Abstract
The rise in the frequency of fungal infections and the increased resistance noted to the widely employed azole antifungals make the development of new antifungals imperative for human health. The sterol biosynthetic pathway has been exploited for the development of several antifungal agents (allylamines, morpholines, azoles), but additional potential sites for antifungal agent development are yet to be fully investigated. The sterol methyltransferase gene (ERG6) catalyzes a biosynthetic step not found in humans and has been shown to result in several compromised phenotypes, most notably markedly increased permeability, when disrupted in Saccharomyces cerevisiae. The Candida albicans ERG6 gene was isolated by complementation of a S. cerevisiae erg6 mutant by using a C. albicans genomic library. Sequencing of the Candida ERG6 gene revealed high homology with the Saccharomyces version of ERG6. The first copy of the Candida ERG6 gene was disrupted by transforming with the URA3 blaster system, and the second copy was disrupted by both URA3 blaster transformation and mitotic recombination. The resulting erg6 strains were shown to be hypersusceptible to a number of sterol synthesis and metabolic inhibitors, including terbinafine, tridemorph, fenpropiomorph, fluphenazine, cycloheximide, cerulenin, and brefeldin A. No increase in susceptibility to azoles was noted. Inhibitors of the ERG6 gene product would make the cell increasingly susceptible to antifungal agents as well as to new agents which normally would be excluded and would allow for clinical treatment at lower dosages. In addition, the availability of ERG6 would allow for its use as a screen for new antifungals targeted specifically to the sterol methyltransferase.
The frequency of occurrence of human fungal infections has been increasing over the past decade in response to a combination of factors (12) which include advances in invasive surgical techniques which allow for opportunistic pathogen access, immunosuppression employed in transplantation or resulting from chemotherapy, and disease states such as AIDS. The threat to human health is further compounded by the increased frequency with which resistance to the commonly employed antifungal agents is appearing.
The most prevalently utilized antifungal agents include the polyenes and the azoles. The polyenes are effective by binding to ergosterol, the fungal membrane sterol, and inducing lethal cell leakage (7). Polyenes often have negative side effects, and resistance has been reported (15, 28). The azoles function by inhibition of the cytochrome P-450-mediated removal of the C-14 methyl group from the ergosterol precursor, lanosterol (32). The azoles are fungistatic drugs and are thus subject to the accumulation of resistant phenotypes due, in part, to the need to continuously administer the drug to patients who are immunocompromised. Resistance has been reported in Candida albicans (8, 30, 31, 37, 38) as well as in other species of Candida (24, 26). In addition, other fungal pathogens, including species of Histoplasma (36), Cryptococcus (19, 33), and Aspergillus (9), have been the subjects of recent reports on azole resistance. The increase in infections coupled with the reduced efficacy of the currently available drugs makes the discovery and development of new antifungals an urgent matter.
The pathway for fungal sterol biosynthesis has provided an excellent target for antifungal development, but there remain additional sites in the pathway that have not been thoroughly investigated. The sterol methyltransferase gene (ERG6) represents a particularly good example because this step is not found in cholesterol biosynthesis, thus avoiding some elements of possible side effects. Saccharomyces cerevisiae erg6 mutants have been available for some time (23), and the ERG6 gene was isolated and disrupted several years ago (11). Although the absence of the ERG6 gene product was not lethal, it did result in several severely compromised phenotypes.
erg6 mutants have been shown to have diminished growth rates as well as limitations on utilizable energy sources (21), reduced mating frequency (11), altered membrane structural features (18, 20), and low transformation rates (11). In addition, several lines of evidence have indicated that erg6 mutants have severely altered permeability characteristics. This has been demonstrated by using dyes (3), cations (3), and spin labels used in electron paramagnetic resonance studies (18). These early observations have been corroborated recently by the cloning of the LIS1 gene (35), mutants of which were selected on the basis of hypersensitivity to sodium and lithium; sequencing of LIS1 has indicated identity to ERG6. This study demonstrated that while the rate of cation uptake was increased three- to fourfold in the mutant strain, the rate of cation efflux was indistinguishable from that of the wild type. In addition, studies using the Golgi inhibitor brefeldin A have routinely employed erg6 mutant strains because of their permeability by this compound (34). Since the absence of a functional sterol methyltransferase would make the cell hypersensitive to exogenous compounds, blocks in ERG6 gene product function could increase the effectiveness of new or existing antifungals. Thus, we have utilized an S. cerevisiae erg6 mutant to isolate the C. albicans ERG6 gene, disrupted both copies in the latter organism, and characterized the resulting phenotype of the C. albicans erg6 mutant.
MATERIALS AND METHODS
Strains and plasmids.
C. albicans CAI4 (Δura3::imm434/Δura3::imm434), received from W. Fonzi (10), was used for disruption of both copies of ERG6. The S. cerevisiae erg6 deletion strain BKY48-5C (α leu2-3 ura3-52 erg6Δ::LEU2) was used as the recipient strain for transformation with the Candida genomic library (13). Escherichia coli DH5α was used as the host strain for all plasmid constructions. Plasmid pRS316 was obtained from P. Heiter, and Bluescript plasmid was obtained from Stratagene, La Jolla, Calif.
Media.
CAI4 was grown on YPD complete medium containing 1% yeast extract (Difco), 2% Bacto Peptone (Difco), and 2% glucose. Complete synthetic medium (CSM) was used for transformation experiments and contained 0.67% yeast nitrogen base (Difco), 2% glucose, and 0.8 g of a mixture of amino acids plus adenine and uracil (Bio 101) per liter. CSM dropout medium contained the same ingredients as CSM, but without uracil. Uridine was added at 80 mg per liter to ensure growth of CAI4. CSM containing uridine and 5-fluoroorotic acid (5-FOA) at 1 g/liter was used to regenerate the ura3 genetic marker as outlined by Fonzi and Irwin (10). All experiments were carried out at 30°C unless otherwise indicated.
Cloning of ERG6.
Transformation of S. cerevisiae BKY48-5C by using the Candida gene library was carried out by a lithium acetate-modified protocol developed by Gaber et al. (11) for erg6 transformations. The C. albicans ERG6 gene was cloned by transforming a S. cerevisiae erg6 deletion strain (BKY48-5C) with a Candida genomic DNA library obtained from S. Scherer at the University of Minnesota (13). Transformants containing putative Candida ERG6 DNA were subcloned into the Saccharomyces vector pRS316 for complementation analyses and DNA sequencing. All Candida transformations for disruption experiments were carried out essentially in accordance with the procedures of Sanglard et al. (30). Plasmid p5921, obtained from Fonzi (10), was the source of the URA3 blaster for Candida ERG6 disruption experiments.
Approximately 1,250 transformants were obtained by plating on a uracil dropout medium that ensured the presence of the plasmid. These transformants were then screened on medium containing 0.06 μg of cycloheximide per ml. S. cerevisiae erg6 strains are nystatin resistant and cycloheximide sensitive. Transformants that were resistant to this level of cycloheximide (Cyhr) were further tested for the presence of intracellular ergosterol. Sterols extracted from the S. cerevisae erg6 strains and the transformants were analyzed by UV spectrophotometry and gas chromatography-mass spectrometry (GC-MS) to confirm the sterol profile.
DNA sequencing of the Candida ERG6 gene.
Both strands of the plasmid insert containing the ERG6 gene were sequenced by the Sanger dideoxy chain termination method. Initially, T3 and T7 primers were used, and as DNA sequence became available, primers were generated from sequenced DNA.
PCR.
PCR analyses were used to verify disruptions of both Candida ERG6 genes. Primers P1, P2, and P3 were used to distinguish disrupted ERG6 genes on the basis of size and are in the ERG6 gene itself. Primer 4 is in the hisG region of the URA3 blaster. P1 was 5′-CACATGGGTGAAATTAG-3′ and could be used with all other primers. P2 was 5′-CTCCAGTTCAATTAGCAG-3′, P3 was 5′-TGTGCGTGTACAAAGCAC-3′, and P4 was 5′ GATAATACCGAGATCGAC-3′. PCR buffers and Taq polymerase were obtained from Promega. The buffer composition was 10 mM Tris-HCl (pH 9) and 2 mM MgCl2, and reactions mixtures contained 0.2 mM deoxynucleoside triphosphates and 0.5 U of polymerase. Conditions for amplification were as follows: the first cycle was denaturation at 94°C for 5 min; this was followed by 40 cycles of annealing at 50°C for 2 min, elongation at 72°C for 3 min, and denaturation at 94°C for 1 min. A final elongation step at 72°C for 20 min completed the reaction. The protocols used for preparation of the Candida template DNA described by Ausubel et al. (1).
Sterol analyses.
Nonsaponifiable sterols were isolated as described previously (23). UV analysis of sterols in extracts was accomplished by scanning wavelengths from 200 to 300 nm with a Beckman DU 640 spectrophotometer. GC analyses of nonsaponifiable sterols were conducted on a HP5890 series II equipped with the Hewlett-Packard Chemstation software package. The capillary column (HP-5) was 15 m by 0.25 mm by 0.25 mm (film thickness) and was programmed to increase from 195 to 300°C (3 min at 195°C and then increased at 5.5°C/min until the final temperature of 300°C was reached and held for 4 min). The linear velocity was 30 cm/s with nitrogen as the carrier gas, and all injections were run in the splitless mode. GC-MS analyses were done with a Varian 3400 GC interfaced to a Finnigan MAT SSQ 7000 MS. The GC separations were done on a DB-5 fused-silica column (15 m by 0.32 mm by 0.25 mm [film thickness]) programmed to increase from 50 to 250°C at 20°C/min after a 1-min hold at 50°C. The oven temperature was then held at 250°C for 10 min before the temperature was increased to 300°C at 20°C/min. Helium was the carrier gas, with a linear velocity of 50 cm/s in the splitless mode. The MS was in the electron impact ionization mode at an electron energy of 70 eV, an ion source temperature of 150°C, and scanning from 40 to 650 atomic mass units at 0.5-s intervals.
Drug susceptibility testing in C. albicans.
Drug susceptibilities of C. albicans wild-type and erg6 strains were conducted by using cells harvested from overnight YPD plates grown at 37°C. Cells were suspended in YPD medium to a concentration of 107 (optical density at 660 nm of 0.5) cells per ml. Cells were plated by transferring 5 μl of the original suspension (100) plus 10−1 and 10−2 dilutions onto YPD plates containing the drug to be tested. The plates were incubated for 48 h at 37°C and observed for growth. Clotrimazole, brefeldin A, cerulenin, cycloheximide, nystatin, and fluphenazine were obtained from Sigma, St. Louis, Mo. Fenpropiomorph and tridemorph were obtained from Crescent Chemical Co., Hauppage, N.Y. Ketoconazole was obtained from ICN, Costa Mesa, Calif. Terbinafine was a gift from D. Kirsch (American Cyanamid, Princeton, N.J.). Stock solutions of terbinafine, tridemorph, brefeldin A, and cerulenin were prepared in ethanol. Clotrimazole, ketoconazole, and fenpropiomorph stocks were prepared in dimethyl sulfoxide, and fluphenazine and cycloheximide stocks were prepared in water. Nystatin was dissolved in N,N-dimethyl formamide (Sigma).
Nucleotide sequence accession numbers.
The GenBank accession number for the C. albicans ERG6 gene is AF031941. GenBank accession numbers for the previously determined nucleotide sequences of ERG6 from S. cerevisiae, Arabidopsis thaliana, and Triticum ativum are X74249, U71400, and U60755, respectively.
RESULTS
Cloning of the C. albicans ERG6 gene.
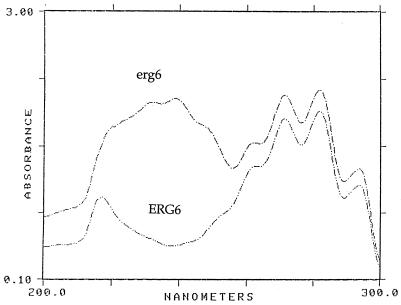
Four Saccharomyces erg6 transformants which grew on cycloheximide were analyzed for sterol content. erg6 mutants which fail to synthesize ergosterol due to defects in the C-24 transmethylase gene accumulate principally zymosterol, cholesta-5,7,24-trien-3β-ol, and cholesta-5,7,22,24-tetraen-3β-ol (23). UV scans of the sterols obtained from a Saccharomyces erg6 strain as well as an erg6 transformant containing the Candida ERG6 gene are shown in Fig. 1. Sterols giving the erg6 spectrum contain absorption maxima at 262, 271, 282, and 293 nm as well as maxima at 230 and 238 nm. The latter two absorption maxima are due to conjugated double bonds which occur in the sterol side chain (cholesta-5,7,22,24-tetra-en-3β-ol). The ERG6 transformed strain does not have a conjugated double bond in the side chain and gives absorption maxima only at 262, 271, 282, and 293 nm. The remaining three transformants yielded similar profiles. Additionally, GC analysis of the erg6 mutant and the ERG6 transformants confirmed the presence of ergosterol in the latter strains (data not shown). These results were confirmed by MS.
FIG. 1.
UV scan of nonsaponifiable sterols in which erg6 sterols containing a conjugated double bond in the sterol side chain show absorption maxima at 230 and 238 nm. Wild-type erg6 transformants containing the Candida ERG6 gene do not have the conjugated double-bond system in the sterol side chain.
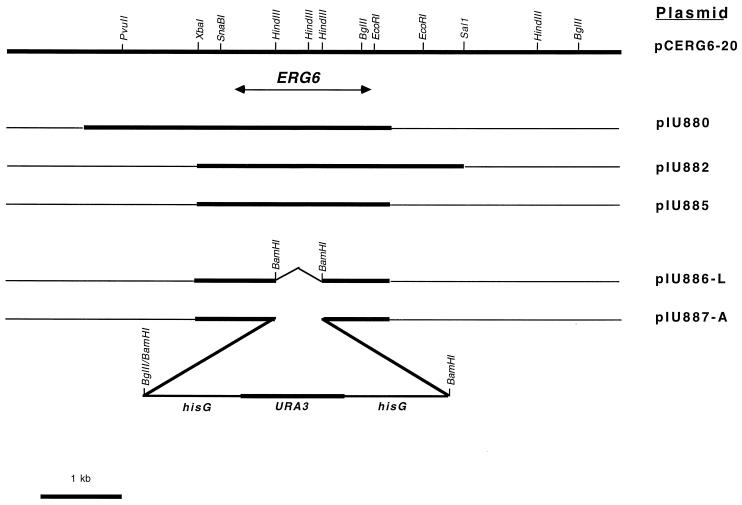
Of the four transformants restoring the ability of the erg6 mutant to synthesize ergosterol, there were two different types, designated pCERG6-20 and pCERG6-9, with insert sizes of 8 and 14 kb, respectively (Fig. 2). The pCERG6-9 insert contained the entire 8-kb DNA fragment of pCERG6-20, suggesting that the ERG6 gene resided within the 8-kb fragment. Growth of ergosterol-producing transformants on media containing 5-FOA resulted in the loss of the transforming plasmid, which restored the BKY48-5c strain back to the erg6 phenotype; this indicated that ergosterol production of the pCERG6-20 and -9 transformants was plasmid mediated. To locate the ERG6 gene within the plasmid insert, an approximately 4-kb subclone of the left arm of pCERG6-20 was inserted into the Saccharomyces vector pRS316, yielding plasmid pIU880, which was able to complement erg6 (Fig. 2). Plasmid pIU882, which contains a 2.4-kb overlap with pIU880, also complemented erg6, suggesting that the Candida ERG6 gene lies within this 2.4-kb fragment. A 2.4-kb XbaI-EcoRI subclone of pIU880 inserted into pRS316 resulted in pIU885 containing the entire ERG6 gene.
FIG. 2.
A C. albicans ERG6 genomic clone (pCERG6-20) with restriction sites and three complementing subclones, pIU880, pIU882, and pIU885. Deletion of a 0.7-kb HindIII fragment within pIU885, filling in of cohesive ends, addition of BamHI linkers (pIU886-L), and subsequent insertion of the URA3 blaster into this site as shown (pIU887-A) are represented.
DNA sequencing of the Candida ERG6 gene.
The 2.4-kb XbaI-EcoRI DNA insert of pIU885 (Fig. 2) was selected for sequencing. The DNA and amino acid sequences are presented in Fig. 3. The Candida ERG6 gene encodes the sterol methyltransferase, which contains 377 amino acids and is 66% identical to the Saccharomyces enzyme. Figure 4 shows the sequence alignment between the Candida, Saccharomyces, Arabidopsis, and Triticum sterol methyltransferases, and the levels of identity of Candida to the latter two are 40 and 49%, respectively. A 9-amino-acid region (Fig. 4; amino acids 127 to 135 in the C. albicans sequence) represents the highly conserved S-adenosylmethionine binding site (6).
FIG. 3.
The DNA and amino acid sequences of the C. albicans ERG6 gene. The S-adenosylmethionine binding site is indicated by underlining.
FIG. 4.
Alignment of the amino acid sequences of the sterol methyltransferases from C. albicans, S. cerevisiae, A. thaliana, and T. ativum. Shaded areas indicate regions of sequence identity.
Creation of a C. albicans ERG6 heterozygote.
Disruption of the Candida ERG6 gene to derive a sterol methyltransferase-deficient strain was made more difficult since Candida, unlike Saccharomyces, is diploid and, thus, both copies of the ERG6 gene must be disrupted. To accomplish this, the URA3 blaster system developed by Fonzi (10) was used. The URA3 blaster contains ∼3.8 kb comprised of repeat elements of hisG (derived from Salmonella) flanking the Candida URA3 gene. The plasmid pIU887-A containing the URA3 blaster inserted into the ERG6 gene is shown in Fig. 2. The 2.4-kb XbaI-EcoRI ERG6 DNA fragment was cloned into the pBluescript vector KS(+) in which a HindIII site was filled in with the Klenow fragment of DNA polymerase I (pIU886). pIU886-L was subsequently derived by deleting a 0.7-kb HindIII fragment within the ERG6 coding sequence, filling in this site with Klenow fragment, followed by the addition of BamHI linkers. Plasmid 5921, containing the URA3 blaster, was digested with SnaBI and StuI, both blunt-cutting enzymes, followed by religation. This resulted in a deletion of 6 bp in one of the hisG regions and destruction of these two sites. The modified 5921 plasmid was then digested with BamHI and BglII to release the 3.8-kb URA3 blaster, which was then ligated into pIU886-L that had been digested with BamHI to generate pIU887-A.
C. albicans CAI4 was transformed by using the 5.3-kb BglII-SnaBI fragment containing the URA3 blaster and ERG6 flanking recombinogenic ends of 0.8 and 0.9 kb. Transformants containing the single disrupted ERG6 allele resulting in heterozygosity for ERG6 were confirmed by using PCR after selection for loss of the URA3-hisG region. Intrachromosomal recombination between the linear hisG sequences resulted in the loss of one of these hisG repeats and the URA3, thus permitting reuse of the URA3 blaster for the subsequent disruption of the ERG6 gene on the homologous chromosome. Selection for colonies on medium containing 5-FOA resulted in growth of only uridine-requiring strains (5).
Creation of C. albicans erg6 strains.
The creation of a Candida erg6 mutant strain in which both alleles were disrupted was accomplished in two different ways. The ERG6 heterozygote was placed onto plates containing high concentrations of nystatin (15 μg/ml), and nystatin-resistant colonies appeared after 3 days. We surmised that mitotic recombination resulted in homozygous ERG6 and erg6 segregants and that these nystatin-resistant colonies might be the erg6 homozygotes. When colony purified, these resistant colonies indeed turned out to be erg6 homozygotes (see below). The second method used to generate erg6 homozygotes was to transform the ERG6 heterozygote with the URA3 blaster. Two kinds of transformants were obtained, wild-type and slow-growing colonies. Both types of colonies were tested for resistance to nystatin, and only the slower-growing colonies were nystatin resistant.
Confirmation of erg6 homozygosity by sterol analyses.
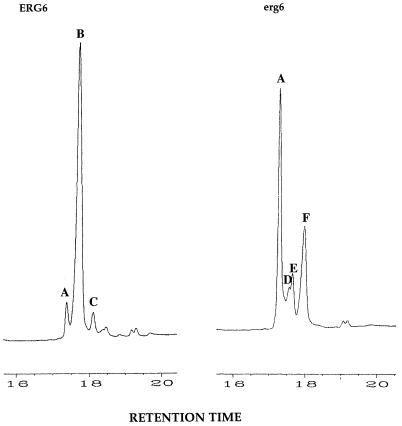
The sterols isolated from wild-type and putative erg6 homozygotes were analyzed by UV spectrophotometry and GC-MS. All of our putative erg6 homozygotes contained erg6-like UV scans similar to the S. cerevisiae erg6 scan shown in Fig. 1. Additionally, GC-MS of erg6 mutant sterols confirmed that only cholesterol-like (C-27) sterols accumulate since the side chain cannot be methylated. Figure 5 shows a GC profile demonstrating that the putative erg6 mutants accumulate C-27 sterols and are deficient in side chain transmethylation. Whereas the predominant sterol in the CAI4 wild type is ergosterol (peak B, 76%), the principal sterols in erg6 mutants are zymosterol (peak A, 43%), cholesta-5,7,24-trien-3β-ol (peak D, 6%), cholesta-7,24-dien-3β-ol (peak E, 9%), and cholesta-5,7,22,24-tetraen-3β-ol (peak F, 29%).
FIG. 5.
GC of the sterols of the wild type and an erg6 strain of C. albicans. Peak A, zymosterol; peak B, ergosterol; peak C, fecosterol; peak D, cholesta-5,7,24-trien-3β-ol; peak E, cholesta-7,24-dien-3β-ol; peak F, cholesta-5,7,22,24-tetraen-3β-ol.
PCR confirmation of homozygous disruptions.
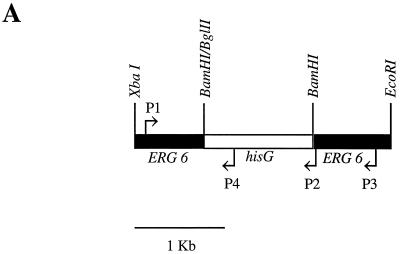
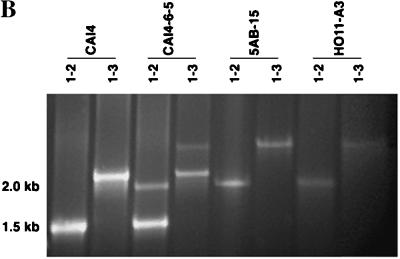
Confirmation of the disruption of both copies of the C. albicans ERG6 gene by mitotic recombination of the heterozygote and by a second transformation using the URA3 blaster was performed by using four PCR primers. The URA3 blaster containing a 3.8-kb region of hisG-URA3-hisG replaced 0.7 kb of ERG6 DNA (Fig. 6A). This was followed by deletion of the hisG-URA3 sequence such that, in effect, the remaining 1.2-kb hisG sequence replaces a 0.7-kb ERG6 deletion. The expected PCR amplifications of CAI4 using primer pair P1-P2 or P1-P3 are 1.5 and 2.15 kb, respectively (Fig. 6B, lanes 1 and 2). The expected products from P1-P2 amplification of the heterozygote CAI-4-6-5 are 1.5 kb (wild-type allele) and 2.01 kb (disrupted ERG6 allele), and the expected products from amplification using the P1-P3 primers are 2.15 kb (wild type) and 2.65 kb (disrupted ERG6); these products are visible in Fig. 6B, lanes 3 and 4. Primer pair P1-P4 gives a 1.1-kb band, demonstrating the presence of hisG within the ERG6 sequence (data not shown). The erg6 homozygotes 5AB-15, obtained by mitotic recombination, and HO11-A3, obtained by URA3 blaster disruption, yield identical amplification products with primer pairs P1-P2 (2.01 kb) and P1-P3 (2.65 kb), as shown in Fig. 6B, lanes 5 to 8.
FIG. 6.
(A) URA3 blaster disruption of the ERG gene showing location of PCR primers; (B) agarose gel electrophoresis confirmation of heterozygote and homozygote disruptants of the ERG6 gene. Lanes (left to right): 1 and 2, CAI4 (wild type); 3 and 4, CA14-6-5 (heterozygote); 5 and 6, 5AB-15 (homozygote derived from URA3 blaster transformation followed by mitotic recombination); 7 and 8, HO11-A3 (homozygote derived from two rounds of URA3 blaster transformation). The PCR primer pairs used are indicated at the tops of the lanes (e.g., 1-2 is P1-P2). The image was captured on disc and the photograph was generated by using Photoshop on Macintosh.
Drug susceptibilities of C. albicans erg6 strains.
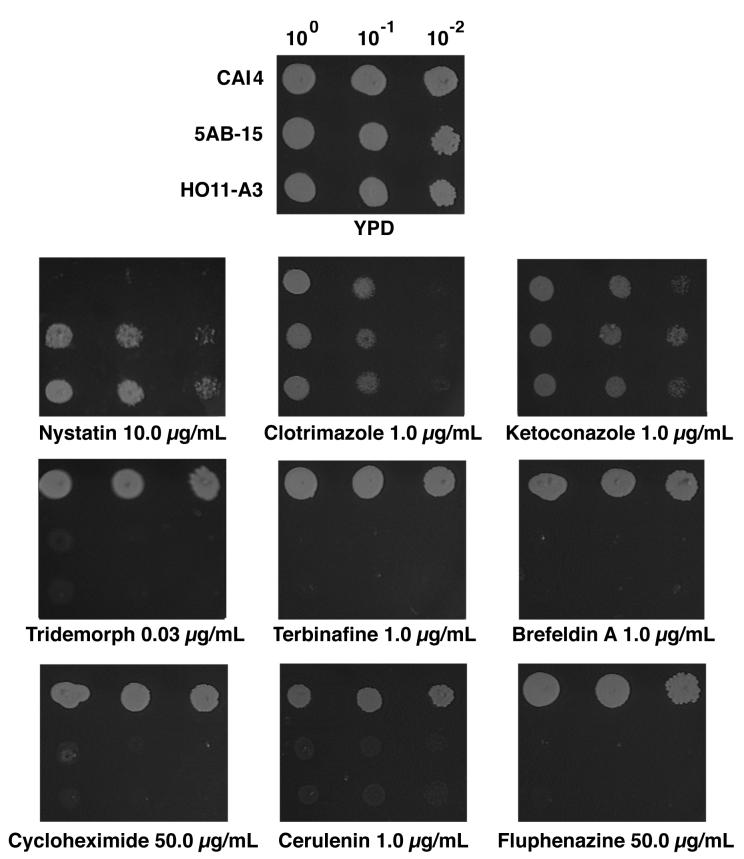
The susceptibilities of the erg6 strains as compared to that of wild-type C. albicans were determined by using a number of antifungal compounds and general cellular inhibitors (Fig. 7). The erg6 strains were shown to be more resistant to nystatin while showing nearly identical sensitivities to the azole antifungals clotrimazole and ketoconazole. Significantly increased susceptibilities of the erg6 strains were noted for tridemorph and fenpropiomorph, inhibitors of sterol Δ14-reductase and Δ8-Δ7 isomerase (2); terbinafine, an allylamine antifungal inhibiting squalene epoxidase (16); brefeldin A, an inhibitor of Golgi function (33); cycloheximide, a common protein synthesis inhibitor; cerulenin, an inhibitor of fatty acid synthesis (25); and fluphenazine, a compound which interferes with the function of calmodulin (14).
FIG. 7.
Growth responses of the wild type (CAI4), a homozygous erg6 strain derived from URA3 blaster transformation (5AB-15), and a homozygous erg6 strain derived from mitotic recombination (HO11-A3) in the presence of sterol biosynthesis inhibitors and metabolic inhibitors. Cells were grown at 37°C to a density of 107 cells/ml, and 5 μl was inoculated at 100, 10−1, and 10−2 dilutions. The image was captured on disc and the photograph was generated by using Photoshop on Macintosh.
The determination of drug concentrations sufficient to completely inhibit growth on plates yielded the data shown in Table 1. The concentration of nystatin required for complete inhibition of the wild type (2.5 μg/ml) is within the normal range for a wild-type strain (23), while the erg6 mutants show a resistance level similar to that noted for erg6 mutants of S. cerevisiae (23). As demonstrated by growth on plates (Fig. 7), the azoles show equal efficacies against both wild-type and erg6 mutant strains. In contrast, the erg6 mutants show significantly increased susceptibilities to other antifungals and metabolic inhibitors. erg6 susceptibilities to cerulenin and fluphenazine were twofold greater, while those for terbinafine and brefeldin A were about 50 times greater, than those of the wild type. Cycloheximide susceptibility was increased about 11-fold in the erg6 mutants, while the greatest increases in susceptibility were shown for the morpholines fenpropiomorph (100-fold) and tridemorph (several thousandfold). The erg6 heterozygote showed essentially the same drug sensitivities as those of the wild type, CAI4, for all inhibitors tested.
TABLE 1.
Susceptibilities of ERG6 and erg6 strains of C. albicans to antifungal agents and metabolic inhibitors
| Drug | Inhibitory concn (μg/ml)a
|
|
|---|---|---|
| ERG6 | erg6 | |
| Nystatin | 2.5 | 15 |
| Clotrimazole | 4 | 4 |
| Ketoconazole | 5 | 5 |
| Terbinafine | >50 | 1 |
| Fenpropiomorph | 0.5 | 0.005 |
| Tridemorph | >90 | 0.03 |
| Brefeldin A | 50 | 1 |
| Cerulenin | 2 | 1 |
| Cycloheximide | >600 | 50 |
| Fluphenazine | 100 | 50 |
Concentration at which no growth appeared after 48 h under the conditions described in the legend to Fig. 7.
DISCUSSION
Strains with mutations in the erg6 gene of S. cerevisiae have been available for many years (23). Since the biosynthetic step that adds the C-24 methyl group is found in fungal but not in human sterol biosynthesis, it was proposed (27) that this step might be essential and that inhibition at this point in the pathway would be lethal. This hypothesis could not be tested until the ERG6 gene could be shown to be completely inactivated, since low levels of leakiness could allow viability. The cloning and disruption of the ERG6 gene (11) provided definitive evidence that the gene is not essential in S. cerevisiae. However, the same study reinforced previous work done with erg6 point mutations that had demonstrated that erg6 mutants have several altered phenotypes (3, 18, 20, 21). Our particular interest is in the alteration of permeability characteristics.
The essential nature of the ERG6 gene in C. albicans has not been reported prior to the work described here. It was possible that this gene could be essential since the ERG11 gene has been shown to be essential in S. cerevisiae but not in C. albicans, indicating that these two species are not identical in their abilities to survive and grow on various sterol intermediates. In addition, it would be of particular interest to assess the permeability of Candida erg6 mutant cells since this characteristic might make them more sensitive to known and new antifungals or might even make them sensitive to compounds previously found not to be effective when ergosterol is present in the cell.
Using a Candida genomic library, we have isolated the Candida ERG6 gene by complementing an erg6 mutant of Saccharomyces. As part of our screen for complementation, sensitivity to nystatin and resistance to cycloheximide were employed. Nystatin functions by binding to membrane ergosterol and causing cell leakage, which leads to cell death (7). Mutants such as erg6 do not produce ergosterol and utilize sterol intermediates in place of membrane ergosterol. Nystatin has lower affinity for sterol intermediates, thus leading to resistance in non-ergosterol-containing strains. Restoration of the ERG6 gene from Candida in Saccharomyces erg6 mutants would restore the nystatin-sensitive phenotype. The wild-type ERG6 gene also reconstitutes the cell permeability barrier to normal levels, thus conferring cycloheximide resistance at low drug concentrations. Cloning of the Candida ERG6 gene was also confirmed by UV analysis of sterol composition and GC-MS analysis of accumulated sterols in Saccharomyces erg6 and transformed strains containing the Candida ERG6 gene. Final confirmation that we had cloned ERG6 was provided by sequencing the Candida ERG6 gene. The Candida sequence showed high identity to the S. cerevisiae ERG6 gene sequence and good agreement with the same gene from Arabidopsis and Triticum. The high homology of the Candida and Saccharomyces sequences accounts for the successful complementation noted in this study.
To determine the essentiality of the ERG6 gene in Candida, the two copies were disrupted by first creating the heterozygote by using the URA3 blaster disruption protocol. The second copy of the ERG6 gene was disrupted either by allowing for mitotic recombination or by a second disruption with the URA3 blaster. In both cases, the resulting erg6 homozygotes were viable, indicating that the ERG6 gene in C. albicans is not essential for viability. Both types of erg6 mutants were confirmed by sterol and PCR analyses of the disruptions.
With the continued increase in resistance to the azole antifungals, new approaches to antifungal chemotherapy are strongly indicated. One approach is to disarm the resistance mechanism. A primary mechanism in C. albicans for azole resistance is the increase in expression of efflux systems which utilize the azoles as substrates. Both the ABC (ATP-binding cassette) transporter gene CDR1 and a gene (BENr) belonging to a major facilitator multidrug efflux transporter have been implicated in this process (31). A report by Sanglard et al. (30) has shown that disruption of the CDR1 gene results in a cell that shows increased susceptibilities to the azole, allylamine, and morpholine antifungals as well as other metabolic inhibitors, including cycloheximide, brefeldin A, and fluphenazine. Although not effective alone, disruptions of BENr were shown to work synergistically with CDR1 with two metabolic inhibitors. The CDR1 system could provide for an assay for drugs not subject to efflux by these transporters or could also be used to select for compounds which could block the action of the transporters directly. Such approaches would avoid or disarm resistance mechanisms, respectively.
In this report, the testing of Candida erg6 mutants for their susceptibility to antifungal and metabolic inhibitors indicated that these mutants had increased sensitivity to a wide variety of compounds. Azoles were an exception in that they showed no difference in efficacy for wild-type and mutant strains. Apparently, the permeability changes are unrelated to the entry mechanism for these compounds. The remainder of the compounds tested, including two other antifungal compounds with different mechanisms of action, are significantly more inhibitory toward the erg6 strain.
These findings have important applicability from several perspectives. First, the results predict that an inhibitor of the ERG6 gene product would result in a fungal organism that is hypersensitive to known compounds or new compounds to which the cell is normally impermeable. Treatment of a cell with both inhibitors would thus produce a synergistic effect. Synergism has been shown (4) by using the experimental sterol methyltransferase inhibitor ZM59620 added simultaneously with allylamine and morpholine antifungals. In these studies, the concentrations of the drugs in the combined treatment were significantly below the individual concentrations necessary for both the inhibition of ergosterol biosynthesis and growth inhibition. Thus, because of the increased drug access produced by inhibitors of the sterol methyltransferase, other inhibitors can be clinically employed at reduced dosages. Second, the availability of the C. albicans ERG6 gene allows it to be used as a screen for the identification of inhibitory compounds that specifically target the ERG6 gene product. This approach has been successfully utilized in cloning of one of the 3-hydroxy-3-methylglutaryl-CoA (HMGCoA) reductase genes (29) as well as the ERG11 (17) and ERG24 (22) genes. In applying this strategy for the purpose of identifying ERG6 gene product inhibitors, the sensitivity of a wild-type strain would be compared to that of a strain carrying additional copies of ERG6 on a high-copy-number plasmid. Inhibition of the wild type but not the multiple-copy strain would identify inhibition specific to the sterol methyltransferase. Treatment of a fungal pathogen with such an inhibitor would result in a metabolically compromised cell that, as in the first application, would be more susceptible to existing antifungals and metabolic inhibitors. Finally, the erg6 system allows for the replacement of in vitro testing of inhibitors by utilizing the increased permeability characteristics inherent in the in vivo mutant system. This will allow characterization of potential inhibitors that normally fail to reach intracellular targets due to a lack of permeability.
Since the erg6 system results in a compromised cell which is highly permeable to a variety of compounds and since selection of new inhibitors using high-copy-number ERG6 plasmids allows for easy identification, we believe that this system has superior potential for the development of new antifungal treatment protocols.
ACKNOWLEDGMENTS
This work was supported by grant DAMD17-95-1-5067 to M.B. and N.D.L. from the Defense Women’s Health Research Program of the U.S. Army.
We thank W. Fonzi for C. albicans CAI4, P. Heiter for plasmid pRS316, and S. Scherer for the C. albicans genomic library. We thank Marilyn Bartlett for advice and discussions on drug susceptibility testing.
REFERENCES
- 1.Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 2.Baloch R I, Mercer E I, Wiggins T E, Baldwin B C. Inhibition of ergosterol biosynthesis in Saccharomyces cerevisiae and Ustilago maydis by tridemorph, fenpropiomorph, and fenpropidin. Phytochemistry. 1984;23:2219–2226. [Google Scholar]
- 3.Bard M, Lees N D, Burrows L A, Kleinhans F W. Differences in crystal violet dye uptake and cation-induced death among yeast sterol mutants. J Bacteriol. 1978;135:1146–1148. doi: 10.1128/jb.135.3.1146-1148.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett-Bee K, Dixon G. Ergosterol biosynthesis inhibition: a target for antifungal agents. Acta Biochim Pol. 1995;42:465–480. [PubMed] [Google Scholar]
- 5.Boeke J D, Truehart J, Natsoulis G, Fink G R. 5-Fluoro-orotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 6.Bouvier-Navé P, Husselstein T, Desprez T, Benveniste P. Identification of cDNAs encoding sterol methyl transferases involved in the second methylation step of plant sterol biosynthesis. Eur J Biochem. 1997;246:518–529. doi: 10.1111/j.1432-1033.1997.t01-1-00518.x. [DOI] [PubMed] [Google Scholar]
- 7.Brajtburg J, Powderley W G, Kobayashi G S, Medoff G. Amphotericin B: current understanding of mechanism of action. Antimicrob Agents Chemother. 1990;34:183–188. doi: 10.1128/aac.34.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark F S, Parkinson T, Hitchcock C A, Gow N A R. Correlation between rhodamine 123 accumulation and azole sensitivity in Candida species: possible role for drug efflux in drug resistance. Antimicrob Agents Chemother. 1996;40:419–425. doi: 10.1128/aac.40.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denning D W, Venkateswarlu K, Oakley K L, Anderson M J, Manning N J, Stevens D A, Warnock D W, Kelly S L. Itraconazole resistance in Aspergillus fumigatus. Antimicrob Agents Chemother. 1997;41:1364–1368. doi: 10.1128/aac.41.6.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaber R F, Copple D M, Kennedy B K, Vidal M, Bard M. The yeast gene ERG6 is required for normal membrane function but is not essential for biosynthesis of the cell-cycle sparking sterol. Mol Cell Biol. 1989;9:3447–3456. doi: 10.1128/mcb.9.8.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georgopapadakou N H, Walsh T J. Human mycoses: drugs and targets for emerging pathogens. Science. 1994;264:371–373. doi: 10.1126/science.8153622. [DOI] [PubMed] [Google Scholar]
- 13.Goshorn A K, Grindle S M, Scherer S. Gene isolation by complementation in Candida albicans and applications to physical and genetic mapping. Infect Immun. 1992;60:876–884. doi: 10.1128/iai.60.3.876-884.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hait W N, Gesmonde J F, Lazo J S. Effect of anti-calmodulin drugs on the growth and sensitivity of C6 rat glioma cells to bleomycin. Anticancer Res. 1993;14:1711–1721. [PubMed] [Google Scholar]
- 15.Hebeka E K, Solotorovsky M. Development of resistance to polyene antibiotics in Candida albicans. J Bacteriol. 1965;89:1533–1539. doi: 10.1128/jb.89.6.1533-1539.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jandrositz A, Turnowski F, Hogenaur G. The gene encoding squalene epoxidase from Saccharomyces cerevisiae: cloning and characterization. Gene. 1991;107:155–160. doi: 10.1016/0378-1119(91)90310-8. [DOI] [PubMed] [Google Scholar]
- 17.Kalb V F, Loper J C, Dey C R, Woods C W, Sutter T R. Isolation of a cytochrome P-450 gene from Saccharomyces cerevisiae. Gene. 1986;45:237–245. doi: 10.1016/0378-1119(86)90021-1. [DOI] [PubMed] [Google Scholar]
- 18.Kleinhans F W, Lees N D, Bard M, Haak R A, Woods R A. ESR determination of membrane permeability in a yeast sterol mutant. Chem Phys Lipids. 1979;23:143–154. doi: 10.1016/0009-3084(79)90042-2. [DOI] [PubMed] [Google Scholar]
- 19.Lamb D C, Baldwin B C, Kwon-Chung K J, Kelly S L. Stereoselective interaction of the azole antifungal agent SCH39304 with the cytochrome P-450 monooxygenase system isolated from Cryptococcus neoformans. Antimicrob Agents Chemother. 1997;41:1465–1467. doi: 10.1128/aac.41.7.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lees N D, Bard M, Kemple M D, Haak R A, Kleinhans F W. ESR determination of membrane order parameter in yeast sterol mutants. Biochim Biophys Acta. 1979;553:469–475. doi: 10.1016/0005-2736(79)90302-x. [DOI] [PubMed] [Google Scholar]
- 21.Lees N D, Lofton S L, Woods R A, Bard M. The effects of varied energy source and detergent on the growth of sterol mutants of Saccharomyces cerevisiae. J Gen Microbiol. 1980;118:209–214. [Google Scholar]
- 22.Marcireaux D, Guyonnet D, Karst F. Construction and growth properties of a yeast strain defective in sterol 14-reductase. Curr Genet. 1992;22:267–272. doi: 10.1007/BF00317919. [DOI] [PubMed] [Google Scholar]
- 23.Molzhan S W, Woods R A. Polyene resistance and the isolation of sterol mutants in Saccharomyces cerevisiae. J Gen Microbiol. 1972;72:339–348. doi: 10.1099/00221287-72-2-339. [DOI] [PubMed] [Google Scholar]
- 24.Moran G P, Sullivan D J, Henman M C, McCreary C E, Harrington B J, Shanley D B, Coleman D C. Antifungal drug susceptibilities of oral Candida dubliniensis isolates from human immunodeficiency virus (HIV)-infected and non-HIV-infected subjects and generation of stable fluconazole-resistant derivatives in vitro. Antimicrob Agents Chemother. 1997;41:617–623. doi: 10.1128/aac.41.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morisaki N, Funabashi H, Shimazawa R, Furukawa J, Kawaguchi A, Okuda S, Iwasaki S. Effect of side-chain structure on inhibition of yeast fatty-acid synthase by cerulenin analogues. Eur J Biochem. 1993;211:111–115. doi: 10.1111/j.1432-1033.1993.tb19876.x. [DOI] [PubMed] [Google Scholar]
- 26.Parkinson T, Falconer D J, Hitchcock C A. Fluconazole resistance due to energy-dependent drug efflux in Candida glabrata. Antimicrob Agents Chemother. 1995;39:1696–1699. doi: 10.1128/aac.39.8.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinto W J, Nes W D. Stereochemical specificity for sterols in Saccharomyces cerevisiae. J Biol Chem. 1983;258:4472–4476. [PubMed] [Google Scholar]
- 28.Powderley W G, Kobayashi G S, Herzig G P, Medoff G. Amphotericin B-resistant yeast infection in severely immunocompromised patients. Am J Med. 1988;84:826–832. doi: 10.1016/0002-9343(88)90059-9. [DOI] [PubMed] [Google Scholar]
- 29.Rine J, Hansen W, Hardeman E, Davis R W. Targeted selection of recombinant clones through gene dosage effects. Proc Natl Acad Sci USA. 1983;80:6750–6754. doi: 10.1073/pnas.80.22.6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanglard D, Ischer F, Monod M, Bille J. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob Agents Chemother. 1996;40:2300–2305. doi: 10.1128/aac.40.10.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanglard D, Kuchler K, Ischer F, Pagani J L, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanden Bossche H, Willemsens G, Marichal P. Anti-Candida drugs—the biochemical basis for their activity. Crit Rev Microbiol. 1987;15:57–72. doi: 10.3109/10408418709104448. [DOI] [PubMed] [Google Scholar]
- 33.Venkateswarlu K, Taylor M, Manning N J, Rinaldi M G, Kelly S L. Fluconazole tolerance in clinical isolates of Cryptococcus neoformans. Antimicrob Agents Chemother. 1997;41:748–751. doi: 10.1128/aac.41.4.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogel J P, Lee J N, Kirsch D R, Rose M D, Sztul E S. Brefeldin A causes a defect in secretion in Saccharomyces cerevisiae. J Biol Chem. 1993;268:3040–3043. [PubMed] [Google Scholar]
- 35.Welihinda A A, Beavis A D, Trumbly R J. Mutations in LIS1 (ERG6) gene confer increased sodium and lithium uptake in Saccharomyces cerevisiae. Biochim Biophys Acta. 1994;1193:107–117. doi: 10.1016/0005-2736(94)90339-5. [DOI] [PubMed] [Google Scholar]
- 36.Wheat J, Marichal P, Vanden Bossche H, Le Monte A, Connolly P. Hypothesis on the mechanism of resistance to fluconazole in Histoplasma capsulatum. Antimicrob Agents Chemother. 1997;41:410–414. doi: 10.1128/aac.41.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White T. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother. 1997;41:1482–1487. doi: 10.1128/aac.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White T. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14α demethylase in Candida albicans. Antimicrob Agents Chemother. 1997;41:1488–1494. doi: 10.1128/aac.41.7.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]