Abstract
A new mutant of Arabidopsis designated bus1-1 (for bushy), which exhibited a bushy phenotype with crinkled leaves and retarded vascularization, was characterized. The phenotype was caused by an En-1 insertion in the gene CYP79F1. The deduced protein belongs to the cytochrome P450 superfamily. Because members of the CYP79 subfamily are believed to catalyze the oxidation of amino acids to aldoximes, the initial step in glucosinolate biosynthesis, we analyzed the level of glucosinolates in a CYP79F1 null mutant (bus1-1f) and in an overexpressing plant. Short-chain glucosinolates derived from methionine were completely lacking in the null mutant and showed increased levels in the overexpressing plant, indicating that CYP79F1 uses short-chain methionine derivatives as substrates. In addition, the concentrations of indole-3-ylmethyl-glucosinolate and the content of the auxin indole-3-acetic acid and its precursor indole-3-acetonitrile were increased in the bus1-1f mutant. Our results demonstrate for the first time that the formation of glucosinolates derived from methionine is mediated by CYP79F1 and that knocking out this cytochrome P450 has profound effects on plant growth and development.
INTRODUCTION
Glucosinolates (mustard oil glycosides) are plant secondary metabolites that are found almost exclusively in plants of the order Capparales (Rodman, et al., 1996; Selmar, 1999). These compounds are sulfonated thioglucosides, which are stored in the vacuole and are hydrolyzed upon cell damage by thioglucosidases known as myrosinases (Bones and Rossiter, 1996). The resulting aglycones rearrange to form a variety of products that are toxins and deterrents to many pathogens and herbivores and that are believed to serve in plant defense (Louda and Mole, 1991). On the basis of their toxic properties and pungent taste, glucosinolates are often classified as antinutritional compounds. For example, rapeseed meal, a by-product of oil extraction from crushed rapeseed, is an excellent feed concentrate whose nutritional value is impaired by a high concentration of glucosinolates. However, glucosinolates are also of substantial benefit to humans as flavoring agents in a variety of cruciferous vegetables (e.g., broccoli and cauliflower) and spices (e.g., mustard). Furthermore, the consumption of such vegetables seems to reduce the risk of developing cancer (Jongen, 1996; Verhoeven et al., 1997). A hydrolytic degradation product of the methionine-derived 4-methylsulfinylbutyl (4MSOB) glucosinolate, 4MSOB isothiocyanate, was found to be a potent anticarcinogen (Zhang et al., 1992). Therefore, glucosinolates are considered to be excellent candidates for use in creating “functional food” designed to reduce the risk of cancer.
Although significant progress in understanding glucosinolate biosynthesis has been made in recent years, the characteristics of the enzymes involved are still imperfectly known (Halkier and Du, 1997; Halkier, 1999). The first step in glucosinolate biosynthesis appears to be the oxidation of amino acids to their corresponding aldoximes, which is analogous to the first step in cyanogenic glycoside formation. After donation of a thiol moiety (probably from cysteine), sequential transfer of glucose and sulfate residues completes the basic skeleton. At present, there is considerable controversy surrounding the nature of the enzymes that catalyze aldoxime formation. For example, in rapeseed, oxidation of phenylalanine to its corresponding aldoxime has been attributed to a flavin-containing monooxygenase (Bennett et al., 1993), whereas in white mustard, aldoxime formation from tyrosine appears to be mediated by a cytochrome P450 monooxygenase (Du et al., 1995). Furthermore, peroxidases have been proposed to be involved in aldoxime production from tryptophan in Chinese cabbage (Ludwig-Müller and Hilgenberg, 1988).
The model plant Arabidopsis, belonging to the Brassicaceae family in the order Capparales, contains more than 30 different glucosinolates (Hogge et al., 1988; Haughn et al., 1991). Therefore, this species is well suited to the examination of glucosinolate biosynthesis and its physiological roles. The glucosinolates in Arabidopsis originate principally from phenylalanine, tryptophan, and chain-elongated methionine derivatives. To date, three genes involved in aldoxime formation have been cloned from this species (Hull et al., 2000; Mikkelsen et al., 2000; Wittstock and Halkier, 2000). The deduced proteins (CYP79A2, CYP79B2, and CYP79B3) all belong to the P450 superfamily. Heterologous expression studies have shown that these enzymes are able to convert phenylalanine (CYP79A2) and tryptophan (CYP79B2 and CYP79B3) to their corresponding aldoximes. In addition, transgenic Arabidopsis plants overexpressing CYP79A2 contained higher concentrations of benzylglucosinolate than did wild-type plants, indicating that CYP79A2 uses phenylalanine as substrate in vivo (Wittstock and Halkier, 2000).
Here, we describe the isolation and characterization of the Arabidopsis mutant bus1-1 (for bushy), a bushy, semisterile plant with crinkled leaves. The phenotype was caused by an En-1 insertion in the gene CYP79F1 encoding a cytochrome P450 monooxygenase. The expression of CYP79F1 in apical plant parts was restricted to the vascular tissues. Analysis of a null mutant (bus1-1f) revealed the complete lack of short-chain methionine-derived glucosinolates, demonstrating that CYP79F1 is involved in the biosynthesis of aliphatic glucosinolates. Furthermore, overexpression of CYP79F1 in Arabidopsis led to an increased level of short-chain methionine-derived glucosinolates. Additionally, the bus1-1f mutant contained an increased level of indole-3-ylmethyl-glucosinolate together with increased indole-3-acetic acid (IAA) and indole-3-acetonitrile (IAN) levels, which could be responsible for the observed developmental phenotypes. These findings might indicate that alterations in the metabolism of methionine-derived glucosinolates may directly or indirectly affect auxin metabolism and plant morphogenesis.
RESULTS
Phenotype Analysis of an En-1–Induced bus1-1 Mutant
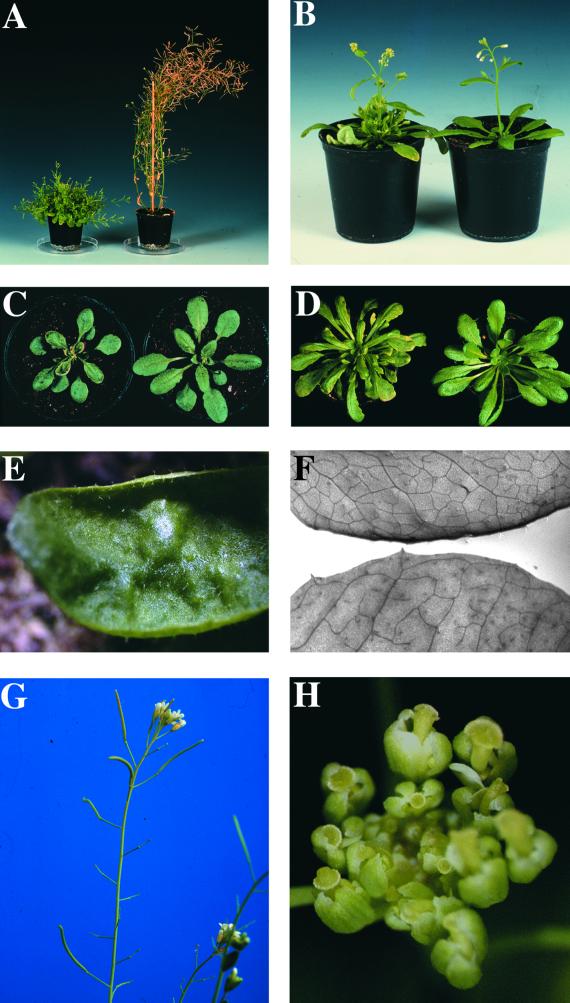
A collection of Arabidopsis plants mutagenized with the maize En-1 transposon (Wisman et al., 1998) was screened for plants with changed shoot growth at maturity. The bus1-1 mutant was chosen for further characterization because of the extremely bushy phenotype (Figure 1A). Segregation analysis revealed that the parent plant (collection number 4AAJ108) was heterozygous and that the phenotype was caused by a recessive mutation.
Figure 1.
Phenotype of the bus1-1 Mutant.
(A) to (D) show bus1-1 at left and wild type at right.
(A) The mature bus1-1 mutant exhibits a small bushy phenotype.
(B) At flowering time, the first side shoot reaches the height of the main inflorescence (5 weeks old).
(C) Under normal greenhouse conditions, the bus1-1 mutant has smaller rosette leaves (3 weeks old).
(D) Under short-day conditions, the rosette leaves are normal in size (6 weeks old).
(E) Fully developed rosette leaves of bus1-1 are crinkled.
(F) bus1-1 leaves (bottom) have fewer veins than do wild-type leaves (top).
(G) The bus1-1 mutant is semisterile, with siliques that vary in size or do not develop at all.
(H) Most mutant pistils are already visible when floral buds are still closed, so the anthers cannot reach the stigmas during dehiscence.
Under normal greenhouse conditions, the bus1-1 mutant was characterized by (1) small crinkled leaves, (2) early formation of secondary rosette leaves, (3) semisterility, and (4) an enhanced number of lateral shoots. The first differences noted between the mutant and wild-type plants were evident in the cotyledons of 5-day-old seedlings. At this stage, the vascularization of cotyledons was restricted to the primary vein, whereas wild-type plants had already formed secondary veins. Retardation of vascular strand formation occurred at all developmental stages but was most pronounced in fully developed rosette leaves (Figure 1F). The defective reticularization of the vascular system might be the reason for the observed leaf shape. Juvenile rosette leaves were strongly curled upward, and the two halves of the blade had the appearance of being hinged along the midrib, resembling the leaves of a Venus flytrap. Later in development, the leaves expanded, although the margins remained curled. Adult leaves were crinkled (Figure 1E). Rosettes of the mutant were smaller compared with those of the wild type (Figure 1C). Furthermore, secondary leaves that developed in the rosette leaf axils were larger and more numerous than those in the wild type.
The onset of the main inflorescence and the flowering time were not affected in the mutant (Figure 1B). Analyses of flower morphology revealed that the number of floral organs varied strongly in the mutant. Flowers had three to four sepals (the wild type had four), one to five petals (wild type, four), and three to five stamens (wild type, six). No differences could be observed in the sizes of these organs. The number of pistils was not affected, but their length varied strongly. Many pistils were already visible at a stage at which floral buds were still closed (Figure 1H). A variable amount of pollen was released on the stigma, depending on whether the anthers could reach the stigma during dehiscence. As a consequence, fertilization was often reduced, and developing siliques of bus1-1 were variable in size and seed content and sometimes did not develop at all (Figure 1G).
At the time of flowering, neither the length of the main inflorescence nor the number of lateral shoots differed between the bus1-1 mutant and the wild type. However, differences were visible in the length of the lateral shoots. In the mutant, the shoot closest to the shoot apex had already reached the height of the main inflorescence (Figure 1B). After flowers began to open, the main inflorescence of the mutant stopped growing or grew much slower compared with that of the wild type. Additionally, secondary inflorescences that emerged from the rosette leaf axils were increased in number and length. The secondary inflorescences also stopped growing after floral transition. The reduced growth, together with the enhanced secondary shoot formation, caused the bushy phenotype observed at maturity (Figure 1A).
All of these characteristics of the bus1-1 phenotype were also observed under short-day conditions, except for the difference in leaf size. Under these conditions, the bus1-1 mutant developed leaves that were normal in size (Figure 1D). When the root development of seedlings grown in vitro was analyzed, no differences in the length or the number of lateral roots were noted.
Molecular Characterization of Different bus Mutants
To determine if the gene of interest in the bus1-1 mutant was still tagged by an En-1 insertion, we screened offspring populations for plants that had reverted to the wild-type phenotype. Germinal reversion was detected at high frequencies (20 to 70%), indicating that the observed phenotype was caused by transposon insertion. Nevertheless, due to the high En-1 copy number (10 to 15), several methods used to isolate the corresponding flanking region failed. To reduce the number of En-1 insertions in the genome, the mutant was crossed to the wild type. In the F2 generation, different lines segregating for the bus1-1 phenotype were identified, but DNA gel blot analysis still revealed a copy number corresponding to at least 10 En-1 insertions. Additionally, germinal reversion could not be observed in the offspring of the F2–bus1-1 mutants, indicating that excision of En-1 during the crossing procedure probably caused a stable footprint mutation. This footprint was confirmed later (see below).
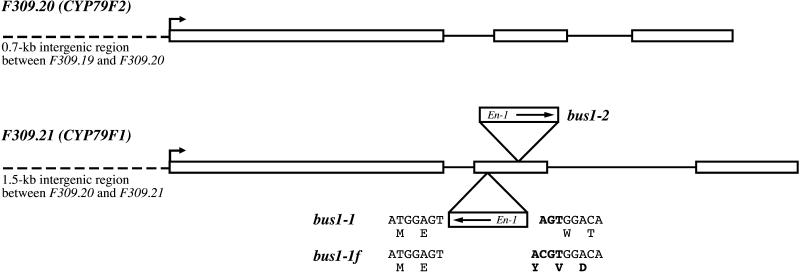
To identify new mutants with fewer En-1 insertions, we rescreened the Arabidopsis En-1 collection for plants exhibiting the characteristic bus phenotype. Three additional independent mutants were found, and segregation analysis of the parent plants (collection numbers 4L9-1, 6N18, and 6AAQ19) revealed that all of them were heterozygous. We chose plant 4L9-1, which had approximately four En-1 insertions, for further characterization and designated the mutant allele bus1-2. Combined phenotypic and genetic analysis of the 4L9-1 progeny revealed an En-1 fragment cosegregating with the mutant phenotype (see Methods). The flanking region of this En-1 insertion was isolated by a polymerase chain reaction (PCR)–based strategy and sequenced. Database searches identified this sequence in the bacterial artificial chromosome (BAC) clone F309 (GenBank accession number AC006341). The En-1 transposon was found to be inserted in gene F309.21 located on chromosome 1 (Figure 2).
Figure 2.
Genomic Organization of CYP79F1 and CYP79F2.
The coding regions of both genes comprise three exons (white boxes). Translation initiation sites are indicated by arrows, and intergenic regions that have been used for GUS expression studies are indicated by dashed lines. In both identified bus alleles (bus1-1 and bus1-2), the En-1 transposon is located in the second exon, although in different orientations (arrows). Insertion of En-1 in bus1-1 caused a 3-bp target duplication (AGT in boldface). The stable footprint mutant bus1-1f contains four additional bases, causing a frameshift (changed bases and amino acids are shown in boldface).
On the basis of this gene sequence, we reanalyzed the original bus1-1 mutant. PCR reactions with gene-specific primers in combination with different En-1 primers revealed that bus1-1 also contained an En-1 insertion in gene F309.21 (Figure 2). This insertion could no longer be detected in the F2–bus1-1 mutants derived from the crosses with the wild type (see above). Amplification with two F309.21-specific primers that flank the original insertion site revealed a sequence containing four additional bases, causing a frameshift (Figure 2). This stable null mutant has been designated bus1-1f (for footprint).
Identification of CYP79F1 and CYP79F2
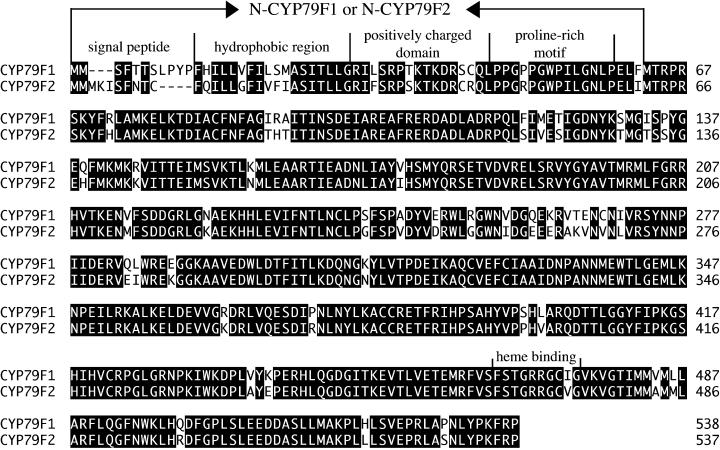
Detailed sequence analysis of the BAC clone F309 led to the identification of the gene F309.20 1.5 kb upstream of F309.21, with 86% homology to F309.21 (Figure 2). On the basis of these sequences, primers comprising the entire coding regions were designed for reverse transcription (RT)–PCR amplification of the cDNAs. Sequence comparison revealed that each gene contains three exons spanning ∼2.1 kb of genomic DNA (Figure 2). The predicted proteins have a calculated molecular mass of ∼62 kDa and share 89% identity at the amino acid level (Figure 3). The deduced amino acid sequences exhibited homology with conserved motifs found in cytochrome P450 enzymes, including the proline-rich domain (consensus, [P/I]PGPX[G/P]XP) and the heme binding domain (consensus, FXXGXXXCXG) (Chapple, 1998) (Figure 3). The greatest level of identity, ∼45%, was found to the members of the CYP79 family. According to the nomenclature for the cytochrome P450 superfamily, the proteins were thus designated CYP79F1 (gene F309.21) and CYP79F2 (gene F309.20) (http://drnelson.utmem.edu/CytochromeP450.html).
Figure 3.
Amino Acid Comparison of CYP79F1 and CYP79F2.
The deduced proteins share 89% identical amino acids (black). At the N termini, the four motifs characteristic of most cytochrome P450 enzymes are indicated. The N-terminal regions fused with GFP (named N-CYP79F1 and N-CYP79F2) are marked by arrows. The heme binding domain is located in the C-terminal part of the proteins.
Subcellular Localization of CYP79F1 and CYP79F2
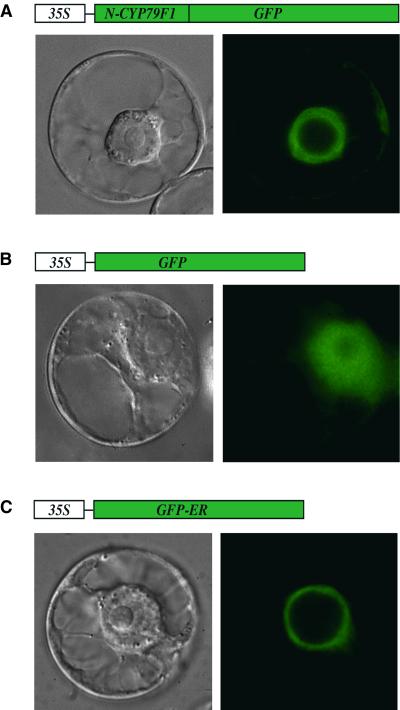
CYP79F1 and CYP79F2 are very similar, but significant differences occur in their extreme N-terminal regions (Figure 3). Several motifs were identified in the N-terminal regions that are conserved in other cytochrome P450s: a short N-terminal signal peptide for targeting to the endoplasmic reticulum (ER); a stretch of hydrophobic amino acids constituting the transmembrane helix, which anchors the protein in the ER; a positively charged domain; and a proline-rich motif (Chapple, 1998) (Figure 3). Consistent with these motifs, computer analyses of CYP79F1 and CYP79F2 suggested a localization in the ER (program TargetP at http://www.cbs.dtu.dk/services). However, because of the observed amino acid differences at the N termini of the two proteins, it seemed important to prove ER localization experimentally. The first 62 amino acids of either CYP79F1 or CYP79F2 (named N-CYP79F1 and N-CYP79F2), which include the motifs mentioned above (Figure 3), were fused to green fluorescent protein (GFP). As controls, we used either GFP alone, which is localized to the cytoplasm, or a GFP variant that is targeted to the ER (Haselhoff et al., 1997). All constructs were expressed in tobacco BY2 cells. Comparison of the fluorescence patterns between the controls and the fusion proteins revealed that the GFP fusions of CYP79F1 and CYP79F2 were both located in the ER (Figure 4).
Figure 4.
Heterologous Expression in BY2 Cells.
Different GFP constructs (at top) were expressed under the control of the cauliflower mosaic virus (CaMV) 35S promoter in tobacco BY2 cells. Transformed cells were analyzed either by differential interference contrast (at bottom left) or by fluorescence microscopy (at bottom right).
(A) Expression of N-CYP79F1 (see Figure 3) fused to GFP. Green fluorescence was observed at the cortical and perinuclear ER. The N-CYP79F2–GFP fusion protein exhibited the same fluorescence pattern (not shown).
(B) Expression of GFP alone. Green fluorescence was observed in the cytoplasm.
(C) Expression of a GFP variant targeted to the endoplasmic reticulum (GFP-ER). Green fluorescence was restricted to the ER.
Studies on the Expression of CYP79F1 and CYP79F2
Due to the great similarity of CYP79F1 and CYP79F2, it was not possible either to generate the specific DNA probes needed for RNA gel blot analyses and in situ hybridization studies or to synthesize the specific peptides needed for antibody production and subsequent protein gel blot analysis. Therefore, we performed promoter analysis for each gene in transgenic Arabidopsis plants. Both promoters (PCYP79F1 and PCYP79F2) were amplified by PCR and fused to the uidA reporter gene. PCYP79F1 corresponds to the entire intergenic region between gene F309.20 and F309.21 (1.5 kb), whereas PCYP79F2 comprises the region between gene F309.19 and F309.20 (0.7 kb; Figure 2). The constructs were transformed into Arabidopsis, and tissue-specific expression patterns were analyzed over three generations. Histochemical staining revealed variations in β-glucuronidase (GUS) activity in different transgenic lines of both PCYP79F1 and PCYP79F2. Such variability is common and probably reflects the positional effects of the insertion site on transgene expression. Nevertheless, all lines exhibited the same qualitative staining pattern.
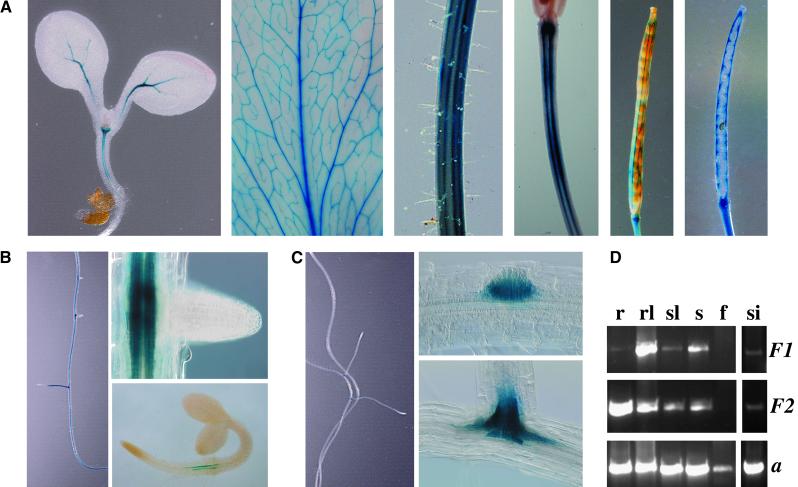
No differences in the activity patterns of the two promoters were found in apical plant parts. Microscopic analysis of whole organs revealed promoter activities exclusively in the vascular bundles of hypocotyls, leaves, stems, and mature siliques (Figure 5A). The highest level of promoter activity was observed in fully expanded rosette leaves, and the lowest level was seen in stems and young stem leaves. No activity was found in flowers.
Figure 5.
Expression Studies of CYP79F1 and CYP79F2 by GUS Staining and RT-PCR.
(A) Promoter activity of PCYP79F1 in apical plant parts. Expression in hypocotyls, leaves, stems, silique stems, and siliques (left to right) is restricted to the vascular tissue. PCYP79F2 exhibited the same staining pattern (not shown).
(B) and (C) Differences in the expression patterns are visible in roots.
(B) PCYP79F2 is active in the vascular tissue throughout the entire root (left) except for the root tips (right, top) and is first detectable in 3-day-old seedlings at the basal end of the main root (right, bottom).
(C) PCYP79F1 is not active in the main root (left) and is detected only in lateral root primordia (right, top) and at the basal ends of lateral roots (right, bottom).
(D) RT-PCR reactions using poly(A)+ RNA from roots (r), rosette leaves (rl), stem leaves (sl), stems (s), flowers (f), and siliques (si) with gene-specific primers for CYP79F1 (F1), CYP79F2 (F2), and actin (a).
Striking differences in the activity of PCYP79F1 and PCYP79F2 were observed in roots. GUS staining from PCYP79F2 activity was found throughout the root, except in the root tips (Figure 5B, left). Activity was restricted to the vascular system and was first detectable 3 days after germination at the basal end of the main root (Figure 5B, right). In contrast, PCYP79F1 was not active in the main root (Figure 5C, left) but was restricted to young lateral root primordia and vascular tissue at the basal ends of lateral roots (Figure 5C, right).
To confirm the results of the GUS expression analysis, RT-PCR was performed with gene-specific primers for CYP79F1 and CYP79F2. Specificity was tested in PCR reactions with genomic DNA, in which mispriming would have given rise to PCR products of different lengths. RT-PCR analyses with poly(A)+ RNA from various tissues of wild-type Arabidopsis plants revealed the following features (Figure 5D): (1) transcription of both genes in all analyzed tissues except flowers; (2) no significant differences in transcription levels between CYP79F1 and CYP79F2 in shoots; and (3) a considerably lower transcription level of CYP79F1 than of CYP79F2 in roots. Together, these results were in full agreement with the results of the GUS expression studies.
Analysis of Glucosinolate Patterns
Members of the Arabidopsis CYP79 family are believed to catalyze the conversion of amino acids to aldoximes, the first step in glucosinolate formation (Bak et al., 1998). Heterologous expression in Escherichia coli of CYP79A2, CYP79B2, and CYP79B3 from Arabidopsis showed that these proteins are able to synthesize aldoximes from phenylalanine (CYP79A2) and tryptophan (CYP79B2 and CYP79B3) (Hull et al., 2000; Mikkelsen et al., 2000; Wittstock and Halkier, 2000). However, given the claims for the involvement of other enzymes in this reaction type, the role of the CYP79 proteins is not fully clear. The availability of the stable footprint mutant bus1-1f now allows assessment of whether or not CYP79F1 is involved in glucosinolate biosynthesis.
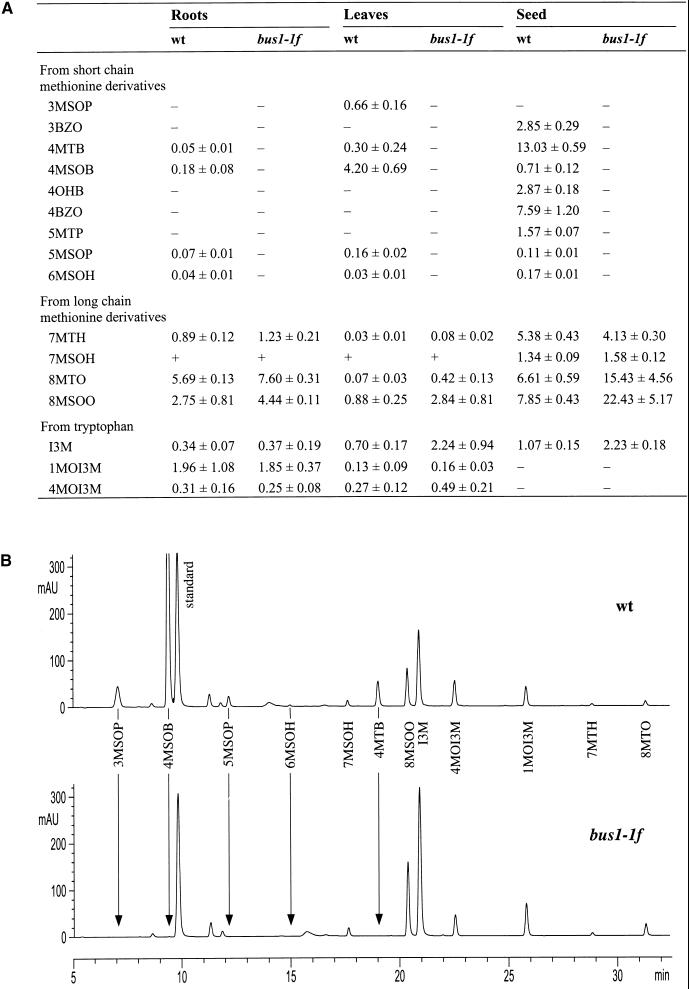
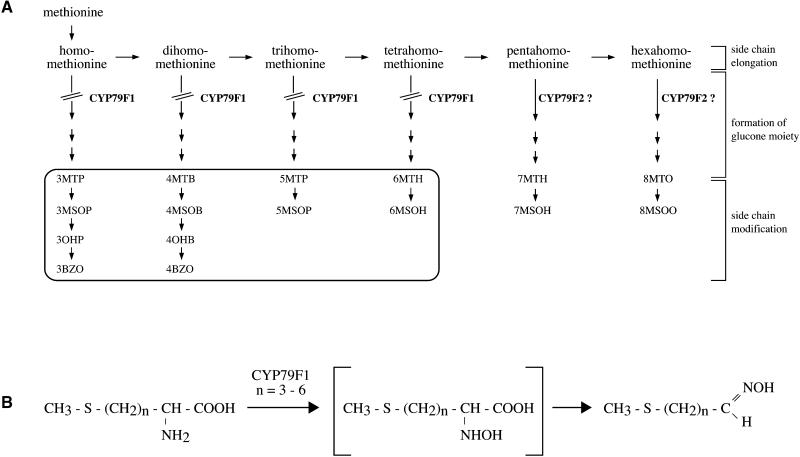
Glucosinolate patterns in roots, leaves, and seed of bus1-1f were compared with those of wild-type plants. HPLC analyses revealed that many glucosinolates derived from methionine were below detectable limits in bus1-1f, suggesting that CYP79F1 is involved in the synthesis of aldoximes from methionine (Figure 6). In fact, all of the major aliphatic glucosinolates of Arabidopsis appear to be derived from chain-elongated derivatives of methionine that are formed by a three-step cycle that yields an amino acid with one to six additional methylene groups in the side chain (Graser et al., 2000). After these methionine derivatives are converted to glucosinolates, subsequent side chain modifications (oxidation, loss of the methylsulfinyl group, and esterification) lead to the formation of the great variety of glucosinolates in Arabidopsis. In the bus1-1f mutant, all of the glucosinolates derived from methionine derivatives extended by one to four methylene residues were completely absent (Figure 6 and Figure 7A). In contrast, all of the glucosinolates derived from methionine derivatives extended by five or six methylene residues were still present. These results suggest that CYP79F1 specifically catalyzes the oxidation of short-chain methionine derivatives and does not use the long-chain derivatives as substrates (Figure 7B).
Figure 6.
Estimation of Glucosinolate Content by HPLC Analysis.
(A) Extracts of roots grown in vitro, rosette leaves, and seed from wild-type plants (wt) and bus1-1f mutants were analyzed by HPLC (see Methods). Concentrations are given in μmol/g dry weight for roots and rosette leaves and μmol/g fresh weight for seed (values shown are means ±sd; n 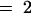 for roots, and n
for roots, and n 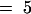 for leaves and seed). (−), not detected, and (+), that the peak was not well resolved from adjacent peaks and could not be quantified in all measurements. 3MSOP, 3-methylsulfinylpropyl; 3BZO, 3-benzoyloxypropyl; 4MTB, 4-methylthiobutyl; 4MSOB, 4-methylsulfinylbutyl; 4OHB, 4-hydroxybutyl; 4BZO, 4-benzoyloxybutyl; 5MTP, 5-methylthiopentyl; 5MSOP, 5-methylsulfinylpentyl; 6MSOH, 6-methylsulfinylhexyl; 7MTH, 7-methylthioheptyl; 7MSOH, 7-methylsulfinylheptyl; 8MTO, 8-methylthiooctyl; 8MSOO, 8-methylsulfinyloctyl; I3M, indole-3-ylmethyl; 1MOI3M, 1-methoxyindole-3-ylmethyl; 4MOI3M, 4-methoxyindole-3-ylmethyl.
for leaves and seed). (−), not detected, and (+), that the peak was not well resolved from adjacent peaks and could not be quantified in all measurements. 3MSOP, 3-methylsulfinylpropyl; 3BZO, 3-benzoyloxypropyl; 4MTB, 4-methylthiobutyl; 4MSOB, 4-methylsulfinylbutyl; 4OHB, 4-hydroxybutyl; 4BZO, 4-benzoyloxybutyl; 5MTP, 5-methylthiopentyl; 5MSOP, 5-methylsulfinylpentyl; 6MSOH, 6-methylsulfinylhexyl; 7MTH, 7-methylthioheptyl; 7MSOH, 7-methylsulfinylheptyl; 8MTO, 8-methylthiooctyl; 8MSOO, 8-methylsulfinyloctyl; I3M, indole-3-ylmethyl; 1MOI3M, 1-methoxyindole-3-ylmethyl; 4MOI3M, 4-methoxyindole-3-ylmethyl.
(B) Representative HPLC chromatograms from leaf extracts of one wild-type plant (wt) and one bus1-1f mutant. Signals are given in arbitrary units (mAU). Peaks that were absent in the bus1-1f mutant are marked by arrows. Abbreviations are as given in (A).
Figure 7.
Proposed Model for the Function of CYP79F1 and CYP79F2.
(A) Scheme of the biosynthetic pathway for methionine-derived glucosinolates. Glucosinolates that are absent in the bus1-1f mutant are boxed. The lack of these glucosinolates is most likely caused by the blocked conversion (interrupted arrows) of short-chain methionine derivatives to their aldoximes due to the absence of CYP79F1. Long-chain methionine derivatives might be converted by CYP79F2, as indicated by the question marks. Glucosinolate abbreviations are defined in the legend to Figure 6.
(B) CYP79F1 is proposed to react exclusively with homo- to tetrahomo-methionine (n 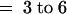 ).
).
Interestingly, the bus1-1f mutant contains higher concentrations of glucosinolates derived from the long-chain methionine derivatives than do wild-type plants (Figure 6). The lack of conversion of short-chain methionine derivatives to their aldoximes probably leads to a buildup of these intermediates, which can then undergo further rounds of chain elongation, resulting in the formation of more glucosinolates with longer side chains (Figure 7A). An increase of the tryptophan-derived indole-3-ylmethyl-glucosinolate was also found in mutant leaves and seed but not in roots.
Auxin Analysis
The biosynthesis of indolyl glucosinolates starts with the conversion of tryptophan to indole-3-acetaldoxime (IAOx) (Halkier, 1999; Hull et al., 2000; Mikkelsen et al., 2000). This reaction also might be the first step in the biosynthesis of the auxin IAA via a tryptophan-dependent pathway involving IAN as an intermediate (Trp→IAOx→IAN→IAA) (Slovin et al., 1999). If IAOx is an intermediate in both IAA and indole glucosinolate biosynthesis, the two pathways might share regulatory features. The increased concentration of indole-3-ylmethyl-glucosinolate in leaves and seed of bus1-1f (Figure 6) raised the possibility that the auxin concentration was changed as well. The morphology of the bus mutant (bushy phenotype and crinkled leaves) also suggested that it was altered in auxin concentration, given the important role that auxin plays in apical dominance and leaf morphology (Fellner, 1999; Procházka and Truksa, 1999). Therefore, we measured the concentrations of free IAA and its precursor IAN in leaves of bus1-1f and wild-type plants. The concentration of both compounds was higher in the mutant than in the wild type: IAA was increased twofold (14 ± 3.5 compared with 7.6 ± 1.2 pg/mg dry weight; n = 3), and IAN was increased fivefold (7.7 ± 2.3 compared with 1.7 ± 0.3 ng/mg dry weight; n = 3). Therefore, it is possible that the observed bus phenotype is caused by an increased concentration of auxin.
Overexpression of CYP79F1 in Arabidopsis
The lack of aliphatic glucosinolates with short carbon chain lengths in the bus1-1f mutant indicated that CYP79F1 oxidizes short-chain methionine derivatives. To further test this hypothesis, we determined the glucosinolate content of wild-type plants overexpressing CYP79F1. We postulated that if the expression of the CYP79F1 gene results in the synthesis of rate-limiting amounts of enzyme catalyzing the N-hydroxylation of short-chain methionine derivatives, overexpression would increase the levels of the corresponding glucosinolates.
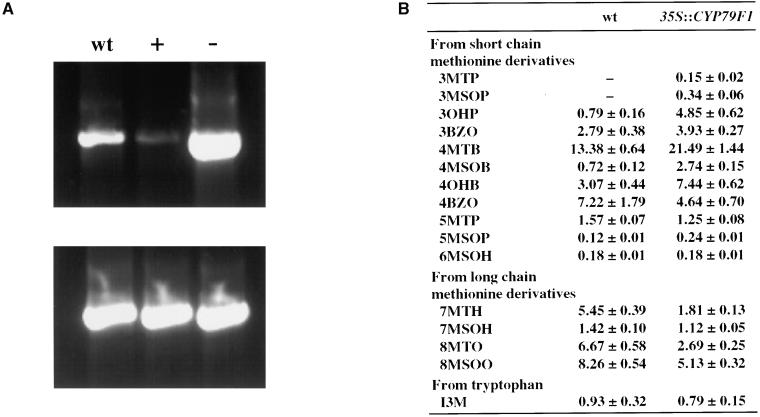
When the CYP79F1 cDNA was expressed under the control of the constitutive cauliflower mosaic virus (CaMV) 35S promoter in Arabidopsis wild-type plants, 90% of the transgenic plants (32 of 35) exhibited a more or less pronounced bus phenotype, indicating different degrees of silencing of the CYP79F1 gene. The remaining transgenic plants did not show any morphological changes compared with wild-type plants. RT-PCR analysis confirmed that the transcript level of CYP79F1 was reduced in leaves of transgenic plants exhibiting the bus phenotype, whereas the transcript level was increased in leaves of transgenic plants exhibiting the wild-type phenotype (Figure 8A). By segregation and DNA gel blot analyses, we identified a homozygous, CYP79F1-overexpressing line carrying a single T-DNA insertion. Seed of this line had a higher content of short-chain methionine-derived glucosinolates compared with that of wild-type seed. The aliphatic glucosinolates with the shortest carbon chain lengths were especially enriched (Figure 8B). For example, 3-methylthiopropyl and 3-methylsulfinylpropyl, which were not detected in the seed of wild-type plants, were present in the transgenic line. On the other hand, the concentration of methionine-derived glucosinolates with long carbon side chains was reduced in the overexpressing line compared with the wild type. Overexpression of CYP79F1 led to an enhanced conversion of short-chain methionine derivatives to their aldoximes and consequently to a higher concentration of glucosinolates with short carbon chain lengths (the only exception was a decrease of 4-benzoyloxybutyl). The more rapid use of short-chain methionine derivatives probably leaves less substrate for further chain elongation and thus results in lower concentrations of aliphatic glucosinolates with long side chains.
Figure 8.
Overexpression of CYP79F1 in Transgenic Arabidopsis Plants.
The CYP79F1 cDNA was expressed in Arabidopsis under the control of the CaMV 35S promoter. wt, wild type.
(A) RT-PCR reactions using poly(A)+ RNA from rosette leaves. Transgenic plants exhibiting the bus phenotype (+) had lower transcription levels of CYP79F1 than did wild-type plants, whereas transgenic plants without the bus phenotype (−) had higher transcription levels.
(B) Glucosinolate pattern in seed of the homozygous plant 35S::CYP79F1 overexpressing CYP79F1 compared with the wild type. Concentrations are given in μmol/g (means ±sd; n 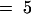 ). Glucosinolate abbreviations are defined in the legend to Figure 6.
). Glucosinolate abbreviations are defined in the legend to Figure 6.
DISCUSSION
Cytochrome P450s and Glucosinolate Biosynthesis
Glucosinolate biosynthesis is initiated by the oxidation of amino acids to their corresponding aldoximes. Several different types of enzymes have been implicated in this conversion: plasma membrane–bound peroxidases in the formation of indolyl glucosinolates from tryptophan in Chinese cabbage (Ludwig-Müller and Hilgenberg, 1988), flavin-containing monooxygenases in the formation of aromatic and aliphatic glucosinolates from homophenylalanine and methionine derivatives (Bennett et al., 1993, 1996), and cytochrome P450s in the formation of aromatic glucosinolates from phenylalanine and tyrosine in white mustard, nasturtium, and papaya (Du et al., 1995; Du and Halkier, 1996; Halkier and Du, 1997). This study unambiguously demonstrated the involvement of the cytochrome P450 CYP79F1 in the formation of aliphatic glucosinolates from chain-elongated methionine derivatives. The lack of short-chain aliphatic glucosinolates in the null mutant and the increased levels in the overexpressing plant indicated that CYP79F1 uses short-chain methionine derivatives as substrates. This is further supported by heterologous expression studies of CYP79F1 in E. coli, which revealed that recombinant CYP79F1 is able to convert dihomo- and trihomomethionine to their corresponding aldoximes (Hansen et al., 2000). Together with the recent cloning of CYP79B2 and CYP79B3 involved in indolyl (Hull et al., 2000; Mikkelsen et al., 2000) and CYP79A2 involved in aromatic (Wittstock and Halkier, 2000) glucosinolate biosynthesis, our results suggest that in Arabidopsis the cytochrome P450s of the family CYP79 perform the initial aldoxime formation for all types of glucosinolates.
CYP79F1 Expression Pattern and Substrate Specificity
The expression pattern of CYP79F1 provides valuable information regarding the site of glucosinolate biosynthesis in Arabidopsis. Glucosinolates in this species accumulate in all tissues that have been analyzed so far, including roots, leaves, and seed (Haughn et al., 1991) (Figure 6). Inspection of the glucosinolate content of various organs revealed a much higher concentration of short-chain methionine-derived glucosinolates in leaves than in roots. This difference correlates well with the higher expression level of CYP79F1 we found in leaves as opposed to roots (Figure 5D).
In other species (white mustard and rapeseed), it has been demonstrated that one of the major sites of glucosinolate biosynthesis is the silique wall (Toroser et al., 1995a, 1995b; Du and Halkier, 1998). Glucosinolates are transferred from the walls to the seed, where they accumulate during seed maturation. The expression of CYP79F1 in the vascular strands of silique walls and seed funicles suggests a similar mode of glucosinolate synthesis and deposition in Arabidopsis.
The involvement of CYP79F1 in the biosynthesis of aliphatic glucosinolates was clearly shown by the complete absence of short-chain methionine-derived glucosinolates in the null mutant and their increased concentration in CYP79F1-overexpressing plants. However, the metabolic function of the neighboring gene, CYP79F2, with 89% identity to CYP79F1 at the amino acid level, is still uncertain. CYP79F2 does not appear to catalyze the same reactions as CYP79F1 because it fails to assume the function of CYP79F1 in the null mutant bus1-1f, although it is expressed in the same cells of apical plant parts. It might be postulated that CYP79F2 converts long-chain methionine derivatives to their corresponding aldoximes (Figure 7). This hypothesis would explain the striking differences in the expression levels of CYP79F1 and CYP79F2 in roots: the low expression of CYP79F1 coincides with a low content of short-chain methionine-derived glucosinolates in these organs, and the high expression of CYP79F2 coincides with a high content of long-chain methionine-derived glucosinolates (Figure 6). To study this problem in more detail, the preparation of knockout mutants for CYP79F2 is currently under way.
The bus Phenotype
The bus phenotype may result from either an altered content of methionine-derived glucosinolates and their biosynthetic intermediates or from an increase of indolyl glucosinolates and IAA concentrations. There are several lines of evidence that suggest a role for auxin in the production of the bus phenotype. First, the changes in indolyl glucosinolate concentrations observed could result in changes in IAA concentrations, given the postulated metabolic links between these two groups of substances (Ludwig-Müller and Hilgenberg, 1988). Second, the intimate relations between auxin and branching, vascularization, and leaf shape, reported in the literature for many years, suggest the involvement of auxin in any mutant with abnormalities in these traits (Davies, 1995; Fellner, 1999; Procházka and Truksa, 1999). Although enhanced shoot branching is traditionally correlated with decreased auxin concentrations, the bus1-1f mutant actually contained increased auxin levels. However, during the last decade, it became clear that changes in hormone concentrations in mutant plants often do not correlate well with the degree of branching (Napoli et al., 1999). For example, the pea mutant rms-2 that displays a highly branched phenotype has an increased instead of a decreased auxin level (Beveridge et al., 1994).
Alternately, the modified profile of chain-elongated methionine derivatives in the bus1-1f mutant may result in changes in methionine metabolism that affect plant growth and development. Besides its role as a structural component of proteins, methionine serves as the direct precursor of both S-adenosylmethionine, the principal methyl donor in trans-methylation reactions, and the hormone ethylene (Ravanel et al., 1998). In addition, methionine is an immediate progenitor of S-methylmethionine, a major sulfur transport form (Bourgis et al., 1999). The genetic lesion in CYP79F1 in bus1-1f may lead to the buildup of chain-elongated analogs of methionine, which may in turn interfere with methionine synthesis, utilization, or transport, leading to changes in gross morphology. Arabidopsis mutants that are deficient in methionine (Ravanel et al., 1998; Kim and Leustek, 2000) or that overaccumulate methionine (Bartlem et al., 2000) show reduced growth compared with wild-type plants.
Glucosinolates in Crop Improvement and Food Design
The nutritional value of rapeseed meal as animal feed is reduced by high glucosinolate content. For this reason, over 40 years ago plant breeders initiated a search for rapeseed that is low in glucosinolates. In 1968, seeds from plants of the Polish Brassica napus variety Bronowski were found to be low in glucosinolates (Josefsson and Appelqvist, 1968). This genetic source was then incorporated into high-yielding varieties. However, the molecular basis of the low glucosinolate content in this variety is still poorly understood and does not appear to result from an obvious biosynthetic block. The identification of genes encoding cytochrome P450s of the family CYP79 as key enzymes in the glucosinolate biosynthesis pathway provides additional tools to modulate glucosinolate profiles.
Manipulation of glucosinolate composition could also improve the pest resistance of rapeseed, because glucosinolates influence the behavior of insect, avian, and molluscan herbivores and are powerful antibacterial and antifungal agents (Louda and Mole, 1991). For example, surveys of herbivore damage on rapeseed lines with different glucosinolate contents have shown that increased glucosinolate levels reduce feeding by pigeons and slugs but increase feeding by specialist insects such as flea beetles (Giamoustaris and Mithen, 1995). However, compositional changes, such as increasing the length of the side chain of methionine-derived aliphatic glucosinolates, decreased flea beetle damage. Hence, altering the chain length of aliphatic glucosinolates, as was achieved in this study by producing a null mutant of CYP79F1 in Arabidopsis, may have agricultural value.
Recent interest in glucosinolates also derives from their role as potential anticancer agents in human diets (Jongen, 1996; Verhoeven et al., 1997). For example, 4MSOB isothiocyanate, a hydrolysis product of 4MSOB found in broccoli, was demonstrated to be a potent anticarcinogen (Zhang et al., 1992). Hence, increasing the concentration of the short- chain methionine-derived glucosinolate 4MSOB, which was achieved in this study by overexpressing CYP79F1 in Arabidopsis, may increase the anticancer properties of glucosinolate-containing foods. Additional experiments are necessary to determine if this strategy can be applied successfully to other species.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana ecotype Columbia was the wild type used for crossing and transformation. The bus mutants have been identified in an En-1–mutagenized collection of Arabidopsis ecotype Columbia (Wisman et al., 1998).
Plants were grown in soil either in a greenhouse at 20°C with a 16-hr light period or in a growth chamber at 22°C with an 8-hr light period (short-day conditions). For in vitro culture, seed were surface-sterilized and plated on agar-solidified half-strength Murashige and Skoog (1962) (MS) medium (2.15 g/L MS salts, 1% sucrose, pH 5.8, with KOH). Plates were grown in a culture chamber using 16-hr light periods at 150 μE m−2 sec−1, 75% humidity, at 21°C. For Arabidopsis root liquid cultures, ∼20 sterilized seed were grown in 40 mL of MS medium (4.3 g/L MS salts, 3% sucrose, pH 5.8, with KOH) in Erlenmeyer flasks and agitated at 90 rpm for 3 weeks. Tobacco BY2 cells were grown according to Nagata et al. (1981) and subcultured weekly.
Segregation Analysis of Plant 4L9-1 and Isolation of Flanking Genomic DNA
The F1 progeny of plant 4L9-1 segregated 3:1 for the bus phenotype. The genotype of F1 plants exhibiting the wild-type phenotype (either heterozygous for the bus1-2 allele or wild type) was determined by phenotypical analysis of the F2 generation. Individual plants with defined genotypes were used in DNA gel blot experiments to determine if one of the four En-1 insertions cosegregates with the bus phenotype. Isolation of genomic DNA and DNA gel blot analysis were performed as described (Sambrook et al., 1989). DNA was digested with Csp6I. Hybridization was performed using the 5′ end of En-1 (∼200 bp) as a probe. Analysis revealed an 800-bp Csp6I fragment (600 bp of En-1 plus 200 bp of flanking genomic DNA) cosegregating with the phenotype. Flanking genomic DNA was isolated from a homozygous mutant plant by ligation-mediated polymerase chain reaction (PCR) (Varotto et al., 2000) and used as a probe on the same DNA gel blot described above. Hybridizing Csp6I fragments of the wild type and the mutant allele were different in length because of the Csp6I restriction site in En-1. The results confirmed that plants homozygous for the bus phenotype were homozygous for the insertion in CYP79F1, plants heterozygous for the bus phenotype were heterozygous for the insertion, and plants with the wild-type phenotype did not contain an insertion in CYP79F1. Cosegregation analysis revealed that none of the other En-1 insertions could be responsible for the phenotype. The possibility that the phenotype was caused by a stable footprint after En-1 excision from another closely linked gene was excluded based on the identification of plants that had reverted to wild type in offspring populations of homozygous bus1-2 mutants.
Genetic Analysis of bus1-1 and Identification of bus1-1f
The En-1 element in bus1-1 was identified in PCR reactions by using genomic DNA as a template (10 ng; for conditions, see Reverse Transcription–PCR Analysis below) and the following primer combinations: the CYP79F1 forward primer 5′-ATGATCATGTGATGTTTTGTC-ACAGGG-3′ (genomic region 64,929 to 64,955 of bacterial artificial chromosome [BAC] clone F309) and the En-1 primer 5′-GCTCCAATG-ACCCACCAACAGAATG-3′ as well as the CYP79F1 reverse primer 5′-CCACAAACATAATCAAAGACTACTCTGT-3′ (genomic region 65,613 to 65,640) and the En-1 primer 5′-AGAAGCACGACGGCT-GTAGAATAGGA-3′.
The bus1-1 mutant was backcrossed to Arabidopsis wild-type plants for two generations. Plants were allowed to self-propagate, and their progeny were tested for the segregation of the bus phenotype. In PCR reactions with the primer combinations mentioned above, the En-1 insertion could not be detected. Amplification with the two CYP79F1-specific primers (see above) and subsequent sequencing led to the identification of bus1-1f.
Reverse Transcription–PCR Analysis
Tissue-Specific Expression Studies
Poly(A)+-RNA was isolated from various tissues by using oligo(dT)25 Dynabeads and following the manufacturer's instructions (Dynal, Oslo, Norway). Roots were harvested from liquid cultures grown in vitro. All other tissues were harvested from plants grown in the greenhouse. Fifty nanograms of poly(A)+ RNA was reverse transcribed with 200 units of Superscript II reverse transcriptase (Life Technologies, Grand Island, NY) in a 50-μL reaction containing 1 × first strand buffer (Life Technologies), 0.5 μg of oligo(dT)12-18 (Amersham, Uppsala, Sweden), 0.2 μmol of DTT, and 25 nmol of each deoxynucleotide triphosphate, according to the manufacturer's instructions (Life Technologies). cDNA samples (5 μL) were used for subsequent PCR amplifications. The CYP79F1 cDNA fragment (1.6 kb) was amplified with the forward primer 5′-TCCATGGCATCAATCACTCTACTGG-3′ (genomic region 63,931 to 63,955 of BAC clone F309) and the reverse primer 5′-AAGCGTTGAAGAAGATAACAGA-3′ (genomic region 66,179 to 66,200). The CYP79F2 cDNA fragment (0.9 kb) was amplified with the forward primer 5′-TCAACTTTCCATCGTAGAGTCCATT-3′ (genomic region 60,584 to 60,608) and the reverse primer 5′-GGTATCTTGACGGGCAACATGAGGT-3′ (genomic region 61,607 to 61,631). The actin-2 cDNA fragment (1.1 kb) was amplified with the forward primer 5′-TTCCTCAATCTCATCTTCTTCC-3′ and the reverse primer 5′-GACCTGCCTCATCATACTCG-3′. PCR was performed in 1 × buffer (Hoffmann-La Roche, Mannheim, Germany) containing 1.5 mM MgCl2, 0.05 mM each deoxynucleotide triphosphate, 1 unit of Taq DNA polymerase (Hoffmann-La Roche), and 20 pmol of each primer, for 35 cycles at an annealing temperature of 58°C.
Cloning of CYP79F1 and CYP79F2 cDNAs
Reverse transcription (RT)–PCR reactions with poly(A)+ RNA isolated from rosette leaves were performed as described above. CYP79F1 cDNA was amplified with the forward primer 5′-ATACATCATGATGAGCTTTACC-3′ (genomic region 63,864 to 63,885 of BAC clone F309) and the reverse primer 5′-AAGCGTTGAAGAAGATAACAGA-3′ (genomic region 66,179 to 66,200), and CYP79F2 cDNA was amplified with the forward primer 5′-TCAAACAAAAATACAAACATTA-3′ (genomic region 60,206 to 60,227) and the reverse primer 5′-ACAAAGCGTCGAAACACATC-3′ (genomic region 63,318 to 63,337). The forward primers contained an additional XbaI restriction site and the reverse primers an additional BamHI restriction site. CYP79F1 cDNA was subcloned into pUC18, and CYP79F2 cDNA was subcloned into pBluescript SK−. Both cDNAs were sequenced by the DNA core facility at the Max-Planck-Institut für Züchtungsforschung (Cologne, Germany) on PE-Applied Biosystems (Weiterstadt, Germany) Abi Prism 377 and 3700 sequencers by using BigDye terminator chemistry.
Expression of N-Terminal Green Fluorescent Protein Fusions in BY2
The 5′ region of CYP79F1 was isolated from the cloned cDNA (see Reverse Transcription–PCR Analysis) by RcaI digestion (the RcaI site is just upstream of the start ATG). The resulting 180-bp fragment was subcloned into the NcoI site of the vector pCATSgfp (kindly provided by Guido Jach, Max-Planck-Institut für Züchtungsforschung). It contains a codon-optimized green fluorescent protein (GFP) S65C mutant gene (Heim et al., 1995) under the control of the cauliflower mosaic virus (CaMV) 35S promoter and an additional translation-enhancing element of tobacco etch virus (the basic vector is described in Carrington and Freed, 1990). The 5′ region of CYP79F2 was isolated by PCR using genomic DNA as a template (10 ng; for conditions, see RT-PCR Analysis) and the following primer combination: forward, 5′-TTAAGATGATGATGAAGATT-3′ (genomic region 60,235 to 60,254 of BAC clone F309, with an additional EcoRI site at the 5′ end), and reverse, 5′-AATTGTCTCCAATGGACTCT-3′ (genomic region 60,599 to 60,618). The PCR product (400 bp) was digested with EcoRI and RcaI, leading to a 180-bp fragment that was subcloned into the vector pCATSgfp digested with EcoRI and NcoI, thereby replacing the translation-enhancing element. As a control for endoplasmic reticulum (ER) localization, the GFP variant mGFP5ER (Haselhoff et al., 1997), which has been cloned into the vector pBT4 (Feldbrügge et al., 1994), was used. As a control for cytoplasmic localization, the vector pCATSgfp (see above) was used. Transient expression in tobacco BY2 protoplasts and microscopic analysis were performed as described by Bischoff et al. (2000).
Construction of Binary Vectors
PCYP79F1 and PCYP79F2 were isolated by PCR using genomic DNA as a template (10 ng; for conditions, see Reverse Transcription–PCR Analysis). PCYP79F1 was amplified with the forward primer 5′-ACTGTTGTTTTTAGACTATCCA-3′ (genomic region 62,359 to 62,380 of BAC clone F309) and the reverse primer 5′-GATGATGTATGTATGCGTGTAG-3′ (genomic region 63,852 to 63,873; the start ATG has been mutated to ATC corresponding to GAT in the complementary strand). PCYP79F2 was amplified with the forward primer 5′-ATTATAAGTGACTGTCTCATCT-3′ (genomic region 59,606 to 59,627) and the reverse primer 5′-ATCTTAATGTTTGTATTTTTGT-3′ (genomic region 60,220 to 60,241; the primer ends just before the start ATG). The forward primers also contained an XbaI restriction site, and the reverse primers contained a BamHI site. PCYP79F2 was cloned as an XbaI-BamHI fragment into pBluescript SK− and subsequently as an SstI-BamHI fragment into the binary vector pVKH35SGUSpA (Reintanz, 1997; see below). PCYP79F1 contains an internal BamHI site and therefore was cloned in several steps into the binary vector. First, the 5′ 800-bp XbaI-BamHI fragment was subcloned into pBluescript SK− and then cloned as an SstI-BamHI fragment into pVKH35SGUSpA. Subsequently, the 3′ 700-bp BamHI fragment was ligated directly into pVKH35SGUSpA. This binary vector allows plant selection on hygromycin and contains the uidA gene under the control of the CaMV 35S promoter. This promoter was excised by digestion with SacI and BamHI and replaced by PCYP79F1 and PCYP79F2, respectively.
The CYP79F1 cDNA was isolated from pUC18 (see Reverse Transcription–PCR Analysis) and cloned into the binary vector pVKH35SpA1 (Reintanz, 1997). This binary vector allows plant selection on hygromycin and contains a CaMV 35S–poly(A) cassette with a polylinker in between. For cloning, HindIII and XbaI restriction sites in the polylinker were used. The vector was digested with HindIII, overhangs were filled in with T4 DNA polymerase, and the vector was redigested with XbaI. Subsequently, CYP79F1 cDNA was ligated as an XbaI-SmaI fragment.
Generation of Transgenic Arabidopsis Plants
Arabidopsis plants were transformed by dipping developing flowers into Agrobacterium cultures that had been grown overnight (Clough and Bent, 1998). Seed were collected and screened for hygromycin resistance (half-strength MS medium supplemented with 15 μg/mL hygromycin). After 2 weeks, 35 resistant seedlings (T1 generation) were transferred to the greenhouse. The T2 generation was screened for 3:1 (resistant:nonresistant) segregation. To identify homozygous plants, the seed of 10 resistant plants from two T2 plant lines were screened for 100% resistance (T3 generation).
Histochemical Assay of β-Glucuronidase Expression
Intact seedlings grown in vitro or excised organs of mature plants were vacuum infiltrated with β-glucuronidase (GUS) staining solution (100 mM Na2HPO4, pH 7, 0.1% Triton-X 100, 2 mM K3Fe[CN]6, 2 mM K4Fe[CN]6, and 0.5 mg/mL 5-bromo-4-chloro-3-indolyl-β-glucuronic acid) two times for 10 min each. After incubation overnight at 37°C, tissue was cleared by treatment with 70% ethanol and analyzed with the Leica microscope DM R (Leica, Wetzlar, Germany).
Glucosinolate Analysis
Rosette leaves were harvested from 6-week-old plants grown under short-day conditions (there were no differences in the sizes of the leaves between bus1-1f and wild-type plants; see Figure 1D). For each sample, all rosette leaves from one plant were collected; five samples were measured. Roots were isolated from liquid cultures grown in vitro, each culture containing ∼20 plants (one culture per sample; two samples were measured). Roots and leaves were lyophilized before measurement. For seed analysis of either the bus1-1f mutant or the transgenic line 35S::CYP79F1, five samples were measured (each 25 mg).
Samples were immersed in 4 mL of boiling water containing 100 μL of 0.3 M Pb(OAc)2/0.3 M Ba(OAc)2 solution and 0.5 μmol of internal standard (allyl glucosinolate) for 15 min. After 30 min of gentle shaking at room temperature, samples were centrifuged at 4000g for 10 min, and the supernatant was loaded onto a small (100 mg) column of DEAE Sephadex A-25. The column was rinsed with 67% (aqueous) methanol and water. After capping, 50 μL of sulfatase solution (Graser et al., 2000) was loaded and incubated overnight. The resulting desulfoglucosinolates were eluted from the column with 0.8 mL of 60% (aqueous) methanol and 0.8 mL of water and pooled. Separation of desulfoglucosinolates was achieved by HPLC on a Hewlett-Packard (Palo Alto, CA) HP 1100 series system with autosampler, column oven, and diode array detector. The procedure used a C18 fully end-capped reverse phase column (LiChrospher RP-18, 250 × 4.6 mm i.d., 5-μm particle size; Chrompack, Raritan, NJ) operated at 1 mL/min and 25°C. The system was equipped with a C18 LiChrospher (75 × 4.6 mm i.d., 5-μm particle size; Chrompack) reverse phase guard column. The injection volume was 20 μL. Elution was accomplished with a gradient (solvent A, water; solvent B, acetonitrile) of 1.5 to 5% solvent B (6 min), 5 to 7% solvent B (2 min), 7 to 21% solvent B (10 min), 21 to 29% solvent B (5 min), and 29 to 57% solvent B (14 min), followed by a cleaning cycle (57 to 93% solvent B for 2 min, 5 min of hold, 93 to 1.5% solvent B for 3 min, and 6 min of hold). Eluting compounds were monitored at 229 nm, and peaks were identified by matching retention times and UV spectra with those of desulfoglucosinolate standards. Concentrations of glucosinolates were calculated in relation to the internal standard by applying response factors established for single desulfoglucosinolates (P.D. Brown and J. Gershenzon, unpublished data).
Indole-3-Acetic Acid (IAA) and Indole-3-Acetonitrile (IAN) Analysis
Rosette leaves were harvested from 3-week-old plants grown under short day conditions (there were no differences in the sizes of the leaves between mutant and wild-type plants). For each sample, all rosette leaves from four plants were pooled, frozen in liquid nitrogen, and homogenized; three samples representing 12 plants were analyzed.
Between 10 and 50 mg per sample was incubated with 1 mL of extraction buffer (50 mM Na2HPO4, pH 7, and 0.02% diethyldithiocarbamic acid) and internal standards (250 pg of 13C6IAA and 20 ng of 13C1IAN per sample) for 6 hr at 4°C. Samples were centrifuged for 10 min at 14,000g. The supernatants were acidified with 1 M HCl to pH 2.7, and 60 mg of Amberlite XAD-7 was added. After 1 hr of incubation, supernatants were removed, XAD-7 resin was washed with 1% acetic acid and vacuum dried, and absorbed compounds were eluted twice with 1.5 mL of dichloromethane. Eluates were combined, vacuum-dried, and methylated with diazomethane. Methylated samples were dried, dissolved in 15 μL of heptane, and silylated with 5 μL of N,O-bis-(trimethylsilyl)-trifluoracetamide/0.1% trimethyl chlorosilane for 20 min at 70°C, and 1 μL of each sample was splitless injected into a gas chromatography–mass spectrometry system (Edlund et al., 1995).
The column used for analysis was a CP-SIL 24CB (30 m × 0.25 mm; Varian, Inc., Lexington, MA), and the injector temperature was set to 270°C. After injection, the column temperature was held at 60°C for 2 min, increased at a rate of 20°C/min to 200°C, and then increased again at a rate of 4°C/min to 270°C. Column effluent was introduced into the ion source of a JEOL JMS SX102/102 mass spectrometer. The ion source and a gas chromatography interface were held at 270°C, and ions were generated with 70 electron volts at an ionization current of 300 μA. The mass spectrometer operated in selected reaction monitoring mode with an acceleration voltage of −10 kV and the ion multiplier set to −1.5 kV. The precursor ions were selected by magnetic switching; the daughter ions were selected by simultaneous switching of the magnetic and electrostatic fields. For IAA analysis, the monitored reactions were between mass-to-charge ratio (m/z) = 261.118 and m/z = 202.105 (endogenous IAA) or between m/z = 267.137 and m/z = 208.125 (internal standard). IAN measurement was based on reactions monitored between m/z = 228.108 and m/z = 213.085 (endogenous IAN) or between m/z = 229.112 and m/z = 214.088 (internal standard) (Ilic et al., 1996). Peak integration and additional data processing were performed using JEOL XMass software.
Acknowledgments
We thank Dr. O. Leyser for helpful comments on the manuscript, our service group ADIS (Automated DNA Isolation and Sequencing) for DNA sequencing, ZIGIA (Center for Functional Genomics in Arabidopsis) for the En lines and support, and members of our laboratory, particularly A. Szyroki, L. Vahlkamp, and Dr. P. Wolff, for critically reading the manuscript. This work was funded by the European Community BIOTECH program and the International Coop-eration Copernicus program. We also acknowledge support by the European Community V Frame work program (Grant No. QLRT-1999-01209).
References
- Bak, S., Nielsen, H.L., and Halkier, B.A. (1998). The presence of CYP79 homologues in glucosinolate-producing plants shows evolutionary conservation of the enzymes in the conversion of amino acid to aldoxime in the biosynthesis of cyanogenic glucosides and glucosinolates. Plant Mol. Biol. 38, 725–734. [DOI] [PubMed] [Google Scholar]
- Bartlem, D., Lambein, I., Okamoto, T., Itaya, A., Uda, Y., Kijima, F., Tamaki, Y., Nambara, E., and Naito, S. (2000). Mutation in the threonine synthase gene results in an over-accumulation of soluble methionine in Arabidopsis. Plant Physiol. 123, 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, R., Donald, A., Dawson, G., Hick, A., and Wallsgrove, R. (1993). Aldoxime-forming microsomal enzyme systems involved in the biosynthesis of glucosinolates in oilseed rape (Brassica napus) leaves. Plant Physiol. 102, 1307–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, R.N., Kiddle, G., Hick, A.J., Dawson, G.W., and Wallsgrove, R.M. (1996). Distribution and activity of microsomal NADPH-dependent monooxygenases and amino acid decarboxylases in cruciferous and non-cruciferous plants and their relationship to foliar glucosinolate content. Plant Cell Environ. 19, 801–812. [Google Scholar]
- Beveridge, C., Ross, J., and Murfet, I. (1994). Branching mutant rms-2 in Pisum sativum: Grafting studies and endogenous indole-3-acetic acid levels. Plant Physiol. 104, 953–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff, F., Vahlkamp, L., Molendijk, A., and Palme, K. (2000). Localization of AtROP4 and AtROP6 and interaction with the guanine nucleotide dissociation inhibitor AtRhoGDI1 from Arabidopsis. Plant Mol. Biol. 42, 515–530. [DOI] [PubMed] [Google Scholar]
- Bones, A.M., and Rossiter, J.T. (1996). The myrosinase-glucosinolate system, its organisation and biochemistry. Physiol. Plant. 97, 194–208. [Google Scholar]
- Bourgis, F., Roje, S., Nuccio, M.L., Fisher, D.B., Tarczynski, M.C., Li, C., Herschbach, C., Rennenberg, H., Pimenta, M.J., Shen, T.-L., Gage, D.A., and Hanson, A.D. (1999). S-Methylmethionine plays a major role in phloem sulfur transport and is synthesized by a novel type of methyltransferase. Plant Cell 11, 1485–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C., and Freed, D.D. (1990). Cap-independent enhancement of translation by a plant potyvirus 5′ nontranslated region. J. Virol. 64, 1590–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple, C. (1998). Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 311–343. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Davies, P.J. (1995). Plant Hormones, Physiology, Biochemistry and Molecular Biology. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Du L., and Halkier, B.A. (1996). Isolation of a microsomal enzyme system involved in glucosinolate biosynthesis from seedlings of Tropaeolum majus L. Plant Physiol. 111, 831–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., and Halkier, B.A. (1998). Biosynthesis of glucosinolates in the developing silique walls and seeds of Sinapis alba. Phytochemistry 48, 1145–1150. [Google Scholar]
- Du L., Lykkesfeldt, J., Olsen, C.E., and Halkier, B.A. (1995). Involvement of cytochrome P450 in oxime production in glucosinolate biosynthesis as demonstrated by an in vitro microsomal enzyme system isolated from jasmonic acid-induced seedlings of Sinapis alba L. Proc. Natl. Acad. Sci. USA 92, 12505–12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund, A., Eklöf, S., Sundberg, B., Moritz, T., and Sandberg, G. (1995). A microscale technique for gas chromatography–mass spectrometry measurements of picogram amounts of indole-3-acetic acid in plant tissue. Plant Physiol. 108, 1043–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldbrügge, M., Sprenger, M., Dinkelbach, M., Yazaki, K., Harter, K., and Weisshaar, B. (1994). Functional analysis of a light-responsive plant bZIP transcriptional regulator. Plant Cell 6, 1607–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellner, M. (1999). Research on mechanisms of auxin action: Isolation and characterization of auxin-related mutants. In Advances in Regulation of Plant Growth and Development, M. Strnad, P. Pec, and E. Beck, eds (Prague: Peres), pp. 139–155.
- Giamoustaris, A., and Mithen, R. (1995). The effect of modifying the glucosinolate content of leaves of oilseed rape (Brassica napus ssp. oleifera) on its interaction with specialist and generalist pests. Ann. Appl. Biol. 126, 347–363. [Google Scholar]
- Graser, G., Schneider, B., Oldham, N.J., and Gershenzon, J. (2000). The methionine chain elongation pathway in the biosynthesis of glucosinolates in Eruca sativa (Brassicaceae). Arch. Biochem. Biophys. 378, 411–419. [DOI] [PubMed] [Google Scholar]
- Halkier, B.A. (1999). Glucosinolates. In Naturally Occurring Glycosides, R. Ikan, ed (New York: John Wiley), pp. 193–223.
- Halkier, B.A., and Du, L.C. (1997). The biosynthesis of glucosinolates. Trends Plant Sci. 2, 425–431. [DOI] [PubMed] [Google Scholar]
- Hansen, C.H., Wittstock, U., Olsen, C.E., Hick, A.J., Pickett, J.A., and Halkier, B.A. (2000). Cytochrome P450 CYP79F1 from Arabidopsis catalyzes the conversion of dihomomethionine and trihomomethionine to the corresponding aldoximes in the biosynthesis of aliphatic glucosinolates. J. Biol. Chem., in press. [DOI] [PubMed]
- Haselhoff, J., Siemering, K.R., Prasher, D.C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn, G.W., Davin, L., Giblin, M., and Underhill, E.W. (1991). Biochemical genetics of plant secondary metabolites in Arabidopsis thaliana: The glucosinolates. Plant Physiol. 97, 217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim, R., Cubitt, A.B., and Tsien, R.Y. (1995). Improved green fluorescence. Nature 373, 663–664. [DOI] [PubMed] [Google Scholar]
- Hogge, L.R., Reed, D.W., and Underhill, E.W. (1988). HPLC separation of glucosinolates from leaves and seeds of Arabidopsis thaliana and their identification using thermospray liquid chromatography/mass spectrometry. J. Chromatogr. Sci. 26, 551–556. [Google Scholar]
- Hull, A.K., Vij, R., and Celenza, J.L. (2000). Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. USA 97, 2379–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic, N., Normanly, J., and Cohen, J.D. (1996). Quantification of free plus conjugated indoleacetic acid in Arabidopsis requires correction for the nonenzymatic conversion of indolic nitriles. Plant Physiol. 111, 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongen, W.M.F. (1996). Glucosinolates in Brassica: Occurrence and significance as cancer-modulating agents. Proc. Nutr. Soc. 55, 433–446. [DOI] [PubMed] [Google Scholar]
- Josefsson, E., and Appelqvist, L.-A. (1968). Glucosinolates in seed of rape and turnip rape as affected by variety and environment. J. Sci. Found. Agric. 19, 564–570. [Google Scholar]
- Kim, J., and Leustek, T. (2000). Repression of cystathione-γ-synthase in Arabidopsis thaliana produces partial methionine auxotrophy and developmental abnormalities. Plant Sci. 151, 9–18. [Google Scholar]
- Louda, S., and Mole, S. (1991). Glucosinolates: Chemistry and ecology. In Herbivores: Their Interaction with Secondary Plant Metabolites. Vol. 1. The Chemical Participants, 2nd ed, G.A. Rosenthal and M.R. Berenbaum, eds (San Diego, CA: Academic Press), pp. 123–164.
- Ludwig-Müller, J., and Hilgenberg, W. (1988). A plasma membrane-bound enzyme oxidizes l-tryptophan to indole-3-acetaldoxime. Physiol. Plant. 74, 240–250. [Google Scholar]
- Mikkelsen, D.M., Hansen, C.H., Wittstock, U., and Halkier, B.A. (2000). Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J. Biol. Chem. 275, 33712–33717. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nagata, T., Okada, K., Tabeke, I., and Mastui, C. (1981). Delivery of tobacco mosaic virus RNA into plant protoplasts mediated by reverse-phase evaporation vesicles (liposomes). Mol. Gen. Genet. 184, 161–165. [Google Scholar]
- Napoli, C.A., Beveridge, C.A., and Snowden, K.C. (1999). Reevaluating concepts of apical dominance and the control of axillary bud outgrowth. Curr. Top. Dev. Biol. 44, 127–169. [DOI] [PubMed] [Google Scholar]
- Procházka, S., and Truksa, M. (1999). Phytohormones and shoot apical dominance. In Advances in Regulation of Plant Growth and Development, M. Strnad, P. Pec, and E. Beck, eds (Prague: Peres), pp. 221–231.
- Ravanel, S., Gakiere, B., Job, D., and Douce, R. (1998). The specific features of methionine biosynthesis and metabolism in plants. Proc. Natl. Acad. Sci. USA 95, 7805–7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reintanz, B. (1997). Funktionelle Charakterisierung der Kaliumkanal a-Untereinheit AtKC1 aus Arabidopsis thaliana. Ph.D. Thesis (Cologne, Germany: University of Cologne).
- Rodman, J.E., Karol, K.G., Price, R.A., and Sytsma, K.J. (1996). Molecules, morphology, and Dahlgren's expanded order Capparales. Syst. Bot. 21, 289–307. [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Selmar, D. (1999). Biosynthesis of cyanogenic glycosides, glucosinolates and non protein amino acids. In Biochemistry of Plant Secondary Metabolism, M. Wink, ed (Boca Raton, FL: CRC Press), pp. 79–150.
- Slovin, J.P., Bandurski, R.S., and Cohen, J.D. (1999). Auxin. In Biochemistry and Molecular Biology of Plant Hormones, P.J.J. Hooykaas, M.A. Hall, and K.R. Libbenga, eds (Dordrecht, The Netherlands: Elsevier Science), pp. 273–331.
- Toroser, D., Griffiths, H., Wood, C., and Thomas, D.R. (1995. a). Biosynthesis and partitioning of individual glucosinolates between pod walls and seeds and evidence for the occurrence of PAPS: Desulphoglucosinolate sulphotransferase in seeds of oilseed rape (Brassica napus L.). J. Exp. Bot. 46, 1753–1760. [Google Scholar]
- Toroser, D., Wood, C., Griffiths, H., and Thomas, D.R. (1995. b). Glucosinolate biosynthesis in oilseed rape (Brassica napus L.): Studies with 35SO42− and glucosinolate precursors using oilseed rape pods and seeds. J. Exp. Bot. 46, 787–794. [Google Scholar]
- Varotto, C., Peraesi, P., Meurer, J., Oelmüller, R., Steiner-Lange, S., Salamini, F., and Leister, D. (2000). Disruption of the Arabidopsis photosystem I gene psaE1 affects photosynthesis and impairs growth. Plant J. 22, 115–124. [DOI] [PubMed] [Google Scholar]
- Verhoeven, D.T.H., Verhagen, H., Goldbohm, R.A., Van Den Brandt, P.A., and Van Poppel, G. (1997). A review of mechanisms underlying anticarcinogenicity by brassica vegetables. Chem.-Biol. Interact. 103, 79–129. [DOI] [PubMed] [Google Scholar]
- Wisman, E., Hartmann, U., Sagasser, M., Baumann, E., Palme, K., Hahlbrock, K., Saedler, H., and Weisshaar, B. (1998). Knock-out mutants from an En-1 mutagenized Arabidopsis thaliana population generated phenylpropanoid biosynthesis phenotypes. Proc. Natl. Acad. Sci. USA 95, 12432–12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittstock, U., and Halkier, B.A. (2000). Cytochrome P450 CYP79A2 from Arabidopsis thaliana L. catalyzes the conversion of l-phenylalanine to phenylacetaldoxime in the biosynthesis of benzylglucosinolate. J. Biol. Chem. 275, 14659–14666. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Talalay, P., Cho, C.-G., and Posner, G.H. (1992). A major inducer of anticarcinogenic protective enzymes from broccoli: Isolation and elucidation of structure. Proc. Natl. Acad. Sci. USA 89, 2399–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]