prediction of DNA-binding regions
Searching with the NLS/DNA motifs, we predicted a relatively small number of DNA-binding proteins in eukaryotes, ranging from 419 in human to 67 in yeast (Table III). However, this was 2–9 times higher than the number of proteins in the respective organism for which SWISS-PROT annotated DNA binding or for which we could infer DNA binding through homology (Table III). Thus, we predicted a new potential DNA-binding region for >800 proteins in all four eukaryotes.
Table III. DNA-binding regions in genomes.
| Genomea | Nprotb | Nprot bind-DNA predictedc | Nprot bind-DNA knownd |
|---|---|---|---|
| Human | 13 933 | 419 | 141 |
| Drosophila | 14 219 | 300 | 37 |
| C. elegans | 16 232 | 251 | 10 |
| Yeast | 6307 | 67 | 10 |
| E. coli | 4286 | 13 | 3 |
aSee Table II.
bTotal number of proteins in entire genome.
cNumber of proteins for which we predict DNA binding using NLS motifs.
dNumber of proteins for which DNA binding is annotated, or can be inferred by homology to a protein for which binding is annotated (note, family relations taken from Liu and Rost, 2000).
Availability of data set and program
Our data set and method are available at: http://cubic.bioc.columbia.edu/predictNLS. The program also allows experimentalists to test accuracy and coverage for new NLS motifs they may find or suspect. This feature has already helped to unravel experimentally a novel NLS in the hairless protein (K. Djabali and A. Christiano, submitted). Finally, we added a form enabling experimentalists to add new NLSs. Every NLS added may help to speed up the next experiment!
METHODS
Collecting the initial set of NLS data from the literature. We searched ∼250 papers and reviews for experimentally determined NLSs. Our main criteria for ‘accepting’ NLSs were that the signal was proven sufficient to mediate the nuclear transport of a non-nuclear protein to the nucleus and that deleting the NLS prevented the nuclear import. Technically, some motifs taken at this step comprised simple protein sequences, others regular expressions.
Sets of nuclear and non-nuclear proteins. We retrieved all proteins in SWISS-PROT release 38.0 (Bairoch and Apweiler, 1999) with annotations of subcellular localization (ignoring PUTATIVE, POTENTIAL, BY SIMILARITY). Finally, we sorted all remaining proteins into two sets: (i) nuclear proteins (true positives, 3142 proteins) and (ii) non-nuclear proteins (true negatives, 5910 proteins). Note, the set of nuclear proteins corresponded to 618 structural families (Rost, 1999).
Extending experimental NLSs through homology. For each experimental NLS protein, we found homologues in SWISS-PROT with PredictProtein (Rost, 1996). For pairs with >80% identical residues, we extended the initial set of experimental NLSs by adding the sequence corresponding to the experimental NLS in the homologues.
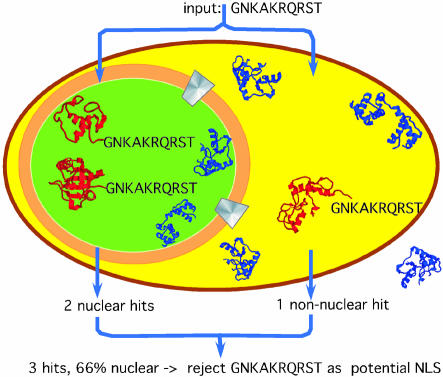
Testing experimental NLSs. We tested the validity of all motifs found in the literature and their homologues by monitoring the matches of any motif in the sets of nuclear and non-nuclear proteins (Figure 3). The rationale was to find all NLSs that matched exclusively in nuclear proteins.
Fig. 3. Scheme for the concept of ‘in silico mutagenesis’. We started the search with the hypothetical motif GNKAKRQRST. We searched the data sets of proteins known to be nuclear and proteins known to be non-nuclear for the presence of this motif. In this particular example, two nuclear and one non-nuclear protein matched. Requiring 100% accuracy for all motifs, we did not include GNKAKRQRST into our data set of potential motifs. Note, the particular example was one of many failed attempts to generalize an experimental NLS.
In silico mutagenesis. Given the list of sustained NLS motifs (experimental and homologues), we increased the number of potential NLSs by ‘in silico mutagenesis’: we changed or removed some residues in the given motifs and monitored the resulting true (nuclear) and false (non-nuclear) matches. Obviously, allowing alternative residues at particular positions increased the number of nuclear proteins found. However, often this also increased the number of matching non-nuclear proteins. For example, the experimentally determined motif GKKRSKA was present in two nuclear proteins. We could infer that the amino acid type at the positions of serine (S) and alanine (A) was not crucial for the NLS motif since GKKRxK found 11 nuclear proteins. For example, KKRxK matched 105 proteins, only 69% of which were nuclear. Thus, we rejected this generalization. In general, while trying to increase our coverage by our extended NLS list, we dropped any NLS present in any non-nuclear protein, i.e. 100% accuracy. Furthermore, we required the motif to be present in at least two distinct protein families. We tried all possible generalizations for the NLS motifs in our initial set through ‘educated-guess trial-and-error’. Finally, we compiled the coverage, i.e. the fraction of the known nuclear proteins correctly detected by our final expert database of NLS motifs.
NLS and DNA-binding regions. We explored two ways of testing whether or not NLS motifs overlapped with known DNA-binding sites. First, we looked at proteins for which the NLS and the three-dimensional structures are experimentally known. Towards this end, we investigated 22 examples of proteins of known structure [PDB codes: 1a02, 1an2, 1an4, 1akh, 1au7, 1b8i, 1cdw, 1fos, 1hlo, 1hry, 1hwt, 1lat, 2lef, 1mdy, 1nk2, 1nk3, 1oct, 1pdn, 1pue, 1tgh; 1ftz, 1ign (Berman et al., 2000)]. Secondly, we compared the DNA-binding regions annotated in SWISS-PROT with the NLS matching in our extended data set (1115 proteins in total).
Supplementary data. Supplementary data to this paper (an appendix of experimentally verified NLS motifs) are available in Embo reports Journal Online.
Supplementary Material
Acknowledgments
ACKNOWLEDGEMENTS
Thanks to Jinfeng Liu (Columbia University) for computer assistance and collection of the genome data sets; to Barry Honig for his valuable comments on DNA binding, to Amos Bairoch (SIB, Geneva), Rolf Apweiler (EBI, Hinxton) and their crews for maintaining the excellent databases SWISS-PROT and TrEMBL. Last but not least, thanks to all those who enabled this analysis by depositing experimental information about NLSs.
REFERENCES
- Bairoch A. and Apweiler, R. (1999) The SWISS-PROT protein sequence data bank and its supplement TrEMBL in 1999. Nucleic Acids Res., 27, 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N. and Bourne, P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifaci N., Moroianu, J., Radu, A. and Blobel, G. (1997) Karyopherin β2 mediates nuclear import of a mRNA binding protein. Proc. Natl Acad. Sci. USA, 94, 5055–5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulikas T. (1993) Nuclear localization signals (NLS). Crit. Rev. Eukaryot. Gene Expr., 3, 193–227. [PubMed] [Google Scholar]
- Boulikas T. (1994) Putative nuclear localization signals (NLS) in protein transcription factors. J. Cell. Biochem., 55, 32–58. [DOI] [PubMed] [Google Scholar]
- Chen L., Glover, J.N., Hogan, P.G., Rao, A. and Harrison, S.C. (1998) Structure of the DNA-binding domains from nfat, fos and jun bound specifically to DNA. Nature, 392, 42–48. [DOI] [PubMed] [Google Scholar]
- Conti E., Uy, M., Leighton, L., Blobel, G. and Kuriyan, J. (1998) Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell, 94, 193–204. [DOI] [PubMed] [Google Scholar]
- Devos D. and Valencia, A. (2000) Practical limits of function prediction. Proteins, 41, 98–107. [PubMed] [Google Scholar]
- Hertz G.Z. and Stormo, G.D. (1999) Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics, 15, 563–577. [DOI] [PubMed] [Google Scholar]
- Hofmann K., Bucher, P., Falquet, L. and Bairoch, A. (1999) The PROSITE database, its status in 1999. Nucleic Acids Res., 27, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J.C., Shimizu, Y., Minoshima, S., Shimizu, N., Haussler, C.A., Jurutka, P.W. and Haussler, M.R. (1998) Novel nuclear localization signal between the two DNA-binding zinc fingers in the human vitamin D receptor. J. Cell. Biochem., 70, 94–109. [PubMed] [Google Scholar]
- Irie Y., Yamagata, K., Gan, Y., Miyamoto, K., Do, E., Kuo, C.H., Taira, E. and Miki, N. (2000) Molecular cloning and characterization of Amida, a novel protein which interacts with a neuron-specific immediate early gene product arc, contains novel nuclear localization signals, and causes cell death in cultured cells. J. Biol. Chem., 275, 2647–2653. [DOI] [PubMed] [Google Scholar]
- Jonassen I. (1997) Efficient discovery of conserved patterns using a pattern graph. Comp. Appl. Biol. Sci., 13, 509–522. [DOI] [PubMed] [Google Scholar]
- LaCasse E.C. and Lefebvre, Y.A. (1995) Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic acid-binding proteins. Nucleic Acids Res., 23, 1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. and Rost, B. (2000) Analysing all proteins in entire genomes. CUBIC, Columbia University, Department of Biochemistry and Molecular Biophysics, http://cubic.bioc.columbia.edu/genomes
- Mattaj I.W. and Englmeier, L. (1998) Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem., 67, 265–306. [DOI] [PubMed] [Google Scholar]
- Minor D.L.J. and Kim, P.S. (1996) Context-dependent secondary structure formation of a designed protein sequence. Nature, 380, 730–734. [DOI] [PubMed] [Google Scholar]
- Moede T., Leibiger, B., Pour, H.G., Berggren, P. and Leibiger, I.B. (1999) Identification of a nuclear localization signal, RRMKWKK, in the homeodomain transcription factor PDX-1. FEBS Lett., 461, 229–234. [DOI] [PubMed] [Google Scholar]
- Rost B. (1996) PHD: predicting one-dimensional protein structure by profile based neural networks. Methods Enzymol., 266, 525–539. [DOI] [PubMed] [Google Scholar]
- Rost B. (1999) Twilight zone of protein sequence alignments. Protein Eng., 12, 85–94. [DOI] [PubMed] [Google Scholar]
- Sayle R.A. and Milner-White, E.J. (1995) RASMOL: biomolecular graphics for all. Trends Biochem. Sci., 20, 37. [DOI] [PubMed] [Google Scholar]
- Tinland B., Koukolikova-Nicola, Z., Hall, M.N. and Hohn, B. (1992) The T-DNA-linked VirD2 protein contains two distinct functional nuclear localization signals. Proc. Natl Acad. Sci. USA, 89, 7442–7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truant R. and Cullen, B.R. (1999) The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin β-dependent nuclear localization signals. Mol. Cell. Biol., 19, 1210–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K. (1998) Importins and exportins: how to get in and out of the nucleus [published erratum appears in Trends Biochem Sci., 1998, 23, 235]. Trends Biochem Sci., 23, 185–189. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.