Abstract
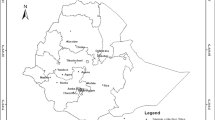
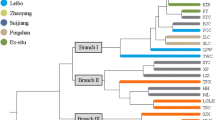
As an ecologically and economically important endemic bamboo species, moso (Phyllostachys edulis) has been widely distributed in Southern China. In the paper, 20 fluorescently labeled microsatellite markers were used to evaluate the genetic structure of Ph. edulis including 34 representative populations (803 individuals) covering the geographic range in China. Moderate genetic diversity (H = 0.376) and differentiation (Gst = 0.162) were detected at the species level, with the majority of genetic diversity occurring within populations (84.55%). Bayesian model-based structure analysis and sNMF/ANLS-AS method revealed the presence of two and three clusters. When K = 2, majority of populations (except SX) were clustered together (C1). It implied that SX, known as an introduced and isolated founder population, significantly differed from other populations for distinct environmental selection and allele mutation with the proof of scarce outcrossing and relatively high frequency of private allele. While K = 3, two subgroups (C1a of 18 populations and C1b of 14 populations) were detected within C1. The C1b displayed as a belt-shape region with an east-west direction. It coincided with the extremely high artificial selection in C1b (lower genetic diversity than that of C1a) due to the intensive plantation in the last four decades. Our results implied that the population protection and germplasm collection of moso bamboo should be not only from representative populations in Zhejiang, Jiangxi, Fujian, and other places with intensive cultivation in the east of China, but from populations with high level of gene and genotypic diversity in the west (e.g., HN5, GD1, GZ2, YN1, and SX).





Similar content being viewed by others
References
Alexander DH, Lange K (2011) Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinf 12(1):246–246. https://doi.org/10.1186/1471-2105-12-246
Bohonak AJ (2002) IBD (Isolation By Distance): a program for analyses of isolation by distance. J Hered 93(2):153–154. https://doi.org/10.1093/jhered/93.2.153
Brown AHD, Feldman MW, Nevo E (1980) Multilocus structure of natural populations of HORDEUM SPONTANEUM. Genetics 96(2):523–536
Chen YY, Han QX, Cheng Y, Li ZZ, Li W (2010) Genetic variation and clonal diversity of the endangered aquatic fern Ceratopteris pteridoides as revealed by AFLP analysis. Biochem Syst Ecol 38(6):1129–1136. https://doi.org/10.1016/j.bse.2010.12.016
Chung MY, López-Pujol J, Chung MG (2013) Low genetic diversity in marginal populations of Bletilla striata (Orchidaceae) in southern Korea: insights into population history and implications for conservation. Biochem Syst Ecol 46:88–96. https://doi.org/10.1016/j.bse.2012.09.019
Clark-Tapia R, Alfonso-Corrado C, Eguiarte LE, Molina-Freaner F (2005) Clonal diversity and distribution in Stenocereus eruca (Cactaceae), a narrow endemic cactus of the Sonoran Desert. Am J Bot 92(2):272–278. https://doi.org/10.3732/ajb.92.2.272
De Witte LC, Armbruster GJ, Gielly L, Taberlet P, Stocklin J (2012) AFLP markers reveal high clonal diversity and extreme longevity in four key arctic-alpine species. Mol Ecol 21(5):1081–1097. https://doi.org/10.1111/j.1365-294X.2011.05326.x
Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS (1991) ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res 19(14):4008. https://doi.org/10.1093/nar/19.14.4008
Doyle JJ, Doyle JL (1991) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14(8):2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10(3):564–567. https://doi.org/10.1111/j.1755-0998.2010.02847.x
Francois O (2016) Running structure-like population genetic analyses with R. R Tutorials in Population Genetics U Grenoble-Alpes:pages 1–9
Frichot E, Francois O (2015) LEA:AnRpackage for landscape and ecological association studies. Methods Ecol Evol 6(8):925–929. https://doi.org/10.1111/2041-210X.12382
Frichot E, Mathieu F, Trouillon T, Bouchard G, Francois O (2014) Fast and efficient estimation of individual ancestry coefficients. Genetics 196(4):973–983. https://doi.org/10.1534/genetics.113.160572
Fu J (2001) Chinese moso bamboo: its importance. Bamboo 22:5–7
Grunwald NJ, Goodwin SB, Milgroom MG, Fry WE (2003) Analysis of genotypic diversity data for populations of microorganisms. Phytopathology 93(6):738–746. https://doi.org/10.1094/PHYTO.2003.93.6.738
Hamrick J, Godt M (1989) Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS (eds) Plant population genetics, breeding and genetic resources. Sinauer Associates, Sunderland, pp 43–63
Hamrick J, Godt M (1996) Effects of life history traits on genetic diversity in plant species. Philos Trans R Soc Lond Ser B Biol Sci 351(1345):1291–1298. https://doi.org/10.1098/rstb.1996.0112
Hamrick JL, Godt MJW, Sherman-Broyles SL (1992) Factors influencing levels of genetic diversity in woody plant species. In: Population genetics of forest trees. New Forests 6:95–124
Isagi Y, Oda T, Fukushima K, Lian C, Yokogawa M, Kaneko S (2016) Predominance of a single clone of the most widely distributed bamboo species Phyllostachys edulis in East Asia. J Plant Res 129(1):21–27. https://doi.org/10.1007/s10265-015-0766-z
Janzen DH (1976) Why bamboos wait so long to flower. Annu Rev Ecol Syst 7(1):347–391. https://doi.org/10.1146/annurev.es.07.110176.002023
Jay F et al (2012) Forecasting changes in population genetic structure of alpine plants in response to global warming. Mol Ecol 21(10):2354–2368. https://doi.org/10.1111/j.1365-294X.2012.05541.x
Jiang WX, Zhang WJ, Ding YL (2013) Development of polymorphic microsatellite markers for Phyllostachys edulis (Poaceae), an important bamboo species in China. Appl Plant Sci 1(7). https://doi.org/10.3732/apps.1200012
Kamvar ZN, Tabima J, Grunwald NJ (2014) Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2:e281. https://doi.org/10.7717/peerj.281
Kim H, Park H (2007) Sparse non-negative matrix factorizations via alternating non-negativity-constrained least squares for microarray data analysis. Bioinformatics 23(12):1495–1502. https://doi.org/10.1093/bioinformatics/btm134
Kitamura K, Kawahara T (2009) Clonal identification by microsatellite loci in sporadic flowering of a dwarf bamboo species, Sasa cernua. J Plant Res 122(3):299–304. https://doi.org/10.1007/s10265-009-0220-1
Kitamura K, Saitoh T, Matsuo A, Suyama Y (2009) Development of microsatellite markers for the dwarf bamboo species Sasa cernua and Sasa kurilensis (Poaceae) in northern Japan. Mol Ecol Resour 1470
Lai C, Hsiao J (1997) Genetic variation of Phyllostachys pubescens (Bambusoideae, Poaceae) in Taiwan based on DNA polymorphisms Botanical Bulletin of Academia Sinica 38
Li Y (1995) Bamboo garden with the great achievement of “South Bamboo Transfer to North” Shanxi. Forestry 06:29–30
Lin Y, Lu JJ, Wu MD, Zhou MB, Fang W, Ide Y, Tang DQ (2014) Identification, cross-taxon transferability and application of full-length cDNA SSR markers in Phyllostachys pubescens. Springer Plus 3:486
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21(9):2128–2129. https://doi.org/10.1093/bioinformatics/bti282
Miyazaki Y, Ohnishi N, Hirayama K, Nagata J (2009) Development and characterization of polymorphic microsatellite DNA markers for Sasa senanensis (Poaceae: Bambuseae). Conserv Genet 10(3):585–587. https://doi.org/10.1007/s10592-008-9575-4
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci 70(12):3321–3323. https://doi.org/10.1073/pnas.70.12.3321
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28(19):2537–2539. https://doi.org/10.1093/bioinformatics/bts460
Peng Z, Lu T, Li L, Liu X, Gao Z, Hu T, Yang X, Feng Q, Guan J, Weng Q, Fan D, Zhu C, Lu Y, Han B, Jiang Z (2010) Genome-wide characterization of the biggest grass, bamboo, based on 10,608 putative full-length cDNA sequences. BMC Plant Biol 10(1):116. https://doi.org/10.1186/1471-2229-10-116
Powell W, Machray GC, Provan J (1996) Polymorphism revealed by simple sequence repeats. Trends Plant Sci 1(7):215–222. https://doi.org/10.1016/S1360-1385(96)86898-0
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Rodriguez M, Rau D, O’Sullivan D, Brown AH, Papa R, Attene G (2012) Genetic structure and linkage disequilibrium in landrace populations of barley in Sardinia. Theor Appl Genet 125(1):171–184. https://doi.org/10.1007/s00122-012-1824-8
Ruan X, Lin X, Lou Y, Guo X, Fang W, Chen C (2008) Genetic diversity of Phyllostachys heterocycla var. pubescens provenances by AFLP and ISSR. J Zhejiang For Sci Technol 28:29–33
Shannon CE (2001) A mathematical theory of communication. ACM SIGMOBILE Mobile Computing and Communications Review 5:3–55
Simpson EH (1949) Measurement of diversity. Nature 163(4148):688–688. https://doi.org/10.1038/163688a0
Slatkin M, Barton NH (1989) A comparison of three indirect methods for estimating average levels of gene flow. Evolution 43(7):1349–1368. https://doi.org/10.1111/j.1558-5646.1989.tb02587.x
Stoddart JA, Taylor JF (1988) Genotypic diversity: estimation and prediction in samples. Genetics 118(4):705–711
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739. https://doi.org/10.1093/molbev/msr121
Tang DQ, JJ L, Fang W, Zhang S, Zhou MB (2010) Development, characterization and utilization of GenBank microsatellite markers in Phyllostachys pubescens and related species. Mol Breed 25(2):299–311. https://doi.org/10.1007/s11032-009-9333-4
Tian B, Yang HQ, Wong KM, Liu AZ, Ruan ZY (2012) ISSR analysis shows low genetic diversity versus high genetic differentiation for giant bamboo, Dendrocalamus giganteus (Poaceae: Bambusoideae), in China populations. Genet Resour Crop Evol 59(5):901–908. https://doi.org/10.1007/s10722-011-9732-3
Watanabe M, Ito M, Kurita S (1994) Chloroplast DNA phylogeny of Asian bamboos (Bambusoideae, Poaceae) and its systematic implication. J Plant Res 107(3):253–261. https://doi.org/10.1007/BF02344252
Wright S (1978) Evolution and the genetics of populations: a treatise in four volumes. Vol. 4, Variability within and among natural populations. University of Chicago Press
Yang HQ, An MY, ZJ G, Tian B (2012) Genetic diversity and differentiation of Dendrocalamus membranaceus (Poaceae: Bambusoideae), a declining bamboo species in Yunnan, China, as based on inter-simple sequence repeat (ISSR) analysis. Int J Mol Sci 13(12):4446–4457. https://doi.org/10.3390/ijms13044446
Yeh F, Yang R, Boyle T, Ye Z, Xiyan J (2000) PopGene32, Microsoft Windows-based freeware for population genetic analysis, version 1.32
Zhang W, Ma N (1990) Vitality of bamboo pollens and natural pollination in bamboo plants. For Res 3:250–255
Zhang SF, Ma QX, Ding YL (2007) RAPD analysis for the genetic diversity of Phyllostachys edulis China forestry. Sci Technol 21:3
Zhou FC (1998) Cultivation and utilization of bamboo. Bamboo Res 1:11–20
Acknowledgements
This work was supported by the Special Fund for Forest Scientific Research in the Public Welfare from State Forestry Administration of China (no. 201504106). We wish to thank Xiaoting Shen, Pei Hu, Yi Fan, Jie Wang and Dr. Liang Chen for sampling assistance and Dr. Tao Xia for experimental suggestion. We thank the public and private owners for access to the sampling sites. The authors are also thankful to the anonymous reviewers whose comments and suggestions have greatly improved the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by W.-W. Guo
Data archiving statement
All the 20 SSR primers used in this work are available in GenBank. The corresponding GenBank accession was provided in supplemental Table S1.
Rights and permissions
About this article
Cite this article
Jiang, W., Bai, T., Dai, H. et al. Microsatellite markers revealed moderate genetic diversity and population differentiation of moso bamboo (Phyllostachys edulis)—a primarily asexual reproduction species in China. Tree Genetics & Genomes 13, 130 (2017). https://doi.org/10.1007/s11295-017-1212-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-017-1212-2