Abstract
Infection of cells by picornaviruses of the rhinovirus, aphthovirus, and enterovirus groups results in the shutoff of host protein synthesis but allows viral protein synthesis to proceed. Although considerable evidence suggests that this shutoff is mediated by the cleavage of eukaryotic translation initiation factor eIF4G by sequence-specific viral proteases (2A protease in the case of coxsackievirus), several experimental observations are at variance with this view. Thus, the cleavage of other cellular proteins could contribute to the shutoff of host protein synthesis and stimulation of viral protein synthesis. Recent evidence indicates that the highly conserved 70-kDa cytoplasmic poly(A)-binding protein (PABP) participates directly in translation initiation. We have now found that PABP is also proteolytically cleaved during coxsackievirus infection of HeLa cells. The cleavage of PABP correlated better over time with the host translational shutoff and onset of viral protein synthesis than did the cleavage of eIF4G. In vitro experiments with purified rabbit PABP and recombinant human PABP as well as in vivo experiments with Xenopus oocytes and recombinant Xenopus PABP demonstrate that the cleavage is catalyzed by 2A protease directly. N- and C-terminal sequencing indicates that cleavage occurs uniquely in human PABP at 482VANTSTQTM↓GPRPAAAAAA500, separating the four N-terminal RNA recognition motifs (80%) from the C-terminal homodimerization domain (20%). The N-terminal cleavage product of PABP is less efficient than full-length PABP in restoring translation to a PABP-dependent rabbit reticulocyte lysate translation system. These results suggest that the cleavage of PABP may be another mechanism by which picornaviruses alter the rate and spectrum of protein synthesis.
Picornaviruses are important pathogens of both human (e.g., poliovirus, coxsackievirus, and hepatitis A virus) and animals (e.g., foot-and-mouth disease virus and encephalomyocarditis virus). Their genomes consist of single-stranded, plus-sense RNAs which act as templates for both translation and replication. On entering the host cell, the RNA directs the translation of a single polyprotein, which is cleaved during translation by virus-encoded proteases into functional proteins (58). To gain advantage over the host cell in the competition for ribosomes and other translational factors, picornaviruses cause a shutoff of host protein synthesis under conditions which favor viral protein synthesis.
Translation initiation in eukaryotes involves RNA-protein and protein-protein interactions at both the 5′ and 3′ ends of the mRNA (61). Eukaryotic cytoplasmic mRNAs contain a 5′-terminal m7G cap, which enhances the efficiency of initiation (57). The joining of the 43S initiation complex to mRNA requires the eIF4 group of initiation factors (48). This group includes eIF4E, a 25-kDa cap-binding protein; eIF4A, a 46-kDa DEAD-box protein which has ATP-dependent RNA helicase activity; eIF4B, a 70-kDa RNA-binding protein which stimulates the helicase activity of eIF4A and also catalyzes RNA annealing; and eIF4G, a 154-kDa protein which contains binding sites for eIF4E, eIF4A, and eIF3 (41, 46). The 43S initiation complex then scans the 5′-untranslated region of the mRNA until it encounters the initiation codon, whereupon the 60S ribosomal subunit joins and the 80S initiation complex is formed (48).
Picornaviruses, on the other hand, utilize a different mode of translation initiation, namely, internal initiation, which involves the direct entry of the 43S initiation complex at an internal ribosomal entry sequence on the viral RNA (32, 55). The switch to viral translation in cells infected with enteroviruses, rhinoviruses, and aphthoviruses is thought to be caused by the cleavage of eIF4G. The first indication of this was the observation that cleavage of eIF4G coincides with the shutoff of host protein synthesis (18). Considerable evidence has been obtained in support of an indirect mechanism whereby viral proteases activate cellular proteases (73). More recently, it was found that the virally encoded 2A protease (rhinoviruses and enteroviruses) or L protease (aphthoviruses) cleaves eIF4G directly (36, 40, 44), although this process is relatively inefficient (9). Regardless of the mechanism, cleavage of eIF4G separates it into two domains, one that binds eIF4E and one that binds eIF3 and eIF4A (41, 46). Cleavage of eIF4G in vitro causes drastic inhibition of translation of capped mRNAs, whereas internal initiation is either unaffected (cardioviruses) or even stimulated (enteroviruses and rhinoviruses) (8, 44, 51).
There is, however, a significant body of evidence which indicates that eIF4G cleavage is not responsible, or is only partially responsible, for the shutoff of host protein synthesis. Various treatments (guanidine, drugs, ionophores, and temperature shifts) can prevent the host translational shutoff but not eIF4G cleavage (6, 30, 56). In vivo expression of 2A protease activates viral mRNA translation independently of its role in the host translational shutoff (26, 45). Complete cleavage of Xenopus oocyte eIF4G by injected 2A protease results in only a modest reduction of endogenous protein synthesis (34). Expression of poliovirus 2A protease in COS-1 cells has a much greater inhibitory effect on transcription by RNA polymerase II than on translation (17). Finally, a cleavage-resistant variant of eIF4G is able to restore some but not all of cap-dependent translation in a 2A protease-treated rabbit reticulocyte lysate (RRL) translation system (42). These studies suggest that eIF4G cleavage is not solely responsible for the host shutoff.
Most eukaryotic mRNAs contain a 3′-terminal poly(A) tract. Some of the known functions of the poly(A) tract include nucleocytoplasmic transport of mRNA (28), stabilization of cytoplasmic mRNA (12), and enhancement of mRNA translation (21, 31). A long 3′-poly(A) tract stimulates initiation synergistically with the 5′ cap (20–22, 49). During vertebrate embryonic development, changes in poly(A) length are the basis for translational control of developmentally regulated mRNAs (2, 25). The poly(A) tract binds a poly(A)-binding protein (PABP, also referred to as PABP I) of ∼70 kDa, which is representative of a large family of eukaryotic RNA-binding proteins. The primary sequence of PABP is highly conserved among the Xenopus, mouse, and human species (>94% identity). The N-terminal two-thirds of the protein consists of four tandemly arranged RNA recognition motifs (RRMs), each of which is ∼90 amino acid residues in length (1, 60). A single RRM is sufficient for poly(A) binding, and the affinity of a truncated PABP containing only the first two RRMs is similar to that of the full-length protein (38, 60). The presence of multiple RRM domains enables PABP to transfer between poly(A) strands (60). The C-terminal one-third of PABP permits homodimerization on RNA and creation of higher-order PABP-poly(A) structures (3, 38).
The role of the poly(A) tract in translation is thought to be mediated by PABP. The poly(A) tracts of only those mRNAs that are actively being translated in polyribosomes are associated with PABP (27). Some human immunodeficiency virus type 1 mRNAs that are translationally inactive in the absence of the Rev protein are also devoid of bound PABP (11). PABP from the ribosomal fraction of embryo axes of pea seeds stimulates the translation of poly(A)-containing mRNAs in vitro (63). Translation of capped, polyadenylated mRNAs is inhibited by addition of poly(A) to an RRL translational system (49). This inhibition can be relieved by the addition of excess PABP, suggesting that the added poly(A) sequesters endogenous PABP (23). Addition of monoclonal antibodies to PABP inhibits the translation of uncapped, polyadenylated mRNA, but this is restored by the addition of recombinant PABP (68). Various experimental approaches suggest that PABP acts in the joining of the 40S ribosomal subunit to the mRNA (68) and the recruitment of the 60S ribosomal subunit (59). The association of PABP with the N terminus of eIF4G and with eIF4B in yeast and wheat germ suggests that poly(A)-dependent translation involves the canonical initiation factors (43, 67). This idea is strengthened by the demonstration that synergism between the cap and the poly(A) tract, mediated by eIF4E and PABP, respectively, also requires the N terminus of eIF4G (68, 69). Recent evidence shows that in mammalian cells, PABP interacts with eIF4A via a novel protein, PAIP-1 (PABP-interacting protein-1), which stimulates protein synthesis (16).
In the present study, we demonstrate that PABP, like eIF4G, is cleaved by 2A protease during coxsackievirus infection of HeLa cells, providing another potential mechanism by which picornavirus infection may affect host cell translation.
MATERIALS AND METHODS
Materials.
[3H]Leu (60 Ci/mmol) was purchased from DuPont-NEN, and [35S]Met (>1,000 Ci/mmol) was purchased from both DuPont-NEN and Amersham. Horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG) and alkaline phosphatase-conjugated horse anti-mouse IgG were obtained from Vector Laboratories, Burlingame, Calif. Immobilon P was purchased from Millipore, Bedford, Mass. Glutathione-Sepharose 4B and protein A-agarose were obtained from Pharmacia and Pierce, respectively. Female frogs (Xenopus laevis) were purchased from Xenopus I, Madison, Wis. Cyanogen bromide-activated Sepharose 4B and poly(A) were purchased from Sigma.
Cells and cell culture.
HeLa cells (a gift from S. A. Huber, University of Vermont, Burlington, Vt.) and COS-1 cells (American Type Culture Collection, Rockville, Md.) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco BRL, Gaithersburg, Md.) with high glucose (4.5 g/liter) supplemented with 10% heat-inactivated fetal bovine serum, 1 mM sodium pyruvate, 2 mM l-glutamine, 100 μg of streptomycin per ml, and 100 U of penicillin G per ml all from Irvine Scientific, Santa Ana, Calif.) at 37°C under 5% CO2.
Virus and viral assays.
A plasmid containing the full-length, infectious cDNA of coxsackievirus B3 (37) was transfected into COS-1 cells. At 72 h posttransfection, the supernatant was transferred to HeLa cells. When the cytopathic effect was complete, cells were lysed and the virus was passaged once in HeLa cells for amplification. The lysate was subjected to three cycles of freeze-thawing for release of intracellular virus. The supernatant was then subjected to titer determination by a plaque-forming assay with HeLa cells as the titering cell line (29).
Infection of HeLa cells with coxsackievirus and pulse-labeling.
HeLa cells (2 × 106) were plated on a 90-mm dish and allowed to adhere overnight. Cells were infected with coxsackievirus B3 at a multiplicity of infection of 2 in 1 ml of DMEM with 2% heat-inactivated fetal bovine serum. After 30 min of incubation, new medium (containing l-Met) was added to the cells. At 30 min prior to harvest, the medium was removed and the cells were washed three times with Met-deficient DMEM. Then Met-deficient medium was added to each plate, followed by 50 μCi of [35S]Met, and the cells were returned to the incubator for 30 min. At 0 h (mock infected) and at various times postinfection, the cells were washed once with phosphate-buffered saline, trypsinized, and pelleted by centrifugation at 300 × g for 10 min. The cells were then resuspended in 100 ml of lysis buffer with 1 mM dithiothreitol (DTT; Clontech, Palo Alto, Calif.) and allowed to stand for 15 min at 4°C. After removal of insoluble material, the supernatant was stored in aliquots at −20°C.
Preparation of 2A protease.
Plasmid pET8c/CVB4 2A, a gift from Tim Skern, University of Vienna, encodes a fusion protein consisting of 13 amino acids of the T7 gene 10 protein, 34 amino acids of coxsackievirus B4 VP1, and 150 amino acids of coxsackievirus B4 2A protease. This plasmid was used to produce recombinant 2A protease in Escherichia coli (44). The enzyme was purified and stored at 4°C at a concentration of 2.9 mg/ml in buffer A (50 mM Tris-HCl [pH 8.0], 50 mM NaCl, 1 mM EDTA, 5 mM DTT, 50% glycerol).
Preparation of PABP.
Poly(A)-Sepharose was prepared by immobilization of poly(A) on cyanogen bromide-activated Sepharose 4B (53). For the preparation of rabbit PABP (rPABP), 40 ml of the postribosomal supernatant of RRL (15) was made 100 mM with respect to KCl and passed through 10 ml of poly(A)-Sepharose in a column as described previously (19). The bound PABP was eluted with buffer B (50 mM Tris-HCl [pH 7.6], 2 M LiCl). Fractions containing PABP (typically 0.5 ml) were identified by the Bradford assay (10), confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% polyacrylamide) with Coomassie blue staining, dialyzed overnight against 10 mM Tris-HCl (pH 7.6)–50 mM KCl at 4°C, and concentrated fourfold in a Microcon-30 apparatus (Amicon). This procedure typically yielded 50 μg of PABP.
A plasmid encoding recombinant human PABP fused at the N terminus to glutathione S-transferase (GST-hPABP) in the pGEX-2T vector (Pharmacia Biotech) was provided by Jnanankur Bag (University of Guelph, Canada) (4). The GST-hPABP was expressed in E. coli XL1-Blue cells (Stratagene) by induction with 0.1 mM isopropyl-β-d-thiogalactopyranoside and purified on glutathione-Sepharose (4). Bound GST-hPABP was eluted with 10 mM Tris-HCl (pH 8.0)–10 mM reduced glutathione.
hPABP was enriched from extracts (500 μl) of coxsackievirus-infected HeLa cells by the addition of 50 μl of a 50% (vol/vol) slurry of oligo(dT)-cellulose followed by 50 μl of a 50% (vol/vol) slurry of poly(A)-Sepharose. To the slurry was added 600 μl of 20 mM HEPES (pH 7.0)–100 mM KCl–250 mM NaCl–2 mM magnesium acetate–0.1 mM EDTA–0.5 mM DTT. The slurry was rotated at 4°C for 1 h and centrifuged at 500 × g. The supernatant was discarded, and the resin was washed once with the same buffer. The bound material was eluted by heating in a twice-concentrated Laemmli sample buffer and subjected to SDS-PAGE (39).
Preparation of protein fragments for sequencing.
For preparation of the 55-kDa N-terminal cleavage product of rabbit PABP (rPABPN), 20 ml of the postribosomal supernatant from RRL was treated overnight with 110 μl of 2A protease (final concentration, 15 μg/ml) at room temperature. The solution was passed over a poly(A)-Sepharose column, and the bound material (rPABPN) was eluted with buffer B, as described for purification of intact rPABP. The protein sample was concentrated in a Microcon-30 apparatus and electrophoretically transferred to a Teflon membrane at Kendrick Laboratories (Madison, Wis.), and the C-terminal amino acid was determined by sequencing on a Hewlett-Packard G1009A C-terminal sequencer at Argo BioAnalytica, Inc. (Morris Plains, N.J.).
For preparation of the 15-kDa C-terminal cleavage product of human PABP (hPABPc), 658 μg of GST-hPABP was treated with 13 μl of 2A protease (final concentration, 50 μg/ml) in a total volume of 0.75 ml of buffer A (without glycerol) at 30°C for 4 h. A slurry of glutathione-Sepharose (50% [vol/vol]; 0.65 ml) was added, and the incubation was continued for 1 h at room temperature with constant rotation. The slurry was then poured into a column, and the flowthrough fraction, containing both hPABPc and 2A protease, was collected. (GST-hPABPN remained bound to the glutathione-Sepharose.) To resolve hPABPc from 2A protease, an aliquot of the flowthrough fraction (∼5%) was applied to a 0.46- by 25-cm C5 reverse-phase column (Phenomenex, Inc., Torrance, Calif.) equilibrated in 0.1% aqueous trifluoroacetic acid, and the column was developed with a linear gradient of acetonitrile (40). Fractions were analyzed by SDS-PAGE to identify hPABPc. The pure hPABPc (0.8 μg) was subjected to automated Edman degradation with an Applied Biosystems model 470A Sequenator at the University of Kentucky Macromolecular Structure Analysis Facility.
Expression and cleavage of PABP in Xenopus oocytes.
Capped, polyadenylated RNA encoding Xenopus PABP with an N-terminal Myc epitope tag (Myc-xPABP) was synthesized in vitro (47) from plasmid pSDM (provided by Peter Good, Louisiana State University Medical Center) by using SP6 polymerase (Promega Biotech). An ovary was surgically removed from female Xenopus laevis and rinsed with modified Barth’s saline. Stage VI oocytes were isolated manually, sorted, and injected with RNA (34). The oocytes were cultured for 12 h at room temperature, injected with recombinant 2A protease, cultured further for 2.5 h, and frozen in groups of 17. The oocytes were homogenized at 4°C in 0.5 ml of 30 mM HEPES (pH 7.5)–70 mM NaCl–7 mM 2-mercaptoethanol–0.1% Triton X-100–0.01% SDS–10 μg of RNase A per ml–0.5 μg of leupeptin per ml–1 mM phenylmethylsulfonyl fluoride and centrifuged at 25,000 × g for 5 min. Supernatants were diluted to 1 ml with the same buffer, and Myc-xPABP was immunoprecipitated with 1 μl of anti-Myc tag monoclonal 9E10 antibody (provided by Kelly Tatchell, Louisiana State University Medical Center). Incubation was carried out for 1 h at 4°C with gentle agitation. Immune complexes were immobilized on protein A-agarose and washed three times with the same buffer at room temperature. Protein was eluted from the beads with twice-concentrated Laemmli sample buffer at 100°C for 5 min. The immunoprecipitates were subjected to immunoblotting as described below.
Immunoblotting.
eIF4G, eIF4E and PABP were separated by SDS-PAGE and electrophoretically transferred to a polyvinylidene difluoride (PVDF) membrane with a Bio-Rad Mini Trans-Blot cell. The membranes were incubated with antibodies against either eIF4E, eIF4G, PABP, or Myc tag. Anti-PABP antibody was provided by Dan Schoenberg (Ohio State University, Columbus, Ohio). Immunoblotting for eIF4G was performed with antiserum which recognizes amino acid residues 327 to 342 in cpN (76). Myc-xPABP was detected with the anti-Myc tag antibody described above. eIF4E was purified from human erythrocytes by affinity chromatography on m7GTP-Sepharose (71) and reverse-phase high-pressure liquid chromatography (HPLC) on a C4 column (33) and used as a source of antigen for production of eIF4E antiserum in New Zealand rabbits. Immunoreactive species were visualized with goat anti-mouse secondary antibody conjugated to alkaline phosphatase (for Myc) or goat anti-rabbit secondary antibody conjugated to either horseradish peroxidase or alkaline phosphatase.
In vitro translation.
In vitro protein synthesis rates in a micrococcal nuclease-treated RRL translation system were measured by incorporating [3H]Leu into newly synthesized globin as described previously (13), except that the potassium acetate concentration was 100 mM instead of 150 mM. Translation reaction mixtures of 25 μl were programmed with total rabbit globin mRNA (10 μg/ml) and incubated at 30°C for the times indicated in the figures. Aliquots of 4 μl were withdrawn and precipitated in 10% trichloroacetic acid, and the radioactivity was determined by scintillation spectrometry.
RESULTS
Cleavage of PABP by 2A protease in RRL.
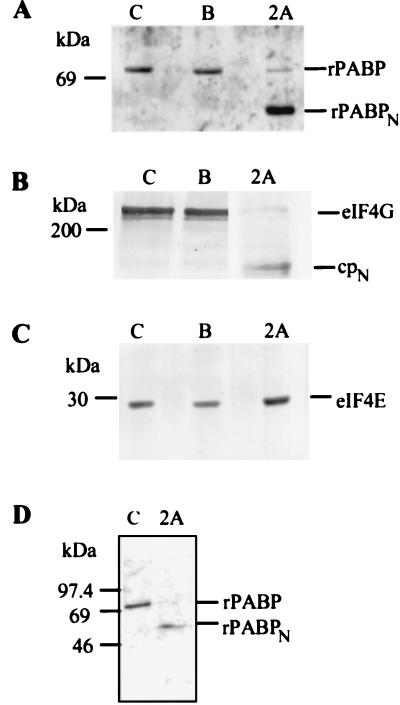
A number of published studies have used recombinant 2A protease (from rhinovirus or coxsackievirus) or L protease (from foot-and-mouth disease virus) to study the effects of cleaving eIF4G on cap-dependent and cap-independent translation in vitro (7, 8, 42, 44, 51, 52). Under conditions that are typically used for eIF4G cleavage in an RRL translation system, PABP was also cleaved (Fig. 1A). The 70-kDa PABP remained intact for 10 min of incubation in the control translation reaction mixtures (lane C) or in reaction mixtures to which only buffer was added (lane B) but was cleaved to a ∼55-kDa product by 2A protease addition (lane 2A). eIF4G also remained intact in the control and buffer-containing translation reactions but was cleaved to cpN by 2A protease (Fig. 1B). (The antigen used to produce the anti-eIF4G antiserum was a peptide located in the N-terminal portion; hence, only full-length eIF4G and cpN are reactive.) The eIF4E in these lysates remained intact under all three conditions (Fig. 1C).
FIG. 1.
Effect of 2A protease on PABP, eIF4G, and eIF4E in vitro. (A to C) An RRL translation system was incubated for 10 min at 30°C with no additions (lanes C), with 2A protease at a final concentration of 300 μg/ml (lanes 2A), or with an equivalent volume of buffer A in which the 2A protease is stored (lanes B). Samples were subjected to SDS-PAGE on either 10% (A and C) or 6% (B) polyacrylamide gels and electroblotted onto PVDF membranes. Immunodetection was carried out either with anti-PABP (A), anti-eIF4G (B), or anti-eIF4E (C) primary antibodies. (D) rPABP (100 μg/ml) from the postribosomal supernatant of RRL was incubated alone (lane C) or was treated with 50 μg of 2A protease per ml (lane 2A) at 30°C for 4 h. Samples were subjected to SDS-PAGE on a 10% gel, stained with Coomassie blue R-250, and destained with 40% methanol.
The reduction in size of the 70-kDa PABP to ∼55 kDa suggests that an ∼15-kDa fragment was removed. Such a fragment, however, was not detected in immunoblots, due to the absence of an antigenic site. (The antigen used for the production of the anti-PABP antiserum was a peptide derived from the central proline-rich region of PABP.) To test for its presence, we synthesized [35S]Met-labeled Myc-xPABP in an RRL translation system. The reaction mixture was incubated with 100 μg of 2A protease per ml overnight, and the cleavage products were subjected to SDS-PAGE on 15% polyacrylamide gels followed by autoradiography. Radioactive products of ∼55 and ∼15 kDa were present in the samples treated with 2A protease but not in control samples incubated with buffer A alone (data not shown).
Several studies have provided evidence that a latent cellular protease becomes activated by 2A protease and is responsible for the cleavage of eIF4G during poliovirus infection (reviewed in reference 73). Other studies have indicated that eIF4G is a substrate for cellular calcium-dependent proteases (74) and requires initiation factor eIF3 for efficient 2A protease-induced cleavage (75). It was therefore of interest to test whether PABP was a direct substrate for 2A protease. rPABP was purified to homogeneity from RRL (Fig. 1D, lane C) and incubated with the homogeneous, recombinant 2A protease. Appearance of the 55-kDa cleavage product (lane 2A) indicates that neither secondary proteases nor ancillary proteins are required.
Cleavage of Myc-xPABP by 2A protease in Xenopus oocytes.
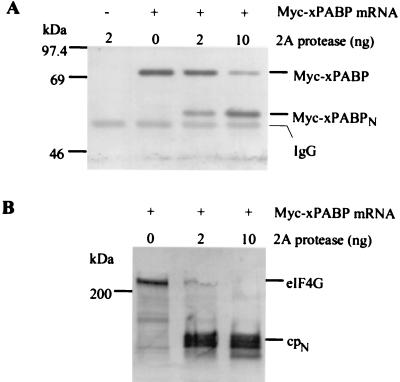
The cleavage of PABP by 2A protease was also studied in Xenopus oocytes. An in vitro-synthesized mRNA encoding Xenopus PABP containing an N-terminal Myc epitope tag was microinjected into oocytes, and the Myc-xPABP was allowed to accumulate. Doses of 2 and 10 ng of 2A protease were subsequently microinjected into the oocytes, and the incubation was continued. Myc-xPABP was immunoprecipitated from oocyte extracts with an anti-Myc tag antibody, and the products were analyzed by immunoblotting with the same antibody (Fig. 2A). PABP was partially cleaved at 2 ng of protease per oocyte and almost completely cleaved at 10 ng. The same extracts were also analyzed for the cleavage of endogenous eIF4G (Fig. 2B). The eIF4G was completely cleaved at both 2 and 10 ng of protease per oocyte, indicating that eIF4G is more sensitive to cleavage by 2A protease than is PABP. Also, the detection of the 55-kDa band by the anti-Myc antibody identifies it as the N-terminal cleavage product of PABP (henceforth referred to as PABPN) and establishes that the C-terminal ∼20% of the molecule (PABPc) is removed by 2A protease. Finally, these results indicate that PABP from evolutionarily distant organisms (rabbit and frog) are similarly cleaved.
FIG. 2.
Effect of 2A protease on PABP and eIF4G in vivo. Xenopus oocytes were microinjected with 15 ng of in vitro-synthesized mRNA for Myc-xPABP (+) or not injected (−) and incubated for 12 h at room temperature. Then oocytes were injected with 2 or 10 ng of 2A protease per oocyte or not injected, as indicated, and incubation was continued for 2.5 h. (A) Myc-xPABP was immunoprecipitated from the oocyte extracts with anti-Myc tag antibody. The immunoprecipitates were resolved by SDS-PAGE on a 6% polyacrylamide gel, transferred to a PVDF membrane, and probed with the same antibody. The additional band seen in all lanes represents the Ig heavy chain (IgG). (B) Equal amounts of total protein from each extract were resolved by SDS-PAGE on a 6% polyacrylamide gel, transferred to PVDF, and probed for endogenous eIF4G with anti-eIF4G antibody.
Cleavage of PABP in coxsackievirus-infected HeLa cells.
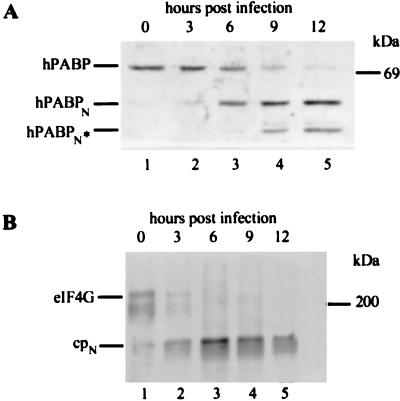
The foregoing experiments establish that PABP can be cleaved in vitro (Fig. 1) and in vivo (Fig. 2) by 2A protease, but they do not indicate whether the levels of 2A protease achieved during picornavirus infection of mammalian cells are sufficient for PABP cleavage. This was investigated by analyzing PABP and eIF4G at various times after infection of HeLa cells with coxsackievirus B3 (Fig. 3). PABP cleavage was more than 50% complete by 6 h postinfection and was essentially complete by 9 h (Fig. 3A). In addition to the 55-kDa PABPN, a faster-migrating, secondary cleavage product, PABPN*, appeared by 9 h postinfection, which could indicate the presence of a second cleavage site for the 2A protease on PABP. However, since secondary cleavage was not observed in reactions with only purified PABP and 2A protease (Fig. 1D), it is more likely that secondary cleavage in HeLa cells was due to another protease. Cleavage of eIF4G in infected HeLa cells was extensive by 3 h and was complete by 6 h (Fig. 3B).
FIG. 3.
Coxsackievirus infection of HeLa cells results in cleavage of PABP. HeLa cells were infected with coxsackievirus B3, and aliquots were withdrawn at various times postinfection. (A) PABP was enriched from samples taken at the indicated times as described in Materials and Methods, subjected to SDS-PAGE on a 10% polyacrylamide gel, and detected by immunoblotting. (B) The same infected cell extracts as in panel A were directly subjected to SDS-PAGE on 6% polyacrylamide gels, and eIF4G was detected by immunoblotting.
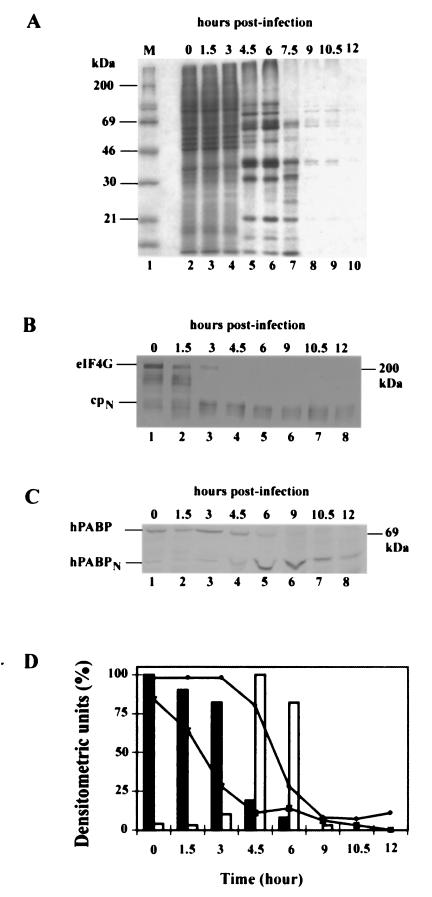
To more precisely determine the kinetics of PABP cleavage relative to those of eIF4G cleavage and also to correlate these cleavages with the progress of viral infection and host shutoff, another experiment was performed in which more frequent time points were used and total protein synthesis was measured by pulse-labeling with [35S]Met for 30 min before each time point. Host protein synthesis continued for 3 h, after which viral protein synthesis predominated until 7.5 h (Fig. 4A). Cleavage of eIF4G was ∼75% complete by 3 h and was ∼100% complete by 4.5 h (Fig. 4B). PABP cleavage was detectable by 4.5 h, ∼75% complete by 6 h, and essentially complete by 9 h (Fig. 4C). PABP cleavage therefore lagged behind eIF4G cleavage by ∼3 h, suggesting that PABP is less sensitive to 2A protease than is eIF4G. This is consistent with the dose-response results obtained with Xenopus oocytes (Fig. 2).
FIG. 4.
Temporal relationships among shutoff of host proteins synthesis, viral protein synthesis, eIF4G cleavage, and PABP cleavage. HeLa cells were infected with coxsackievirus B3 and pulse-labeled with [35S]Met 30 min before being harvested as described in Materials and Methods. Aliquots were withdrawn at various times postinfection. (A) Total protein (10 μg) from each cell extract was resolved by SDS-PAGE on a 10% polyacrylamide gel, dried, and subjected to autoradiography. (B) Cell extracts (1 μg of total protein) were subjected to immunoblotting for eIF4G as in Fig. 3. (C) PABP was detected by immunoblotting as in Fig. 3. (D) The autoradiogram and immunoblots were scanned on a densitometer, and the percentages (expressed in arbitrary densitometric units) of intact PABP (—•—), intact eIF4G (—×—), synthesis of a representative cellular protein (∼46 kDa; solid bars), and synthesis of a representative a viral protein (∼35 kDa; open bars) are shown as a function of time.
All four parameters are quantified in Fig. 4D. Interestingly, at 3 h, host protein synthesis had decreased only ∼20% and viral protein synthesis had just become detectable at a time when eIF4G cleavage was ∼75% complete, indicating a lack of correlation between the translational switch and eIF4G cleavage. At 4.5 and 6 h, by contrast, PABP was partially cleaved, host protein synthesis had been almost completely shut off, and viral protein synthesis was at maximal levels. The subsequent decline in viral synthesis correlated with a further decline in the amount of intact PABP.
Determination of the cleavage site in PABP.
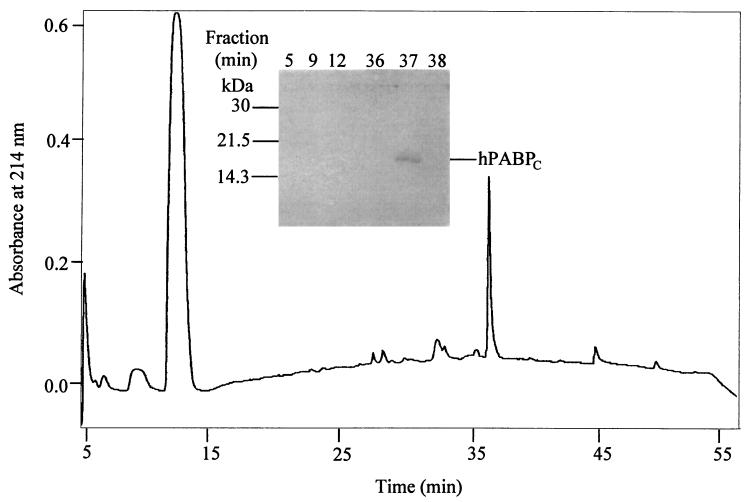
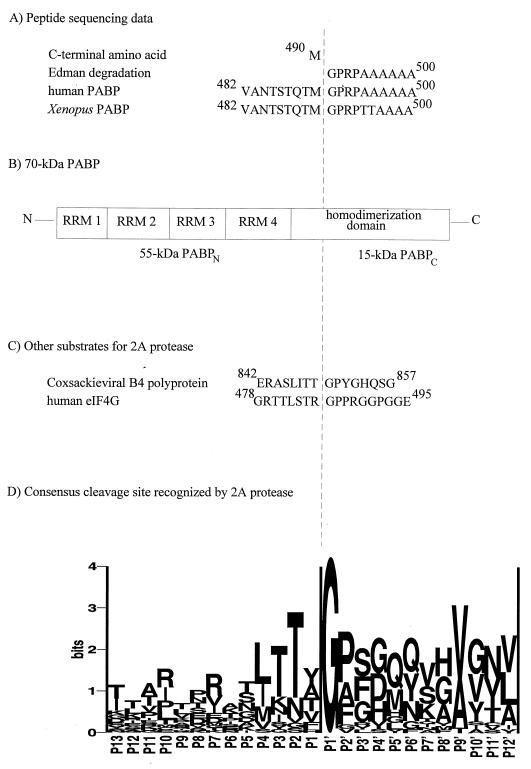
We next determined the cleavage site of 2A protease in PABP. Recombinant GST-hPABP was digested with 2A protease in vitro, and hPABPc was separated from GST-hPABPN on glutathione-Sepharose. hPABPc was further purified by reverse-phase HPLC on a C5 column (Fig. 5), and fractions were analyzed by SDS-PAGE (inset). The hPABPc peak was subjected to automated Edman degradation for determination of its N-terminal sequence (Fig. 6A). Ten residues were unambiguously assigned, and their sequence exactly matched that of hPABP beginning at Gly-491. These results indicate that the cleavage by 2A protease occurs at 490M↓G491 in hPABP. The sequences of hPABP and xPABP are identical in this region (Fig. 6A). The sequence of rPABP does not appear in public databases. However, amino acid sequences derived from human and mouse cDNAs are 99.5% identical, and those derived from human and Xenopus cDNAs are 93.7% identical. Thus, it is likely that PABP from all four species is cleaved at the same site. This is consistent with the fact that the apparent molecular masses of cleavage products, derived from electrophoretic mobility, are the same for rPABP, xPABP, and hPABP (after correction for N-terminal tags) (Fig. 1–4).
FIG. 5.
Isolation of hPABPc by reverse-phase HPLC. Purified GST-hPABP was cleaved in vitro by 2A protease and loaded onto a glutathione-Sepharose column as described in Materials and Methods. The flowthrough fraction containing hPABPc was loaded directly onto a C5 column. Fractions with absorbance at 214 nm were subjected to SDS-PAGE on a 15% polyacrylamide gel followed by Coomassie blue staining (inset).
FIG. 6.
Determination of the in vitro cleavage site of 2A protease in PABP. (A) rPABPN was isolated as described in Materials and Methods and subjected to C-terminal amino acid analysis. hPABPc was isolated as described in the legend to Fig. 5 and subjected to sequential Edman degradation. The results of each analysis are compared with a portion of the sequences of hPABP and xPABP. (B) Schematic diagram showing the position of the cleavage site on PABP. (C) Known cleavage sites for coxsackievirus 2A protease on other naturally occurring proteins. (D) Sequence logo of 2A protease cleavage sites. The size of the letters representing amino acid residues denotes the frequency of their occurrence in 22 enterovirus and rhinovirus polyproteins relative to the 2A protease cleavage site. Thus, Gly is found exclusively in the position immediately C-terminal to the cleavage site (P1′), etc. Panel D reprinted from reference 5 with permission of the authors and publisher.
Cleavage by 2A protease is highly dependent on the amino acid sequence of the substrate (64, 65), but it was possible that it cleaved PABP at two nearby sites, thus removing a short peptide between PABPN and PABPc. To verify that a single cleavage occurs, we prepared rPABPN and subjected it to C-terminal sequencing (see Materials and Methods). Since C-terminal sequencing is much less processive than N-terminal sequencing, it was possible to assign only the C-terminal amino acid residue with confidence. The assignment of Met matched the sequence of both hPABP and xPABP upstream of Gly-491 (Fig. 6A), supporting the existence of only a single cleavage site. Further evidence comes from the observation that the molecular masses for hPABPN and hPABPc estimated from electrophoretic mobility (55 and 15 kDa, respectively) are in excellent agreement with the calculated masses for cleavage at 490M↓G491 (55.2 and 15.5 kDa, respectively).
Cleavage of PABP by 2A protease diminishes its ability to stimulate translation in vitro.
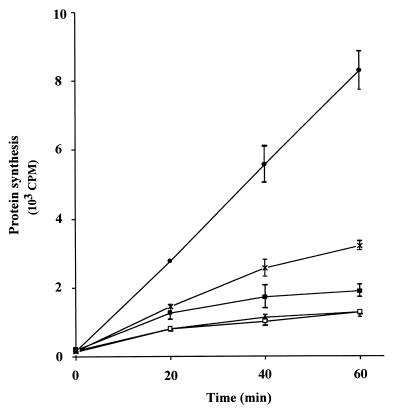
The effect of PABP cleavage on in vitro translation was determined in an RRL translation system programmed with exogenous rabbit globin mRNA. The system was preincubated for 5 min at room temperature with the cap analog m7GTP and with low levels of poly(A) to make it less dependent on cap-mediated translation (69) and more dependent on exogenous PABP (23), respectively. The addition of both m7GTP and poly(A) inhibited translation synergistically: 100 μM m7GTP alone caused 15% inhibition, 8 μg of poly(A) per ml alone caused 58% inhibition, but the combination of 100 μM m7GTP and 8 μg of poly(A) per ml caused 95% inhibition (data not shown). Inhibition by m7GTP was relatively weak because of the low potassium acetate concentration (14). These results are consistent with earlier published studies showing that the cap and poly(A) are functionally redundant for mRNA recruitment (21, 68), although this has not previously been demonstrated in RRL.
Equimolar amounts (0.6 nM) of either GST-hPABP or GST-hPABPN were added to this PABP-dependent system to measure restoration of translation (Fig. 7). GST-hPABP was more effective than GST-hPABPN in restoring translation (28% restoration of mRNA-stimulated protein synthesis after 60 min compared with 9% for PABPN). At 0.4 and 0.2 nM GST-hPABP and GST-hPABPN, the extent of restoration was lower, but GST-hPABP was consistently more effective than GST-hPABPN (data not shown). These results indicate that cleavage of PABP impairs poly(A)-dependent translation in vitro.
FIG. 7.
Effect of PABP cleavage on in vitro translation. Protein synthesis was measured by the addition of [3H]Leu and globin mRNA (10 μg/ml) to a micrococcal nuclease-treated RRL system (•). The system was made dependent on added PABP by preincubation with 8 μg of poly(A) per ml (23) and 100 μM m7GTP (69) for 5 min at room temperature. Equimolar amounts (0.6 nM) of GST-hPABP (×) or GST-hPABPN (■) were then added, and translation was initiated. Reaction mixtures containing no mRNA and no inhibitors (□) or mRNA plus poly(A) plus m7GTP but no PABP (▴) served as controls. Similar results were obtained in two other experiments with GST-hPABP and GST-hPABPN at the same concentrations (data not shown).
DISCUSSION
The data presented here identify a previously unreported event that occurs during infection with coxsackievirus, namely, the cleavage of PABP, a factor involved in several aspects of mRNA metabolism. We have shown that the 2A protease separates a 55-kDa N-terminal fragment, containing the four RRMs, from the C-terminal homodimerization domain (Fig. 6B). The C-terminal domain does not contribute to RNA affinity or selectivity but cooperates with RRMs 3 and 4 to achieve a poly(A)-organizing activity, whereby multiple copies of PABP can assemble on poly(A) to form a repetitive, higher-order complex with a repeat RNA length of 27 nucleotides (3, 38). Yeast extracts containing PABP with a C-terminal truncation have a 10-fold reduction in their ability to translate uncapped polyadenylated mRNAs relative to wild-type extracts (35). This is consistent with our observation that GST-hPABPN is less able to restore translation to a PABP-dependent translation system than is intact GST-hPABP (Fig. 7). Similarly, in mature Xenopus oocytes, PABP lacking the C terminus and RRM 4 is not able to protect mRNAs from deadenylation (72). RRM 2 of PABP is required for interaction with eIF4G in yeast (35), but the domains on PABP that potentially interact with PAIP-1, eIF4B, or 40S or 60S ribosomal subunits have not yet been identified. Thus, it is possible that the loss of the C-terminus alters the interaction of PABP with PAIP-1 and/or some component of the translational machinery, making it unavailable for initiation or reinitiation. Alternatively, the C terminus of PABP may not be involved in “direct” interactions with other protein synthesis components but may function by homodimerization; the C termini of two PABP molecules would homodimerize, allowing the N terminus of one PABP molecule to bind poly(A) and the N terminus of the other PABP molecule to bind to PAIP-1 or other proteins.
The role of PABP cleavage in coxsackievirus infection is unclear. The picornavirus polyprotein is cleaved by virus-encoded proteases into 11 functional proteins during translation (58). One of these proteases, 2A, plays at least two roles in the picornavirus life cycle. One role is to cleave the capsid precursor protein P1 from the nascent polypeptide, a step that is necessary for continued efficient translation (70). Another role is to cleave eIF4G, either directly or indirectly, after which host protein synthesis shuts down and viral protein synthesis proceeds (36, 40, 41, 44, 73). The absence of competition with host mRNAs may contribute to the increase in viral protein synthesis, but in the case of enteroviruses and rhinoviruses, there is also a stimulation by the C-terminal eIF4G cleavage product, cpc (8, 50, 52). Given that both the 5′-cap and the 3′-poly(A) tract have a positive effect on translation initiation rates (61), the cleavage of PABP may enable the virus to more effectively shut down protein synthesis.
Alternatively, the cleavage of PABP may affect the translation of picornavirus RNA. The poly(A) tracts of picornavirus RNAs are quite heterogeneous in length. The function of these tracts is not known, but poliovirus RNA molecules with short poly(A) tracts have a lower specific infectivity (62, 66). It is possible, by analogy to host cellular mRNAs, that PABP plays a positive role in the translation of picornavirus RNAs. Cleavage of PABP by 2A protease may be related to the switch from translation of picornavirus RNA to replication. Increasing evidence supports a model for picornavirus RNA amplification that initiates with the elongation of a pUpU-VPg primer derived from the viral protein 3AB, which in turn primes the transcription of polyadenylated viral RNA (54). The virus may conceivably cleave PABP to eliminate higher-order PABP-poly(A) structures (3) that favor translation, thereby allowing the poly(U)-VPg primer to anneal to poly(A) and permitting viral replication to occur.
The substrate specificity of 2A protease has been used to predict cellular proteins that may be potential targets for the protease. Based on cleavage of a series of peptides in vitro, a consensus sequence of I/L-X-T-X↓G-P (where X is any amino acid) was proposed (64, 65). More recently, the sequence specificity of 2A protease has been characterized by using a neural network algorithm and graphical visualization technique (5). An analysis of 22 naturally occurring 2A protease cleavage sites in enterovirus and rhinovirus polyproteins indicated that there is an absolute requirement for Gly at position P1′, a strong preference for Thr at P2 and Pro at P2′, and a variety of less highly conserved preferences (Fig. 6D). The experimentally determined 2A protease cleavage sites in the coxsackievirus B4 polyprotein and eIF4G adhere to this specificity (Fig. 6C). The cleavage site on the coxsackievirus B3 polyprotein has not been deduced experimentally, but comparison with other enteroviruses and rhinoviruses predicts the site to be TTMTNT↓GAFGQ. The cleavage site within PABP, determined in the present study, also conforms to this specificity (compare Fig. 6A and D). The invariant Gly is present at P1′, and Ala, Thr, and Ser, are present at P9′, P4, and P5, respectively, each of which is the second most favorable residues at that position (Fig. 6D). Interestingly, an inducible form of PABP has been found in activated T lymphocytes which is 79% identical to the noninducible form but which lacks the consensus sequence for cleavage by 2A protease (77).
It is surprising that the neural network algorithm did not predict PABP as a potential cellular target (5). However, the authors of this study removed from the list of potential cellular targets those proteins in which the cleavage sites were predicted to be topologically inaccessible to the protease. This raises the interesting possibility that the cleavage site in PABP is normally inaccessible but becomes exposed upon binding to poly(A), PAIP-1, or eIF4G. This, in fact, occurs in the case of eIF4G upon binding to eIF4E (24).
ACKNOWLEDGMENTS
We are indebted to Nadejda Korneeva for synthesizing the poly(A)-Sepharose resin and for her input and expert advice. We thank Kelly Tatchell and Dan Schoenberg for mouse anti-Myc tag and rabbit anti-PABP antibodies, respectively. We also thank Jnanankur Bag, Peter Good, and Tim Skern for expression vectors encoding GST-hPABP, myc-xPABP, and 2A protease, respectively. Finally, we thank S. A. Huber for HeLa cells.
This work was supported by grant GM20818 from the National Institutes of Health, grant 96-303 from the American Heart Association, grant S96-38 from the UCSD Biotechnology Star project, and grant Ba1668/1-1 from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Adam S A, Nakagawa T, Swanson M S, Woodruff T K, Dreyfuss G. mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol Cell Biol. 1986;6:2932–2943. doi: 10.1128/mcb.6.8.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachvarova R F. A maternal tail of poly(A)—the long and the short of it. Cell. 1992;69:895–897. doi: 10.1016/0092-8674(92)90606-d. [DOI] [PubMed] [Google Scholar]
- 3.Baer B W, Kornberg R D. The protein responsible for the repeating structure of cytoplasmic poly(A)-ribonucleoprotein. J Cell Biol. 1983;96:717–721. doi: 10.1083/jcb.96.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bag J, Wu J. Translational control of poly(A)-binding protein expression. Eur J Biochem. 1996;273:143–152. doi: 10.1111/j.1432-1033.1996.0143n.x. [DOI] [PubMed] [Google Scholar]
- 5.Blom N, Hansen J, Blaas D, Brunak S. Cleavage site analysis in picornaviral polyproteins: discovering cellular targets by neural networks. Protein Sci. 1996;5:2203–2216. doi: 10.1002/pro.5560051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonneau A-M, Sonenberg N. Proteolysis of the p220 component of the cap-binding protein complex is not sufficient for complete inhibition of host cell protein synthesis after poliovirus infection. J Virol. 1987;61:986–991. doi: 10.1128/jvi.61.4.986-991.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borman A M, Bailly J-L, Girard M, Kean K M. Picornavirus internal ribosome entry segments: comparison of translation efficiency and the requirements for optimal internal initiation of translation in vitro. Nucleic Acids Res. 1995;23:3656–3663. doi: 10.1093/nar/23.18.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borman A M, Kirchweger R, Ziegler E, Rhoads R E, Skern T, Kean K M. eIF4G and its proteolytic cleavage products: effect on initiation of protein synthesis from capped, uncapped, and IRES-containing mRNAs. RNA. 1997;3:186–196. [PMC free article] [PubMed] [Google Scholar]
- 9.Bovee M L, Lamphear B J, Rhoads R E, Lloyd R E. Direct cleavage of eIF4G by poliovirus 2A protease is inefficient in vitro. Virology. 1998;245:241–249. doi: 10.1006/viro.1998.9172. [DOI] [PubMed] [Google Scholar]
- 10.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 11.Campbell L H, Borg K T, Haines R T, Moon R T, Schoenberg D R, Arrigo S J. Human immunodeficiency virus type 1 is required for binding of poly(A)-binding protein to rev-dependent RNAs. J Virol. 1994;68:33–38. doi: 10.1128/jvi.68.9.5433-5438.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caponigro G, Parker R. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol Rev. 1996;60:233–249. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu L Y, Rhoads R E. Inhibition of cell-free messenger ribonucleic acid translation by 7-methylguanosine 5′-triphosphate: effect of messenger ribonucleic acid concentration. Biochemistry. 1980;19:184–191. doi: 10.1021/bi00542a028. [DOI] [PubMed] [Google Scholar]
- 14.Chu L-Y, Rhoads R E. Translational recognition of the 5′-terminal 7-methylguanosine of globin messenger RNA as a function of ionic strength. Biochemistry. 1978;17:2450–2455. doi: 10.1021/bi00605a032. [DOI] [PubMed] [Google Scholar]
- 15.Clemens M J. Translation of eukaryotic messenger RNA in cell-free extracts. In: Hames B D, Higgins S L, editors. Transcription and translation. A practical approach. Washington, D.C: IRL Press; 1984. pp. 231–270. [Google Scholar]
- 16.Craig A W B, Haghighat A, Yu A T K, Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 17.Davies M V, Pelletier J, Meerovitch K, Sonenberg N, Kaufman R J. The effect of poliovirus proteinase 2Apro expression on cellular metabolism. Inhibition of DNA replication, RNA polymerase II transcription, and translation. J Biol Chem. 1991;266:14714–14720. [PubMed] [Google Scholar]
- 18.Etchison D, Milburn S C, Edery I, Sonenberg N, Hershey J W B. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem. 1982;257:14806–14810. [PubMed] [Google Scholar]
- 19.Fukami H, Itano H A. In vitro translation of globin: effect of proteins purified by affinity chromatography on polyadenylate-sepharose. Biochemistry. 1976;15:3529–3535. doi: 10.1021/bi00661a021. [DOI] [PubMed] [Google Scholar]
- 20.Galili G, Kawata, E. E, Smith L D, Larkins B A. Role of the 3′-poly(A) sequence in translational regulation of mRNAs in Xenopus laevis oocytes. J Biol Chem. 1988;263:5764–5770. [PubMed] [Google Scholar]
- 21.Gallie D R. The cap and poly(A) tail function synergistically to regulate mRNA translation efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 22.Gallie D R, Tanguay R. Poly(A) binds to initiation factors and increases cap-dependent translation in vitro. J Biol Chem. 1994;269:17166–17173. [PubMed] [Google Scholar]
- 23.Grossi de Sa M F, Standart N, Martins de Sa C, Akhayat O, Huesca M, Scherrer K. The poly(A)-binding protein facilitates in vitro translation of poly(A)-rich mRNA. Eur J Biochem. 1988;176:521–526. doi: 10.1111/j.1432-1033.1988.tb14309.x. [DOI] [PubMed] [Google Scholar]
- 24.Haghighat A, Svitkin Y, Novoa I, Kuechler E, Skern T, Sonenberg N. The eIF4G-eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. J Virol. 1996;70:8444–8450. doi: 10.1128/jvi.70.12.8444-8450.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hake L E, Richter J D. Translational regulation of maternal mRNA. Biochim Biophys Acta. 1997;1332:M31–M38. doi: 10.1016/s0304-419x(96)00039-x. [DOI] [PubMed] [Google Scholar]
- 26.Hambidge S J, Sarnow P. Translational enhancement of the poliovirus 5′ noncoding region mediated by virus-encoded polypeptide-2A. Proc Natl Acad Sci USA. 1992;89:10272–10276. doi: 10.1073/pnas.89.21.10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hensold J O, Barth-Baus D, Stratton C A. Inducers of erythroleukemic differentiation cause messenger RNAs that lack poly(A)-binding protein to accumulate in translationally inactive, salt-stable 80S ribosomal complexes. J Biol Chem. 1996;271:23246–23254. doi: 10.1074/jbc.271.38.23246. [DOI] [PubMed] [Google Scholar]
- 28.Huang Y, Carmichael G C. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol Cell Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huber S A. VCAM-1 is a receptor for encephalomyocarditis virus on murine vascular endothelial cells. J Virol. 1994;68:3453–3458. doi: 10.1128/jvi.68.6.3453-3458.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irurzun A, Sanchez-Palomino S, Novoa I, Carrasco L. Monensin and nigericin prevent the inhibition of host translation by poliovirus, without affecting p220 cleavage. J Virol. 1995;69:7453–7460. doi: 10.1128/jvi.69.12.7453-7460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson R J, Standart N. Do the poly(A) tail and 3′ untranslated region control mRNA translation? Cell. 1990;62:15–24. doi: 10.1016/0092-8674(90)90235-7. [DOI] [PubMed] [Google Scholar]
- 32.Jang S K, Krausslich H G, Nicklin M J, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′-nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joshi B, Cai A-L, Keiper B D, Minich W B, Mendez R, Beach C M, Stepinski J, Stolarski R, Darzynkiewicz E, Rhoads R E. Phosphorylation of eukaryotic protein synthesis initiation factor 4E at Ser-209. J Biol Chem. 1995;270:14597–14603. doi: 10.1074/jbc.270.24.14597. [DOI] [PubMed] [Google Scholar]
- 34.Keiper B D, Rhoads R E. Cap-independent translation initiation in Xenopus oocytes. Nucleic Acids Res. 1997;25:395–403. doi: 10.1093/nar/25.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kessler S H, Sachs A. RNA recognition motif 2 of yeast pabp1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:51–57. doi: 10.1128/mcb.18.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirchweger R, Ziegler E, Lamphear B J, Waters D, Liebig H-D, Sommergruber W, Sobrino F, Hohenadl C, Blaas D, Rhoads R E, Skern T. Foot-and-mouth-disease virus leader proteinase: purification of the Lb form and determination of its cleavage site on eIF-4γ. J Virol. 1994;68:5677–5684. doi: 10.1128/jvi.68.9.5677-5684.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knowlton K U, Jeon E S, Berkley N, Wessely R, Huber S A. A mutation in the puff region of VP2 attenuates the myocarditic phenotype of an infectious cDNA of the Woodruff variant of coxsackievirus B3. J Virol. 1996;70:7811–7818. doi: 10.1128/jvi.70.11.7811-7818.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuhn U, Pieler T. Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interaction. J Mol Biol. 1996;256:20–30. doi: 10.1006/jmbi.1996.0065. [DOI] [PubMed] [Google Scholar]
- 39.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 40.Lamphear B, Yan R, Yang F, Waters D, Liebig H-D, Klump H, Kuechler E, Skern T, Rhoads R E. Mapping the cleavage site in protein synthesis initiation factor eIF-4γ of the 2A proteases from human Coxsackievirus and rhinovirus. J Biol Chem. 1993;268:19200–19203. [PubMed] [Google Scholar]
- 41.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 42.Lamphear B J, Rhoads R E. A single amino acid change in protein synthesis initiation factor 4G renders cap-dependent translation resistant to picornaviral 2A proteases. Biochemistry. 1996;35:15726–15733. doi: 10.1021/bi961864t. [DOI] [PubMed] [Google Scholar]
- 43.Le H, Tanguay R L, Balasta M L, Wei C-C, Browning K S, Metz A M, Goss D J, Gallie D R. Translation initiation factors eIF-iso4G and eIF4B interact with the poly(A)-binding protein and increase its RNA binding activity. J Biol Chem. 1997;272:16247–16255. doi: 10.1074/jbc.272.26.16247. [DOI] [PubMed] [Google Scholar]
- 44.Liebig H-D, Ziegler E, Yan R, Hartmuth K, Klump H, Kowalski H, Blaas D, Sommergruber W, Frasel L, Lamphear B, Rhoads R E, Kuechler E, Skern T. Purification of two picornaviral 2A proteinases: interaction with eIF-4γ and influence on in vitro translation. Biochemistry. 1993;32:7581–7588. doi: 10.1021/bi00080a033. [DOI] [PubMed] [Google Scholar]
- 45.Macadam A, Ferguson G, Fleming T, Stone D, Almond J, Minor P. Role for poliovirus protease 2A in cap independent translation. EMBO J. 1994;13:924–927. doi: 10.1002/j.1460-2075.1994.tb06336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melton D A, Krieg P A, Rebagliati M R, Maniatis T, Zinn K, Green M R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984;12:7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merrick W C, Hershey J W B. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 31–69. [Google Scholar]
- 49.Munroe D, Jacobson A. mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol Cell Biol. 1990;10:3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohlmann T, Pain V M, Wood W, Rau M, Morley S J. The proteolytic cleavage of eukaryotic initiation factor (eIF) 4G is prevented by eIF4E binding protein (PHAS-I; 4E-BP1) in the reticulocyte lysate. EMBO J. 1997;16:844–855. doi: 10.1093/emboj/16.4.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohlmann T, Rau M, Morley S J, Pain V M. Proteolytic cleavage of initiation factor eIF-4G in the reticulocyte lysate inhibits translation of capped mRNAs but enhances that of uncapped mRNAs. Nucleic Acids Res. 1995;23:334–340. doi: 10.1093/nar/23.3.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohlmann T, Rau M, Pain V M, Morley S J. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 1996;15:1371–1382. [PMC free article] [PubMed] [Google Scholar]
- 53.Ovchinnikov L P, Seriakova T A, Avaneskov A T, Alzhanova A T, Radzhabov H M, Spirin A S. RNA-binding proteins of rabbit reticulocytes. Eur J Biochem. 1978;90:517–525. doi: 10.1111/j.1432-1033.1978.tb12631.x. [DOI] [PubMed] [Google Scholar]
- 54.Paul A V, van Boom J H, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 55.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 56.Perez L, Carrasco L. Lack of direct correlation between p220 cleavage and the shut-off of host translation after poliovirus infection. Virology. 1992;189:178–186. doi: 10.1016/0042-6822(92)90693-j. [DOI] [PubMed] [Google Scholar]
- 57.Rhoads R E. The cap structure of eukaryotic messenger RNA and its interaction with cap-binding protein. Prog Mol Subcell Biol. 1985;9:104–155. [Google Scholar]
- 58.Rueckert R R. Picornaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Vol. 1. New York, N.Y: Lippincott-Raven Press; 1996. pp. 609–654. [Google Scholar]
- 59.Sachs A B, Davis R W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 60.Sachs A B, Davis R W, Kornberg R D. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol Cell Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 62.Sarnow P. Role of 3′-end sequences in infectivity of poliovirus transcripts made in vitro. J Virol. 1989;63:467–470. doi: 10.1128/jvi.63.1.467-470.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sieliwanowicz B. The influence of poly(A)-binding proteins on translation of poly(A)+ RNA in a cell-free system from embryo axes of dry pea seeds. Biochim Biophys Acta. 1987;908:54–59. [Google Scholar]
- 64.Sommergruber W, Ahorn H, Klump H, Seipekt J, Zoephel A, Fessl F, Krystek E, Blaas D, Kuechler E, Liebig H-D, Skern T. 2A proteinases of coxsackie- and rhinovirus cleave peptides derived from eIF-4γ via a common recognition motif. Virology. 1994;198:741–745. doi: 10.1006/viro.1994.1089. [DOI] [PubMed] [Google Scholar]
- 65.Sommergruber W, Ahorn H, Zoephel A, Maurer-Fogy I, Fessl F, Schnorrenberg G, Liebig H-D, Blaas D, Kuechler E, Skern T. Cleavage specificity on synthetic peptide substrates of human rhinovirus 2 proteinase 2A. J Biol Chem. 1992;267:22639–22644. [PubMed] [Google Scholar]
- 66.Spector D H, Baltimore D. Requirement of 3′-terminal poly (adenylic acid) for the infectivity of poliovirus RNA. Proc Natl Acad Sci USA. 1974;71:2983–2987. doi: 10.1073/pnas.71.8.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tarun S Z, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 68.Tarun S Z J, Sachs A B. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 69.Tarun S Z J, Wells S E, Deardorff J A, Sachs A B. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci USA. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Toyoda H, Nicklin M J H, Murray M G. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell. 1986;45:761–770. doi: 10.1016/0092-8674(86)90790-7. [DOI] [PubMed] [Google Scholar]
- 71.Webb N R, Chari R V J, DePillis G, Kozarich J W, Rhoads R E. Purification of the messenger RNA cap-binding protein using a new affinity medium. Biochemistry. 1984;23:177–181. doi: 10.1021/bi00297a001. [DOI] [PubMed] [Google Scholar]
- 72.Wormington M, Searfoss A M, Hurney C A. Overexpression of poly(A) binding protein prevents maturation-specific deadenylation and translational inactivation in Xenopus oocytes. EMBO J. 1996;15:900–909. [PMC free article] [PubMed] [Google Scholar]
- 73.Wyckoff E E. Inhibition of host cell protein synthesis in poliovirus-infected cells. Semin Virol. 1993;4:209–215. [Google Scholar]
- 74.Wyckoff E E, Croall D E, Ehrenfeld E. The p220 component of eukaryotic initiation factor 4F is a substrate for multiple calcium-dependent enzymes. Biochemistry. 1990;29:10055–10061. doi: 10.1021/bi00495a007. [DOI] [PubMed] [Google Scholar]
- 75.Wyckoff E E, Hershey J W B, Ehrenfeld E. Eukaryotic initiation factor 3 is required for poliovirus 2A protease-induced cleavage of the p220 component of eukaryotic initiation factor 4F. Proc Natl Acad Sci USA. 1990;87:9529–9533. doi: 10.1073/pnas.87.24.9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yan R, Rychlik W, Etchison D, Rhoads R E. Amino acid sequence of the human protein synthesis initiation factor eIF-4γ. J Biol Chem. 1992;267:23226–23231. [PubMed] [Google Scholar]
- 77.Yang H, Duckett C S, Lindsten T. iPABP, an inducible poly(A)-binding protein detected in activated human T cells. Mol Cell Biol. 1995;15:6770–6776. doi: 10.1128/mcb.15.12.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]