Abstract
Platelet-derived growth factor (PDGF) is a potent mitogen for many cell types. The PDGF receptor (PDGFR) is a receptor tyrosine kinase that mediates the mitogenic effects of PDGF by binding to and/or phosphorylating a variety of intracellular signaling proteins upon PDGF-induced receptor dimerization. We show here that the Na+/H+ exchanger regulatory factor (NHERF; also known as EBP50), a protein not previously known to interact with the PDGFR, binds to the PDGFR carboxyl terminus (PDGFR-CT) with high affinity via a PDZ (PSD-95/Dlg/Z0-1 homology) domain-mediated interaction and potentiates PDGFR autophosphorylation and extracellular signal-regulated kinase (ERK) activation in cells. A point-mutated version of the PDGFR, with the terminal leucine changed to alanine (L1106A), cannot bind NHERF in vitro and is markedly impaired relative to the wild-type receptor with regard to PDGF-induced autophosphorylation and activation of ERK in cells. NHERF potentiation of PDGFR signaling depends on the capacity of NHERF to oligomerize. NHERF oligomerizes in vitro when bound with PDGFR-CT, and a truncated version of the first NHERF PDZ domain that can bind PDGFR-CT but which does not oligomerize reduces PDGFR tyrosine kinase activity when transiently overexpressed in cells. PDGFR activity in cells can also be regulated in a NHERF-dependent fashion by stimulation of the β2-adrenergic receptor, a known cellular binding partner for NHERF. These findings reveal that NHERF can directly bind to the PDGFR and potentiate PDGFR activity, thus elucidating both a novel mechanism by which PDGFR activity can be regulated and a new cellular role for the PDZ domain-containing adapter protein NHERF.
Receptor tyrosine kinases (RTKs) are a large family of transmembrane proteins that transduce signals from the extracellular environment to the cell interior. RTKs are typically activated by ligand-induced dimerization or oligomerization, which leads to stimulation of their intrinsic tyrosine kinase activity. Platelet-derived growth factor (PDGF) activates an RTK known as the PDGF receptor (PDGFR), which can comprise α and/or β subunits. Following PDGF-induced dimerization, the PDGFR autophosphorylates and then associates via its large intracellular carboxyl terminus (CT) with a variety of intracellular proteins in order to mediate its effects on cell growth, motility, and proliferation (20).
Nearly a decade ago, it was reported that removal of the last several dozen amino acids from the PDGFR-β CT could result in a significant decrease in receptor autophosphorylation and signaling (40). Such a minimal truncation of the CT would not be expected to block the interaction of the PDGFR with most known PDGFR-associated proteins except possibly for phospholipase Cγ (39, 40). Since point mutations that block phospholipase Cγ binding to the PDGFR do not reduce PDGFR tyrosine kinase activity (39), however, the reduction in the tyrosine kinase activity of minimally truncated PDGFR has remained an unexplained finding.
We recently described an interaction between the CT of the β2-adrenergic receptor (β2AR) and an intracellular protein called the Na+/H+ exchanger regulatory factor (NHERF) and demonstrated that this interaction plays a role in β2AR regulation of Na+/H+ exchange (17). NHERF contains two PSD-95/Dlg/ZO-1 homology (PDZ) domains, which are protein-protein interaction domains known to associate with specific CT motifs on target proteins (15). NHERF binds avidly to the motif D(S/T)XL (17, 18, 47), which is found at the CT of the β2AR (β2AR-CT) as well as at those of a small number of other proteins, including the PDGFR. In the experiments described here, we examined (i) whether NHERF might indeed associate via its PDZ domains with the PDGFR and (ii) whether this interaction might help to explain the apparent importance of the distal PDGFR-CT in regulation of receptor activity.
MATERIALS AND METHODS
Fusion protein preparation and overlays.
Hexahistidine- and S-tagged NHERF fusion proteins, for both full-length NHERF and various NHERF truncations, were created via insertion of PCR products derived from a rabbit NHERF cDNA into pET-30A (Novagen), followed by expression and purification. β2AR-CT (last 80 amino acids of the human β2AR) as well as PDGFR-CT (last 45 amino acids of human PDGFR-β) were expressed as glutathione S-transferase (GST) fusion proteins. The GST fusion proteins were expressed using the pGEX-2TK vector (Pharmacia), which places a consensus protein kinase A (PKA) phosphorylation site at the start of each fusion protein. This site was radiolabeled with PKA (Calbiochem) and [32P]ATP (DuPont-NEN) for experiments where PDGFR-CT–GST was labeled with 32P, cleaved from the GST with thrombin (Novagen), and then overlaid onto unlabeled PDGFR-CT–GST in the presence and absence of NHERF fusion proteins. For all other experiments, however, the GST fusion proteins were not radiolabeled.
NHERF binding to the receptor-CT–GST fusion proteins was assayed via a far-Western blot overlay technique. The GST fusion proteins (5 μg per lane) were run on sodium dodecyl sulfate (SDS)–4 to 20% polyacrylamide gels (Novex, San Diego, Calif.), blotted, and overlaid with NHERF in 2% milk and 0.1% Tween 20 in phosphate-buffered saline (PBS) (blot buffer) for 1 h at room temperature. The blots were then washed three times with blot buffer, incubated for 1 h at room temperature with a horseradish peroxidase-conjugated anti-S-tag antibody (Novagen) in blot buffer, washed three more times with blot buffer, and visualized via enhanced chemiluminescence. To perform saturation-binding curves, equal amounts of PDGFR-CT–GST and control GST were loaded into multiple alternating lanes on SDS–4 to 20% polyacrylamide gels, and the resultant blots were cut into two-lane strips. The strips were incubated with increasing concentrations of NHERF(1-151) fusion protein, and the amount of binding for each strip was quantified via scanning laser densitometry; results are shown as means ± standard errors of the means (SEM). The specific binding of NHERF(1-151) to PDGFR-CT–GST was defined as the total amount of binding to PDGFR-CT–GST minus the binding of NHERF(1-151) to control GST on the same strip of blot. Binding of PDGFR-CT to NHERF-2, also known as SIP-1, E3KARP, and TKA-1 (18, 37, 51), was also examined via the overlay assay; NHERF-2 fusion proteins were prepared as previously described (18).
Cell culture and transfection.
CHO cells were maintained in Ham's F-12 medium, and COS-7 cells were maintained in Dulbecco's modified Eagle medium (GibcoBRL). Both media were supplemented with 10% fetal calf serum and 1% penicillin (10 U/ml)-streptomycin (10 μg/ml) at 37°C in a humidified 5% CO2 atmosphere. Transient transfections of CHO cells with hemagglutinin epitope (HA)-tagged rabbit NHERF cDNAs or Flag-tagged human β2AR, or of COS-7 cells with human PDGFR-β cDNA, were performed using 5 μl of Lipofectamine per μg of cDNA transfected. Transfections were performed over 2 h in 100-mm-diameter plates with a total of 10 μg of cDNA transfected per plate. All cells were serum starved for 12 to 16 h before experimentation by incubation in either Ham's F-12 medium or Dulbecco's modified Eagle medium supplemented with 0.5% bovine serum albumin, 0.5% penicillin-streptomycin, and 10 mM HEPES. All assays described were performed 48 h posttransfection. The level of PDGFR expression was quantified via binding studies with radiolabeled PDGF, following a protocol described previously (41). For the majority of experiments described here, the amount of DNA used in transfection was 4 μg of full-length NHERF cDNA per 100-mm-diameter plate of cells (referred to as “moderate” overexpression of NHERF in Results). For the NHERF truncation constructs NHERF(1-151) and NHERF(1-121), 9 μg of cDNA was transfected per 100-mm-diameter plate, as expression of these truncated proteins was somewhat lower than for full-length NHERF per unit of DNA transfected.
Some experiments, as indicated in the text, were performed on AP1-CHO cells stably transfected with either wild-type (WT) β2AR or mutant L413A β2AR or and PKA− β2AR in the vector pBK-CMV. The stable transfections were performed in the same way as the transient transfections, but the cells were then selected for 2 to 3 weeks by the addition of G418 (1 mg/ml) to the F-12–calf serum medium. Cellular expression of WT and mutant β2AR was confirmed by radioligand binding of 125I-cyanopindolol. The relative levels of expression of the WT, L413A, and PKA− β2AR variants were comparable for the three stable cell lines (0.50, 0.55, and 0.34 pmol of receptor per mg of protein, respectively).
Phospho-ERK assay.
Serum-starved cells were either unstimulated or treated with PDGF-BB homodimer (Calbiochem) at 37°C. To generate the PDGF-BB dose-response series, the exposure time for each dose was 5 min. For single-dose agonist stimulations, 200 pM PDGF-BB was applied for 5 min. Agonist-stimulated cell monolayers were lysed in Laemmli sample buffer, lysates resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene difluoride (PVDF) membrane (NEN Life Sciences) by electroblotting. Activated extracellular signal-regulated kinase (ERK) was detected using anti-phospho-ERK antibodies (New England Biolabs), visualized by enhanced chemiluminescence (Amersham), and quantified by scanning laser densitometry. Blots were then stripped and reprobed with an anti-ERK1/2 antibody (Upstate Biotechnology, Inc.) in order to normalize the amount of phosphorylated ERK to the amount of total ERK.
PDGFR immunoprecipitation and covalent cross-linking.
After either no stimulation or stimulation with 200 pM PDGF-BB, cell monolayers in 100-mm-diameter plates were washed at 4°C with PBS followed by lysis in radioimmunoprecipitation assay (RIPA) buffer. To covalently cross-link extracellular domains of the PDGFR-β, 2 mM BS3 (Pierce Laboratories) was applied to the washed monolayers in PBS supplemented with 10 mM HEPES and slowly agitated at 4°C for 30 min. Cross-linking was terminated by aspiration of the BS3-containing PBS solution followed by two washes of the monolayers with PBS containing 0.1 M Tris. After such cross-linking, monolayers were lysed with RIPA buffer as described previously (32). For cross-linking or non-cross-linking experiments, cell lysates were then clarified by centrifugation and normalized for protein content. Immunoprecipitation was performed with 5 μg of an anti-PDGFR-β rabbit polyclonal antibody (Upstate Biotechnology) plus 50 μl of a 50% slurry of protein G-plus-protein A–agarose (Calbiochem), which was agitated for 4 h at 4°C. Immunocomplexes were washed twice with ice-cold RIPA buffer, washed once with ice-cold PBS, denatured in Laemmli sample buffer, resolved by SDS-PAGE, and then transferred to a PVDF membrane. Tyrosine phosphorylation of the PDGFR immunocomplexes was detected using a horseradish peroxidase-conjugated antiphosphotyrosine monoclonal antibody, PY20H (Signal Transduction Laboratories). Nonspecific antibody interaction with the membrane was prevented by incubation of the PVDF membrane in 4% bovine serum albumin–Tris-buffered saline (0.05% Tween 20, 0.05% NP-40, 150 mM NaCl, 10 mM Tris). Immunocomplexes on PVDF membranes were visualized by enhanced chemiluminescence and quantified by scanning laser densitometry. Blots were then stripped and reprobed with the anti-PDGFR antibody in order to normalize the amount of phosphorylated receptor to the amount of total receptor.
Coimmunoprecipitation of PDGFR and NHERF.
CHO cells transfected with 4 μg of cDNA encoding WT PDGFR-β or both the PDGFR and 2 μg of HA-tagged NHERF were stimulated with 200 pM PDGF for 5 min as previously described, lysed, and clarified by centrifugation. HA-NHERF was immunoprecipitated by addition of a 20:1 dilution of HA affinity matrix slurry (BAbCo) with agitation at 4°C for 4 h. Anti-HA immunocomplexes were washed twice in RIPA lysis buffer and once with ice-cold PBS. Whole-cell lysates and HA affinity matrix immunocomplexes were transferred to PVDF membranes for immunodetection of HA-NHERF and the PDGFR. The PDGFR was detected with a 1:1,000 dilution of rabbit anti-PDGFR polyclonal antibody (Upstate Biotechnology) and a 1:7,000 dilution of a horseradish peroxidase-conjugated anti-rabbit polyclonal antibody (Jackson Immunochemicals). HA-tagged NHERF was detected with a 1:4,000 dilution of mouse anti-HA monoclonal antibody 12CA5 (Boehringer Mannheim) and a 1:5,000 dilution of a horseradish peroxidase-conjugated anti-mouse polyclonal antibody (Jackson Immunochemicals).
Creation of L1106A PDGFR-β full-length and CT fusion protein.
WT human PDGFR-β cDNA in the pUC13 vector was the generous gift of A. Kazlauskas. The PDGFR-β cDNA was subcloned into the pBK-CMV vector (Stratagene) using EcoRI and XbaI restriction sites. The L1106A mutant was created using custom-designed oligonucleotides containing the desired mutation in the last codon (CTG to GCG) with flanking NcoI and XbaI sites. The 380-bp fragment was gel purified, restriction enzyme digested, and then subcloned into the pUC13 vector containing the WT PDGFR-β cDNA, which was digested with NcoI and XbaI. The full-length mutant gene was subcloned into pBK-CMV using the EcoRI and XbaI sites. The single point mutation was confirmed by ABI sequencing. When either WT or L1106A PDGFR was overexpressed in COS-7 cells, cells were stimulated with 40 pM PDGF-BB (fivefold lower than for experiments done with endogenous receptors in CHO cells) to avoid reaching maximum levels of receptor autophosphorylation or ERK activation. To create a GST fusion protein corresponding to the CT of L1106A PDGFR, oligonucleotide primers bracketing the final 135 nucleotides of the mutant receptor and containing a 5′ BamHI site and a 3′ EcoRI site were used in a 25-cycle PCR using the full-length mutant receptor as template. The PCR product was gel purified, enzyme digested, and subcloned into the expression vector pGEX-2T, using the BamHI and EcoRI restriction sites. The point mutation was confirmed by ABI sequencing.
RESULTS
PDGFR-CT binds NHERF with high affinity.
The CTs of both the α and β forms of the PDGFR end in DSFL, a sequence consistent with the D(S/T)XL motif preferred for binding by NHERF PDZ domains (17, 18, 47). It has therefore been proposed (18) that the PDGFR-CT might bind with high affinity to NHERF. This possibility was tested and confirmed in the blot overlay experiments shown in Fig. 1A.
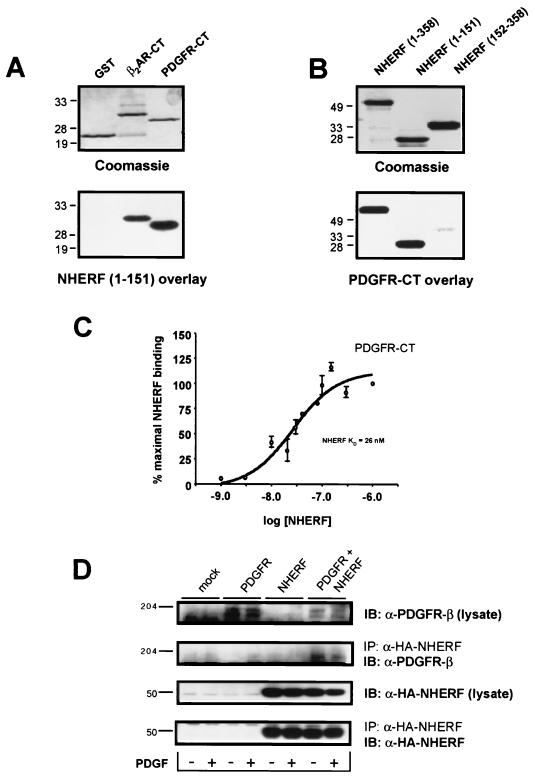
FIG. 1.
PDGFR-CT binds NHERF. (A) NHERF(1-151) binds to immobilized PDGFR-CT. PDGFR-CT, expressed as a GST fusion protein, was run on an SDS-polyacrylamide gel alongside equal amounts of control GST or the β2AR-CT expressed as a GST fusion protein (top). These samples were blotted to nitrocellulose and overlaid with 25 nM NHERF(1-151), which bound equally well to the PDGFR-CT and β2AR-CT but did not bind to control GST (bottom). (B) PDGFR-CT binds to immobilized NHERF. Equal amounts of NHERF(1-358) (full-length NHERF), NHERF(1-151), and NHERF(152-358) were run on an SDS-polyacrylamide (top), then transferred to nitrocellulose, and overlaid with 25 nM PDGFR-CT. PDGFR-CT bound strongly to full-length NHERF and NHERF(1-151) but only weakly to NHERF(152-358). The numbers adjacent to each panel represent relative molecular masses in kilodaltons. (C) PDGFR-CT binds NHERF(1-151) with high affinity. Increasing concentrations (1 to 1,000 nM) of NHERF(1-151) were overlaid onto immobilized PDGFR-CT, and binding was expressed as a percentage of maximal specific binding. The KD of the interaction was estimated at 26 nM. Points and error bars represent the mean ± SEM for three independent experiments. (D) Full-length PDGFR expressed in CHO cells coimmunoprecipitates with NHERF. Whole-cell lysates and anti-HA immunocomplexes were probed for either the PDGFR (top two panels) or HA-NHERF (lower two panels). Only in CHO cells transiently transfected with cDNAs encoding WT PDGFR and HA-NHERF was PDGFR immunoreactivity detected in anti-HA immunocomplexes. The figures adjacent to each panel represent relative molecular masses in kilodaltons. These experiments were performed in both the absence (−) and presence (+) of stimulation with 200 pM PDGF for 5 min. Stimulation with PDGF had no apparent effect on the association of PDGFR with NHERF. The data shown are representative of six independent experiments. IB, immunoblotting; IP, immunoprecipitation.
NHERF(1-151), a truncated version of full-length NHERF that encompasses the first PDZ domain of NHERF, has previously been shown to bind the β2AR-CT (17, 18). NHERF(1-151) binds as well to immobilized PDGFR-CT (the carboxyl-terminal 45 amino acids of the PDGFR-β, expressed as a GST fusion protein) as to immobilized β2AR-CT. In the reverse overlay experiment, PDGFR-CT binds avidly to both immobilized full-length NHERF and immobilized NHERF(1-151) but only weakly to immobilized NHERF(152-358), which encompasses the second NHERF PDZ domain (Fig. 1B). The estimated KD for the interaction of NHERF(1-151) with PDGFR-CT is 26 nM (Fig. 1C), a value similar to that estimated previously for the affinity of the interaction of NHERF(1-151) with β2AR-CT (KD = 18 nM) (18). The affinity of the second NHERF PDZ domain for PDGFR-CT is too low to accurately measure in saturation binding overlay studies. The interaction between NHERF and the PDGFR was also assessed in CHO cells expressing WT PDGFR and HA-tagged NHERF. PDGFR was detected in anti-HA-NHERF immunoprecipitates in an agonist-independent manner (Fig. 1D).
NHERF potentiates cellular PDGFR activity.
We next examined whether NHERF might alter PDGFR signaling in cells. As shown in Fig. 2A, transient overexpression of HA-tagged NHERF in CHO cells led to a potentiation of ERK phosphorylation induced by PDGF stimulation of endogenous PDGFR. This potentiation was not due to increased levels of cellular PDGFR, as assessed by radiolabeled ligand binding studies (data not shown). The magnitude of the potentiation was highly sensitive to the level of cellular HA-NHERF overexpression: excessive levels of NHERF overexpression failed to yield the potentiation of PDGF-induced ERK phosphorylation observed at more moderate levels (Fig. 2B).
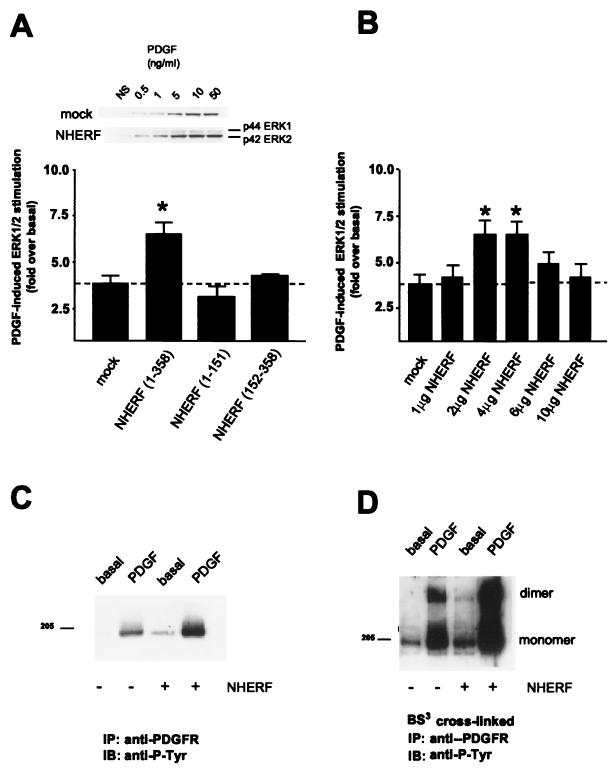
FIG. 2.
NHERF potentiates PDGFR signaling. (A) Overexpression of NHERF in CHO cells enhances PDGF-induced ERK1/2 activation. Columns and error bars represent the mean ± SEM for six independent experiments. NS, nonstimulated. (B) Overexpression of NHERF has a biphasic effect on PDGF-induced ERK1/2 activation. The level of NHERF expression is indicated as micrograms of cDNA encoding HA-tagged NHERF transfected per 100-mm-diameter plate of CHO cells. ∗, significant to P < 0.05. (C) Overexpression of NHERF enhances both basal and PDGF-induced PDGFR autophosphorylation. Endogenous PDGFRs from CHO cells that were either unstimulated or stimulated with 200 pM PDGF were immunoprecipitated (IP), resolved by SDS-PAGE, immunoblotted (IB), and probed with an antiphosphotyrosine antibody (anti-P-Tyr). Compared to cells not transfected with NHERF, NHERF overexpression elevated the basal and PDGF-induced tyrosine phosphorylation of the receptor 1.8 ± 0.3-fold (n = 4) and 2.4 ± 0.4-fold (n = 4), respectively. (D) Overexpression of NHERF enhances both basal and PDGF-induced PDGFR dimerization. Endogenous PDGFR in CHO cells was covalently cross-linked with BS3, either under basal conditions or following stimulation with 200 pM PDGF. The receptors were then immunoprecipitated, resolved by SDS-PAGE, blotted, and probed with an antiphosphotyrosine antibody. PDGF stimulation resulted in an increase in tyrosine phosphorylation of both PDGFR monomer and dimer. Compared to cells not transfected with NHERF, NHERF transfection resulted in 2.7 ± 0.1-fold (n = 4) and 2.9 ± 0.3-fold (n = 4), respectively, increases in basal and agonist-stimulated dimer tyrosine phosphorylation. The 205-kDa band is indicated on the left.
Moderate overexpression of NHERF not only enhanced PDGF-induced ERK activation but also potentiated agonist-induced autophosphorylation of endogenous PDGF receptors (Fig. 2C). This potentiation was similar in magnitude to the NHERF-induced potentiation of PDGF-stimulated ERK phosphorylation and, like the potentiation of ERK phosphorylation, was blunted at the highest levels of NHERF overexpression (data not shown). As an additional measure of the effect of NHERF overexpression on PDGFR function, both basal and PDGF-stimulated PDGFR dimerization were assessed via nonhydrolyzable receptor cross-linking. As was seen for both PDGF-induced receptor autophosphorylation and subsequent ERK activation, both basal and PDGF-induced levels of PDGFR dimerization were promoted severalfold by moderate overexpression of NHERF (Fig. 2D).
The cell lines used in this study, CHO and COS-7, contain significant levels of endogenous NHERF (17). To test the possibility that PDGFR activity in these cells is potentiated by endogenous NHERF, we prepared a point mutant of the PDGFR, L1106A, in which the terminal leucine of the PDGFR polypeptide was changed to alanine. A similar mutation of β2AR-CT, L413A, abolishes NHERF binding (17). The L1106A point mutation in the PDGFR carboxyl-terminal tail prevented NHERF binding in vitro when the mutant PDGFR-CT was expressed as a GST fusion protein (Fig. 3A). The full-length L1106A mutant and WT PDGFR were next expressed separately in COS-7 cells, which contain significantly lower levels of endogenous PDGFR than CHO cells (data not shown). In this context, L1106A PDGFR was impaired relative to the similarly overexpressed WT PDGFR with respect to PDGF-induced receptor autophosphorylation (Fig. 3B) and downstream activation of ERK (Fig. 3C).
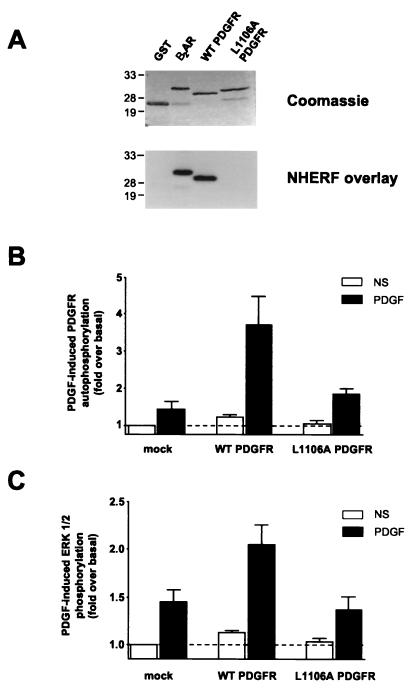
FIG. 3.
The PDGFR L1106A mutant does not bind to NHERF and is less active than WT PDGFR. (A) L1106A PDGFR-CT does not bind NHERF(1-151). L1106A PDGFR-CT, which is identical to WT PDGFR-CT except that its terminal leucine is mutated to alanine, was expressed as a GST fusion protein and run on an SDS-polyacrylamide gel alongside equal amounts of control GST, β2AR-CT, and WT PDGFR-CT (top). These samples were overlaid with 25 nM NHERF(1-151), which bound well to WT PDGFR-CT but did not bind specifically to either control GST or L1106A PDGFR-CT (bottom). The positions of molecular weight markers are shown on the left in kilodaltons. (B) Full-length L1106A PDGFR exhibits less PDGF-induced autophosphorylation than WT PDGFR in COS-7 cells. WT and L1106A PDGFR were separately transfected into COS-7 cells, and their levels of expression were comparable as assessed by radiolabeled PDGF binding. The differentially transfected cells were then not stimulated (NS) or stimulated with PDGF (40 pM) and assayed for PDGFR autophosphorylation. Bars and error bars represent the mean ± SEM for five independent experiments. (C) L1106A PDGFR exhibits reduced ability to activate ERK relative to WT PDGFR. The low level of endogenous PDGFR in COS-7 cells mediates an approximately 1.4-fold increase in ERK activation following stimulation with 40 pM PDGF (left pair of bars). Overexpression of WT PDGFR enhances PDGF-induced ERK activation (middle pair of bars), whereas overexpression of L1106A PDGFR does not (right pair of bars). The level of ERK activation under all conditions is expressed as fold over the level observed in the absence of transfected PDGFR and the absence of PDGF stimulation. Bars and error bars represent the mean ± SEM for four independent experiments.
NHERF oligomerizes when bound to PDGFR-CT.
We next explored the mechanism by which NHERF might potentiate PDGFR activation and signaling events. We hypothesized that NHERF might act as an adapter protein to facilitate a closer spatial association of two PDGFR monomers, thereby reducing the energy required to dimerize the PDGFR and activate its intrinsic kinase activity. This hypothesis was investigated by examining the ability of NHERF to facilitate the physical association of PDGFR-CTs in blot overlay studies. Radiolabeled PDGFR-CT did not detectably bind to nitrocellulose-immobilized PDGFR-CT in overlay assays, although it bound very well to immobilized NHERF in the same experiments (Fig. 4A and B). When these studies were repeated in the presence of a molar excess of NHERF in the overlay solution, radiolabeled PDGFR-CT bound to the immobilized PDGFR-CT (Fig. 4C). NHERF(1-151), the fusion protein encompassing the first PDZ domain, was also able to bridge radiolabeled PDGFR-CT to immobilized PDGFR-CT (data not shown). This suggests that the second PDZ domain, which as shown earlier binds only very weakly to the PDGFR-CT, is not required for NHERF to facilitate the association of PDGFR-CTs. Since crystallographic studies of peptides complexed with PDZ domains suggest that it is unlikely that a single PDZ domain can bind multiple carboxyl-terminal tails (6, 13), the ability of NHERF(1-151) to cross-link PDGFR-CTs in the overlay studies was somewhat surprising.
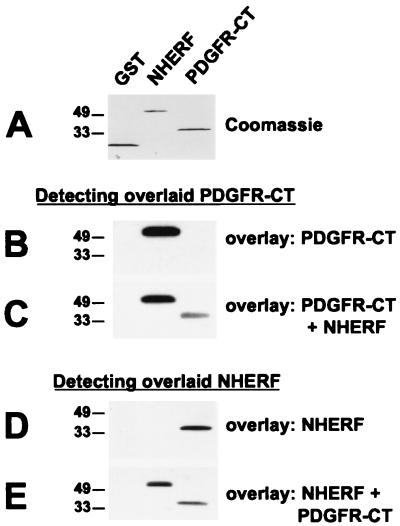
FIG. 4.
NHERF can facilitate oligomerization of PDGFR-CT, and PDGFR-CT can facilitate oligomerization of NHERF. (A) Equal amounts (2 μg) of GST, NHERF, and PDGFR-CT were run on SDS-polyacrylamide gels. (B) The samples were blotted and overlaid with 25 nM radiolabeled PDGFR-CT, which bound to immobilized NHERF but not to immobilized GST or PDGFR-CT. (C) When the experiment was repeated in the presence of 50 nM NHERF, binding of radiolabeled PDGFR-CT to immobilized PDGFR-CT was detectable. (D) The same samples were overlaid with 25 nM NHERF, which was detected via a far-Western blot approach. The overlaid NHERF bound to immobilized PDGFR-CT but not to immobilized GST or NHERF. (E) When the experiment was repeated in the presence of 250 nM PDGFR-CT, binding of overlaid NHERF to immobilized NHERF was detectable, while binding of overlaid NHERF to immobilized PDGFR-CT was reduced. These data are representative of four to six independent experiments each. The positions of molecular weight standard markers (in kilodaltons) are shown on the left of each panel.
Since some individual PDZ domains can oligomerize (3, 4, 23, 44, 50), we examined the possibility that NHERF could oligomerize to facilitate PDGFR-CT association. Initial experiments revealed that overlaid NHERF did not bind detectably to immobilized NHERF in overlay assays, although it bound very well to immobilized PDGFR-CT (Fig. 4D). To explore whether binding of PDGFR-CT to NHERF might enhance NHERF-NHERF association, we repeated the NHERF overlay experiments in the presence of a molar excess of PDGFR-CT in the overlay solution. Under these conditions, overlaid NHERF bound very well to the immobilized NHERF (Fig. 4E).
NHERF(1-121) can act as a dominant negative of cellular PDGFR activity.
The findings described above demonstrate that NHERF can oligomerize, but that it does so efficiently only when one of its PDZ domains is actively engaged with a carboxyl-terminal ligand such as the PDGFR-CT. To further characterize the phenomenon of CT-induced NHERF oligomerization, truncations of NHERF were examined for the ability to bind each other. Immobilized full-length NHERF, NHERF(1-151), and NHERF(152-358) all bound to NHERF(1-151) overlaid in the presence of a molar excess of PDGFR-CT (Fig. 5A). Further successive truncations of NHERF(1-151) shed light on the determinants of NHERF PDZ domain oligomerization. Truncation of 30 amino acids from the amino-terminal side resulted in loss of PDGFR-CT binding as well as loss of NHERF-NHERF oligomerization, consistent with previous reports of the importance of the amino-terminal portions of PDZ domains in binding to carboxyl-terminal ligands (4, 50). Removal of 30 or 50 residues from the carboxyl-terminal end, resulting in NHERF(1-121) and NHERF(1-101), respectively, eliminated NHERF-NHERF oligomerization without altering PDGFR-CT binding. Removal of 70 amino acids, resulting in NHERF(1-81), prevented both PDGFR-CT binding and NHERF-NHERF oligomerization.
FIG. 5.
NHERF(1-121) binds PDGFR-CT, does not oligomerize, and inhibits PDGFR autophosphorylation in cells. (A) Truncations of NHERF have differential effects on PDGFR-CT binding versus PDGFR-CT-induced NHERF oligomerization. Equal amounts of various truncated versions of NHERF expressed as fusion proteins were run on SDS-polyacrylamide gels and then blotted to nitrocellulose. The schematic diagram depicts the NHERF truncations and summarizes their abilities to bind overlaid 25 nM PDGFR-CT and to bind overlaid 25 nM NHERF(1-151) in the presence of 250 nM PDGFR-CT. +++, amount of specific binding observed with full-length NHERF; ++, somewhat less binding than observed with full-length NHERF; +, barely detectable binding, −, no specific binding detectable. These data are representative of four to eight independent experiments. (B) EGF-induced EGFR autophosphorylation is unaffected by overexpression of full-length NHERF, NHERF(1-151), or NHERF(1-121) in CHO cells. Human EGFR was transfected into CHO cells and stimulated with 160 pM EGF. The bars and error bars representative the mean ± SEM of three independent experiments. (C) NHERF(1-121) acts as a dominant negative for PDGF-induced PDGFR autophosphorylation in CHO cells. The endogenous PDGFR in CHO cells was stimulated with 100 pM PDGF, and the PDGFR was then immunoprecipitated, run on an SDS-polyacrylamide gel, blotted, and probed with an antiphosphotyrosine antibody. The level of PDGF-induced PDGFR autophosphorylation observed in cells transfected with either full-length NHERF, NHERF(1-151), or NHERF(1-121) is expressed as a percentage of that observed in untransfected cells. Bars and error bars represent the mean ± SEM for four independent experiments. ∗∗, significant to P < 0.01.
The in vitro association data with the truncated mutants suggested that NHERF(1-121) might be a useful reagent for testing the hypothesis that NHERF potentiation of PDGFR activity in cells depends in part on CT-induced NHERF oligomerization, since NHERF(1-121) would bind to the same intracellular partners as endogenous NHERF without being able to facilitate NHERF oligomerization. We therefore examined the effects of overexpression of full-length NHERF, NHERF(1-151), or NHERF(1-121) on autophosphorylation of endogenous PDGFR or overexpressed epidermal growth factor (EGF) receptor (EGFR) in CHO cells. EGF-induced EGFR autophosphorylation was unaffected by overexpression of NHERF or the truncated versions of NHERF (Fig. 5B), revealing that NHERF does not have a general effect on cellular RTK activity. PDGF-induced autophosphorylation of endogenous PDGFR, as described above, was consistently enhanced twofold by overexpression of full-length NHERF, while overexpression of NHERF(1-151) had no significant effect. Overexpression of NHERF(1-121), in contrast, decreased PDGF-induced autophosphorylation of endogenous PDGFR by nearly 50% (Fig. 5C).
Activation of β2AR can influence cellular PDGFR activity in a NHERF-dependent manner.
It has previously been shown that NHERF binds in an agonist-promoted fashion to the β2AR and that this association in cells imparts to the β2AR the ability to regulate Na+/H+ exchanger type 3 activity (17). One might therefore infer that the β2AR would also be able to inhibit the activity of the cellular PDGFR in an agonist-dependent fashion, since the two receptors should compete for binding to cellular NHERF. This simplistic prediction needs to be tempered, however, by the potential for the existence of a NHERF-independent stimulatory coupling between the β2AR and PDGFR. It has been shown for many G-protein-coupled receptors that agonist activation can induce cellular mitogenic signals that are mediated via transactivation of an RTK. Transactivation of growth factor receptors, such as the PDGFR (21, 29) or EGFR (11, 12, 28, 32, 46), has often been reported to be a step necessary for mitogenic signaling by G-protein-coupled receptors. If stimulation of the β2AR leads to such a transactivation of the PDGFR, this obviously would complicate studies aimed at examining the potential for NHERF-mediated interactions between the two receptors. Thus, before examining whether the β2AR can inhibit PDGFR function by competing for binding to intracellular NHERF, we investigated the potential for β2AR-mediated transactivation of the PDGFR. These studies were carried out with CHO cells, which express fairly high levels of endogenous PDGFR.
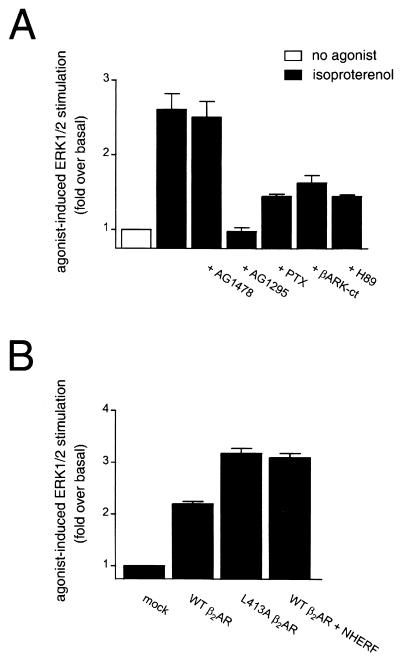
The β2AR has previously been shown to activate ERK in HEK-293 and COS-7 cells via a Gi/Gβγ-dependent pathway (10, 32), with switching of the G-protein coupling of the β2AR from Gs to Gi occurring via PKA phosphorylation of the receptor (10). CHO cells transfected with WT β2AR exhibited robust ERK activation in response to stimulation with the β-adrenergic agonist isoproterenol (Fig. 6A, first and second bars). We next examined whether this response involved RTK transactivation. CHO cells are known to express little or no endogenous EGFR (30), and indeed the β2AR-mediated activation of ERK reported here was completely insensitive to preincubation with the specific EGFR antagonist AG1478 (Fig. 6A, third bar). In contrast, the specific PDGFR antagonist AG1295 potently inhibited isoproterenol-induced increases in ERK phosphorylation (Fig. 6A, fourth bar), revealing an essential role for PDGFR kinase activity in β2AR mitogenic signaling in CHO cells. The isoproterenol-mediated ERK activation in CHO cells, like that in HEK-293 cells, was attenuated by pretreatment of the cells with pertussis toxin, by overexpression of the Gβγ sequestrant βARK-ct, and by preincubation with the specific PKA inhibitor H89 (Fig. 6A, final three bars). Thus, the β2AR in CHO cells employs a Gi/Gβγ/PKA-dependent pathway to stimulate ERK via PDGFR transactivation.
FIG. 6.
β2AR stimulates ERK phosphorylation via PDGFR transactivation. (A) Isoproterenol-induced ERK activation in CHO cells transfected with WT β2AR is inhibited by PDGFR-specific tyrosine kinase inhibitors and is mediated by a Gi/Gβγ-dependent pathway. Application of 10 μM isoproterenol for 2 min resulted in activation of ERK. This effect was not blocked by preincubation with the EGFR-specific tyrphostin AG1478 (100 nM for 10 min) but was blocked by preincubation with the PDGFR-specific tyrphostin AG1295 (10 μM for 40 min), pertussis toxin (PTX; 100 ng/ml for 16 h), or the PKA inhibitor H89 (10 μM for 10 min) and by coexpression of the Gβγ sequestrant βARK-ct. The data points represent the mean ± SEM for four separate experiments. (B) L413A β2AR exhibits a greater capacity to activate ERK than WT β2AR. Isoproterenol stimulation of the point-mutated L413A β2AR, which is incapable of sequestering cellular NHERF, resulted in more robust activation of ERK than did isoproterenol stimulation of WT β2AR. Overexpression of NHERF with WT β2AR resulted in a potentiation of the ERK activation induced by stimulation of WT β2AR. The data points represent the mean ± SEM for five separate experiments.
We next examined whether, in addition to this G-protein-dependent transactivation of the PDGFR by the β2AR, there might also be a simultaneous inhibitory form of cross talk between the two receptors due to competition for binding to endogenous NHERF. For these studies, we used a point-mutated β2AR that would be unable to sequester NHERF from the PDGFR. This mutant has the final residue of the WT β2AR-CT, Leu413, changed to alanine. This mutant receptor (L413A β2AR) does not bind NHERF yet is identical to the WT β2AR with respect to agonist binding and G-protein coupling (17). As would be expected if β2AR binding of NHERF leads to diminished PDGFR activity, removing the ability of the β2AR to bind endogenous NHERF resulted in an enhanced capacity of L413A β2AR to activate ERK compared to WT β2AR (Fig. 6B, first three bars). These data suggest that mutation of the final residue of the β2AR alleviates a negative regulatory effect of WT β2AR activation on PDGFR function. If this regulatory effect is indeed due to competition for NHERF binding, then it should also be attenuated by overexpression of NHERF. This prediction is borne out by the observation that NHERF overexpression enhances the ability of the WT β2AR to activate ERK via PDGFR transactivation (Fig. 6B, fourth bar).
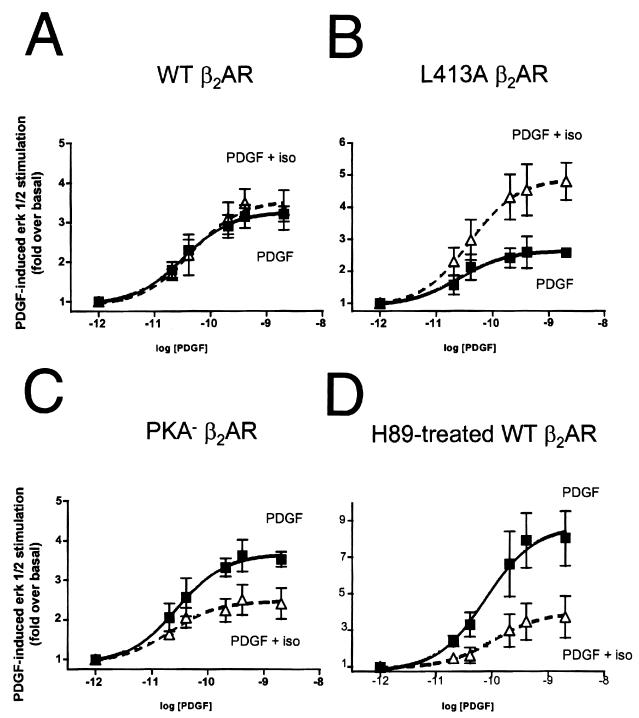
To more definitively establish the presence of two opposing forms of β2AR cross talk to the PDGFR, we studied the effect of β2AR prestimulation on PDGF-induced ERK activation. It might be predicted that in cells stably expressing WT β2AR, a combination of G-protein-mediated transactivation and NHERF-mediated inhibition of PDGFR signaling may occur. The data shown in Fig. 7A demonstrate that isoproterenol stimulation of WT β2AR before determination of the level of ERK activation at various PDGF concentrations did not cause a significant alteration in PDGFR signaling to ERK. In contrast, prestimulation of L413A β2AR, which cannot sequester NHERF from the PDGFR, resulted in a significant enhancement of subsequent PDGF-mediated ERK activation (Fig. 7B). Thus, L413A β2AR retains the capacity to transactivate the PDGFR and in fact transactivates the PDGFR more strongly than does WT β2AR. Conversely, prestimulation of the mutant receptor PKA− β2AR, which can sequester NHERF but which lacks PKA phosphorylation sites and thus cannot engage the Gi/Gβγ-dependent transactivation pathway (10), yielded no PDGFR transactivation. Instead, isoproterenol prestimulation of PKA− β2AR resulted in an attenuation of subsequent PDGF-mediated ERK activation (Fig. 7C).
FIG. 7.
Dissection of G-protein-mediated transactivation from NHERF-mediated inhibition of PDGFR signaling by the β2AR. (A) Prestimulation of WT β2AR stably expressed in AP1-CHO cells does not significantly alter subsequent ERK activation by PDGF. Cells pretreated with isoproterenol (iso) were incubated with 10 μM isoproterenol for 10 min before determination of the levels of ERK activation induced by increasing concentrations of PDGF. (B) Prestimulation of L413A β2AR, stably expressed in AP1-CHO cells, results in an elevation of PDGF-induced ERK activation. This set of experiments reveals that removing the capacity of the β2AR to sequester NHERF relieves an inhibitory action of the β2AR on PDGFR function. (C) Prestimulation of PKA− β2AR, stably expressed in AP1-CHO cells, results in an attenuation of PDGF-induced ERK activation. This set of experiments indicates that without the ability to couple to Gi and transactivate the PDGFR, PKA− β2AR can still bind NHERF and sequester it from the PDGFR, thus inhibiting PDGFR function. (D) Prestimulation of WT β2AR under conditions of reduced PKA activity results in an attenuation of PDGFR function. AP1-CHO cells stably transfected with WT β2AR were pretreated with the PKA inhibitor H89 and then stimulated with increasing concentrations of PDGF in the absence or presence of isoproterenol pretreatment. Inhibition of PKA prevents β2AR transactivation of the PDGFR and uncovers an inhibitory action of the WT β2AR on PDGFR signaling. This set of experiments reveals the capacity of WT β2AR to inhibit PDGFR function, presumably by sequestering cellular NHERF from the PDGFR.
Since prestimulation of WT β2AR exhibited little effect on PDGFR function in the experiments described above, it is likely that the G-protein-mediated transactivation and NHERF-mediated inhibition roughly negated each other under our experimental conditions. One might predict, however, that under conditions of impaired PKA activation, PDGFR transactivation by WT β2AR would be suppressed and NHERF-mediated inhibition would predominate. To mimic conditions where PKA is inactive or unavailable, and PDGFR transactivation by the β2AR is thus minimized, we performed experiments in which cells were incubated with the PKA inhibitor H89. Pretreatment with H89 blocked isoproterenol-induced increases in ERK phosphorylation, consistent with previous findings (10), but did not attenuate PDGF-induced activation of ERK (data not shown). However, isoproterenol prestimulation of the WT β2AR in H89-treated cells profoundly inhibited PDGF-induced ERK activation (Fig. 7D).
DISCUSSION
The findings described here reveal that NHERF can associate with the most distal portion of the PDGFR-CT and can potentiate PDGFR function. It has previously been reported that truncation of the last 74 amino acids from the PDGFR-β significantly decreases receptor autophosphorylation and signaling (40). The only protein previously shown to associate with the last 74 amino acids of the PDGFR is phospholipase Cγ (39, 40), but this interaction has been demonstrated to have no effect on PDGFR kinase activity (39). Thus, no satisfactory explanation has been offered for the observed reduction in the tyrosine kinase activity of slightly truncated PDGF receptors. The present results suggest a potential resolution to this mystery, since truncation of even a single amino acid from the PDGFR-CT would result in a loss of NHERF binding to the PDGFR and a consequent reduction in PDGFR activity.
The studies of truncated murine PDGFR by Seedorf et al. (40) also examined PDGFRs with 80 and 115 amino acids removed from the CT. Like the −74 truncation, the −115 truncation exhibited a substantial reduction in receptor activity. The −80 truncation, in contrast, was nearly as active as the WT receptor. Moreover, stimulation of the −80 truncated receptor resulted in tyrosine phosphorylation of a set of cellular proteins distinct from the set of proteins phosphorylated following stimulation of WT PDGFR. Interestingly, removal of 80 amino acids from the CT of the murine PDGFR-β results in the creation of a carboxyl-terminal motif (ESDN) that might exhibit some affinity for binding to the NHERF PDZ domains, even though Asn at the terminal position is less optimal than Leu (17, 18, 47). Alternatively, since there are many known PDZ proteins with slightly different binding preferences, the carboxyl-terminal motif created by the −80 truncation might facilitate association with PDZ proteins other than NHERF. Such a scenario might help to explain the anomalous gain in activity of the −80 truncated receptor relative to the −74 truncated receptor. Furthermore, this scenario might help to explain the unusual pattern of tyrosine-phosphorylated proteins observed following stimulation of the −80 truncated receptor, since association with a novel scaffolding or adapter protein might alter the set of substrates targeted by the PDGFR.
The effects of carboxyl-terminal truncations on PDGFR activity have also been studied by Mori et al. (33). Three truncations of the human PDGFR-β were made for these studies: −98, −141, and −155 amino acids. The −141 and −155 truncations both exhibited substantial reductions in receptor activity, but the −98 truncation was nearly as active as the WT receptor. Such a finding might seem to be at odds with our findings that the distal PDGFR-CT is important for modulating receptor activity. However, it should be noted that removal of 98 amino acids from the human PDGFR results in the creation of a carboxyl-terminal motif (SSVL) that is very likely to bind to NHERF with a reasonable affinity, given the known binding preferences of the NHERF PDZ domains (17, 18, 47). Thus, it is possible that the −98 truncated receptor created by Mori et al. exhibits little reduction in activity because it can still bind to NHERF almost as well as WT PDGFR. Such a scenario calls attention to the importance of examining the last few amino acids of any truncated protein, since the unintentional creation of short motifs capable of associating with PDZ domains or other modular protein binding domains is a very real possibility.
The conservative Leu-to-Ala mutation in the final residue of the PDGFR L1106A mutant described here is unlikely to interfere with the binding of any PDGFR-associated proteins other than NHERF or a closely related protein. The identified binding sites for other proteins that associate with the PDGFR are considerably removed from the distal CT (20). NHERF-2, a close relative of NHERF that is also called SIP-1, E3KARP, and TKA-1 (18, 37, 51), binds to carboxyl-terminal tails with a specificity similar to that of NHERF (18) and thus may also be a cellular binding partner for PDGFR. Indeed, a database entry by Seedorf and Ullrich identifies NHERF-2 as a binding partner for the PDGFR-CT (K. Seedorf and A. Ullrich, unpublished observations; GenBank accession number Z50150), and we have found that NHERF-2 binds the PDGFR-CT at least as well as NHERF when the two are directly compared in single-concentration overlay experiments (data not shown). Thus, the decreased activity of the L1106A mutant PDGFR in COS-7 cells is likely to be a result of a loss of binding to endogenous NHERF family proteins, although the relative contributions to this effect of endogenous NHERF and NHERF-2 are unknown.
Although NHERF enhances PDGFR activity, it is clear from both the aforementioned truncation experiments (33, 40) and the present study that NHERF is by no means required for PDGFR signaling, since truncated PDGFR as well as the L1106A mutant receptor can still autophosphorylate in response to agonist. We offer the hypothesis that NHERF can aid the formation and stabilization of active PDGFR dimers by creating oligomeric complexes that keep individual PDGFR monomers in close proximity to one another. This NHERF-mediated regulation of RTK activity is likely to be confined to the PDGFR, since the NHERF PDZ domains recognize specific carboxyl-terminal motifs at the ends of target proteins (17, 18, 47), and the present work demonstrates that EGFR activity is not enhanced by NHERF overexpression. The broader phenomenon of RTK regulation by PDZ domain-containing adapter proteins, however, may be quite general. Members of the Eph family of RTKs have been shown to interact with and be clustered by PDZ proteins such as AF6, GRIP, and PICK1 (5, 22, 45), and the Caenorhabditis elegans RTK LET-23 has been shown to associate with the PDZ protein LIN-7 in a physiologically relevant manner (43).
As shown in Fig. 6 and 7, the ability of NHERF to potentiate PDGFR signaling imparts a specialized mechanism for inhibiting PDGFR function to the β2AR, since the β2AR binds NHERF in agonist-dependent fashion (17). However, NHERF-mediated inhibition of cellular PDGFR activity by the β2AR is offset under our experimental conditions by G-protein-mediated PDGFR transactivation. These two opposing mechanisms of PDGFR regulation can be clearly teased apart only by the use of point-mutated versions of the β2AR: (i) L413A β2AR, which can transactivate the PDGFR but cannot bind NHERF, and (ii) PKA− β2AR, which can bind NHERF but cannot transactivate the PDGFR. It is not clear why the β2AR should have this potential for bidirectional regulation of PDGFR function. The two receptors are expressed together in a variety of cell types, but their potential for cross talk and mutual regulation has not been examined in native cells under conditions where neither receptor is overexpressed.
The interaction of the PDGFR with NHERF might have physiological consequences beyond potentiation of PDGFR signaling. PDGF potently regulates Na+/H+ exchange in various cell types (8, 31), and the mechanisms underlying this regulation are only partially understood. Since the association of NHERF with the β2AR allows a specialized β2-adrenergic regulation of Na+/H+ exchange in some cells (17), it is possible that the association of NHERF with the PDGFR might play a role in PDGFR regulation of Na+/H+ exchange. Moreover, since NHERF is also known as EBP50 (ezrin-binding protein of 50 kDa [38]) due to its ability to bind the actin-associated MERM (merlin, ezrin, radixin, and moesin) family proteins (35, 38), the PDGFR may be functionally linked to the actin cytoskeleton via its association with NHERF.
The experiments reported in Fig. 5 reveal that NHERF(1-121), a truncated version of NHERF that can bind to the PDGFR-CT but which cannot oligomerize, inhibits PDGFR activity when overexpressed in cells. The difference in the effects of NHERF(1-121) and NHERF(1-151) on PDGFR activity reveals that the inhibition mediated by NHERF(1-121) is not due simply to the loss of the second PDZ domain or the carboxyl-terminal domain of NHERF that binds to MERM family proteins (35, 38). The intermediate effect of NHERF(1-151) relative to full-length NHERF and NHERF(1-121) presumably reflects the fact that NHERF(1-151) is still somewhat competent to oligomerize. This truncated protein is therefore not as effective as NHERF(1-121) at inhibiting oligomerization of endogenous full-length NHERF, nor is it as effective as full-length NHERF at facilitating PDGFR dimerization. These data suggest that CT-induced NHERF oligomerization is important for NHERF-mediated potentiation of PDGFR activity, since a fragment of NHERF that binds PDGFR but does not oligomerize can act as a dominant negative with respect to cellular PDGFR activity.
NHERF oligomerization is dependent on structural determinants in the carboxyl-terminal flanking region of the NHERF PDZ domains, consistent with reports for other PDZ domains (4, 50). However, oligomerization of other PDZ domains has been reported to be either inhibited (3, 4, 23) or unaffected (50) by PDZ domain association with the CTs of other proteins. The present findings, in contrast, indicate that oligomerization of the NHERF PDZ domains is markedly enhanced by association with the PDGFR-CT. The concept of CT-induced oligomerization of NHERF PDZ domains may be important for understanding other physiological roles of NHERF, such as its ability to regulate NHE3 (17, 48, 49, 51) and its association with MERM proteins (35, 38). Moreover, PDZ domains other than those found in NHERF may also exhibit CT-induced oligomerization, and thus the phenomenon reported here may have broad implications for a variety of cellular signaling pathways.
The observation that NHERF potentiates PDGFR activity raises the possibility that NHERF may play a role in tumor formation and development, since PDGF is a potent mitogen known to correlate with the development of several kinds of tumors, especially breast cancer tumors (1, 2, 9, 42). In this context, it is of interest to note that the gene for human NHERF (GenBank accession number NM_004252) is found on chromosome 17q25.1, a frequently reported locus of allelic aberrations in breast cancer tumors (16, 24, 25, 36). Moreover, recent studies aimed at elucidating the mechanisms by which estrogen can promote cell proliferation and breast cancer tumor growth (26) have revealed that estrogen profoundly regulates the expression of cellular NHERF (14). Our data suggest that estrogen-induced increases in cellular NHERF expression would potentiate PDGFR function, thus providing a potential mechanism by which estrogen might regulate cell proliferation and breast tumor growth through alterations in the levels of cellular NHERF. Such a scenario is highly speculative at present but may be worthy of further investigation.
Previous studies have focused on the physiological consequences of NHERF association with the β2AR (7, 17). The present findings demonstrate that association of NHERF with a different type of receptor, the PDGFR, also has physiological consequences. The PDZ domains of NHERF associate with the PDGFR-CT and with each other to potentiate PDGFR activity by apparently stabilizing the formation of active PDGF-receptor complexes. Thus, the expression and availability of NHERF in a given cell may affect the responsiveness of the cell to PDGF. While cellular responses to PDGF are known to be regulated by PDGFR phosphorylation (19) and dephosphorylation (27), as well as by ligand-induced PDGFR internalization and degradation (34), regulation of PDGFR activity through receptor association with NHERF provides cells with an additional level of control over the potent mitogenic and proliferative effects induced by PDGF.
ACKNOWLEDGMENTS
We thank Shirish Shenolikar and Ed Weinman for providing NHERF cDNAs and advice, Rusty Williams and Andrius Kazlauskas for providing PDGFR cDNAs and antibodies, and Julie Pitcher, Richard Premont, and Rusty Williams for discussion and for critical readings of the manuscript. We also thank Millie McAdams and Judy Phelps for DNA sequencing and Donna Addison and Mary Holben for help in preparation of the manuscript.
This work was supported in part by NIH grants HL16037 (to R.J.L.) and HL64713 (to R.A.H.). R.J.L. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Anan K, Morisaki T, Katano M, Ikubo A, Kitsuki H, Uchiyama A, Kuroki S, Tanaka M, Torisu M. Vascular endothelial growth factor and platelet-derived growth factor are potential angiogenic and metastatic factors in human breast cancer. Surgery. 1996;119:333–339. doi: 10.1016/s0039-6060(96)80120-6. [DOI] [PubMed] [Google Scholar]
- 2.Ariad S, Seymour L, Bezwoda W R. Platelet-derived growth factor (PDGF) in plasma of breast cancer patients: correlation with stage and rate of progression. Breast Cancer Res Treat. 1991;20:11–17. doi: 10.1007/BF01833352. [DOI] [PubMed] [Google Scholar]
- 3.Brenman J E, Chao D S, Gee S H, McGee A W, Craven S E, Santillano D R, Wu Z, Huang F, Xia H, Peters M F, Froehner S C, Bredt D S. Interaction of nitric oxide syntahse with the postsynaptic density protein PSD-95 and α1-syntrophin mediated by PDZ domains. Cell. 1996;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- 4.Brenman J E, Christopherson K S, Craven S E, McGee A W, Bredt D S. Cloning and characterization of postsynaptic density 93, a nitric oxide synthase interacting protein. J Neurosci. 1996;16:7407–7415. doi: 10.1523/JNEUROSCI.16-23-07407.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchert M, Schneider S, Meskenaite V, Adams M T, Canaani E, Baechi T, Moelling K, Hovens C M. The junction-associated protein AF-6 interacts and clusters with specific Eph receptor tyrosine kinases at specialized sites of cell-cell contact in the brain. J Cell Biol. 1999;144:361–371. doi: 10.1083/jcb.144.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabral J H M, Petosa C, Sutcliffe M J, Raza S, Byron O, Poy F, Marfatia S M, Chishti A H, Liddington R C. Crystal structure of a PDZ domain. Nature. 1996;382:649–652. doi: 10.1038/382649a0. [DOI] [PubMed] [Google Scholar]
- 7.Cao T T, Deacon H W, Reczek D, Bretscher A, Von Zastrow M. A kinase-regulated PDZ-domain interaction controls endocytic sorting of the β2-adrenergic receptor. Nature. 1999;401:286–290. doi: 10.1038/45816. [DOI] [PubMed] [Google Scholar]
- 8.Cassel D, Rothenberg P, Zhuang X, Deuel T F, Glaser L. Platelet-derived growth factor stimulates Na+/H+ exchange and induces cytoplasmic alkalinization in NR6 cells. Proc Natl Acad Sci USA. 1983;80:6224–6228. doi: 10.1073/pnas.80.20.6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coltrera M D, Wang J, Porter P L, Gown A M. Expression of platelet-derived growth factor B-chain and the platelet-derived growth factor receptor subunit in human breast tissue and breast carcinoma. Cancer Res. 1995;55:2703–2708. [PubMed] [Google Scholar]
- 10.Daaka Y, Luttrell L M, Lefkowitz R J. Switching of the coupling of the β2-adrenergic receptor to different G proteins by protein kinase A. Nature. 1997;390:88–91. doi: 10.1038/36362. [DOI] [PubMed] [Google Scholar]
- 11.Daub H, Weiss F U, Wallasch C, Ullrich A. Role of transactivation of the EGF receptor in signalling by G-protein-coupled receptors. Nature. 1996;379:557–560. doi: 10.1038/379557a0. [DOI] [PubMed] [Google Scholar]
- 12.Daub H, Wallasch C, Lankenau A, Herrlich A, Ullrich A. Signal characteristics of G protein-transactivated EGF receptor. EMBO J. 1997;16:7032–7044. doi: 10.1093/emboj/16.23.7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doyle D A, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Crystal structure of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ domains. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 14.Ediger T R, Kraus W L, Weinman E J, Katzenellenbogen B S. Estrogen receptor regulation of the Na+/H+ exchange regulatory factor. Endocrinology. 1999;140:2976–2982. doi: 10.1210/endo.140.7.6885. [DOI] [PubMed] [Google Scholar]
- 15.Fanning A S, Anderson J M. PDZ domains: fundamental building blocks in the organization of protein complexes at the plasma membrane. J Clin Investig. 1999;103:767–772. doi: 10.1172/JCI6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukino K, Iido A, Teramoto A, Sakamoto G, Kasumi F, Nakamura Y, Emi M. Frequent allelic loss at the TOC locus on 17q25.1 in primary breast cancers. Genes Chromosomes Cancer. 1999;24:345–350. doi: 10.1002/(sici)1098-2264(199904)24:4<345::aid-gcc8>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 17.Hall R A, Premont R T, Chow C-W, Blitzer J T, Pitcher J A, Claing A, Stoffel R H, Barak L S, Shenolikar S, Weinman E J, Grinstein S, Lefkowitz R J. The β2-adrenergic receptor interacts with the Na+/H+ exchanger regulatory factor (NHERF) to control Na+/H+ exchange. Nature. 1998;392:626–630. doi: 10.1038/33458. [DOI] [PubMed] [Google Scholar]
- 18.Hall R A, Ostedgaard L S, Premont R T, Blitzer J T, Rahman N, Welsh M J, Lefkowitz R J. A C-terminal motif found in the β2-adrenergic receptor, P2Y1 receptor and cystic fibrosis transmembrane conductance regulator determines binding to the Na+/H+ exchanger regulatory factor family of PDZ proteins. Proc Natl Acad Sci USA. 1998;95:8496–8501. doi: 10.1073/pnas.95.15.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen K, Johnell M, Siegbahn A, Rorsman C, Engstrom U, Wernstedt C, Heldin C-H, Ronnstrand L. Mutation of a Src phosphorylation site in the PDGF beta-receptor leads to increased PDGF-stimulated chemotaxis but decreased mitogenesis. EMBO J. 1996;15:5299–5313. [PMC free article] [PubMed] [Google Scholar]
- 20.Heldin C-H, Ostman A, Ronnstrand L. Signal transduction via platelet-derived growth factor receptors. Biochim Biophys Acta. 1998;378:F79–F113. doi: 10.1016/s0304-419x(98)00015-8. [DOI] [PubMed] [Google Scholar]
- 21.Herrlich A, Daub H, Knebel A, Herrlich P, Ullrich A, Schultz G, Gudermann T. Ligand-independent activation of platelet-derived growth factor receptor is a necessary intermediate in lysophosphatidic acid-stimulated mitogenic activity in L cells. Proc Natl Acad Sci USA. 1998;95:8985–8990. doi: 10.1073/pnas.95.15.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hock B, Bohme B, Karn T, Yamamoto T, Kaibuchi K, Holtrich U, Holland S, Pawson T, Rubsamen-Waigmann H, Strebhardt K. PDZ-domain-mediated interaction of the Eph-related receptor tyrosine kinase EphB3 and the ras-binding protein AF6 depends on the kinase activity of the receptor. Proc Natl Acad Sci USA. 1998;95:9779–9784. doi: 10.1073/pnas.95.17.9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaffrey S R, Snowman A M, Eliasson M J, Cohen N A, Snyder S H. CAPON: a protein associated with neuronal nitric oxide synthase that regulates its interactions with PSD-95. Neuron. 1998;20:115–124. doi: 10.1016/s0896-6273(00)80439-0. [DOI] [PubMed] [Google Scholar]
- 24.Kalikin L M, Qu X, Frank T S, Caduff R F, Svoboda S M, Law D J, Petty E M. Detailed deletion analysis of sporadic breast tumors defines an interstitial region of allelic loss on 17q25. Genes Chromosomes Cancer. 1996;17:64–68. doi: 10.1002/(SICI)1098-2264(199609)17:1<64::AID-GCC10>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 25.Kalikin L M, Frank T S, Svoboda-Newman S M, Wetzel J C, Cooney K A, Petty E M. A region of interstitial 17q25 allelic loss in ovarian tumors coincides with a defined region of loss in breast tumors. Oncogene. 1997;14:1991–1994. doi: 10.1038/sj.onc.1201013. [DOI] [PubMed] [Google Scholar]
- 26.Katzenellenbogen B S, Montano M M, Ekena K, Herman M E, McInerney E M. Antiestrogens: mechanisms of action and resistance in breast cancer. Breast Cancer Res Treat. 1997;44:23–38. doi: 10.1023/a:1005835428423. [DOI] [PubMed] [Google Scholar]
- 27.Klinghoffer R A, Kazlauskas A. Identification of a putative Syp substrate, the PDGF-β receptor. J Biol Chem. 1994;270:22208–22217. doi: 10.1074/jbc.270.38.22208. [DOI] [PubMed] [Google Scholar]
- 28.Li X, Lee J W, Graves L M, Earp H S. Angiotensin II stimulates ERK via two pathways in epithelial cells—protein kinase C suppresses a G-protein coupled receptor EGF receptor transactivation pathway. EMBO J. 1998;17:2574–2583. doi: 10.1093/emboj/17.9.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linesman D, Benjamin C W, Jones D A. Convergence of angiotensin II and platelet-derived growth factor receptor signaling cascades in vascular smooth muscle cells. J Biol Chem. 1995;270:12563–12568. doi: 10.1074/jbc.270.21.12563. [DOI] [PubMed] [Google Scholar]
- 30.Livneh E, Prywes R, Kashles O, Reiss N, Sasson I, Mory Y, Ullrich A, Schlessinger J. Reconstitution of human epidermal growth factor receptor and its deletion mutants in cultured hamster cells. J Biol Chem. 1986;261:12490–12497. [PubMed] [Google Scholar]
- 31.Ma Y H, Reusch H P, Wilson E, Escobedo J A, Fantl W J, Williams L T, Ives H E. Activation of Na+/H+ exchange by platelet-derived growth factor involves phosphatidylinositol 3′-kinase and phospholipase C gamma. J Biol Chem. 1994;269:30734–30739. [PubMed] [Google Scholar]
- 32.Maudsley S, Pierce K L, Zamah A M, Miller W E, Ahn S, Daaka Y, Lefkowitz R J, Luttrell L M. The β2-adrenergic receptor mediates extracellular signal-regulated kinase activation via assembly of a multireceptor complex with the epidermal growth factor receptor. J Biol Chem. 2000;275:2239–2245. doi: 10.1074/jbc.275.13.9572. [DOI] [PubMed] [Google Scholar]
- 33.Mori S, Claesson-Welch L, Heldin C-H. Identification of a hydrophobic region in the carboxyl terminus of the platelet-derived growth factor β-receptor which is important ligand-mediated endocytosis. J Biol Chem. 1991;266:21158–21164. [PubMed] [Google Scholar]
- 34.Mori S, Heldin C-H, Claesson-Welch L. Ligand-induced ubiquitination of the platelet-derived growth factor β-receptor plays a negative regulatory role in its mitogenic signaling. J Biol Chem. 1993;268:577–583. [PubMed] [Google Scholar]
- 35.Murthy A, Gonzalez-Agosti C, Cordero E, Pinney D, Candia C, Solomon F, Gusella J, Ramesh V. NHE-RF, a regulatory co-factor for Na+/H+ exchange, is a common interactor for Merlin and ERM (MERM) proteins. J Biol Chem. 1998;273:1273–1276. doi: 10.1074/jbc.273.3.1273. [DOI] [PubMed] [Google Scholar]
- 36.Orsetti B, Courjal F, Cuny M, Rodriguez C, Theillet C. 17q21-q25 aberrations in breast cancer: combined allelotyping and CGH analysis reveals 5 regions of allelic imbalance among which two correspond to DNA amplification. Oncogene. 1999;18:6262–6270. doi: 10.1038/sj.onc.1203006. [DOI] [PubMed] [Google Scholar]
- 37.Poulat F, de Santa Barbara P, Desclozeaux M, Soullier S, Moniot B, Bonneaud N, Boizet B, Berta P. The human testis determining factor SRY binds a nuclear factor containing PDZ protein interaction domains. J Biol Chem. 1997;272:7167–7172. doi: 10.1074/jbc.272.11.7167. [DOI] [PubMed] [Google Scholar]
- 38.Reczek D, Berryman M, Bretscher A. Identification of EBP50: a PDZ-containing phosphoprotein that associates with members of the ezrin-radixin-moesin family. J Cell Biol. 1997;139:169–179. doi: 10.1083/jcb.139.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ronnstrand L, Mori S, Arridsson A K, Eriksson A, Wernstedt C, Hellman U, Claesson-Welsh L, Heldin C-H. Identification of two C-terminal autophosphorylation sites in the PDGF beta-receptor: involvement in the interaction with phospholipase C-gamma. EMBO J. 1992;11:3911–3919. doi: 10.1002/j.1460-2075.1992.tb05484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seedorf K, Millauer B, Kostka G, Schlessinger J, Ullrich A. Differential effects of carboxyl-terminal sequence deletions on platelet-derived growth factor receptor signaling activities and interactions with cellular substrates. Mol Cell Biol. 1992;12:4347–4356. doi: 10.1128/mcb.12.10.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seifert R A, Hart C E, Phillips P E, Forstrom J W, Ross R, Murray M J, Bowen-Pope D F. Two different subunits associate to create isoform-specific platelet-derived growth factor receptors. J Biol Chem. 1989;264:8771–8778. [PubMed] [Google Scholar]
- 42.Seymour L, Bezwoda W R. Positive immunostaining for platelet-derived growth factor (PDGF) is an adverse prognostic factor in patients with advanced breast cancer. Breast Cancer Res Treat. 1994;32:229–233. doi: 10.1007/BF00665774. [DOI] [PubMed] [Google Scholar]
- 43.Simske J S, Kaech S M, Harp S A, Kim S K. LET-23 receptor localization by the cell junction protein LIN-7 during C. elegans vulval induction. Cell. 1996;85:195–204. doi: 10.1016/s0092-8674(00)81096-x. [DOI] [PubMed] [Google Scholar]
- 44.Srivastava S, Osten P, Vilim F S, Khatri L, Inman G, States B, Daly C, DeSouza S, Abagyan R, Valtschanoff J G, Weinberg R J, Ziff E B. Novel anchorage of GluR2/3 to the postsynaptic density by the AMPA receptor-binding protein ABP. Neuron. 1998;21:581–591. doi: 10.1016/s0896-6273(00)80568-1. [DOI] [PubMed] [Google Scholar]
- 45.Torres R, Firestein B L, Dong H, Staudinger J, Olson E N, Huganir R L, Bredt D S, Gale N W, Yancopoulos G D. PDZ proteins bind, cluster, and synaptically colocalize with Eph receptors and their ephrin ligands. Neuron. 1998;21:1453–1463. doi: 10.1016/s0896-6273(00)80663-7. [DOI] [PubMed] [Google Scholar]
- 46.Tsai W, Morielli A D, Peralta E G. The m1 muscarinic acetylcholine receptor transactivates the EGF receptor to modulate ion channel activity. EMBO J. 1997;16:4597–4605. doi: 10.1093/emboj/16.15.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang S, Raab R W, Schatz P J, Guggino W B, Li M. Peptide binding consensus of the NHE-RF PDZ1 domain matches the C-terminal sequence of cystic fibrosis transmembrane conductance regulator (CFTR) FEBS Lett. 1998;427:103–108. doi: 10.1016/s0014-5793(98)00402-5. [DOI] [PubMed] [Google Scholar]
- 48.Weinman E J, Steplock D, Wang Y, Shenolikar S. Characterization of a protein cofactor that mediates protein kinase A regulation of the renal brush border membrane Na+-H+ exchanger. J Clin Investig. 1995;95:2143–2149. doi: 10.1172/JCI117903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weinman E J, Steplock D, Tate K, Hall R A, Spurney R F, Shenolikar S. Structure-function of recombinant Na/H exchanger regulatory factor (NHE-RF) J Clin Investig. 1998;101:2199–2206. doi: 10.1172/JCI204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu X-Z S, Choudhury A, Li X, Montell C. Coordination of an array of signaling proteins through homo- and heteromeric interactions between PDZ domains and target proteins. J Cell Biol. 1998;142:545–555. doi: 10.1083/jcb.142.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yun C H C, Oh S, Zizak M, Steplock D, Tsao S, Tse C-M, Weinman E J, Donowitz M. cAMP-mediated inhibition of the epithelial brush border Na+/H+ exchanger, NHE3, requires an associated regulatory protein. Proc Natl Acad Sci USA. 1997;94:3010–3015. doi: 10.1073/pnas.94.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]