Abstract
In Escherichia coli, the MarA protein controls expression of multiple chromosomal genes affecting resistance to antibiotics and other environmental hazards. For a more-complete characterization of the mar regulon, duplicate macroarrays containing 4,290 open reading frames of the E. coli genome were hybridized to radiolabeled cDNA populations derived from mar-deleted and mar-expressing E. coli. Strains constitutively expressing MarA showed altered expression of more than 60 chromosomal genes: 76% showed increased expression and 24% showed decreased expression. Although some of the genes were already known to be MarA regulated, the majority were newly determined and belonged to a variety of functional groups. Some of the genes identified have been associated with iron transport and metabolism; other genes were previously known to be part of the soxRS regulon. Northern blot analysis of selected genes confirmed the results obtained with the macroarrays. The findings reveal that the mar locus mediates a global stress response involving one of the largest networks of genes described.
The chromosomal multiple-antibiotic-resistance (mar) locus, first described for Escherichia coli (22), is also present among other enteric bacteria (14). Molecular characterization of this locus has been performed in E. coli (11), Salmonella enterica serovar Typhimurium (51), and more recently Shigella flexneri (T. M. Barbosa and S. B. Levy, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. A-42, p. 9, 1999). In all three genera, the locus consists of two divergently transcribed units, marC and marRAB, which are regulated from independent promoters (PmarI and PmarII, respectively) located in the marC-marR intergenic promoter/operator region. MarC has characteristics of a putative integral inner membrane protein whose function is unknown. marRAB specifies two regulatory proteins, MarR, the repressor of the operon, and MarA, a transcriptional activator. The function of MarB has not yet been defined. Increased expression of the marRAB operon results from mutations in marO or marR or from inactivation of MarR following exposure to different inducing agents, such as salicylate (1, 12). The resultant Mar phenotype includes resistance to structurally unrelated antibiotics (21, 43), organic solvents (6, 54), oxidative stress agents (4), and disinfectant products (40, 42).
The Mar phenotype is achieved through the differential expression of many chromosomal genes within the mar regulon. Regulation by MarA is achieved by its binding to a specific DNA sequence, “marbox,” in the vicinity of the promoters of controlled genes (37) or by other mechanisms yet to be identified.
Considering the broad Mar phenotype, we hypothesized that MarA affected the expression of a much wider collection of genes than is currently known. Using E. coli Panorama gene macroarrays we identified a large number of genes differentially expressed by constitutive expression of MarA, whose products may be involved in the cell's response to different environmental stresses.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
E. coli K-12 strain AG100 (21) was used for the PCR amplification of specific DNA probes. This strain was originally described (21, 22) as Δ(gal-uvrB), but the deletion was never characterized genotypically. Results obtained in the present study and by PCR of genes located in that genomic segment (galT and bioF) have, however, shown that this region is not deleted in AG100 and its derivatives. E. coli AG100Kan, a derivative of AG100 in which a 1.2-kb kanamycin resistance cassette replaces the mar locus from within marC to within marB (36), was used in the experiments described. pAS10 (48), derived from temperature-sensitive pMAK705 (Chlr) (26), carries a 2.5-kb PCR-amplified fragment containing the marCORAB sequence bearing the marR5 mutation, which produces no MarR and thus constitutively expresses MarA.
Bacterial strains were grown in Luria-Bertani media at 30°C with vigorous aeration. E. coli AG100Kan cells were made competent by the standard CaCl2 method (47), and transformants containing plasmid pMAK705 or pAS10 were maintained in the presence of 25 μg of chloramphenicol (Sigma, St. Louis, Mo.) ml−1.
RNA extraction.
Total RNA from bacterial cultures in mid-logarithmic phase (A530 = 0.35 to 0.40) was isolated by a modification of the hot acidic phenol extraction method in accordance with the manufacturer's instructions (Sigma-Genosys Biotechnologies, Inc., The Woodlands, Tex.). Following ethanol precipitation the RNA pellet was resuspended in water and treated with DNase I (Life Technologies Inc., Gaithersburg, Md.). The absence of genomic DNA was confirmed by examining samples of the RNA in nondenaturing agarose gels and by performing PCR on DNase-treated RNA samples using primers known to target the genomic DNA. The RNA concentration was determined spectrophotometrically (47).
Preparation of labeled cDNA and hybridization to the arrays.
Labeled cDNA was prepared using the E. coli cDNA-labeling primers (Sigma-Genosys) by following the manufacturer's instructions. The primers were annealed to 1 μg of total RNA in the presence of 333 μM dATP, dCTP, and dTTP and reverse transcriptase buffer in a final volume of 25 μl at 90°C for 2 min. The mixture was cooled to 42°C, and 50 U of avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim, Indianapolis, Ind.) and 20 μCi of [α-33P]dGTP (2,000 Ci/mmol) (New England Nuclear) were added. Incubation was at 42°C for 2 h 30 min. The unincorporated nucleotides were removed using a NucTrap probe purification column (Stratagene, La Jolla, Calif.).
Hybridization of the purified labeled cDNA to the Panorama E. coli gene arrays (Sigma-Genosys) was performed in roller bottles by following the manufacturer's instructions. Essentially, arrays were prehybridized for 2 h at 65°C in 5 ml of prewarmed hybridization solution. Denatured labeled cDNA in 5 ml of hybridization solution replaced the prehybridization solution, and hybridization proceeded for ∼18 h at 65°C. The arrays were washed three times with 50 ml of wash buffer at room temperature for 3-min intervals and three times with 100 ml of prewarmed (65°C) wash buffer for 20-min intervals. The compositions of the hybridization solution and wash buffer are described by Tao et al. (52). Hybridizing signals were visualized by exposure to Kodak BioMax MR X-ray film and to a Kodak storage phosphorimager screen SO230 (Molecular Dynamics, Sunnyvale, Calif.). Phosphor screens were scanned, after 1 to 3 days of exposure, at 50-μm pixel resolution in a Storm 860 phosphorimaging instrument (Molecular Dynamics). Arrays were stripped by immersing the membranes in a boiling solution of 0.5% (wt/vol) sodium dodecyl sulfate (SDS).
Description and quantification of the arrays.
The Panorama E. coli gene arrays (Sigma-Genosys) contain 4,290 PCR-amplified open reading frames (ORFs) of the E. coli K-12 (MG1655) genome (8), spotted in duplicate (see Tao et al. [52] for a more-detailed description of the arrays).
Quantification of the hybridizing signals in the phosphorimager file was carried out by Sigma-Genosys using the Array Vision&Trade software (Imaging Research, Inc.). The relative pixel values for the duplicate spots of each gene were averaged and normalized by expressing the averaged spot signal as a percentage of the signal from the averaged pixel values of the genomic DNA spots in the respective field where each gene was printed (Fig. 1). The ratio between these values in samples from cells expressing or lacking MarA represented the fold change in gene expression. Background values were determined for each field in each array by averaging the pixel values of the empty spaces located in the same secondary grid as the genomic DNA (Fig. 1). Genes whose averaged pixel values were close to background (less than a twofold difference from background values) in both experimental and control samples were not considered here.
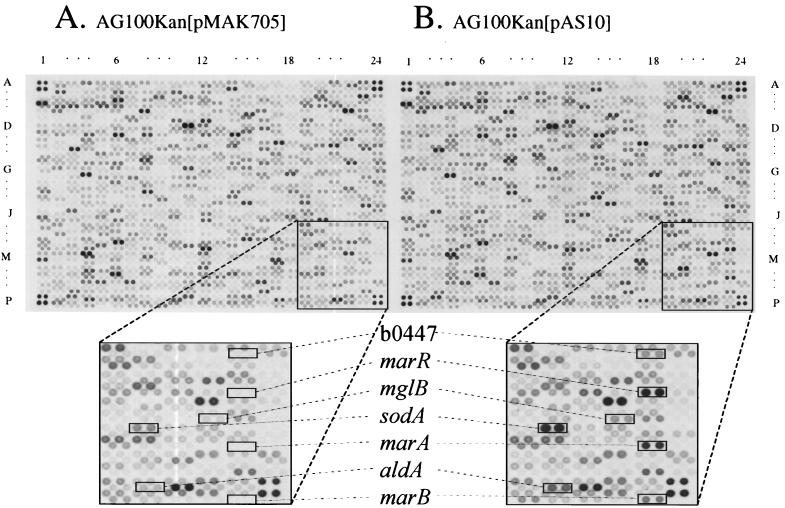
FIG. 1.
Expression profiles of E. coli strains with MarA deleted and constitutively expressing MarA. Identical arrays were probed with labeled 33P-cDNA populations prepared from total RNA from mar-deleted, AG100Kan[pMAK705] (A), and mar-expressing, AG100Kan[pAS10] (B), strains. Columns (1 to 24) and rows (A to P) forming the primary grid in field 1 of the autoradiogram are shown. Fields 2 and 3 are similar in format to field 1 and are not shown. The four spots in the four corners of each field are genomic DNA. Boxes underneath are expanded views of representative areas shown in panels A and B, where changes in expression levels are visible for several genes (seven of the differentially expressed genes are labeled as examples).
Genes identified by computer analysis as being differentially regulated by constitutive expression of MarA (greater than or equal to a twofold change in at least one experiment and with the same regulation trend, i.e., up-regulated or down-regulated, in the other) were confirmed by visual analysis of autoradiograms of the arrays in three independent experiments. Only those genes that satisfied both criteria were considered to be affected by MarA.
Northern blot analysis.
Duplicate samples of DNase I-treated total RNA (5 to 10 μg) were separated on 1 to 1.2% denaturing formaldehyde-agarose gels, and RNA was transferred to nylon membranes (Hybond-N; Amersham Life Science Inc., Arlington Heights, Ill.) using established capillary blotting methods (47). DNA probes were amplified by PCR from E. coli AG100 chromosomal DNA using the appropriate PCR primer pairs (Sigma-Genosys), according to the supplier specifications. Labeling of DNA probes with [32P]dCTP (New England Nuclear) using the room temperature stable (RTS) RadPrime DNA-labeling system (Life Technologies) was carried out according to the manufacturer's instructions. Hybridizations were performed at 65°C, and RNA membranes were washed at 65°C for 15-min intervals, four times in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer–0.1% SDS and two to four times in 0.1× SSC buffer–0.1% SDS. Hybridizing bands were visualized as described above.
DNA manipulations.
Genomic and plasmid DNA was purified from E. coli strains using the QIAamp tissue kit and the QIAprep spin Miniprep kit (Qiagen), respectively, by following the manufacturer's instructions.
RESULTS AND DISCUSSION
Identification of genes affected by constitutive MarA expression.
DNA macroarrays, which contain most of the genomic ORFs of E. coli (8), allowed studies of expression of the complete genome in the presence or absence of MarA. E. coli AG100Kan strain (36) bearing only plasmid pMAK705 represented the control, i.e., a strain deficient in mar expression. Experimental strain AG100Kan(pAS10), containing the pMAK705-derived plasmid pAS10, expresses MarA constitutively (48) and showed the expected increase (∼4- to 20-fold) in resistance to multiple antibiotics (data not shown).
33P-labeled cDNAs prepared from RNA extracted from mar-deleted and mar-expressing strains were hybridized to paired macroarrays, and phosphorimager files and autoradiograms were obtained (Fig. 1). Previously ∼15 genes were known to be regulated by MarA (2). The gene macroarrays identified a total of 62 genes responsive to constitutive expression of MarA in logarithmic phase: 47 induced and 15 repressed (Table 1). The differential regulation of the genes listed in Table 1 was confirmed visually in all three experiments.
TABLE 1.
Genes affected by constitutive expression of MarA
| Gene name | Producta | Fold changeb |
|---|---|---|
| Increased expression | ||
| acnA | Aconitate hydrase 1 | 2.7/5.9 |
| acrA | Acridine efflux pump | 1.9/2.3 |
| aldA | Aldehyde dehydrogenase, NAD linked | 7.4/3.2 |
| b0447 | Putative LRP-like transcriptional regulator | 3.5/4.4 |
| b0853 | Putative sensory transduction regulator | 1.4/4.2 |
| b1448 | Putative resistance protein | 1.8/2.3 |
| b2889 | Putative enzyme | 2.5/5.6 |
| b2948 | ORF; hypothetical protein | 1.4/2.5 |
| cobU | Cobinamide kinase or cobinamide phosphate guanylyltransferase | 1.6/2.2 |
| fumC | Fumarase C (fumarase hydratase class II); isoenzyme | 2.5/2.9 |
| galK | Galactokinase | 1.5/2.0 |
| galT | Galactose-1-phosphate uridylyltransferase | 2.5/2.4 |
| gatA | Galactitol-specific enzyme IIA of phosphotransferase system | 2.0/1.8 |
| gatC | PTS system galactitol-specific enzyme IIC | 3.4/1.6 |
| gltA | Citrate synthase | 2.1/1.9 |
| gshB | Glutathione synthetase | 3.5/5.7 |
| hemB | 5-Aminolevulinate dehydratase | 5.7/5.1 |
| inaA | pH-inducible protein involved in stress response | 5.0/20.2 |
| map | Methionine aminopeptidase | 1.7/2.1 |
| marA | Multiple-antibiotic resistance; activator | 24.0/46.6 |
| marB | Multiple-antibiotic resistance protein | 7.5/16.3 |
| marR | Multiple-antibiotic resistance protein; repressor | 15.9/46.3 |
| mdaA | Modulator of drug activity A | 3.8/8.2 |
| mdaB | Modulator of drug activity B | 5.5/8.2 |
| mglB | Galactose-binding transport protein; receptor for galactose taxis | 5.3/2.6 |
| mtr | Tryptophan-specific transport protein | 1.3/2.2 |
| nfnB | Oxygen-insensitive NADPH nitroreductase | 12.4/20.1 |
| ompX | Outer membrane protein X | 1.6/2.1 |
| pflB | Formate acetyltransferase 1 | 2.1/2.2 |
| pgi | Glucose-6-phosphate isomerase | 2.4/2.1 |
| ribA | GTP cyclohydrolase II | 1.1/2.2 |
| ribD | Bifunctional pyrimidine deaminase or reductase in pathway of riboflavin synthesis | 1.7/2.5 |
| rimK | Ribosomal protein S6 modification protein | 1.6/3.0 |
| sodA | Superoxide dismutase, manganese | 7.0/4.6 |
| srlA2 | PTS system, glucitol/sorbitol-specific IIB component and second of two IIC component | 3.0/2.0 |
| tnaA | Tryptophanase | 7.9/8.4 |
| tnaL | Tryptophanase leader peptide | 1.3/2.1 |
| tolC | Outer membrane channel; specific tolerance to colicin E1; segregation of daughter chromosomes | 3.1/2.8 |
| tpx | Thiol peroxidase | 2.1/1.6 |
| yadG | Putative ATP-binding component of a transport system | 9.2/11.2 |
| yadH | ORF; hypothetical protein | 1.9/2.7 |
| ybjC | ORF; hypothetical protein | 6.7/17.4 |
| ydeA | Putative resistance or regulatory protein | 1.9/3.9 |
| yfaE | ORF; hypothetical protein | 2.5/5.9 |
| yggJ | ORF; hypothetical protein | 3.1/4.2 |
| yhbW | Putative enzyme | 10.6/6.5 |
| zwf | Glucose-6-phosphate dehydrogenase | 2.7/1.8 |
| Decreased expression | ||
| accB | Acetyl coenzyme A carboxylase, BCCP subunit; carrier of biotin | 2.2/2.0 |
| aceE | Pyruvate dehydrogenase (decarboxylase component) | 6.1/5.2 |
| aceF | Pyruvate dehydrogenase (dihydrolipoyltransacetylase component) | 5.1/4.1 |
| ackA | Acetate kinase | 1.8/2.6 |
| b0357 | Putative alpha helix chain | 3.2/2.2 |
| b2530 | Putative aminotransferase | 1.2/2.3 |
| b3469 | Zinc-transporting ATPase | 1.6/2.2 |
| fabB | 3-Oxoacyl-(acyl carrier protein) synthase I | 2.6/3.1 |
| fecA | Outer membrane receptor, citrate-dependent iron transport | 2.5/2.8 |
| glpD | Sn-glycerol-3-phosphate dehydrogenase (aerobic) | 1.4/2.1 |
| guaB | IMP dehydrogenase | 2.9/2.3 |
| ndh | Respiratory NADH dehydrogenase | 5.8/3.8 |
| ompF | Outer membrane protein 1a (Ia, b, F) | 2.7/3.0 |
| purA | Adenylosuccinate synthetase | 2.1/2.1 |
| rplE | 50S ribosomal subunit protein L5 | 3.5/2.0 |
From the E. coli K-12 genome project (http://www.genetics.wisc.edu/). PTS, phosphotransferase system.
Fold changes in gene expression between experimental and control samples obtained from two independent experiments.
The three genes constituting the marRAB operon were easily detected in the cDNA from the mar-expressing strain but not from the mar-deleted strain (Fig. 1). This finding was reassuring given that cDNAs from genes belonging to the family of marA homologues, e.g., soxS and rob, could have caused some level of nonspecific binding (45). Although the fold changes in gene expression for marR and marA were the highest of those for all the genes identified (31- and 35-fold [averaged values], respectively; Table 1), these values cannot be taken as a direct measurement of regulation by MarA, since these genes are deleted in the control strain. Nevertheless, the signal for marB expression (12-fold change in expression [averaged value]) was consistently less than the signals for marR and marA, but the meaning of this observation is unclear. Since the spotted PCR products differ in length (which has an effect on hybridizing intensities [45]) and because the efficiency of reverse transcription varies for different RNAs, the results do not allow comparative analysis between different genes. The expression of the divergent marC (referred to as ydeB in GenBank) was close to background in the experimental sample. Thus it does not appear to be significantly affected by MarA under these conditions, and the results confirm previous reports suggesting that marC is regulated by a promoter different from that which regulates the marRAB operon (2). Also, salicylate has been shown to induce transcription of the marRAB operon but did not affect expression of marC in E. coli (51).
The genes identified in this analysis are dispersed throughout the chromosome and are involved in a wide range of cell functions (Fig. 2, Table 2), some known but others yet uncharacterized. For instance gene b0447 encodes a putative leucine-responsive regulatory protein (LRP)-like transcriptional regulator and yadG encodes a putative ATP-binding component of a transport system, but b1448 and yggJ have no known homologues. It is not clear how all these genes relate to each other in the development of the Mar phenotype. gshB is involved in the synthesis of glutathione, which is part of the cell's antioxidant defenses (27), and, among other functions, is involved in the reduction of OxyR to its normal redox state (9) and in the detoxification of toxic electrophiles (18). The induction of gshB by MarA could help to explain why resistance to oxidative stress is a Mar phenotype.
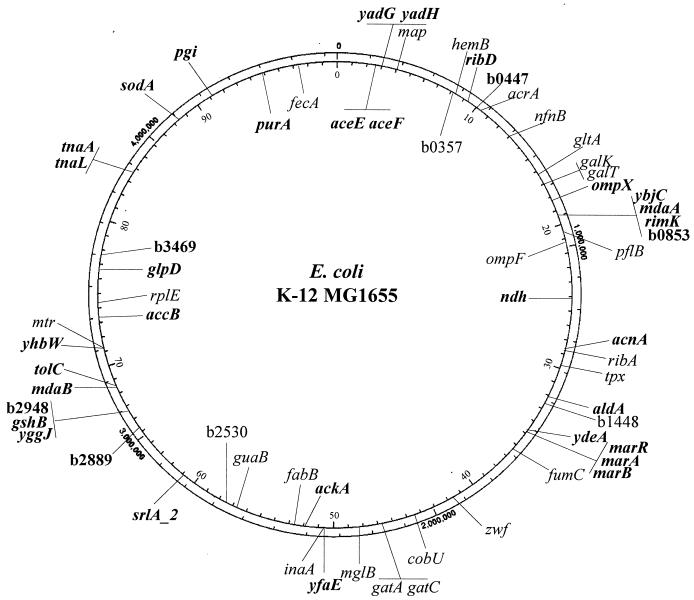
FIG. 2.
Chromosomal distribution and location of the different genes affected by MarA expression. The internal circle represents the chromosome of E. coli K-12 MG1655 divided in intervals of 1 min, while the external circle is divided in intervals of 100,000 nucleotide residues (adapted from Blattner et al. [8]). Genes induced by constitutive expression of MarA are plotted to face the exterior of the chromosome, and genes repressed by MarA are plotted to face the interior of the chromosome. Boldface genes read in the clockwise direction, while lightface genes are on the opposite strand (8). Genes that are in the immediate vicinity of each other are together over the same designation line.
TABLE 2.
Functional classification of genes affected by MarA expression
| Physiological functiona | Genes |
|---|---|
| Energy metabolism, carbon | aceE, aceF, ackA, acnA, aldA, fumC, glpD, gltA, mdaA, ndh, pflB, pgi, zwf |
| Biosynthesis of cofactors, carriers | accB, cobU, hemB, gshB, ribA, ribD |
| Carbon compound catabolism | galK, galT |
| Amino acid biosynthesis and metabolism | tnaA, tnaL |
| Fatty acid biosynthesis | fabB |
| Nucleotide biosynthesis | guaB, purA |
| Adaptation | inaAb |
| Transport/binding proteins | gatA, gatC, fecA, mglB, mtr, srlA2, tolCc, yadG, yadH, ydeA, b3469 |
| Protection responses | acrA, marA, marB, marR, nfnB, sodA, tpx |
| Cell envelope | ompF, ompX |
| Ribosome constituents | rimK, rplE |
| Macromolecule synthesis, modification | map |
| Not classified | b0357, b0447, b0853, mdaB, yhbW |
| Encoding unknown proteins | b1448, b2530, b2889, b2948, ybjC, yfaE, yggJ |
From GenProtEC E. coli genome and proteome database (http://genprotec.mbl.edu/start).
Based on presumptive evidence.
Also involved in cell division and protection responses.
There is the possibility that the differential expression of some of these genes could result from an indirect effect of the constitutive expression of MarA in the experimental strain and/or its absence in the control strain. The absence of MarA could possibly effect a physiological response which causes changes in the deleted strain compared to the wild type, AG100. We believe this to be unlikely since expression from the mar locus is tightly controlled by MarR. We could not detect mRNA from the mar locus in wild-type strains (36, 43). On the other hand the constitutive expression of MarA could produce a stress situation within the cell, with subsequent change in the expression of genes which would compensate for the possible adverse effects. Nevertheless, naturally occurring mar mutants among clinical isolates of E. coli (36, 43) which, like our experimental strain, constitutively express MarA have been reported. Still, differences in the quantity of MarA, i.e., produced with low-copy-number vector pMAK705 versus single-copy marA on the chromosome, may influence the results.
Although no in-depth comparative physiological studies were carried out, no difference in growth rate between the wild type (AG100) and the mar-deleted strain (AG100Kan) was found. Control strain AG100Kan carrying pMAK705 had a growth rate 6% slower than that of AG100 or AG100Kan, and the experimental strain, AG100Kan[pAS10], had a growth rate which was 15% slower than that of the control strain. While this growth difference could possibly affect the expression of some of the reported genes, an effect on the bacterial growth would not be unexpected as an integral part of a stress response system such as mar.
Despite the fact that AG100Kan[pAS10] constitutively expresses both MarA and MarB proteins, we believe the differential regulation of the multiple genes here reported to be associated with MarA, as MarB shows no characteristics of a transcriptional activator. Additionally marB does not appear to be necessary either for basal or inducible expression of the mar regulon or for the selection of mar mutants (39). Nevertheless we cannot rule out the possibility that MarB may indirectly affect the expression of some of these genes, e.g., by triggering a non-mar-regulated response in the cell.
Confirmation of previously identified MarA-regulated genes.
The differential expression of most of the genes previously identified as part of the mar regulon, e.g., inaA, sodA, ompF, zwf, and fumC (4, 25, 30, 46) was confirmed (Table 1). A major role in the Mar phenotype is played by the efflux system acrAB, which acts by pumping toxic compounds out of the cell (42, 44, 54). An increase in the expression of the acrA gene of the acrAB operon was also observed (Table 1); however, the expression values for acrB were not above background. This kind of finding is not fully understood but could arise from differential processing of the polycistronic transcript and/or by differences in transcript stability.
Previous studies suggest coordinate activation of TolC and the AcrAB efflux pump in the development of the Mar phenotype (3, 19). Changes in the expression of outer membrane proteins (e.g., increased OmpX expression and decreased OmpF and LamB expression) in E. coli marR mutants and wild-type strains overexpressing MarA have also been reported (3, 13). Down-regulation of ompF translation is controlled by micF, a regulatory antisense RNA known to be activated by MarA, which binds to the 5′ untranslated region of the former gene mRNA, blocking translation (16). We confirm some of these reports and show for the first time that MarA expression increases the transcription of both tolC and ompX (Table 1). Although we observed a decrease in the levels of ompF, we found no evidence for a similar decrease in lamB expression, suggesting that LamB may not be the underproduced protein identified in the earlier study (3) or that regulation may be posttranscriptional. The micF gene is not spotted on the arrays (which contain only genes coding for ORFs), and therefore we were unable to confirm activation of this gene by MarA. Nevertheless, under this assumption and given the observed down-regulation of ompF, the results indicate that micF is also involved in the destabilization of the ompF mRNA as suggested by others (13, 16).
Transcription of the previously identified mlr1 (b1451) and mlr2 (b0603) genes (48) was increased in the mar-expressing strain in two experiments but appeared to be unaffected in a third experiment, so these genes were not included in Table 1. Expression of the slp gene, previously described as repressed by MarA (48), was so low that any mar-mediated change would have been difficult to detect. This observation may reflect the fact that our experiments were performed on cells in mid-logarithmic phase while slp is a stationary-phase-inducible gene (48).
Relationship between soxRS and mar regulons.
SoxS is the activator of the soxRS regulon (17), which mediates a cellular response to oxidative stress and, like MarA, is a member of the XylS/AraC family of transcriptional activators (20). Many oxidative stress genes, which are known to respond to SoxS, are also responsive to MarA (30, 41). Conversely, SoxS is able to confer a Mar phenotype via activation of genes that are under the control of MarA (4, 25). Genes known to be regulated directly or indirectly by both the MarA and SoxS regulators include zwf, fpr, fumC, micF, nfo, inaA, sodA, and acrAB (4, 25, 30, 46, 54). We confirmed the positive regulation of zwf, fumC, acrA, inaA, and sodA by MarA and also the down-regulation of ompF. However, although binding of MarA to nfo and fpr was shown in cell-free studies (30), no significant change in expression of these two genes was detected using the experimental conditions employed here.
Our findings revealed further overlap between the mar and soxRS regulons. The levels of aconitase (acnA) and GTP cyclohydrolase II (ribA) genes and that of the major oxygen-insensitive nitroreductase gene (nfsA/mdaA), previously known to be under the control of soxRS (15, 31, 33), were increased in mar-expressing strains (Table 1). While NfsA was shown to be the major isoenzyme affected by paraquat (33), the oxygen-sensitive NADPH nitroreductase B gene, nfnB (also designated nfsB), was shown to be slightly induced. We found that nfnB, like nfsA, is under the positive control of MarA (Table 1).
nfsA was initially designated mdaA (modulator of drug activity), as one of two genes associated with bacterial resistance to tumoricidal compounds (10). The other gene, designated mdaB, was also found to be affected by MarA (Table 1). Information about mdaB is very limited, and its function remains unknown. Our findings provide suggestive evidence for a putative physiological role in protection against environmental stresses.
The exact mechanisms for the overlapping regulation by MarA and SoxS are still poorly understood. Multiple-antibiotic resistance encoded by the soxRS locus appeared partly dependent on an intact mar locus; strains overexpressing SoxS showed increased levels of marRAB transcription (41). On the other hand, other work showed that regulation of some genes by mar and by soxRS can occur through independent pathways, e.g., inaA (46). An effect of mar on soxRS has not been detected, and we observed no up-regulation of soxS expression by MarA. Therefore, MarA appears to operate independently of SoxS. A recent report suggests promoter discrimination by the two transcriptional activators dependent on differential binding to the marboxes of the involved genes (38).
Rob, a MarA and SoxS homologue, is also able to bind to promoters of genes belonging to the mar regulon, and overexpression of this protein leads to multiple-antibiotic resistance and organic solvent tolerance in E. coli (5, 29). We found no substantial change in the expression of rob due to MarA.
Effect of constitutive expression of MarA on operons and cotranscribed units.
Some of the genes affected by constitutive expression of MarA were clustered in discrete regions, as part of documented or predicted operons (Fig. 2). Interestingly, we observed considerable variability in the levels of expression of different genes from the same operon, and therefore only some of these genes were eligible for listing in Table 1. For example, the fold increase in expression of the three genes in the tryptophanase operon (tnaLAB; 83.8 min) was 1.7 for tnaL and 8.1 for tnaA (averaged values), while tnaB was unclear; it gave background values in one experiment but was clearly up-regulated in the other two experiments.
Differential expression of genes within MarA-regulated operons could arise as a result of other factors besides regulation of transcriptional initiation, e.g., differences in mRNA stability or the presence of regulatory secondary structures in the intercistronic regions of the operon. For example, the β-methylgalactoside (mgl) transport operon is composed of three ORFs, mglBAC. Northern analysis showed the presence of two transcripts, a polycistronic mglBAC mRNA and a smaller transcript which corresponds to the first gene in the operon, mglB (28). This finding may result from 3′-to-5′ nuclease degradation of the larger mRNA and from protection of the smaller transcript by a repetitive extragenic palindrome sequence located at its 3′ end (28). In agreement, our findings showed the smaller transcript at a much higher level than the larger one (Fig. 3).
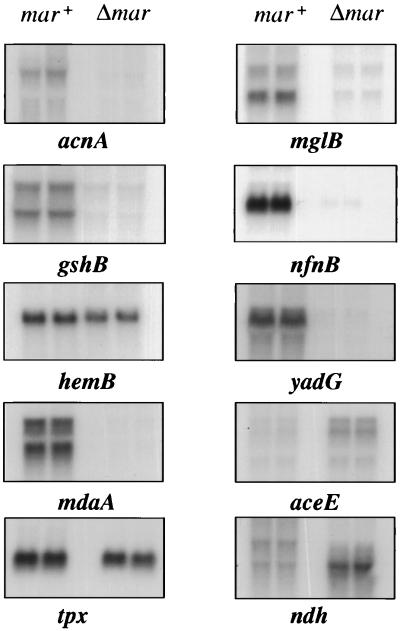
FIG. 3.
Northern blot analysis of genes affected by constitutive expression of MarA. Eight genes up-regulated by MarA, acnA, gshB, hemB, mdaA, tpx, mglB, nfnB, and yadG, and two genes down-regulated by MarA, aceE and ndh, were selected from those listed in Table 1. Samples were prepared and run in duplicate from mar-expressing (mar+) and mar-deleted (Δmar) cells. RNA samples were transferred to nylon membranes and hybridized to 32P-labeled PCR-amplified probes of the genes in the study.
From all the genes listed in Table 1 only five (acrA, pflB, ompF, marA, and mtr) appear to have a paralog in the E. coli genome. However, with the possible exception of mtr versus tnaB, none of the paralogs for these genes was identified as being affected by MarA, and therefore artifacts of cross-hybridization with other genes having substantial sequence homology (45) do not appear to account for the observed findings.
We also observed a MarA effect on neighboring genes which are not part of previously documented operons (Table 1 and Fig. 2). Up-regulation of gshB (min 66.6) expression by MarA was routinely observed; moreover, yggJ, whose function remains unknown and which is located immediately upstream from gshB, and the ORF downstream from gshB, yqgE (b2948), were also affected by constitutive MarA expression. There are only 13 bp between the end of yggJ and the beginning of gshB and 37 bp between gshB and yqgE, a situation which does not allow for the presence of promoter sequences in the respective intergenic regions. Our results support the annotation of these three genes as a “predicted operon” (8).
Transcription of the gene ybjC, a small ORF immediately upstream from nfsA, also seems to be affected by constitutive MarA expression. A promoter sequence internal to ybjC and near its start codon has been proposed for nfsA (55). Thus, nfsA could be transcribed independently from this promoter, but the resulting transcript would hybridize to both genes in the array. On the other hand, the E. coli genome sequence suggests that these two genes may form an operon (8). Expression of the two genes downstream from nfsA, rimK and b0853, is also increased in the presence of MarA. A putative transcriptional terminator in the intergenic region of nfsA and rimK has been identified (55). Nevertheless, a certain level of read-through transcription would explain the coexpression of this complex of genes.
Relationship between the mar regulon and iron.
Some of the genes affected by MarA expression are associated with iron, e.g., hemB, fumC, fecA, acnA, and sodA. Some of the encoded proteins contain iron-sulfur clusters, which play a major role in sensing O2 and iron and in regulatory functions (7). Iron is an essential element for the bacterial cell, and iron acquisition from the host is important in bacterial pathogenesis (34). However, iron can also be harmful, as it catalyzes the production of hydroxyl ions via the Fenton reaction, which may damage all cellular components and even lead to cell death (56). Some genes known to be regulated by Fur (ferric uptake regulator) are also responsive to SoxS, MarA, and other regulators, e.g., acnA and sodA (15, 50). This coregulation would allow the cell to deal with the iron-associated oxidative stress and provides a suggested role for mar in bacterial pathogenesis.
Northern blot analysis of selected genes.
The altered expression of 10 genes newly identified by the macroarrays was confirmed by Northern blot analysis, which showed changes in the expression of mono- or polycistronic transcripts (Fig. 3). The magnitude of these changes, not unexpectedly, differed somewhat from that obtained for the macroarrays. Regulation of gshB, mdaA, and aceE genes involved alteration in the levels of multiple transcripts as expected based on reported or predicted involvement of these genes in polycistronic elements (8).
Conclusions.
The transcriptional activator MarA may control the expression of genes directly or indirectly. It could activate intermediate activator or inhibitor regulatory proteins, which then could up- or down-regulate the expression of other genes in the regulon. An example is the MarA regulation of ompF. MarA activates micF, an antisense RNA which negatively affects the translation of ompF, leading to decreased outer membrane porin OmpF (13). Furthermore, transcriptional activators can act also as repressor proteins, depending on the position of the regulator binding site at the exclusive zone of repression (23).
We only report genes whose expression trends were consistent in three experiments. It is therefore likely that the size of the mar regulon is underestimated. Some of the genes containing putative marboxes in their promoter regions (37) were not shown to be part of the mar regulon under the conditions used here. Moreover, a large number of genes were expressed at background level or responded to MarA expression with small changes that were below the threshold applied in this study and therefore were not included. Under a different set of experimental conditions, such as examining cells in a different stage of the growth phase or grown in different media, it is possible that the magnitude of these changes would increase or that new genes would be affected. Certainly small and transient changes in gene expression could have important implications in the cell's response to external stresses.
Observed differences in global expression analysis between experiments have been seen and extensively addressed by other authors (45, 52). The authors observed that, among other factors, the signal intensities of some genes were significantly different between experiments when different batches of RNA were used. We hope to have addressed this problem in part by performing the study in triplicate; two experiments were quantified, and the MarA-affected genes were judged visually in all three. Changes detected by the gene array method should also be confirmed by other available molecular and biochemical techniques, such as Northern blot analysis (as was done for selected genes) and promoter fusion studies, using cells with a different genetic background.
E. coli is a natural inhabitant of different ecosystems and hosts. In order to successfully survive in such diverse conditions, this bacterium has presumably developed regulatory loci which control adaptational responses to the different environmental stresses to which it is exposed (e.g., fluctuations in temperature and pH, oxidative stress and oxygen limitation, antibiotics, and starvation). Regulatory systems, such as SoxRS, OxyR, Mar, SOS, and Fur, share the capacity to produce a global response by activating or repressing multiple genes in the bacterial chromosome. While some genes are members only of one regulon, others can be regulated by different transcriptional factors.
Some members of the mar regulon are known to be directly or indirectly controlled by other transcriptional regulators. For example, acnA expression can be activated by cyclic AMP receptor protein (CRP), FruR, Fur, and SoxRS and repressed by ArcA and FNR (15); aldA is repressed by the ArcA system and induced by an inducer-regulator complex and by CRP (32); ndh can be repressed by FNR and integration host factor (IHF) and activated by Arr (24); sodA is regulated not only by SoxS and MarA but also by FNR, ArcAB, IHF, and Fur (50); and fumC is regulated by SoxS, MarA, ArcAB and ςs (50). Both MarA and MarR have been identified as members of the heat shock stimulon (45). Additionally, pflB and guaB, which are repressed in response to heat shock, are also part of the mar regulon. A fine-tuning of the cross talk between these global regulators and the genes which they control may provide the cell with the required machinery to enhance its fitness in the new environments which it encounters.
E. coli global responses involving multiple genes include the heat shock stimulon (119 genes) (45), the Fnr modulon with over 70 genes (35), the SOS regulon with over 20 unlinked genes (49), and the soxRS and oxyR regulons, which comprise approximately 15 and 12 genes, respectively (17, 50, 56). Use of two-dimensional gel electrophoresis also identified 16 proteins as being induced upon cold shock of the E. coli cell, although only 12 of them have been identified (53). Despite the caveat that some of the changes observed may result from constitutive rather than induced expression of MarA, it seems reasonable to conclude that mar is one of the largest E. coli regulons known to date.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM51661.
We thank Laura M. McMurry and Michael N. Alekshun for helpful comments in the preparation of the manuscript.
REFERENCES
- 1.Alekshun M A, Levy S B. Alteration of the repression activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J Bacteriol. 1999;181:4669–4672. doi: 10.1128/jb.181.15.4669-4672.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alekshun M N, Levy S B. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother. 1997;41:2067–2075. doi: 10.1128/aac.41.10.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aono R, Tsukagoshi N, Yamamoto M. Involvement of outer membrane protein TolC, a possible member of the mar-sox regulon, in maintenance and improvement of organic solvent tolerance of Escherichia coli K-12. J Bacteriol. 1998;180:938–944. doi: 10.1128/jb.180.4.938-944.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ariza R R, Cohen S P, Bachhawat N, Levy S B, Demple B. Repressor mutations in the marRAB operon that activate oxidative stress genes and multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1994;176:143–148. doi: 10.1128/jb.176.1.143-148.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ariza R R, Li Z, Ringstad N, Demple B. Activation of multiple antibiotic resistance and binding of stress-inducible promoters by Escherichia coli Rob protein. J Bacteriol. 1995;177:1655–1661. doi: 10.1128/jb.177.7.1655-1661.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asako H, Nakajima K, Kobayashi K, Kobayashi M, Aono R. Organic solvent tolerance and antibiotic resistance increased by overexpression of marA in Escherichia coli. Appl Environ Microbiol. 1997;63:1428–1433. doi: 10.1128/aem.63.4.1428-1433.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beinert H, Kiley P J. Fe-S proteins in sensing and regulatory functions. Curr Opin Chem Biol. 1999;3:152–157. doi: 10.1016/S1367-5931(99)80027-1. [DOI] [PubMed] [Google Scholar]
- 8.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 9.Chater K F, Nikaido H. Cell regulation. Maintaining integrity and efficiency in microbial cells. Curr Opin Microbiol. 1999;2:121–125. [Google Scholar]
- 10.Chatterjee P K, Sternberg N L. A general genetic approach in Escherichia coli for determining the mechanism(s) of action of tumoricidal agents: application to DMP 840, a tumoricidal agent. Proc Natl Acad Sci USA. 1995;92:8950–8954. doi: 10.1073/pnas.92.19.8950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen S P, Hachler H, Levy S B. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol. 1993;175:1484–1492. doi: 10.1128/jb.175.5.1484-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen S P, Levy S B, Foulds J, Rosner J L. Salycilate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J Bacteriol. 1993;175:7856–7862. doi: 10.1128/jb.175.24.7856-7862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen S P, McMurry L M, Levy S B. marA locus causes decreased overexpression of OmpF porin in multiple-antibiotic-resistance (Mar) mutants of Escherichia coli. J Bacteriol. 1988;170:5416–5422. doi: 10.1128/jb.170.12.5416-5422.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen S P, Yan W, Levy S B. A multidrug resistance regulatory chromosomal locus is widespread among enteric bacteria. J Infect Dis. 1993;168:484–488. doi: 10.1093/infdis/168.2.484. [DOI] [PubMed] [Google Scholar]
- 15.Cunningham L, Gruer M J, Guest J R. Transcriptional regulation of the aconitase genes (acnA and acnB) of Escherichia coli. Microbiology. 1997;143:3795–3805. doi: 10.1099/00221287-143-12-3795. [DOI] [PubMed] [Google Scholar]
- 16.Delihas N. Antisense micF RNA and 5′-UTR of the target ompF RNA: phylogenetic conservation of primary and secondary structures. Nucleic Acids Symp Ser. 1997;36:33–35. [PubMed] [Google Scholar]
- 17.Demple B. Redox signaling and gene control in the Escherichia coli soxRS oxidative stress regulon—a review. Gene. 1996;179:53–57. doi: 10.1016/s0378-1119(96)00329-0. [DOI] [PubMed] [Google Scholar]
- 18.Ferguson G P. Protective mechanisms against toxic electrophiles in Escherichia coli. Trends Microbiol. 1999;7:242–247. doi: 10.1016/s0966-842x(99)01510-3. [DOI] [PubMed] [Google Scholar]
- 19.Fralick J A. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J Bacteriol. 1996;178:5803–5805. doi: 10.1128/jb.178.19.5803-5805.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallegos M-T, Schleif R, Bairoch A, Hofmann K, Ramos J L. AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.George A M, Levy S B. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid-determined efflux of tetracycline. J Bacteriol. 1983;155:531–540. doi: 10.1128/jb.155.2.531-540.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.George A M, Levy S B. Gene in the major cotransduction gap of the Escherichia coli K-12 linkage map required for the expression of chromosomal resistance to tetracycline and other antibiotics. J Bacteriol. 1983;155:541–548. doi: 10.1128/jb.155.2.541-548.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gralla J D, Collado-Vides J. Organization and function of transcriptional regulatory elements. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1232–1245. [Google Scholar]
- 24.Green J, Anjum M F, Guest J R. Regulation of the ndh gene of Escherichia coli by integration host factor and a novel regulator, Arr. Microbiology. 1997;143:2865–2875. doi: 10.1099/00221287-143-9-2865. [DOI] [PubMed] [Google Scholar]
- 25.Greenberg J T, Chou J H, Monach P A, Demple B. Activation of oxidative stress genes by mutations at the soxQ/cfxB1/marA locus of Escherichia coli. J Bacteriol. 1991;173:4433–4439. doi: 10.1128/jb.173.14.4433-4439.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hidalgo E, Demple B. Adaptive response to oxidative stress: the soxRS and oxyR regulons. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. R. G. Austin, Tex: Landes Company; 1995. pp. 433–450. [Google Scholar]
- 28.Hogg R W, Voelker C, von Carlowitz I. Nucleotide sequence and analysis of the mgl operon of Escherichia coli K12. Mol Gen Genet. 1991;229:453–459. doi: 10.1007/BF00267469. [DOI] [PubMed] [Google Scholar]
- 29.Jair K-W, Yu X, Skarstad K, Thony B, Fujita N, Ishihama A, Wolf R E., Jr Transcriptional activation of promoters of the superoxide and multiple antibiotic resistance regulons by Rob, a binding protein of the Escherichia coli origin of chromosomal replication. J Bacteriol. 1996;178:2507–2513. doi: 10.1128/jb.178.9.2507-2513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jair K-W, Martin R G, Rosner J L, Fujita N, Ishihama A, Wolf R E., Jr Purification and regulatory properties of MarA protein, a transcriptional activator of Escherichia coli multiple antibiotic resistance and superoxide resistance promoters. J Bacteriol. 1995;177:7100–7104. doi: 10.1128/jb.177.24.7100-7104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koh Y S, Chung W-H, Lee J-H, Roe J-H. The reversed SoxS-binding site upstream of the ribA promoter in Escherichia coli. Mol Gen Genet. 1999;261:374–380. doi: 10.1007/s004380050978. [DOI] [PubMed] [Google Scholar]
- 32.Limon A, Hidalgo E, Aguilar J. The aldA gene of Escherichia coli is under the control of at least three transcriptional regulators. Microbiology. 1997;143:2085–2095. doi: 10.1099/00221287-143-6-2085. [DOI] [PubMed] [Google Scholar]
- 33.Liochev S I, Hausladen A, Fridovich I. Nitroreductase A is regulated as a member of the soxRS regulon of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:3537–3539. doi: 10.1073/pnas.96.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Litwin C M, Calderwood S B. Role of iron in regulation of virulence genes. Clin Microbiol Rev. 1993;6:137–149. doi: 10.1128/cmr.6.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lynch A S, Lin E C C. Responses to molecular oxygen. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1526–1538. [Google Scholar]
- 36.Manneewannakul K, Levy S B. Identification of mar mutants among quinolone-resistant clinical isolates of Escherichia coli. Antimicrob Agents Chemother. 1996;40:1695–1698. doi: 10.1128/aac.40.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin R G, Gillette W K, Rhee S, Rosner J L. Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol Microbiol. 1999;34:431–441. doi: 10.1046/j.1365-2958.1999.01599.x. [DOI] [PubMed] [Google Scholar]
- 38.Martin R G, Gillette W K, Rosner J L. Promoter discrimination by the related transcriptional activators MarA and SoxS: differential regulation by differential binding. Mol Microbiol. 2000;35:623–634. doi: 10.1046/j.1365-2958.2000.01732.x. [DOI] [PubMed] [Google Scholar]
- 39.Martin R G, Nyantakyi P S, Rosner J L. Regulation of the multiple antibiotic resistance (mar) regulon by marORA sequences in Escherichia coli. J Bacteriol. 1995;177:4176–4178. doi: 10.1128/jb.177.14.4176-4178.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McMurry L M, Oethinger M, Levy S B. Overexpression of marA, soxS or acrAB produces resistance to triclosan in Escherichia coli. FEMS Microbiol Lett. 1998;166:305–309. doi: 10.1111/j.1574-6968.1998.tb13905.x. [DOI] [PubMed] [Google Scholar]
- 41.Miller P F, Gambino L F, Sulavik M C, Gracheck S J. Genetic relationship between soxRS and mar loci in promoting multiple antibiotic resistance in Escherichia coli. Antimicrob Agents Chemother. 1994;38:1773–1779. doi: 10.1128/aac.38.8.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moken M C, McMurry L M, Levy S B. Selection of multiple-antibiotic-resistant (Mar) mutants of Escherichia coli by using the disinfectant pine oil: roles of the mar and acrAB loci. Antimicrob Agents Chemother. 1997;41:2770–2772. doi: 10.1128/aac.41.12.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oethinger M, Podglajen I, Kern W V, Levy S B. Overexpression of the marA or soxS regulatory gene in clinical topoisomerase mutants of Escherichia coli. Antimicrob Agents Chemother. 1998;42:2089–2094. doi: 10.1128/aac.42.8.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance phenotype (Mar) mutants. J Bacteriol. 1996;178:306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richmond C S, Glasner J D, Mau R, Jin H, Blattner F R. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999;27:3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosner J L, Slonczewski J L. Dual regulation of inaA by the multiple antibiotic resistance (Mar) and superoxide (SoxRS) stress response systems of Escherichia coli. J Bacteriol. 1994;176:6262–6269. doi: 10.1128/jb.176.20.6262-6269.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 48.Seoane A S, Levy S B. Identification of new genes regulated by the marRAB operon in Escherichia coli. J Bacteriol. 1995;177:530–535. doi: 10.1128/jb.177.3.530-535.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith B T, Walker G C. Mutagenesis and more: umuDC and the Escherichia coli SOS response. Genetics. 1998;148:1599–1610. doi: 10.1093/genetics/148.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Storz G, Imlay J A. Oxidative stress. Curr Opin Microbiol. 1999;2:188–194. doi: 10.1016/s1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 51.Sulavick M C, Dazer M, Miller P F. The Salmonella typhimurium mar locus: molecular and genetic analysis and assessment of its requirement for virulence. J Bacteriol. 1997;179:1857–1866. doi: 10.1128/jb.179.6.1857-1866.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tao H, Bausch C, Richmond C, Blattner F R, Conway T. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J Bacteriol. 1999;181:6425–6440. doi: 10.1128/jb.181.20.6425-6440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thieringer H A, Jones P G, Inouye M. Cold shock and adaptation. Bioessays. 1998;20:49–57. doi: 10.1002/(SICI)1521-1878(199801)20:1<49::AID-BIES8>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 54.White D G, Goldman J D, Demple B, Levy S B. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J Bacteriol. 1997;179:6122–6126. doi: 10.1128/jb.179.19.6122-6126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zenno S, Koike H, Kumar A N, Jayarman R, Tanokura M, Saigo K. Biochemical characterization of NfsA, the Escherichia coli major nitroreductase exhibiting a high amino acid sequence homology to Frp, a Vibrio harveyi flavin oxidoreductase. J Bacteriol. 1996;178:4508–4514. doi: 10.1128/jb.178.15.4508-4514.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng M, Doan B, Schneider T D, Storz G. OxyR and SoxRS regulation of fur. J Bacteriol. 1999;181:4639–4643. doi: 10.1128/jb.181.15.4639-4643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]