Abstract
Background
The WD motif (also known as the Trp-Asp or WD40 motif) is found in a multitude of eukaryotic proteins involved in a variety of cellular processes. Where studied, repeated WD motifs act as a site for protein-protein interaction, and proteins containing WD repeats (WDRs) are known to serve as platforms for the assembly of protein complexes or mediators of transient interplay among other proteins. In the model plant Arabidopsis thaliana, members of this superfamily are increasingly being recognized as key regulators of plant-specific developmental events.
Results
We analyzed the predicted complement of WDR proteins from Arabidopsis, and compared this to those from budding yeast, fruit fly and human to illustrate both conservation and divergence in structure and function. This analysis identified 237 potential Arabidopsis proteins containing four or more recognizable copies of the motif. These were classified into 143 distinct families, 49 of which contained more than one Arabidopsis member. Approximately 113 of these families or individual proteins showed clear homology with WDR proteins from the other eukaryotes analyzed. Where conservation was found, it often extended across all of these organisms, suggesting that many of these proteins are linked to basic cellular mechanisms. The functional characterization of conserved WDR proteins in Arabidopsis reveals that these proteins help adapt basic mechanisms for plant-specific processes.
Conclusions
Our results show that most Arabidopsis WDR proteins are strongly conserved across eukaryotes, including those that have been found to play key roles in plant-specific processes, with diversity in function conferred at least in part by divergence in upstream signaling pathways, downstream regulatory targets and /or structure outside of the WDR regions.
Similar content being viewed by others
Background
The so-called WD-repeat (WDR) proteins comprise an astonishingly diverse superfamily of regulatory proteins, representing the breadth of biochemical mechanisms and cellular processes. These proteins have been found to play key roles in such disparate mechanisms as signal transduction, cytoskeletal dynamics, protein trafficking, nuclear export, and RNA processing, and are especially prevalent in chromatin modification and transcriptional mechanisms. WDR proteins are intimately involved in a variety of cellular and organismal processes, including cell division and cytokinesis, apoptosis, light signaling and vision, cell motility, flowering, floral development, and meristem organization, to name a few. Within the cell, WDR proteins have been found to be components of the cytoplasm or nucleoplasm, linked to the cytoskeleton, or associated with membranes through binding to membrane proteins or through membrane-interacting, ancillary domains. Known WDR proteins range in size from small proteins such as the pleiotropic plant developmental regulator VIP3, to massive (>400-kDa) proteins such as the mammalian protein trafficking factor Lyst.
The common and defining feature of these proteins is the WD (also called Trp-Asp or WD-40) motif, a ~40-amino acid stretch typically ending in Trp-Asp, but exhibiting only limited amino acid sequence conservation [1]. When present in a protein, the WD motif is typically found as several (4–10) tandemly repeated units. In the WDR proteins for which structure has been determined, including a mammalian Gβ subunit of heterotrimeric GTPases, repeated WD units form a series of four-stranded, antiparallel beta sheets [[2]; D.K. Wilson, pers. commun.], which fold into a higher-order structure termed a β-propeller. This structure can be visualized as a short, open cylinder where the strands form the walls [2]. At least four repeats are believed to be required to form a β-propeller [3]. In Gβ, which contains seven WDRs, the first and last (i.e., amino- and carboxyl-terminal) WDRs participate in the same propeller blade, potentially reinforcing the structure (for an extensive discussion of WD motifs and WDR structure, the reader is referred to Smith et al., 1999 [2]).
It is now accepted that WDR domains within proteins act as sites for interaction with other proteins. This characteristic of WDRs allows for three general functional roles. First, WDRs within one protein can provide binding sites for two or more other proteins and foster transient interactions among these other proteins. This type of role is best illustrated by Gβ, which has been the most extensively studied of the WDR proteins. The heterotrimeric GTPases in which Gβ s participate functionally associate with a variety of heptahelical membrane receptors (G protein-coupled receptors or GPCRs) to propagate cellular response to a multitude of extracellular signals. Upon receptor activation by an extracellular ligand, Gβ, along with the tightly bound Gγ peptide, dissociates from the Gα subunit, and both Gβγ and Gα then can interact with a variety of effectors. Gβ associates reversibly with at least 14 other proteins, including phospholipases, adenylate cyclases, and ion channels [4]. Another example of this type of role is the yeast histone acetylase subunit Hat2, which is required for efficient interaction of the catalytic subunit Hat1 with the target histone [5]. In both Gβ and Hat2 (and in many other WDR proteins) nearly all of the protein is composed of WDRs.
A second potential role of WDR proteins is as an integral component of protein complexes. This functional mode is probably best illustrated by the snoRNP U3 particle, involved in splicing of the small subunit ribosomal RNAs. Of the 28 characterized subunits of U3, no less than 7 are WDR proteins [6]. Another example is yeast Pfs2, a protein that is found associated with the poly(A) polymerase Pap1 and several multisubunit factors in a large protein complex required for pre-mRNA 3'-end processing and polyadenylation [7]. Within this large complex, Pfs2 interacts directly with specific subunits of two of the processing factors, suggesting that Pfs2 is important for integrity of the larger complex [8]. Many other WDR proteins have been found in relatively stable complexes, including the nuclear pore complex [9], the general transcription factor TFIID [10, 11], and the yeast SET1 histone methyltransferase complex [12].
A third recognized role of the WDR is to act as a modular interaction domain of larger proteins. The presumed role of the WDR in these cases is to bring the protein and associated ancillary domain(s) into proximity of its target(s). Two examples in plants are the light signaling proteins COP1 and SPA1, which juxtapose carboxyl-terminal WDRs with an amino-terminal ring-finger or kinase-like domain, respectively (below). Other common examples of ancillary domains seen in WDR proteins from yeast, animals or plants include the F-box, SET domain, and bromodomain (not shown).
Many WDR-containing proteins of unknown function have been designated as 'Gβ-like', even in the absence of any sequence-based or functional relationship with Gβ. These misleading annotations suggest that a phylogenetic analysis of this superfamily is needed. Here, we evaluated the extent of the predicted WDR protein superfamily in Arabidopsis, and the sequence-based and functional relationships between these proteins and known or hypothetical proteins from budding yeast (Saccharomyces cerevisiae), fruit fly (Drosophila melanogaster) and humans (Homo sapiens). Our results suggest that most Arabidopsis WDR proteins are strongly conserved across eukaryotes, including those that have been found to play key roles in plant-specific processes.
Results and Discussion
The Arabidopsis WDR protein family
This analysis identified 269 Arabidopsis proteins containing at least one copy of the WD motif. The vast majority of these (237) contained four or more recognizable copies of the motif. We classified these 237 proteins into 143 distinct families, 49 of which contained more than one Arabidopsis member. Approximately 113 of these families or individual proteins showed clear homology with WDR proteins from yeast, fly, and/or human (Table 1 [see Additional file 1]). Where conservation was found, it often extended across all of these organisms, suggesting that many of these proteins are components of basic cellular mechanisms.
The Arabidopsis proteome is apparently lacking counterparts of several WDR proteins that have been extensively studied in other eukaryotes and might have been expected to be conserved. For example, we found no protein related to the cell death initiator Dark (fly)/Apaf-1 (human). This protein is the central scaffolding of the apoptosome, a protein complex that activates specific cellular proteases in response to death signals [13]. Many parallels exist between animal and plant apoptotic pathways, and many other components of the animal pathways have been strongly conserved in plants [14]. Arabidopsis also appears to lack a protein closely related to the intermediate chain of the microtubule motor protein dynein, involved in transporting cellular cargo along microtubules. In mammalian cytoplasmic dynein, the intermediate chain plays a crucial scaffolding role, mediating interactions among the heavy chain and other dynein subunits [15]. Arabidopsis was previously hypothesized to lack the dynein heavy chain, based on nearly-completed genomic sequence [16]. It was suggested that if Arabidopsis did lack functional dynein, this could be compensated for by the relative variety of carboxyl-terminal motor domain kinesins in this species [17].
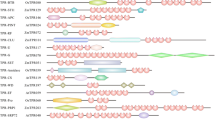
In contrast to these apparently lacking proteins, we found several proteins that were not expected. One example is a protein very closely related to Notchless (Nle), a fly protein that binds to the intracellular domain of the developmental signal receptor Notch and modulates its activity [18] (Fig. 1). Arabidopsis lacks a recognizable Notch, and other components of the associated signaling pathways appear to be absent [19]. In addition, we found two proteins strongly related to the transcription-coupled, DNA-repair (TCR) proteins Rad28 (yeast) and Csa (human), even though plants are not known to undergo TCR (Fig. 1).
Also notable were several cases where Arabidopsis apparently did not participate in the expansion of gene families seen in the other eukaryotes. One example is a conserved component of the transcription factor TFIID, represented by the human TAFII-100 protein. This protein interacts directly with at least three other components of TFIID, and thus probably serves as a scaffolding for construction of the complex [10, 11]. Multiple paralogs of this protein exist in fly, worm, and human (Table 1 [see Additional file 1] and not shown); in flies a form designated Cannonball (Can) appears to operate outside of basal transcription as a key regulator of spermatogenesis [20]. One possibility is that the paralogs within each species act as a interchangeable components of the general transcription machinery to mediate expression of developmentally regulated target genes [21]. The presence of only a single form of this protein in the Arabidopsis proteome suggests that this potential means of expanding the transcriptional repertoire has not evolved in plants. Another example of an evolutionarily stagnant family is Gβ, which exists as only a single form in Arabidopsis (Table 1 [see Additional file 1]; below). In mammals, each of the heterotrimeric G protein subunits, as well as the GPCRs, are encoded by multigene families, and combinatorial interaction among the proteins are believed to modulate much of the diversity of response to extracellular signals. The restriction of this gene family in Arabidopsis would suggest that, if a 'typical' heterotrimeric G protein does exist, it would likely lack the functional complexity seen in mammals. This scenario would be similar to that in yeast, where only a single heterotrimeric G protein, incorporating the Gβ protein Ste4, has a specialized role in transducing mating type signals from heptahelical mating-factor receptors [22].
In contrast, some other Arabidopsis WDR proteins show relatively expanded gene families compared with the other eukaryotes studied. One of the largest Arabidopsis WDR families, consisting of nine members, is orthologous to the conserved Cdc20/Fizzy class of cell cycle regulators including yeast Cdc20 and Cdh1 (Table 1 [see Additional file 1]). These proteins activate the anaphase promoting complex (APC) ubiquitin ligase, which targets downstream cell cycle regulators for proteolysis [23], potentially by mediating interaction of the APC complex with target proteins. Mutation in Cdc20 or Cdh1 affect distinct aspects of the cell cycle, and Cdc20 and Cdh1 coimmunoprecipitate with distinct APC target proteins [24, 25], indicating that these proteins have non-overlapping functions. One explanation for the expansion of this family in Arabidopsis is that the several distinct proteins each specify distinct targets for the APC. Another example of an expanded gene family is the MSI1/RbAp48 group of chromatin-related proteins, which includes five members in Arabidopsis (below), but is represented by only a single form in flies (Table 1 [see Additional file 1]).
Several examples were seen where Arabidopsis WDR proteins have used elements from the inceptive 'molecular toolbox' in original ways. One example is the pleiotropic developmental regulator LEUNIG (LUG) [26]. LUG contains seven, carboxyl-terminal WD motif repeats, internal polyglutamine tracts, and an extended motif termed the single-stranded DNA-binding-protein (SSDP) motif ([27], Fig. 1 and not shown). The SSDP motif was defined in a small family of animal proteins including chicken SSDP, which binds to a single-stranded, polypyrimidine region of the α2(I) collagen promoter [28]. SSDP-like proteins function in transcriptional complexes with LIM homeodomain proteins and LIM-domain-binding proteins (Ldbs) to regulate specific embryonic developmental processes [29]. The arrangement of the SSDP motif with carboxyl-terminal WD repeats appears to be unique to LUG and its orthologs from other plants (not shown). However, the juxtapositioning of polyglutamine tracts with carboxyl-terminal WD repeats, while diverging from the domain structure seen in the SSDPs, resembles that seen in the yeast transcriptional corepressor Tup1 and a related corepressor from fly, Groucho [27]. This, in conjunction with the observation that loss of LUG activity leads to ectopic expression of a floral regulatory gene [30] has led to the speculation that LUG acts as a Tup1/Groucho-like transcriptional corepressor [27]. Intriguingly, it was recently shown that LUG functions in floral development together with SEUSS (SEU), a protein related to the mammalian LIM-domain-binding protein, Ldb1 [26], suggesting the existence of a LUG-SEU transcriptional complex analogous to that involving LIM proteins. Collectively, this information suggests that LUG participates in an evolutionarily distinct mechanism of gene regulation incorporating elements of both Tup1/Groucho and Ldbs.
Linking conserved mechanisms with plant-specific processes: Functional specificity through divergence in regulatory targets
With the exception of LUG, the Arabidopsis WDR proteins that have been functionally characterized are strongly conserved within the WDR regions among yeast, fly and/or human (Table 1 [see Additional file 1]). Most of these proteins have been identified as components of basic cellular machinery in these other eukaryotes, yet have been found to regulate plant-specific processes (Table 2). An interesting question for further consideration is how these proteins have become adapted to their plant roles.
In several cases, the homologous WDR proteins are highly conserved throughout the length of the proteins, and appear to operate in highly analogous mechanisms, with specificity in function conferred by changes in upstream signaling pathways and/or downstream effectors. One case is AGB1, the only clear Arabidopsis ortholog of Gβ [31] (Fig. 1). Loss of AGB1 function leads to developmental pleiotropy including shortened fruits [32] and changes in patterns of cell division in the hypocotyl and root [33]. These phenotypes are associated with the derepression of genes that are normally turned on by auxin, suggesting a role for AGB1 as a negative regulator of auxin signaling [33]. There appears to be one Gα-like protein (GPA1) and two Gγ-like proteins (AGG1 and AGG2) in the Arabidopsis proteome [31], and molecular modeling and yeast two-hybrid studies of potential interactions among AGB1, GPA1 and AGG1 are not inconsistent with the possibility that these could form a heterotrimeric protein [33, 34]. In addition, both AGG1 and AGG2 contain domains expected to recruit AGB1 to membranes [34], and GPA1 has been demonstrated to bind GTP(γ)S [35]. These findings lead to the prediction that AGB1 participates in a prototypical heterotrimeric G protein. However, the Arabidopsis proteome does not contain obvious heptahelical receptors with which a heterotrimeric G protein might interact [31]. One possibility is that the AGB1-containing G protein might be unlinked from a receptor. In animals, several receptor-independent activators of heterotrimeric G proteins are known, including the Ras-related protein Ags1 [36], and AGB1 might function in concert with any of the many Ras-related proteins in Arabidopsis. Alternatively, the Arabidopsis G protein may have evolved an interaction with a distinct type of receptor. Intriguingly, genetic experiments suggested that AGB1 may act in a common mechanism of fruit development with ERECTA (ER), a leucine-rich receptor kinase-like (LRR-RLK) protein, and based on this it was suggested that the two proteins could be functionally associated [32]. Another link between a potential plant heterotrimeric G protein and LRR-RLKs was demonstrated by the observation that gpa1 mutants share the same level of insensitivity to GAs during seed germination as mutants for the LRR-RLK protein BRI1 [33].
An example of downstream functional divergence is shown by CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1). In dark-grown plants, this WDR protein acts as a key repressor of photomorphogenesis; in light, COP1 becomes inactivated, at least partly through gradual nuclear exclusion [37]. The COP1 protein contains a carboxyl-terminal WDR domain, a RING Zn-finger domain near the amino terminus, and a predicted coiled-coil domain (Fig. 1). The WD domains mediate its interaction with, and negative regulation of, HY5 and HYH, closely related bZIP transcription factors [38, 39]. This negative regulation potentially results from the ability of COP1 to target these proteins for ubiquitin-dependent proteolysis, because COP1 has been shown to function in this manner to downregulate a distinct regulatory protein in vivo [40]. Humans and mice have a recognizable COP1 homolog, although yeast or flies do not ([41]; Table 1 [see Additional file 1], Fig. 1). Similar to the relationship between Arabidopsis COP1 and HY5 or HYH, human Cop1 binds to and negatively regulates Jun (and other related bZIP transcription factors) [42]. The mammalian proteins conserve the important structural features of Arabidopsis COP1, including the RING finger, coiled-coil domain, and WDRs. When expressed in plant cells, mammalian Cop1 displays the light-specific nucleo-cytoplasmic partitioning characteristic of Arabidopsis COP1, but fails to rescue the cop1 mutant phenotype [41]. Thus it appears that certain 'upstream' mechanisms regulating COP1 activity are conserved, but that downstream effectors of COP1 are not. This is consistent with the observation that components of the COP9 signalosome, which is required for COP1 localization, are conserved in mammals [43].
FASCIATA2 (FAS2) and the Arabidopsis family of Msi1-related proteins provide more examples of highly conserved WDR proteins involved in plant-specific processes through specificity in downstream targets. The FAS2 protein is the single Arabidopsis homolog of one of the three subunits (Cac1, Cac2 and another WDR protein, Msi1) of chromatin assembly factor 1 (CAF-1) [44], which participates in chromatin assembly following DNA replication or repair [45]. FAS2 physically interacts with the only Arabidopsis ortholog of Cac1 and with AtMSI1, one of five paralogous Msi1-like proteins, and several lines of evidence suggest that Arabidopsis FAS1, FAS2, and at least AtMSI1 comprise a plant CAF-1 [46]. Mutations in FAS2 lead to meristem disorganization at the shoot and root apex, associated with the aberrant expression of two previously identified genes that organize the meristem, WUSCHEL in the shoot and SCARECROW in the root [44].
The conserved WDR protein subfamily represented by Msi1 and its Arabidopsis orthologs also includes human RbAp48 and fly p55 (Table 1 [see Additional file 1], Fig. 1). These latter proteins are CAF-1 subunits [47], but may serve supplemental functions. For example, both RbAp48 and p55 are found in several chromatin-remodeling complexes [48], and RbAp48 has been found tightly bound to the catalytic core subunit of the human histone deacetylase HDAC1 [49]. One possibility is that these Msi1-like proteins link chromatin assembly with the maintenance of gene silencing by recruiting activities that instigate the formation of localized heterochromatin [46]. It is not unlikely that this is the case in Arabidopsis as well. Loss of AtMSI1 function leads to various developmental defects, at least some of these associated with the ectopic expression of the floral homeotic genes AGAMOUS and APETALA2 [50]. Loss of the paralogous AtMSI4 (also called FVE) leads to late flowering, associated with loss of transcriptional repression of the MADS-box flowering inhibitor gene FLC ([51]; J. Martinez-Zapater, pers. comm.). The observation that loss of AtMSI1 or AtMSI4 confer phenotypes that are distinct from those seen in fas2 mutants suggests that these proteins also probably have functions that are not limited to a role in CAF-1. Thus it seems likely that the FAS2 and Msi1 families of proteins are involved in equivalent regulatory mechanisms, with diversity in function due to the identity of the regulated gene(s).
A similar example is FERTILIZATION-INDEPENDENT ENDOSPERM (FIE). FIE is required for repression of endosperm development in the absence of fertilization and is also essential for proper endosperm and embryo development when fertilization occurs [52]. The FIE protein has strong similarity to fly Esc and mammalian Eed, identified originally as polycomb-group repressors of homeotic gene expression [53] (Table 1 [see Additional file 1], Fig. 1). Esc and Eed interact directly with homologous SET-domain E(z) proteins in heterogeneous protein complexes also including the zinc-finger protein Su(z)12 or the histone deacetylase Rpd3 [54, 55]. Esc-E(z) polycomb complexes have been shown to repress gene activity, in part through their histone methyltransferase and histone deacetylase activities [56, 57]. In a highly analogous manner, FIE binds directly to the SET-domain protein MEA, and genetic experiments suggest that a FIE-MEA pair interacts strongly with the Su(z)12-like protein FIS2 [58, 59].
Mutations in FIE also cause misexpression of floral induction genes in the embryo and seedling, and consequent ectopic production of flower-like structures [60], suggesting that FIE has an additional and unrelated role in repression of the flowering program during the vegetative stage. Interestingly, this embryonic flowering resembles the phenotype of a mutant for a Su(z)12-like protein designated EMF2 [61], and potentially FIE and EMF2 are components of a distinct Esc-E(z)-like complex. Although FIE has no apparent paralogs within the Arabidopsis proteome, there are multiple Su(z)12-related and E(z)-related proteins (not shown). Thus there is the potential for FIE to be involved in numerous aspects of growth and development through combinatorial interactions with several distinct complexes. Similarly, Eed, which also appears to be encoded by a single gene, interacts with multiple E(z)-like proteins [62].
Recently it was shown that FIE is required for transcriptional repression of PHERES1, a member the homeotic-function MADS-box gene class [63]. It is notable that FIE and its homologs participate in an evolutionarily conserved mechanism to repress homeotic genes, even as the structure of homeotic genes have greatly diverged between plants (i.e., MADS-box) and animals (i.e., homeobox).
Functional specificity through structural divergence outside of the WDR region
In almost all cases, proteins that we classified as homologous between species showed the highest degree of sequence similarity within the WDR regions, in several instances exhibiting little or no related sequence outside of the WDR region. In at least some of these cases, function of the homologous proteins may nevertheless be conserved. For example, the transcriptional corepressor Tup1 is highly variant outside of the WDRs among different strains of yeast, but is functionally interchangeable among them [64]. In other cases, sequences outside of the WDR may confer a new functional specificity to the protein even as conserved protein interactions are maintained through the WDRs, potentially adapting basic cellular mechanisms to organism-specific processes. One potential example in Arabidopsis is PRL1, a nuclear protein implicated in the response to several stimuli including glucose, sucrose, cold, and multiple phytohormones [65]. PRL1 interacts through its non-conserved amino-terminal domain with yeast Snf1, a conserved protein kinase that plays an important transcriptional role in sugar sensing and response, and with Arabidopsis Snf1-related protein kinases (SnRKs) [66]. The carboxyl-terminal WDR domains of PRL1 exhibit very strong conservation with proteins from yeast (Prp46), fly, and human (Plrg1) (Table 1 [see Additional file 1], Fig. 1 and not shown). These non-plant proteins have not been related to Snf1-associated gene regulation, possibly as a result of divergence with PRL1 in the amino-terminal regions of the proteins. Instead, the yeast and human PRL1 homologs were identified as non-snRNP components of the spliceosome [67, 68], a large pre-mRNA splicing assembly consisting of RNAs, small nuclear ribonucleoproteins (snRNPs) and numerous other proteins. The WDR region of Prp46 and Plrg1 mediate interaction with other spliceosomal subunits that are structurally and functionally conserved in Arabidopsis ([68] and not shown), and therefore it would not be surprising if PRL1 is also found to be associated with the spliceosome. A growing body of evidence points to the physical coordination of transcriptional regulation and downstream events including pre-mRNA processing [69], and thus PRL1 could link conserved spliceosomal components with plant-specific transcriptional regulators.
The flowering regulator FY provides another potential example of this type of recruitment of basic cellular processes for plant-specific functions by WDR proteins. In a developmental role closely related to that of AtMSI1/FVE (above), FY promotes flowering by repressing the flowering inhibitor FLC. FY physically interacts with the plant-specific, RNA-binding-motif protein FCA; this interaction is required for a negative autoregulatory mechanism controlling FCA protein accumulation, where full-length FCA protein promotes premature cleavage and polyadenylation of its own transcript [70]. This autoregulation is evidently disrupted in the shoot apex of plants undergoing the transition to flowering, leading to increased accumulation of active FCA [71]. In accordance with a role in RNA processing, the WDR region of FY exhibits strong homology with that of the aforementioned yeast RNA splicing factor Pfs2 and homologs from higher eukaryotes (Table 1 [see Additional file 1]; Fig. 1). Outside of the WDR region, these proteins diverge, with FY containing an extensive, proline-rich carboxyl-terminal region through which its interaction with FCA is mediated [71]. It was suggested that this non-conserved portion of Pfs2/FY proteins in other eukaryotes might similarly link core RNA-processing machinery with regulatory proteins [71].
In our classification of Arabidopsis WDRs into gene families, we also noted several instances where proteins designated as paralogs diverged significantly outside of the WDR region. These proteins could link a common cellular mechanism with alternative or additional upstream regulators or downstream effectors, potentially in a temporal or spatially restricted manner. One notable case is SPA1, a nuclear-localized, light-dependent repressor of photomorphogenesis proposed to link the phytochrome A-specific branch of light signaling to a COP1-associated mechanism (Hoecker and Quail, 2001). Within its WDR domain, SPA1 is very closely related to COP1, but outside of this region SPA1 substitutes a SER/THR/TYR-protein-kinase-like domain for the RING-finger domain of COP1. SPA1 and COP1 physically interact through coiled-coil regions of both proteins [72]. How the functional relationship between SPA1 and COP1 is related to this molecular linkage is not known, but genetic and biochemical studies suggest that SPA1 may somehow stimulate the ability of residual nuclear COP1 to target proteins for destruction after the onset of illumination [40].
Conclusions
To date, the function of only about 10 members of the Arabidopsis WDR protein superfamily have been described, in spite of the fact that many of these proteins are expected to participate in key cellular and organismal processes. This study provides a useful framework for a functional analysis of unknown Arabidopsis WDR proteins. Because they have the potential to interact with several proteins simultaneously, WDR proteins are attractive subjects for analysis through protein linkage mapping as these techniques become developed for use in plants. This same feature allows WDR proteins to integrate molecular mechanisms and pathways, and so reversed-genetic analyses targeting these proteins should also be highly informative. Studies of these plant proteins may also provide unique insights into the mechanism of pathogenicity of human diseases. Studies in plants have obvious advantages over mammalian disease models, foremost among them being much more powerful and tractable approaches for genetic analyses. To date, six human diseases have been linked to defects in WDR genes, and five of the respective proteins are strongly conserved in Arabidopsis (Table 3). The only of these to be studied to date, the peroxisomal import receptor-associated protein PEX7, appears to be very closely related in function to its yeast and human counterpart [73] (Fig. 1), and this is a preliminary indication that such plants studies will be highly relevant.
Methods
Predicted Arabidopsis proteins containing at least one WD motif were identified using motif-search software maintained by The Arabidopsis Information Resource [74] and current InterPro signatures (Prosite PS50294, PS00678, or PS50082; Pfam PF00400, PRINTS PR00320, or SMART SM0320 [75]). The database used for this analysis, ATH1. pep, was provided by The Institute for Genomic Research (TIGR) and was released Apr 17, 2003. Proteins containing at least four WD motifs were assigned into families using Blastclust (unpublished, available from the National Center for Biotechnology Information [76]), a single linkage clustering tool that uses the BLAST algorithm to determine distance. Blastclust uses these default values for the BLAST: matrix BLOSUM62, gap opening cost 11, gap extension cost 1, no low-complexity filtering, and an Expectation (E)-value cutoff of 1E-6. It is configurable, and accepts several different parameters which can be set to alter the distance calculations and the clustering threshold. Because there was no a priori evidence as to which parameters would yield biologically relevant clusters, we ran the Blastclust software over several iterations, varying two parameters. The L parameter (range: 0.3–0.8) represents the amount of overlap coverage between query and subject, expressed as a ratio. The S parameter (range: 0.7–1.5) is a measure of the information content density of the alignment. As L and S increase, so does the stringency of the match. The analysis presented here used L = 0.3 and S = 0.7. Other protein motifs in WDR-containing proteins were identified using the InterProScan. pl program (Release 3.1) [77] and the Interpro 5.3 database as maintained by the European Bioinformatics Institute, in combination with Pfam Release 7.8 [78] and the PRODOM database (2002.1).
To identify WD motif-containing proteins in S. cerevisiae, D. melanogaster, and H. sapiens, we analyzed previously compiled proteome datasets available from the Saccharomyces Genome Database [79], FlyBase [80], and Ensembl (v. 13.31.1, released Mar 31, 2003)[81] as described above. The sequences utilized can be obtained through HTML links in Table 1 (see Additional file 1]. These sequences were used to query the ATH1. pep dataset using Washington University BLAST (WUBLAST) version 2.0 as maintained by TAIR. An Arabidopsis protein or paralogous group was designated as orthologous if it met the following three criteria: 1) it was the most closely related protein(s) 2) The E value for the match was less than 10E-11, and 3) the protein or all members of the paralogous group were more closely related than the next most significant match by a factor equal to or greater than 10E15.
References
Neer EJ, Schmidt CJ, Nambudripad R, Smith TF: The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994, 371: 297-300. 10.1038/371297a0.
Smith TF, Gaitatzes C, Saxena K, Neer EJ: The WD repeat: a common architecture for diverse functions. Trends Biochem Sci. 1999, 24: 181-185. 10.1016/S0968-0004(99)01384-5.
Chothia C, Hubbard T, Brenner S, Barns H, Murzin A: Protein folds in the all-beta and all-alpha classes. Annu Rev Biophys Biomol Struct. 1997, 26: 597-627. 10.1146/annurev.biophys.26.1.597.
van der Voorn L, Ploegh HL: The WD-40 repeat. FEBS Lett. 1992, 307: 131-134. 10.1016/0014-5793(92)80751-2.
Ruiz-Garcia AB, Sendra R, Galiana M, Pamblanco M, Perez-Ortin JE, Tordera V: HAT1 and HAT2 proteins are components of a yeast nuclear histone acetyltransferase enzyme specific for free histone H4. J Biol Chem. 1998, 273: 12599-12605. 10.1074/jbc.273.20.12599.
Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, Beyer AL, Hunt DF, Baserga SJ: A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002, 417: 967-970. 10.1038/nature00769.
Zhao J, Hyman L, Moore C: Formation of mRNA 3'-ends in eukaryotes: mechanism, regulation and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999, 63: 405-445.
Ohnacker M, Barabino SM, Preker PJ, Keller W: The WD-repeat protein Pfs2p bridges two essential factors within the yeast pre-mRNA 3' end processing complex. EMBO J. 2000, 19: 37-47. 10.1093/emboj/19.1.37.
Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ: Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002, 158: 915-927. 10.1083/jcb.200206106.
Dubrovskaya V, Lavigne AC, Davidson I, Acker J, Staub A, Tora L: Distinct domains of hTAFII100 are required for functional interaction with transcription factor TFIIF beta (RAP30) and incorporation into the TFIID complex. EMBO J. 1996, 15: 3702-3712.
Tao Y, Guermah M, Martinez E, Oelgeschlager T, Hasegawa S, Takada R, Yamamoto T, Horikoshi M, Roeder RG: Specific interactions and potential functions of human TAFII100. J Biol Chem. 1997, 272: 6714-6721. 10.1074/jbc.272.10.6714.
Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF: The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001, 20: 7137-7148. 10.1093/emboj/20.24.7137.
Cain K, Bratton SB, Cohen GM: The Apaf-1 apoptosome: a large caspase-activating complex. Biochimie. 2002, 84: 203-214. 10.1016/S0300-9084(02)01376-7.
Hoeberichts FA, Woltering EJ: Multiple mediators of plant programmed cell death: interplay of conserved cell death mechanisms and plant-specific regulators. Bioessays. 2003, 25: 47-57. 10.1002/bies.10175.
Susalka SJ, Nikulina K, Salata MW, Vaughan PS, King SM, Vaughan KT, Pfister KK: The roadblock light chain binds a novel region of the cytoplasmic dynein intermediate chain. J Biol Chem. 2002, 277: 32939-32946. 10.1074/jbc.M205510200.
Reddy AS, Day IS: Analysis of the myosins encoded in the recently completed Arabidopsis thaliana genome sequence. Genome Biol. 2001, 2: research0024.1-0024.17. 10.1186/gb-2001-2-7-research0024.
Vale RD: The molecular motor toolbox for intracellular transport. Cell. 2003, 112: 467-480.
Royet J, Bouwmeester T, Cohen SM: Notchless encodes a novel WD40-repeat-containing protein that modulates Notch signaling activity. EMBO J. 1998, 17: 7351-7360. 10.1093/emboj/17.24.7351.
Wigge PA, Weigel D: Arabidopsis genome: life without notch. Curr Biol. 2001, 11: R112-4. 10.1016/S0960-9822(01)00043-4.
Hiller MA, Lin TY, Wood C, Fuller MT: Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 2001, 15: 1021-1030. 10.1101/gad.869101.
Verrijzer CP: Transcription factor IID – not so basal after all. Science. 2001, 293: 2010-2011. 10.1126/science.1064980.
Dohlman HG, Thorner JW: Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu Rev Biochem. 2001, 70: 703-754. 10.1146/annurev.biochem.70.1.703.
Visintin R, Prinz S, Amon A: CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997, 278: 460-463. 10.1126/science.278.5337.460.
Hilioti Z, Chung YS, Mochizuki Y, Hardy CF, Cohen-Fix O: The anaphase inhibitor Pds1 binds to the APC/C-associated protein Cdc20 in a destruction box-dependent manner. Curr Biol. 2001, 11: 1347-1352. 10.1016/S0960-9822(01)00399-2.
Schwab M, Neutzner M, Mocker D, Seufert W: Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J. 2001, 20: 5165-5175. 10.1093/emboj/20.18.5165.
Franks RG, Wang C, Levin JZ, Liu Z: SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development. 2002, 129: 253-263.
Conner J, Liu Z: LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development. Proc Natl Acad Sci U S A. 2000, 97: 12902-12907. 10.1073/pnas.230352397.
Bayarsaihan D, Soto RJ, Lukens LN: Cloning and characterization of a novel sequence-specific single-stranded-DNA-binding protein. Biochem J. 1998, 331: 447-452.
Chen L, Segal D, Hukriede NA, Podtelejnikov AV, Bayarsaihan D, Kennison JA, Ogryzko VV, Dawid IB, Westphal H: Ssdp proteins interact with the LIM-domain-binding protein Ldb1 to regulate development. Proc Natl Acad Sci U S A. 2002, 99: 14320-14325. 10.1073/pnas.212532399.
Liu Z, Meyerowitz EM: LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development. 1995, 121: 975-991.
Jones AM: G-protein-coupled signaling in Arabidopsis. Curr Opin Plant Biol. 2002, 5: 402-407. 10.1016/S1369-5266(02)00288-1.
Lease KA, Wen J, Li J, Doke JT, Liscum E, Walker JC: A mutant Arabidopsis heterotrimeric G-protein beta subunit affects leaf, flower, and fruit development. Plant Cell. 2001, 13: 2631-2641. 10.1105/tpc.13.12.2631.
Ullah H, Chen JG, Temple B, Boyes DC, Alonso JM, Davis KR, Ecker JR, Jones AM: The beta-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell. 2003, 15: 393-409. 10.1105/tpc.006148.
Mason MG, Botella JR: Isolation of a novel G-protein gamma-subunit from Arabidopsis thaliana and its interaction with Gbeta. Biochim Biophys Acta. 2001, 1520: 147-153. 10.1016/S0167-4781(01)00262-7.
Wise A, Thomas PG, Carr TH, Murphy GA, Millner PA: Expression of the Arabidopsis G-protein GP alpha1: purification and characterisation of the recombinant protein. Plant Mol Biol. 1997, 33: 723-728. 10.1023/A:1005732423622.
Cismowski MJ, Ma C, Ribas C, Xie X, Spruyt M, Lizano JS, Lanier SM, Duzic E: Activation of heterotrimeric G-protein signaling by a ras-related protein. Implications for signal integration. J Biol Chem. 2000, 275: 23421-23424. 10.1074/jbc.C000322200.
von Arnim AG, Osterlund MT, Kwok SF, Deng XW: Genetic and developmental control of nuclear accumulation of COP1, a repressor of photomorphogenesis in Arabidopsis. Plant Physiol. 1997, 114: 779-788. 10.1104/pp.114.3.779.
Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW: Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell. 1998, 1: 213-222.
Holm M, Ma LG, Qu LJ, Deng XW: Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Devel. 2002, 16: 1247-1259. 10.1101/gad.969702.
Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH: LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature. 2003, 424: 995-999. 10.1038/nature01696.
Wang H, Kang D, Deng XW, Wei N: Evidence for functional conservation of a mammalian homologue of the light-responsive plant protein COP1. Curr Biol. 1999, 9: 711-714. 10.1016/S0960-9822(99)80314-5.
Bianchi E, Denti S, Catena R, Rossetti G, Polo S, Gasparian S, Putignano S, Rogge L, Pardi R: Characterization of human constitutive photomorphogenesis protein 1, a RING finger ubiquitin ligase that interacts with Jun transcription factors and modulates their transcriptional activity. J Biol Chem. 2003, 278: 19682-19690. 10.1074/jbc.M212681200.
Wei N, Tsuge T, Serino G, Dohmae N, Takio K, Matsui M, Deng XW: The COP9 complex is conserved between plants and mammals and is related to the 26S proteasome regulatory complex. Curr Biol. 1998, 8: 919-922.
Kaya H, Shibahara KI, Taoka KI, Iwabuchi M, Stillman B, Araki T: FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell. 2001, 104: 131-142.
Mello JA, Almouzni G: The ins and outs of nucleosome assembly. Curr Opin Genet Dev. 2001, 11: 136-141. 10.1016/S0959-437X(00)00170-2.
van Nocker S: CAF-1 and MSI1-related proteins: linking nucleosome assembly with epigenetics. Trends Plant Sci.
Verreault A: De novo nucleosome assembly: new pieces in an old puzzle. Genes Dev. 2000, 14: 1430-1438.
Wolffe AP, Urnov FD, Guschin D: Co-repressor complexes and remodeling chromatin for repression. Biochem Soc Trans. 2000, 28: 379-386.
Nicolas E, Morales V, Magnaghi-Jaulin L, Harel-Bellan A, Richard-Foy H, Trouche D: RbAp48 belongs to the histone deacetylase complex that associates with the retinoblastoma protein. J Biol Chem. 2000, 275: 9797-9804. 10.1074/jbc.275.13.9797.
Hennig L, Taranto P, Walser M, Schönrock N, Gruissem W: Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development. 2003, 130: 2555-2565. 10.1242/dev.00470.
Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES: The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell. 1999, 11: 445-458. 10.1105/tpc.11.3.445.
Sorensen MB, Chaudhury AM, Robert H, Bancharel E, Berger F: Polycomb group genes control pattern formation in plant seed. Curr Biol. 2001, 11: 277-281. 10.1016/S0960-9822(01)00072-0.
Jacobs JJ, van Lohuizen M: Polycomb repression: from cellular memory to cellular proliferation and cancer. Biochim Biophys Acta. 2002, 1602: 151-161. 10.1016/S0304-419X(02)00052-5.
Tie F, Furuyama T, Prasad-Sinha J, Jane E, Harte PJ: The Drosophila Polycomb Group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development. 2001, 128: 275-286.
Furuyama T, Tie F, Harte PJ: Polycomb group proteins ESC and E(Z) are present in multiple distinct complexes that undergo dynamic changes during development. Genesis. 2003, 35: 114-124. 10.1002/gene.10173.
Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA: Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002, 111: 197-208.
Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V: Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002, 111: 185-196.
Yadegari R, Kinoshita T, Lotan O, Cohen G, Katz A, Choi Y, Katz A, Nakashima K, Harada JJ, Goldberg RB, Fischer RL, Ohad N: Mutations in the FIE and MEA genes that encode interacting polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell. 2000, 12: 2367-2382. 10.1105/tpc.12.12.2367.
Spillane C, MacDougall C, Stock C, Kohler C, Vielle-Calzada JP, Nunes SM, Grossniklaus U, Goodrich J: Interaction of the Arabidopsis polycomb group proteins FIE and MEA mediates their common phenotypes. Curr Biol. 2000, 10: 1535-1538. 10.1016/S0960-9822(00)00839-3.
Kinoshita T, Harada JJ, Goldberg RB, Fischer RL: Polycomb repression of flowering during early plant development. Proc Natl Acad Sci U S A. 2001, 98: 14156-14161. 10.1073/pnas.241507798.
Yoshida N, Yanai Y, Chen L, Kato Y, Hiratsuka J, Miwa T, Sung ZR, Takahashi S: EMBRYONIC FLOWER 2, a novel polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell. 2001, 13: 2471-2481. 10.1105/tpc.13.11.2471.
van Lohuizen M, Tijms M, Voncken JW, Schumacher A, Magnuson T, Wientjens E: Interaction of mouse polycomb-group (Pc-G) proteins Enx1 and Enx2 with Eed: indication for separate Pc-G complexes. Mol Cell Biol. 1998, 18: 3572-3579.
Kohler C, Hennig L, Spillane C, Pien S, Gruissem W, Grossniklaus U: The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 2003, 17: 1540-1553. 10.1101/gad.257403.
Mukai Y, Matsuo E, Roth SY, Harashima S: Conservation of histone binding and transcriptional repressor functions in a Schizosaccharomyces pombe Tup1p homolog. Mol Cell Biol. 1999, 19: 8461-8468.
Nemeth K, Salchert K, Putnoky P, Bhalerao R, Koncz-Kalman Z, Stankovic-Stangeland B, Bako L, Mathur J, Okresz L, Stabel S, Geigenberger P, Stitt M, Redei GP, Schell J, Koncz C: Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev. 1998, 12: 3059-3073.
Bhalerao RP, Salchert K, Bako L, Okresz L, Szabados L, Muranaka T, Machida Y, Schell J, Koncz C: Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc Natl Acad Sci U S A. 1999, 96: 5322-5327. 10.1073/pnas.96.9.5322.
Ohi MD, Gould KL: Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA. 2002, 8: 798-815. 10.1017/S1355838202025050.
Ajuh P, Sleeman J, Chusainow J, Lamond AI: A direct interaction between the carboxyl-terminal region of CDC5L and the WD40 domain of PLRG1 is essential for pre-mRNA splicing. J Biol Chem. 2001, 276: 42370-42381. 10.1074/jbc.M105453200.
Hirose Y, Manley JL: RNA polymerase II and the integration of nuclear events. Genes Dev. 2000, 14: 1415-1429.
Quesada V, Macknight R, Dean C, Simpson GG: Autoregulation of the site of 3' end formation in FCA pre-mRNA prevents precocious flowering. EMBO J. 2003, 22: 3142-3152. 10.1093/emboj/cdg305.
Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C: FY is an RNA 3'end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell. 2003, 113: 777-787.
Hoecker U, Quail PH: The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J Biol Chem. 2001, 276: 38173-38178.
Nito K, Hayashi M, Nishimura M: Direct interaction and determination of binding domains among peroxisomal import factors in Arabidopsis thaliana. Plant Cell Physiol. 2002, 43: 355-366. 10.1093/pcp/pcf057.
The Arabidopsis Information Resource. [http://www.arabidopsis.org/]
Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Barrell D, Bateman A, Binns D, Biswas M, Bradley P, Bork P, Bucher P, Copley RR, Courcelle E, Das U, Durbin R, Falquet L, Fleischmann W, Griffiths-Jones S, Haft D, Harte N, Hulo N, Kahn D, Kanapin A, Krestyaninova M, Lopez R, Letunic I, Lonsdale D, Silventoinen V, Orchard SE, Pagni M, Peyruc D, Ponting CP, Selengut JD, Servant F, Sigrist CJ, Vaughan R, Zdobnov EM: The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res. 2003, 31: 315-318. 10.1093/nar/gkg046.
National Center for Biotechnology Information. [http://www.ncbi.nlm.nih.gov/]
Zdobnov EM, Apweiler R: InterProScan-an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001, 17: 847-848. 10.1093/bioinformatics/17.9.847.
Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL: The Pfam protein families database. Nucleic Acids Res. 2002, 30: 276-280. 10.1093/nar/30.1.276.
Saccharomyces Genome Database. [http://www.yeastgenome.org/]
FlyBase. [http://flybase.bio.indiana.edu/]
Ensembl Human Genome Browser. [http://www.ensembl.org/]
Wall MA, Coleman DE, Lee E, Iniguez-Lluhi JA, Posner BA, Gilman AG, Sprang SR: The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell. 1995, 83: 1047-1058.
Protein Data Bank. [http://www.rcsb.org/pdb/]
Hogue CWV: Cn3D: a new generation of three-dimensional molecular structure viewer. Trends Biochem Sci. 1997, 22: 314-316. 10.1016/S0968-0004(97)01093-1.
Nagle DL, Karim MA, Woolf EA, Holmgren L, Bork P, Misumi DJ, McGrail SH, Dussault BJ, Perou CM, Boissy RE, Duyk GM, Spritz RA, Moore KJ: Identification and mutation analysis of the complete gene for Chediak-Higashi syndrome. Nat Genet. 1996, 14: 307-311.
Introne W, Boissy RE, Gahl WA: Clinical, molecular, and cell biological aspects of Chediak-Higashi syndrome. Mol Genet Metab. 1999, 68: 283-303. 10.1006/mgme.1999.2927.
Bassi MT, Ramesar RS, Caciotti B, Winship IM, De Grandi A, Riboni M, Townes PL, Beighton P, Ballabio A, Borsani G: X-linked late-onset sensorineural deafness caused by a deletion involving OA1 and a novel gene containing WD-40 repeats. Am J Hum Genet. 1999, 64: 1604-1616. 10.1086/302408.
Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R: A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000, 14: 1048-1057.
Henning KA, Li L, Iyer N, McDaniel LD, Reagan MS, Legerski R, Schultz RA, Stefanini M, Lehmann AR, Mayne LV, Friedberg EC: The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell. 1995, 82: 555-564.
Tullio-Pelet A, Salomon R, Hadj-Rabia S, Mugnier C, de Laet MH, Chaouachi B, Bakiri F, Brottier P, Cattolico L, Penet C, Begeot M, Naville D, Nicolino M, Chaussain JL, Weissenbach J, Munnich A, Lyonnet S: Mutant WD-repeat protein in triple-A syndrome. Nat Genet. 2000, 26: 332-335. 10.1038/81642.
van den Brink DM, Brites P, Haasjes J, Wierzbicki AS, Mitchell J, Lambert-Hamill M, de Belleroche J, Jansen GA, Waterham HR, Wanders RJ: Identification of PEX7 as the second gene involved in Refsum disease. Am J Hum Genet. 2003, 72: 471-477. 10.1086/346093.
Purdue PE, Lazarow PB: Peroxisome biogenesis. Annu Rev Cell Dev Biol. 2001, 17: 701-752. 10.1146/annurev.cellbio.17.1.701.
Acknowledgements
This work was supported by a grant from the USDA National Research Initiative Program to SVN (2003-35304-13299) and a Michigan State University Plant Science Graduate Student Recruitment Fellowship to PL.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
SVN conceived the study, carried out sequence manipulation, and drafted the manuscript. PL carried out literature searches and identified information relevant to protein function. Both authors read and approved the final manuscript.
Electronic supplementary material
12864_2003_99_MOESM1_ESM.html
Additional File 1: Conservation of predicted Arabidopsis WDR proteins among budding yeast, fruit fly, and human. Entries are provided with an HTML link to the corresponding genome database entry in The Arabidopsis Information Resource, Saccharomyces Genome Database, FlyBase, or Ensembl Human Genome Browser. WDR proteins from these other eukaryotes that did not exhibit a significant match with any Arabidopsis protein are not listed. Compilation of predicted protein sets, groupings, and homology searches were performed as described in Methods. (HTML 126 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
van Nocker, S., Ludwig, P. The WD-repeat protein superfamily in Arabidopsis: conservation and divergence in structure and function. BMC Genomics 4, 50 (2003). https://doi.org/10.1186/1471-2164-4-50
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-4-50